Abstract
Rho GTPase-activating proteins (GAPs) are responsible for the inactivation of Rho GTPases, which are involved in the regulation of critical biological responses in eukaryotic cells, ranging from cell cycle control to cellular morphogenesis. The genome of fission yeast Schizosaccharomyces pombe contains six genes coding for putative Rho GTPases, whereas nine genes code for predicted Rho GAPs (Rga1 to Rga9). One of them, Rga4, has been recently described as a Cdc42 GAP, involved in the control of cell diameter and symmetry in fission yeast. In this work we show that Rga4 is also a Rho2 GAP that negatively modulates the activity of the cell integrity pathway and its main effector, MAPK Pmk1. The DYRK-type protein kinase Pom1, which regulates both the localization and phosphorylation state of Rga4, is also a negative regulator of the Pmk1 pathway, but this control is not dependent upon the Rga4 role as a Rho2-GAP. Hence, two subsets of Rga4 negatively regulate Cdc42 and Rho2 functions in a specific and unrelated way. Finally, we show that Rga7, another Rho2 GAP, down-regulates the Pmk1 pathway in addition to Rga4. These results reinforce the notion of the existence of complex mechanisms determining the selectivity of Rho GAPs toward Rho GTPases and their functions.
Keywords: Cell/Wall/Yeast, G Proteins, Organisms/Yeast, Signal Transduction, Signal Transduction/Protein Kinases/MAP, GTPase-activating Proteins, Fission Yeast
Introduction
The Rho family GTPases are found in both higher and lower eukaryotic organisms, and they are involved in diverse key biological responses (1, 2). In yeast cells, Rho GTPases are essential in the regulation of the cytoskeleton, morphogenesