Abstract
The PIAS proteins (protein inhibitor of activated STAT) were originally identified as inhibitors of the JAK-STAT pathway. Subsequently, their roles on transcriptional regulation have been identified in modulation of the androgen receptor (AR) and other nuclear hormone receptor-mediated actions. Zimp7, also named Zmiz2, is a novel PIAS-like protein and functions as a transcriptional co-activator. In this study, we demonstrate an interaction between Zimp7 and PIAS proteins with higher preference for PIAS3. A modified mammalian one-hybrid assay showed that the NH2-terminal proline-rich domain of Zimp7 and the region spanning amino acids 321–486 of PIAS3 were the primary interaction segments. The interaction between Zimp7 and PIAS3 proteins was further confirmed by in vitro protein pull-down and co-immunoprecipitation assays with both exogenous and endogenous proteins. Expression of exogenous PIAS3 further enhances Zimp7-mediated augmentation of AR transcription. Knockdown of the endogenous PIAS3 protein using a specific PIAS3 small hairpin RNA reduced the augmentation of Zimp7 on AR-mediated transcription. Co-localization of Zimp7 and PIAS3 proteins was observed in the nuclei of cells by immunostaining. Exogenous PIAS3 expression enhances the stability of the Zimp7 protein. Using chromatin immunoprecipitation assays, we showed that PIAS3 is involved in the AR- and Zimp7-formed protein complex(es) in the AR downstream target promoter to facilitate androgen-induced transcription. Finally, we further demonstrated that loss of Zimp7 significantly impaired PIAS3-mediated enhancement on AR activity in mouse Zimp7 null (zimp7−/−) embryonic fibroblasts. Taken together, these results demonstrate a novel interaction between PIAS and PIAS-like proteins and elucidate a novel regulatory mechanism for PIAS proteins in AR-mediated transcription.
Keywords: DNA/Transcription, Receptors/Steroid/Thyroid, Transcription, Transcription/Coactivators, Transcription/STAT, PIAS Proteins, Zimp Proteins, Androgen Receptor
Introduction
The PIAS2 proteins (protein inhibitor of activated STAT) were originally identified as repressors of STAT transcription factors (1). Recent data have shown that they function as transcriptional co-regulators to modulate the activity of a diverse set of transcription factors, such as p53, Smads, and nuclear hormone receptors (2–5). Members of the PIAS family share a high degree of sequence similarity, and are characterized by the presence of a highly conserved X-SPRING (eXtended SP-RING) domain, also named the Miz (Msx-interacting zinc finger) domain (6). This motif appears to be important for interactions with target proteins and is highly similar to the RING finger domain present in E3 ubiquitin ligases (5). Indeed, numerous studies have implicated a role for PIAS proteins in the ubiquitin-like sumoylation pathway, where they appear to enhance SUMO conjugation of target proteins through the Miz domain (7–10).
Zimp7 and Zimp10, also named zmiz2 and zmiz1, are novel PIAS-like proteins originally identified as androgen receptor (AR) interacting proteins (11, 12). They both share an X-SPRING domain with other PIAS proteins (13). In addition to this domain, the two Zimp proteins contain a strong intrinsic transactivation domain, through which they augment the transcriptional activity of nuclear hormone receptors and other transcriptional factors (12, 14–16). An ortholog of Zimp10 and Zimp7, called tonalli (tna), has been identified in Drosophila, and genetically interacts with the ATP-dependent SWI/SNF and Mediator complexes, suggesting a potential role for the Zimp proteins in chromatin remodeling (13). We have shown that Zimp7 interacts with Brg-1 and BAF57, components of the mammalian SWI/SNF complexes (11).
Previously, we observed that the full-length Zimp proteins, when fused to the DNA-binding domain of GAL4, display a very limited activity compared with truncated mutants containing the COOH-terminal proline-rich domain only (11, 12). Using a series of deletion mutants, we further demonstrated that the NH2-terminal domains of Zimp proteins inhibit the transcriptional activity of the COOH-terminal regions, suggesting a potential mechanism that may be involved in switching the Zimp proteins from an inactive form to an active form. In an attempt to identify a regulatory mechanism for Zimp protein-mediated transcription, we searched for Zimp7 interacting proteins. Interestingly, we identified that the PIAS3 protein was a Zimp7 interacting protein in a yeast two-hybrid screen. Using different in vitro and in vivo approaches, we further demonstrated that Zimp7 physically interacts with PIAS proteins, among which PIAS3 showed the strongest interaction. Through the interaction, PIAS3 augments Zimp7-mediated transcription. The above data elucidate a regulatory role of PIAS proteins in Zimp-mediated transcription.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid System
Yeast two-hybrid experiments were performed as described previously (19). The DNA fragments containing truncated human Zimp10, Zimp7, or LZTS2 were fused in-frame to the GAL4 DNA binding domain (DBD) in the pGBKT7 vector (Clontech). Truncated PIAS3 (amino acids 365–482) was fused to GAL4 TAD in the pVP16 vector (Clontech). The constructs were transformed into the modified yeast strain PJ69-4A (20). A cDNA library from the mouse EML cell line was used for this screening. Transformants were selected on Sabouraud dextrose medium lacking tryptophan, leucine, and/or adenine. The specificity of the interaction with PIAS3 was measured by a liquid β-galactosidase assay (19).
Recombinant DNAs
Full-length and truncated human Zimp7 or Zimp10 were subcloned into pcDNA3-FLAG, pcDNA3-HA, or pM vector, which contains a GAL4 DBD (11). The plasmid constructs containing full-length AR, SMAD2, SMAD3, and p53 in the pM vector used in this study were generated previously (11, 12, 15). The truncated PIAS3 fragments that contain amino acids 321–486 or 486–628 were amplified with the following pairs of primers for cloning into pcDNA-FLAG vector: 5′-TTTAAGATCTCTCCGGGTGTCACTCATGTG-3′ and 5′-ATATCTCGAGGGTCAGGGCTCCTTTGCTTCCA-3′, 5′-TTTAAGATCTACCTCTGGTCACCAGCCATC-3′ and 5′-ATATCTCGAGCCAAAAAGCTCAGTCCAAGG-3′, respectively. The oligonucleotides corresponding to human PIAS3 (5′-GGCTGTCGGTCAGACATCATT-3′) were cloned into the pBS/U6 vector (21). To make lentiviral shRNA constructs, the DNA fragment containing the U6 promoter and the PIAS3 shRNA sequences was transferred into the pLentiSuper vector (Invitrogen). Zimp7 shRNA lentiviral constructs were prepared as described previously (16, 23). The virus production was carried out in human embryonic kidney 293T cells (24). The pcDNA3-FLAG and pM vectors expressing PIAS1, PIAS3, PIASy, PIASxα, PIASxβ, and His-SUMO-1 were generously donated by Dr. Palvimo (25). The reporter plasmid, pPSA7kb-luc, was a kind gift of Dr. Jan Trapman (26).
Cell Cultures, Transient Transfections, and Proliferation Assay
The monkey kidney cell line, CV-1, was maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (FBS) (HyClone, Denver, CO). The human embryonic kidney cell line, HEK293, was grown in 10% FBS/Dulbecco's modified Eagle's medium. For cell proliferation assays, LNCaP cells were cultured in T-medium (Invitrogen) with 5% FBS, and then infected with Zimp7 shRNA or control lentiviruses for 3 days and replated in 96-well dishes. Cell viability was determined over a 6-day time course using the MTS (Cell Titer 96® AQueous Non-Radioactive Cell) Proliferation Assay kit (Promega).
Transient transfections were carried out using a Lipofectamine transfection kit or Lipofectamine 2000 (Invitrogen). Transfection and reporter assays were performed as described previously (11, 15). The relative light units from individual transfections were normalized by β-galactosidase activity in the same samples to eliminate nonspecific factors. Individual transfection experiments were done in triplicate, and the results are reported as mean RLU/β-galactosidase (±S.D.) from representative experiments.
GST Pull-down Assay
Glutathione S-transferase (GST)-PIAS3 fusion proteins were constructed in the pGEX-4T-1 vector (Amersham Biosciences). Expression and purification of GST fusion proteins were performed as described previously (19). Equal amounts of GST fusion proteins coupled to glutathione-Sepharose beads were incubated with HEK293 cell lysates at 4 °C for 2 h in the lysis buffer as described previously (16). Beads were carefully washed three times with binding buffer and then analyzed by SDS-PAGE, followed by Western blot analysis using a Zimp7 antibody (11).
RNA Isolation and Reverse Transcription-PCR
LNCaP cells were infected with shRNA lentiviruses for AR, Zimp7, PIAS3, or a control for 6 h and subsequently cultured in the absence or presence of 1 nm DHT for 12 h. Total RNA was extracted using RNAWiz isolation reagent (Ambion). The reverse transcription-PCR was carried out as described previously (27). The following primers were used in amplification: PSA, 5′-ACCATGTGGGTCCCGGTTGT-3′ and 5′-GAGTTGATAGGGGTGCTCAGG-3′) and GAPDH (5′-CCATGGAGAAGGCTGGGG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′).
Immunoprecipitation and Western Blotting
Whole cell lysates from HEK293 cells were prepared as described previously (11), and incubated with an anti-PIAS3 antibody or normal mouse IgG at 4 °C with gentle rotation overnight. Equilibrated Protein A-Sepharose beads were then added for 1.5 h at 4 °C, and subsequently collected. The beads were washed, proteins were eluted using 2× sample buffer (125 mm Tris-HCl, pH 6.8, 4% SDS, 20% (v/v) glycerol, 0.004% bromphenol blue) and analyzed by Western blot. Detection was performed with ECL reagents according to the manufacturer's protocol using ECL Hyperfilm (Amersham Biosciences). Anti-FLAG antibody (Sigma), anti-HA antibodies (Covance, CA, and Santa Cruz Biotechnology, Santa Cruz, CA), anti-PIAS3 antibody (Santa Cruz Biotechnology), anti-SUMO-1 antibody (Santa Cruz Biotechnology), anti-GAL4 DBD antibody (Santa Cruz Biotechnology), anti-AR antibody (PG-21, Upstate, Lake Placid, NY), and anti-Zimp7 antibody (11) were used in the experiments.
Immunostaining
CV-1 or HEK293 cells were seeded onto chamber slides overnight. The FLAG-PIAS3 plasmid was transfected with pcDNA3-HA-Zimp7 into CV-1 cells using Lipofectamine reagents (Invitrogen). Cells were fixed with 4% paraformaldehyde and immunostained with an anti-FLAG antibody at a 1:1000 dilution and an anti-HA antibody at a 1:500 dilution, or with anti-PIAS3 antibody at a 1:200 dilution and anti-Zimp7 antibody at a 1:500 dilution followed by incubation with species-specific Alexa Fluor 488- and 594-conjugated secondary antibodies. Samples were also counterstained with 1 ng/ml of 4′,6′-diamidino-2-phenylindole. Images were examined with an epifluorescence microscope (Zeiss, Thornwood, NY).
Pulse-Chase Assay
CV-1 cells were transfected with wild-type pcDNA3-FLAG-Zimp7 in the presence or absence of pcDNA3-HA-PIAS3. After 24 h of transfection, the cells were incubated with Dulbecco's modified Eagle's medium without l-methionine and l-cysteine (Invitrogen) for 3 h, and then pulse-labeled with 100 μCi of Tran35S-label (ICN, Irvine, CA) for 30 min. The cells were washed twice with phosphate-buffered saline and then chased by incubating in complete Dulbecco's modified Eagle's medium for various periods of time. The cells were lysed in RIPA buffer (50 mm Tris-Cl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.2 mm Na3VO4, 0.5 mm phenylmethylsulfonyl fluoride). 35S-Labeled Zimp7 protein was immunoprecipitated from the cytosolic fractions using an anti-FLAG monoclonal antibody (Sigma) and analyzed by SDS-PAGE, followed by autoradiography.
Chromatin Immunoprecipitation (ChIP)
LNCaP cells were grown in 5% charcoal-stripped FBS medium for 3 days and then treated with 1 nm DHT or vehicle alone for 2 h. Cells were treated with formaldehyde and subjected to ChIP analysis as described previously (16). The chromatin was sheared to an average size of 800 bp by sonication and diluted 10-fold in ChIP dilution buffer (2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl, pH 8.1, 1% Triton X-100), and then subjected to immunoprecipitation with anti-Zimp7 antibody, anti-AR antibody, anti-PIAS3 antibody, or normal IgG overnight at 4 °C and recovered with Protein A-Sepharose (Amersham Biosciences). ChIP and input DNA were analyzed by PCR using AREI- and AREII-specific primers as described previously (28). The samples were also amplified with GAPDH primers, 5′-CGGTGCGTGCCCAGTTG-3′ and 5′-GCGACGCAAAAGAAGATG-3′, as a control (29).
Mouse Embryonic Fibroblasts
Mice heterozygous for a disrupted allele of the Zimp7 gene by Cre/loxP-mediated recombination were mated, and pregnant females were sacrificed at 16.5 days post-coitus. Embryos were isolated in cold phosphate-buffered saline and then incubated in 500 μl of trypsin (0.05%) for 15 min at 37 °C with intermittent agitation. Embryos were disrupted by pipetting and then added to at least a 3× volume of Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were plated, allowed to adhere, and used for assays. To determine the mouse embryo fibroblasts (MEF) genotype, embryo tails isolated during the dissection were digested, and genomic DNA was extracted and analyzed by PCR with specific primers to mouse zimp7 locus to determine wild-type or deleted zimp7 alleles.
RESULTS
PIAS3 Is a Zimp7 Interacting Protein
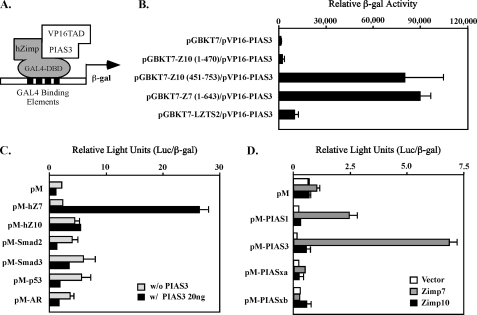
Our previous data suggested that the NH2 termini of Zimp proteins might inhibit the transcriptional activity of the COOH-terminal transactivation domain. To search for potential mechanisms causing Zimp proteins to switch from inactive forms into active forms, we employed a modified yeast two-hybrid screen to identify proteins that potentially interact with the NH2-terminal region of Zimp7 using a bait construct containing Zimp7 amino acids 1–643, which is highly conserved in Zimp10. Of 3.4 × 107 transformants, 123 grew under selective conditions and showed increased adenine and β-galactosidase production in medium. Rescue of the plasmids and sequencing of the inserts revealed that eight cDNAs encoded PIAS proteins. Three of them were PIAS3 and the rest were PIASy. Because Zimp7 and Zimp10 share significant sequence similarity, we examined the interaction between PIAS3 and Zimp proteins in yeast two-hybrid assays (Fig. 1A). The PIAS3 clone was cotransformed with various constructs containing either GAL4 DBD alone or fusion proteins with the NH2-terminal fragment of human Zimp7 (1–643 amino acids), and different truncations of Zimp10 (Fig. 1B). The original bait construct, pGBKT7-hZimp7 (1–643 amino acids), showed a specific interaction with pVP16-PIAS3 (365–482 amino acids) (Fig. 1B). Interestingly, the construct containing the fragment of hZimp10 between amino acids 451 and 753 appeared to interact with pVP16-PIAS3 as well. We then performed mammalian one-hybrid assays to further confirm the interaction between PIAS3 and Zimp proteins. In the experiments, we also included several other transcription factors, such as Smad2, Smad3, AR, and p53, as controls. To eliminate other nonspecific factors and equalize transfection efficiency between samples, we normalized the luciferase activity by measuring expression of co-transfected β-galactosidase in the above experiments. In addition, we also examined expression of the proteins in the above samples by Western blotting (supplemental Fig. S1A). As we observed previously, both full-length Zimp7 and Zimp10 showed very limited activity in the absence of PIAS3 (Fig. 1C). However, a significant induction of luciferase activity was observed when Zimp7 and PIAS3 vectors were co-transfected into cells, suggesting that PIAS3 interacts with Zimp7 and confers its transcriptional activity. Interestingly, only moderate activity was observed in the samples cotransfected with PIAS3 and Zimp10. To assess the interactions between PIAS and Zimp proteins, we extended our experiments to other PIAS proteins. In this set of experiments, we cotransfected pM vectors containing a number of full-length PIAS proteins in the presence or absence of Zimp7 or Zimp10 and examined their expression (supplemental Fig. S1B). As shown in Fig. 1D, Zimp7 showed the strongest interaction with PIAS3. In addition, an interaction between Zimp7 and PIAS1 also appeared. Taken together, the above experiments demonstrate a protein-protein interaction between PIAS3 and Zimp7.
FIGURE 1.
Specific interaction between Zimp7 and PIAS3 proteins. A, a schematic representation of the yeast two-hybrid assay for demonstrating the interaction between PIAS3 and hZimp proteins. B, the pVP16-PIAS3 plasmid containing VP16-TAD and PIAS3 (amino acid 365–482) was co-transformed with pGBKT7 vector or different pGBKT7 fusion constructs. Numbers correspond to amino acid residues. Transformed cells were plated on SD-Ade-Leu-Trp plates and SD-Leu-Trp plates to monitor transformation efficiency. Three independent colonies were inoculated from each transformation experiment for subsequent liquid β-galactosidase assays. The data for the liquid β-galactosidase assays are shown as the mean ± S.D. C, the full-length human Zimp7, Zimp10, AR, SMAD2, SMAD3 or p53 were fused to Gal4 DBD in the pM vector. CV-1 cells were cotransfected with the pM constructs with or without a PIAS3 expression construct, a luciferase reporter construct containing Gal4 binding sites within the chicken myelomonocytic growth factor gene minimal promoter (−41 to +61), and a constitutive β-galactosidase reporter. Data are presented in the RLUs, which were obtained by normalizing the activities of luciferase to those of β-galactosidase. Individual transfection experiments were done in triplicate, and the results reported as the mean ± S.D. D, the full-length PIAS1, PIAS3, PIASxα, or PIASxβ were fused to pM vector, and co-transfected into CV-1 cells with full-length hZimp7 or Zimp10 proteins. The luciferase assay was performed as described above.
Mapping Interacting Regions for Zimp7 and PIAS3
To confirm the NH2-terminal region of Zimp7 was responsible for interaction with PIAS3, we made various truncated constructs of Zimp7 and tested them with full-length PIAS3 protein using the mammalian one-hybrid assay. As shown in Fig. 2A, full-length and two NH2-terminal-truncated Zimp7 mutants (207–892 and 372–892 amino acids) showed very limited activity in the absence of PIAS3 protein. However, a significant induction was observed in the same samples when the full-length PIAS3 protein was added. Further removal of amino acids 372–512 elevated the transcriptional activity of Zimp7 in the absence and presence of PIAS3. Using co-immunoprecipitation assays, we further assessed the interactions between PIAS3 and Zimp7 proteins. FLAG-tagged full-length or truncated Zimp7 and HA-tagged PIAS3 were transfected into HEK293 cells. Nearly equal levels of exogenous Zimp7 proteins were detected in the immunoprecipitates pulled down by anti-FLAG antibodies (Fig. 2B, top panel). Consistent with the above luciferase assay result, PIAS3 protein was only detected in samples co-transfected with full-length or NH2-terminal Zimp7 constructs (Fig. 2B, bottom panel). These data suggest that PIAS3 binds to the NH2-terminal region of Zimp7, which disrupts the intra-molecular interaction to release the autoinhibition of Zimp7 activity.
FIGURE 2.
Mapping the putative interaction regions on hZimp7 and PIAS3 proteins. A, full-length or truncated fragments of Zimp7 were fused to Gal4 DBD in the pM vector. Numbers correspond to amino acid residues. CV-1 cells were cotransfected with pM vectors, luciferase reporter constructs, with or without PIAS3 plasmids. B, HEK293 cells were transfected with pcDNA3-FLAG-tagged full-length or truncated Zimp7 (1 μg) with HA-tagged PIAS3 (1 μg). Equal amounts of the whole cell lysates were used for immunoprecipitation (IP) with an anti-FLAG monoclonal antibody. Immunoprecipitates were then analyzed by immunoblotting (IB) and probed by Zimp7 and PIAS3 antibodies. C, a schematic representation of the domains of human PIAS3 protein. Numbers correspond to amino acid residues. D, equal amounts of GST fusion proteins were used to pull down endogenous Zimp7 protein in HEK293 cells. GST protein alone was used as a negative control. Equal amounts of the above GST proteins were analyzed on SDS-PAGE. Material bound to the GST columns was subjected to SDS-PAGE and analyzed by Western blot with a Zimp7-specific antibody.
The interaction between hZimp7 and PIAS3 was further assessed by GST pull-down experiments. Various truncated PIAS3 fragments were cloned in-frame to generate different truncated GST fusion proteins (Fig. 2C). Using them, we searched for the regions responsible for binding to Zimp7. GST proteins expressed upon isopropyl 1-thio-β-d-galactopyranoside induction were analyzed on SDS-PAGE gels with Coomassie Blue staining (Fig. 2D, bottom panel). To examine the binding of Zimp7 with GST fusion proteins, whole cell lysates isolated from HEK293 cells that express endogenous Zimp proteins were applied to each sample, which was immobilized onto a glutathione-Sepharose matrix for the binding assay. The elution from the above samples was analyzed by SDS-PAGE and Western blot with a human Zimp7 antibody. As shown in Fig. 2D (top panel), the PIAS3 mutant (amino acids 321–486) containing Miz and AD/SIM domains had the strongest binding activity with full-length Zimp7. Deletion of the region between amino acids 321 and 364 covering the full Miz domain significantly reduced the binding. Notably, the GST fusion constructs containing the fragment between amino acids 321 and 386 covering the full Miz domain showed strong binding with Zimp7, whereas the constructs containing the fragments between amino acids 412 and 506 covering the AD/SIM domain and amino acids 486–628 covering the S/T domain showed no binding with Zimp7. Taken together, the above results indicate that the region spanning the Miz and AD/SIM domains is primarily responsible for binding to Zimp7.
Demonstration of the Interaction between Zimp7 and PIAS3 in Cells
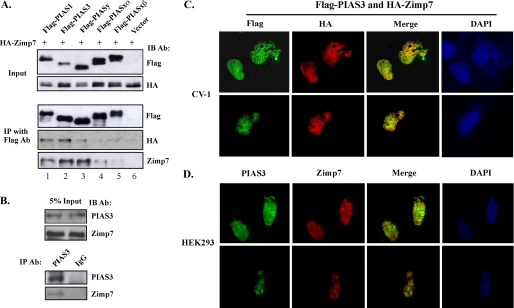
Next, we examined the interactions between Zimp and PIAS proteins in intact cells using co-immunoprecipitation. Both FLAG-tagged PIAS and HA-tagged Zimp7 were expressed in HEK293 cells. Whole cell lysates were prepared from these cells and analyzed by Western blotting. Exogenous PIAS and Zimp7 proteins were detected in transfected cells using FLAG and HA antibodies, respectively (Fig. 3A, top panels). Equal amounts of the above whole cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody. Almost equal levels of exogenous FLAG-tagged PIAS proteins were detected in the immunoprecipitates pulled down by FLAG antibodies (Fig. 3A). Intriguingly, HA-tagged Zimp7 proteins were only detected in immunoprecipitates from cells co-transfected with Zimp7 and PIAS1, PIAS3, or PIASy with either HA or Zimp7 antibody (Fig. 3A). We also examined the existence of endogenous PIAS3 and Zimp7 protein complexes in cells. Both Zimp7 and PIAS3 were detected in whole cell lysates of HKE293 cells with appropriate antibodies (Fig. 3B, top panel). Equal amounts of whole cell lysates were immunoprecipitated by the anti-PIAS3 antibody or normal IgG, a negative control, and subsequently analyzed by immunoblotting with PIAS3 or Zimp7 antibody. Both PIAS3 and Zimp7 proteins were detected in immunoprecipitates pulled by the PIAS3 antibody. These results indicate that Zimp7 complexes with PIAS3, probably as well as PIAS1 and PIASy proteins in mammalian cells.
FIGURE 3.
PIAS3 and Zimp7 interact and co-localize with each other in the nucleus. A, CV-1 cells were transiently co-transfected with pcDNA3-HA-Zimp7 and different pcDNA3-FLAG-PIAS plasmids or pcDNA3-FLAG vector. Equal amounts of whole cell lysates were blotted with HA or FLAG antibody to detect expression levels of the two proteins (input) or subjected to immunoprecipitation (IP) with anti-FLAG antibody. The precipitated fractions were then resolved by SDS-PAGE and analyzed by Western blot (IB) using the anti-HA or anti-Zimp7 antibodies. B, 5% of the total HEK293 cell lysate volume (input) as well as PIAS3 and normal IgG immunoprecipitates were analyzed on SDS-PAGE by either the PIAS3 antibody or Zimp7 antibody (Ab). C, CV-1 cells were transfected with pcDNA3-FLAG-PIAS3 (100 ng) and pcDNA3-HA-Zimp7 (100 ng). Forty-eight hours after transfection, cells were fixed, incubated with the anti-FLAG (1:1000 dilution) monoclonal antibody and the anti-HA (1:500) polyclonal antibody, and then visualized with fluorophore-conjugated secondary antibodies, goat anti-mouse 488 and goat anti-rabbit 594, at a dilution of 1:1000, respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Merge of different stains indicates areas of co-localization. D, HEK293 cells were seeded and immunostained with the rabbit anti-Zimp7 and mouse anti-PIAS3 antibodies followed by incubation with species-specific Alexa Fluor 488- and 594-conjugated secondary antibodies, respectively. Images were examined with an epifluorescence microscope. Nuclei were co-stained with DAPI. Merge of different stains indicates areas of co-localization.
PIAS3 and Zimp7 Proteins Co-localize in Nuclear Granules
Here, we examined whether a dynamic interaction between PIAS3 and hZimp7 exists in cells. Expression vectors for FLAG-tagged PIAS3 and HA-tagged hZimp7 were transfected into CV-1 cells. As shown in Fig. 3C, PIAS3 and hZimp7 displayed speckled patterns of nuclear distribution, which is consistent with previous reports (7, 11). A significant amount of overlay between PIAS3 and Zimp7 proteins appear in cells. Next, we confirmed the co-localization of endogenous PIAS3 and Zimp7 proteins in HEK93 cells with specific antibodies. Both Zimp7 and PIAS3 proteins displayed similar nuclear distributions as observed in the above experiments and there is a significant amount of overlay between these two proteins. Based on the above data, we conclude that Zimp7 and PIAS3 co-localize in the nuclear granules.
PIAS3 Affects the Stability of Zimp7 Protein
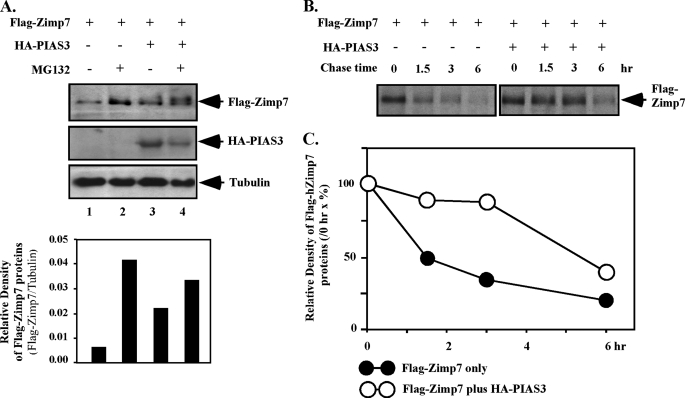
It has been shown that PIAS proteins are able to stabilize SOX9, a transcription factor required for cell differentiation and development (30). We therefore examined whether PIAS3 affects the stability of Zimp7. We first measured cellular levels of Zimp7 protein in the presence or absence of MG132, a proteasome inhibitor. As shown in Fig. 4A, the level of exogenous Zimp7 protein was higher in the presence of MG132 than in the absence of MG132, indicating that the proteasomal degradation pathway contributes to the stability of Zimp7. We then tested whether PIAS3 can affect the stability of Zimp7 in cells. In the absence of MG132, cells co-transfected with Zimp7 and PIAS3 expression vectors showed a higher level of Zimp7 than cells transfected with Zimp7 alone (Fig. 4A, lane 1 versus lane 3). However, in the presence of MG132, co-expression of PIAS3 protein did not significantly increase the level of cellular Zimp7 protein (Fig. 4A, lane 2 versus 4), suggesting that the effect of PIAS3 on Zimp7 stability is a result of increasing the resistance of Zimp7 to the proteasome degradation pathway.
FIGURE 4.
PIAS3 enhances the stability of the Zimp7 protein. A, CV-1 cells were transfected with pcDNA3-FLAG-hZimp7 (1 μg) and/or pcDNA3-HA-PIAS3 (1 μg). After 24 h of transfection, cells were cultured in the presence or absence of MG132, a proteasome inhibitor, at 10−6 m for 12 h. Whole cell lysates were prepared and analyzed by Western blotting with the FLAG or HA antibodies, respectively. The antibody against tubulin was also used as a control for protein loading. Intensities of Zimp7 protein were analyzed by densitometry and levels of Zimp7 protein were shown graphically as a relative percentage normalized by tubulin levels. B, CV-1 cells transfected by Zimp7 and/or PIAS3 expression vectors were pulsed with Tran35S-label and chased with medium containing an excess of cold methionine/cysteine for the indicated times. Cell lysates were immunoprecipitated by FLAG antibody for exogenous Zimp7. C, levels of Zimp7 protein in the above experiments were analyzed by densitometry and normalized by the density of Zimp7 at 0 h. The experiments have been performed more than two times and the result from one set of experiments shown here.
To further demonstrate the effect of PIAS3 on the stability of Zimp7, we performed pulse-chase experiments in CV-1 cells. Cells were pulsed with Tran35S-label and chased in the presence or absence of PIAS3 for a total of 6 h. 35S-Labeled Zimp7 protein was immunoprecipitated from cytosolic fractions and analyzed by SDS-PAGE. As shown in Fig. 4, B and C, PIAS3 enhanced the stability of the Zimp7 protein by increasing its half-life from 1.5 to 6 h. These results provide a direct line of evidence demonstrating that PIAS3 indeed affects the stability of Zimp7.
PIAS3 Enhances Zimp7-mediated Transcription
Zimp7 has been identified as an AR co-activator (11, 14). Therefore, we examined a possible role for PIAS3 in Zimp7-mediated transcription. A luciferase reporter driven by the PSA promoter (PSA-Luc) was co-transfected with expressing vectors for AR, Zimp7, and PIAS3 into CV-1 cells in the absence or presence of DHT (Fig. 5A). In the presence of 1 nm DHT, a nearly 2-fold ligand-dependent induction was observed in cells transfected with AR expression vector alone. Co-transfection of Zimp7 plasmids augmented the ligand-dependent AR activity about 3–4-fold, which is consistent with our previous results (11). Overexpression of the PIAS3 protein further enhanced the androgen-induced AR activity to 7–8-fold. Importantly, the augmentation of AR activity by PIAS3 was only observed in cells co-transfected with Zimp7, suggesting the effect of PIAS3 on AR is dependent on the presence of Zimp7. We next assessed the activity of endogenous PIAS3 on AR-mediated transcription in LNCaP cells using lentiviruses containing a PIAS3-specific shRNA. As shown in Fig. 5B, an induction of PSA luciferase activity was observed in the presence of 1 nm DHT. Overexpression of Zimp7 further augmented the activity of the PSA luciferase reporter ∼2-fold. However, introduction of the specific PIAS3 shRNA into cells reduced the PSA luciferase activity in a dose-dependent manner. There was no significant change of PSA luciferase activity in the samples infected with a control lentivirus (Fig. 5B).
FIGURE 5.
PIAS3 enhances Zimp7-mediated transcription. A, 100 ng of the luciferase reporter driven by the 7-kb PSA promoter, 25 ng of pSV40-β-galactosidase, 5 ng of AR expression vector (pSV-hAR), 0 or 5 ng of pcDNA3-HA-Zimp7, and 0 or 5 ng of pcDNA3-FLAG-PIAS3 were transfected into CV-1 cells. Twenty-four hours after transfection, cells were incubated with or without 1 nm DHT for another 24 h. Cell lysates were then prepared for assessment of luciferase and β-galactosidase activities. B, LNCaP cells were transfected with the PSA-luc reporter (100 ng), pcDNA3-β-galactosidase (25 ng), pcDNA3-FLAG-hZimp7 (20 ng), and the PIAS3 shRNA or control shRNA as indicated. Twenty-four hours after transfection, cells were treated without or with 1 nm DHT for 24 h. Cell lysates were measured for luciferase and β-galactosidase activities as described above. Levels of PIAS3 and tubulin proteins were measured by Western blot assays to determine the knockdown effect in the above correlated samples. C, LNCaP cells were transfected with the PSA-luc reporter (100 ng), pcDNA3-β-galactosidase (25 ng), pcDNA3-FLAG-hZimp7 (20 ng), and three different PIAS3 expression vectors (15 ng) as indicated. Twenty-four hours after transfection, cells were treated without or with 1 nm DHT for 24 h. Cell lysates were measured for luciferase and β-galactosidase activities as described above. The expression of PIAS3 and tubulin was measured by Western blot assays. D, LNCaP cells were infected with shRNA lentiviruses for AR, Zimp7, PIAS3, or a control for 6 h and cultured in the absence or presence of 1 nm DHT for 12 h. Total RNA was isolated and analyzed by reverse transcription-PCR with PSA and GAPDH-specific primers. Levels of PCR products were analyzed by densitometry and normalized by the density of GAPDH. The experiments have been performed more than two times and the result from one set of experiments shown. E, LNCaP cells were cultured in 5% charcoal-stripped FBS for 3 days and then in the absence or presence of 1 nm DHT for 2 h. Cells were then cross-linked with formaldehyde, sheared by sonication, and then immunoprecipitated with anti-AR, anti-Zimp7, anti-PIAS3, or normal rabbit IgG antibody. Primers specific to the AREI and AREII were used to PCR amplify the eluted chromatin. Primers specific to GAPDH were used as a control to monitor immunoprecipitation specificity. F, LNCaP cells were cultured in charcoal-stripped FBS medium for 3 days, and then treated with 1 nm DHT or vehicle only. Cells were cross-linked with 0.5 mm DSP (dithiobis(succinimidylpropionate)) and then the nuclear extracts were isolated and used in immunoprecipitation with anti-Zimp7 antibody or normal rabbit IgG. Five percent of the initial nuclear extracts or immunoprecipitates (IP) were analyzed by Western blotting (IB) with specific antibodies as labeled in the figure.
To further assess augmentation of hZimp7 and PIAS3 on AR signaling, we made several truncated mutants of PIAS3 corresponding to the defined binding regions for Zimp7 as described earlier (Fig. 2, C and D). Using these deletion mutants, we further tested whether the enhancement of PIAS3 on Zimp7 is mediated through the interaction between the proteins. As shown in Fig. 5C, the truncated PIAS3 containing amino acids 321–486 acted as a dominant-negative mutant to attenuate the enhancement of Zimp7 on AR-mediated transcription. However, the truncated PIAS3 protein containing the fragment between amino acids 486 and 628 showed no significant effect in the experiment. The above observations suggest that the region between amino acids 321 and 486 covering the Miz and AD/SIM domains directly interacts with Zimp7, conferring AR transcriptional activity.
Next, we examined the effect of PIAS3 on the expression of PSA, an AR downstream target gene in LNCaP cells using reverse transcription-PCR assays. Approximately 7-fold induction of PSA transcription was observed in the presence of 1 nm DHT (Fig. 5D). Knockdown of AR, Zimp7, or PIAS3 individually reduces endogenous PSA mRNA levels significantly. Intriguingly, knockdown of both Zimp7 and PIAS3 simultaneously almost fully diminishes PSA expression. Using GAPDH as a loading control, we measured the relative level of PSA expression. These results were consistent with the transactivation assays and provide additional evidence to show a collaborative role for Zimp7 and PIAS3 in regulating AR-mediated transcription.
To demonstrate the Zimp7·PIAS3·AR complex formation in LNCaP cells, ChIP assays were performed to detect the occupancy of the above proteins on AR-regulated promoters after androgen induction. LNCaP cells were grown in charcoal-stripped FBS for 3 days followed by treatment with 1 nm DHT. Soluble chromatin was prepared after formaldehyde treatment of the cell cultures, and specific antibodies against AR, hZimp7, or PIAS3 were used to immunoprecipitate AR, Zimp7, or PIAS3-bound genomic DNA fragments, which were then analyzed by PCR using specific primers for amplifying the AREI and AREII sites in the PSA promoter region (28). AR and hZimp7 recruitments were detected in both AREI and AREII sites in the presence of DHT (Fig. 5E), whereas only a weak binding for PIAS3 at the AREII site was detected. To further evaluate whether AR, PIAS3, and Zimp7 are involved in the same protein complex, we performed a co-immunoprecipitation assay using nuclear extracts isolated from LNCaP cells after being cross-linked with 0.5 mm dithiobis(succinimidylpropionate) in the presence or absence of 1 nm DHT. As shown in Fig. 5F, both AR and PIAS3 proteins were detected in the immunoprecipitates pulled down by Zimp7 antibody in the presence of DHT, which provides an additional line of evidence demonstrating that AR, Zimp7, and PIAS3 are involved in the same protein complex(es) upon androgen induction.
PIAS3-mediated Enhancement on AR Activity Is Significantly Impaired in Zimp7 Null Cells
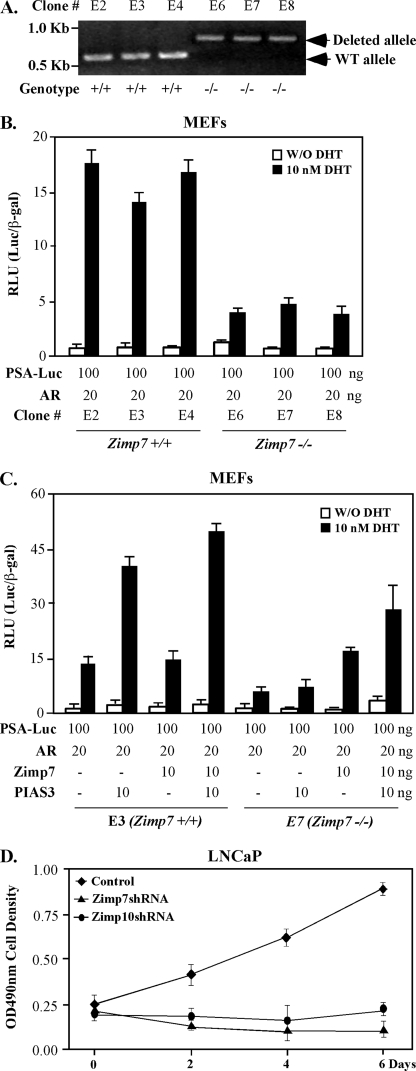
To fully evaluate the cooperative role of PIAS3 in Zimp7-mediated enhancement of AR activity, we made an effort to produce MEFs from embryos that were generated through intercrosses between Zimp7 heterozygous knock-out mice (zimp7+/−). Three E16.5-day-old wild-type and homozygous knock-out (zimp7−/−) embryos were isolated and analyzed by genomic PCR (Fig. 6A), and then used to generate MEFs. We first tested androgen-induced transcriptional activity in the above MEFs. Transfection of the PSA luciferase report and AR expression vectors into three lines of wild-type MEFs showed 12–17-fold induction in the presence of 10 nm DHT. In contrast, only 2–4-fold induction was observed in the zimp7−/− MEFs, confirming a critical role of Zimp7 in AR-mediated transcription. We then further assessed the role of PIAS3 in inducing AR activity using the above wild-type and Zimp7 null MEFs. Overexpression of both PIAS3 and AR proteins showed androgen-induced activity of PSA-luciferase reporter nearly three times higher than expression of AR alone in wild-type MEFs. Addition of Zimp7 into MEFs enhanced the PSA-luciferase activity to another 20–30% in the above experiment. However, in zimp7−/− MEFs, expression of both exogenous AR and PIAS3 only slightly induced PSA-luciferase activity in comparison with expression of AR alone, whereas co-expression of Zimp7 and AR generated a 2-fold induction of PSA-luciferase activity. Most importantly, co-expression of PIAS3, AR, and Zimp7 proteins significantly restored the androgen-induced activity of PSA-luciferase reporter in Zimp7 null MEFs. Therefore, these data not only provide a direct line of evidence that demonstrates the importance of Zimp7 in AR-mediated transcription but also indicate the requirement for Zimp7 in PIAS3-regulated AR activity.
FIGURE 6.
Examination of the effect of PIAS3 on AR transcriptional activity in Zimp7 null MEFs. A, MEFs isolated from 6 embryos at E16.5 were genotyped by PCR with specific primers to the mouse zimp7 locus at chromosome 11. PCR fragments representing wild-type alleles (400 base pair) or deleted alleles (876 bp) were shown on 1% agarose gel. B, three wild-type and Zimp7 null MEFs were transfected with 100 ng of PSA-Luc, 25 ng of CMV-β-galactosidase, and 20 ng of human AR expression vector. Twenty-four hours after transfection, cells were treated with or without 10 nm DHT for 24 h. Cell lysates were measured for luciferase and β-galactosidase activities, and the RLUs were calculated. Bars represent the mean ± S.D. of triplicate determinations. C, wild-type or zimp7−/− MEFs were transfected with 100 ng of PSA-Luc, 25 ng of CMV-β-galactosidase, and 20 ng of human AR expression vector in the presence or absence of 10 ng of Zimp7 or PIAS3 expression vector. Twenty-four hours after transfection, cells were treated without or with 10 nm DHT for 24 h. Then cells lysates were prepared and used for measuring luciferase and β-galactosidase activities. The RLUs were calculated as above. Similar results were obtained from two independent MEF isolations. D, LNCaP cells were infected with shRNA lentiviruses for Zimp7, and Zimp10, as well as control viruses and then used for the MTS cell proliferation assay.
Finally, we examined the promoting role of Zimp7 in prostate cancer cell growth. LNCaP cells were infected with shRNA lentiviruses for Zimp7 and Zimp10, and a control virus. As shown in Fig. 6D, knockdown of Zimp7 significantly reduced LNCaP cell growth, confirming the regulatory role of Zimp7 in cell growth. The similar inhibitory effect by Zimp10 shRNA viruses was also observed in the experiments, which is consistent with our previous observation (31). The above data further confirm a promoting role for Zimp7 and Zimp10 in prostate cancer cell growth.
DISCUSSION
Zimp7 is a novel PIAS-like protein and a transcriptional co-activator (11, 14). Previous studies have shown that Zimp7 and its homolog, Zimp10, interact with the AR and enhance AR-mediated transcription. Subsequent identification of the interaction between Zimp7 and components of the SWI/SNF chromatin remodeling complexes suggests a possible role for Zimp7 in chromatin modification (11). Our previous studies have shown that the carboxyl-terminal regions of Zimp7 and Zimp10 possess a very strong intrinsic transcriptional activation domain that can be inhibited by their amino-terminal domains through intra-molecular interactions (11, 12). However, the molecular mechanisms for these autoinhibitions are currently unclear. Therefore, identification of regulatory mechanisms that can release the inhibition and switch the Zimp proteins from inactive forms into active forms will be extremely important to fully understand the biological roles of the proteins. In this study, we demonstrated that PIAS3 physically interacts with the amino-terminal region of Zimp7 and represses autoinhibition of the protein in resulting activation of its mediated transcription. Our findings provide the first line of evidence demonstrating the inter-regulation between PIAS and PIAS-like proteins.
The PIAS proteins were originally identified as negative regulators of the JAK-STAT cytokine signaling pathway, but have subsequently been shown to regulate the activity of steroid hormone receptors and other transcription factors, such as LEF-1 and p53 (25, 32, 33). Accumulated evidence indicates that the PIAS proteins can either positively or negatively regulate the activity of other transcription factors in a different cell context (5). Of particular relevance to AR signaling, the PIAS family member PIASxα was first identified as AR-interacting protein 3 (ARIP3) (34). Other PIAS proteins have also been found to interact with SUMO-1 and Ubc9 and to mediate sumoylation of nuclear hormone receptors and other transcription factors (7, 8). In this study, we have provided multiple lines of evidence demonstrating that PIAS3 interacts with Zimp7 and participates the augmentation on AR-mediated transcription and androgen-induced cell growth. Identification of an interaction between PIAS3 and Zimp7 and their collaborative roles on AR action provides a new mechanism for PIAS proteins in the regulation of nuclear hormone receptor-mediated transcription.
The PIAS proteins contain several functionally conserved domains. The C3HC4-type RING-finger-like zinc-binding domain or Miz domain is located in the center region of the proteins and is required for the SUMO-E3 ligase activity of PIAS proteins (3, 35, 36). The carboxyl-terminal regions of PIAS proteins contain a putative SUMO1 interacting motif (SIM), a highly acidic region (AD), and a serine/threonine-rich region (S/T) (37, 38). The functional roles of SIM, AD, or S/T domains in PIAS proteins remain to be defined. In this study, we demonstrated that the region spanning the Miz and AD/SIM domains of PIAS3 between amino acids 321 and 486 is mainly responsible for binding to Zimp7. Through this interaction, PIAS3 can further enhance Zimp7 activity on AR-mediated transcription. Using MG132, a proteasome inhibitor, we showed that the level of the Zimp7 protein is regulated through a ubiquitin-dependent degradation pathway. Interestingly, overexpression of exogenous PIAS3 can prolong the half-life of the Zimp7 protein from 1.5 to 6 h in cells. These findings suggest a novel regulatory mechanism between PIAS and PIAS-like proteins in cells, which may involve their regulation of the activity of AR and other transcription factors.
It has been shown that the PIAS proteins possess E3 ligase activity for the ubiquitin-like SUMO pathway, through which the PIAS proteins can modulate AR-mediated transcription (8, 39). Although the precise mechanism by which the PIAS proteins regulate the transcriptional activity of AR is not fully clear, previous studies have shown that sumoylation by the PIAS proteins of co-regulators within the AR transcriptional complex rather than the AR protein itself may play an important role in inducing AR transcriptional activity (8, 10, 37). In this study, we have investigated the possible role of PIAS3 as a SUMO E3-ligase in the sumoylation of Zimp7. Co-expression of PIAS3 and Zimp7 in the presence SUMO-1 in CV-1 cells showed that PIAS3 has no significant effect on the sumoylation of Zimp7 (data not shown). Analysis of the human Zimp7 sequence showed two potential sumoylation sites, Lys-659 and Lys-665, which are conserved among different species. Either single or double mutations within these sites did not affect intrinsic Zimp7 transcriptional activity and AR-mediated transcription (data not shown). Thus, it appears that sumoylation of Zimp7 may not directly contribute to its transcriptional activity through these sites. It would be interest to search for other regulatory mechanisms for PIAS3 to regulate the stability of Zimp7.
Previously, it has been suggested that PIAS proteins can regulate transcription in a SUMO E3 ligase-independent manner (22). PIAS1 interacts with Msx1 and confers Msx1-mediated transcriptional repression by modulating its cellular localization to the nuclear periphery (18). PIASx has also been shown to desumoylate Elk1 in the resulting disassociation with HDAC2 and activation of Elk1-mediated transcription (17). In this study, we have provided multiple lines of evidence showing that PIAS3 interacts with the Zimp7 protein and enhances its stability. Through the interaction, PIAS3 can increase the intrinsic transcriptional activity of Zimp7 in cis and augment AR ligand-dependent activity in trans. Thus, the above data suggest another potential SUMO E3 ligase-independent mechanism for PIAS proteins in modulating transcription.
In this study, we used MEFs generated from Zimp7 knock-out mice to assess the biological significance of Zimp7 and its interaction with PIAS3 in AR-mediated transcription. We demonstrated that loss of Zimp7 expression significantly reduces AR-mediated transcription, and impairs the effect of PIAS3 on AR action. Addition of the exogenous Zimp7 protein partially restored the enhancement of PIAS3 on AR transcriptional activity in Zimp7 null MEFs. These data provide intriguing evidence that demonstrates a crucial role for the interaction between Zimp7 and PIAS3 in inducing AR activity in vivo. Further investigation of the biological roles of PIAS proteins in androgen signaling should be pursued using appropriate animal models.
In conclusion, this study demonstrates an interaction between PIAS3 and Zimp7, a newly identified PIAS-like protein. Through this interaction, PIAS3 can further augment the activity of Zimp7 on AR-mediated transcription. The interaction between PIAS3 and Zimp7 may facilitate the formation of the AR-involved transcriptionally active protein complex(es) and confer ligand-induced AR action. Our results also explore a novel molecular mechanism by which PIAS proteins regulate AR and other nuclear hormone receptor-mediated transcription. Further studies on regulation of the cellular Zimp7 level at normal or disease states, as well as the biological consequences of Zimp7 and PIAS3 interaction in vivo, will further explore biological roles of PIAS and PIAS-like proteins.
Supplementary Material
Acknowledgment
We are especially grateful to Curtis Clark for reading the manuscript.
This work was supported in part by National Institutes of Health Grant CA70297.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PIAS
- protein inhibitor of activated STAT
- STAT
- signal transducer and activator of transcription
- AR
- androgen receptor
- DBD
- DNA binding domain
- shRNA
- small hairpin RNA
- SIM
- SUMO1 interacting motif
- AD
- acidic domain
- S/T
- serine/threonine-rich region
- RLU
- relative luciferase units
- FBS
- fetal bovine serum
- GST
- glutathione S-transferase
- DHT
- dihydroxytestosterone
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HA
- hemagglutinin
- ChIP
- chromatin immunoprecipitation
- MEF
- mouse embryo fibroblast
- Miz
- Msx-interacting zinc finger.
REFERENCES
- 1.Shuai K. (2000) Oncogene 19, 2638–2644 [DOI] [PubMed] [Google Scholar]
- 2.Megidish T., Xu J. H., Xu C. W. (2002) J. Biol. Chem. 277, 8255–8259 [DOI] [PubMed] [Google Scholar]
- 3.Jackson P. K. (2001) Genes Dev. 15, 3053–3058 [DOI] [PubMed] [Google Scholar]
- 4.Kotaja N., Vihinen M., Palvimo J. J., Jänne O. A. (2002) J. Biol. Chem. 277, 17781–17788 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt D., Müller S. (2003) Cell Mol. Life Sci. 60, 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L., Wu H., Ma L., Sangiorgi F., Wu N., Bell J. R., Lyons G. E., Maxson R. (1997) Mech. Dev. 65, 3–17 [DOI] [PubMed] [Google Scholar]
- 7.Kotaja N., Karvonen U., Jänne O. A., Palvimo J. J. (2002) Mol. Cell. Biol. 22, 5222–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishida T., Yasuda H. (2002) J. Biol. Chem. 277, 41311–41317 [DOI] [PubMed] [Google Scholar]
- 9.Kahyo T., Nishida T., Yasuda H. (2001) Mol. Cell 8, 713–718 [DOI] [PubMed] [Google Scholar]
- 10.Kotaja N., Karvonen U., Jänne O. A., Palvimo J. J. (2002) J. Biol. Chem. 277, 30283–30288 [DOI] [PubMed] [Google Scholar]
- 11.Huang C. Y., Beliakoff J., Li X., Lee J., Li X., Sharma M., Lim B., Sun Z. (2005) Mol. Endocrinol. 19, 2915–2929 [DOI] [PubMed] [Google Scholar]
- 12.Sharma M., Li X., Wang Y., Zarnegar M., Huang C. Y., Palvimo J. J., Lim B., Sun Z. (2003) EMBO J. 22, 6101–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutiérrez L., Zurita M., Kennison J. A., Vázquez M. (2003) Development 130, 343–354 [DOI] [PubMed] [Google Scholar]
- 14.Beliakoff J., Sun Z. (2006) Nucl. Recept. Signal 4, e017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Thyssen G., Beliakoff J., Sun Z. (2006) J. Biol. Chem. 281, 23748–23756 [DOI] [PubMed] [Google Scholar]
- 16.Lee J., Beliakoff J., Sun Z. (2007) Nucleic Acids Res. 35, 4523–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S. H., Sharrocks A. D. (2005) EMBO J. 24, 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H., Quinn J. C., Prasanth K. V., Swiss V. A., Economides K. D., Camacho M. M., Spector D. L., Abate-Shen C. (2006) Genes Dev. 20, 784–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F., Li X., Sharma M., Sasaki C. Y., Longo D. L., Lim B., Sun Z. (2002) J. Biol. Chem. 277, 11336–11344 [DOI] [PubMed] [Google Scholar]
- 20.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sui G., Soohoo C., Affar el B., Gay F., Shi Y., Forrester W. C., Shi Y. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5515–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharrocks A. D. (2006) Genes Dev. 20, 754–758 [DOI] [PubMed] [Google Scholar]
- 23.Li T. H., Zhao H., Peng Y., Beliakoff J., Brooks J. D., Sun Z. (2007) Nucleic Acids Res. 35, 2767–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delenda C. (2004) J. Gene Med. 6, Suppl. 1, S125–S138 [DOI] [PubMed] [Google Scholar]
- 25.Kotaja N., Aittomäki S., Silvennoinen O., Palvimo J. J., Jänne O. A. (2000) Mol. Endocrinol. 14, 1986–2000 [DOI] [PubMed] [Google Scholar]
- 26.Cleutjens K. B., van Eekelen C. C., van der Korput H. A., Brinkman A. O., Trapman J. (1996) J. Biol. Chem. 271, 6379–6388 [DOI] [PubMed] [Google Scholar]
- 27.Sun Z., Yergeau D. A., Tuypens T., Tavernier J., Paul C. C., Baumann M. A., Tenen D. G., Ackerman S. J. (1995) J. Biol. Chem. 270, 1462–1471 [DOI] [PubMed] [Google Scholar]
- 28.Shang Y., Myers M., Brown M. (2002) Mol. Cell 9, 601–610 [DOI] [PubMed] [Google Scholar]
- 29.Bates G. J., Nicol S. M., Wilson B. J., Jacobs A. M., Bourdon J. C., Wardrop J., Gregory D. J., Lane D. P., Perkins N. D., Fuller-Pace F. V. (2005) EMBO J. 24, 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hattori T., Eberspaecher H., Lu J., Zhang R., Nishida T., Kahyo T., Yasuda H., de Crombrugghe B. (2006) J. Biol. Chem. 281, 14417–14428 [DOI] [PubMed] [Google Scholar]
- 31.Beliakoff J., Lee J., Ueno H., Aiyer A., Weissman I. L., Barsh G. S., Cardiff R. D., Sun Z. (2008) Mol. Cell. Biol. 28, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R. (2001) Genes Dev. 15, 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson V., Davis G. E., Maxwell S. A. (2001) Apoptosis 6, 221–234 [DOI] [PubMed] [Google Scholar]
- 34.Moilanen A. M., Karvonen U., Poukka H., Yan W., Toppari J., Jänne O. A., Palvimo J. J. (1999) J. Biol. Chem. 274, 3700–3704 [DOI] [PubMed] [Google Scholar]
- 35.Hochstrasser M. (2001) Cell 107, 5–8 [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K., Taira T., Niki T., Seino C., Iguchi-Ariga S. M., Ariga H. (2001) J. Biol. Chem. 276, 37556–37563 [DOI] [PubMed] [Google Scholar]
- 37.Jiménez-Lara A. M., Heine M. J., Gronemeyer H. (2002) FEBS Lett. 526, 142–146 [DOI] [PubMed] [Google Scholar]
- 38.Shuai K. (2006) Cell Res. 16, 196–202 [DOI] [PubMed] [Google Scholar]
- 39.Schmidt D., Müller S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.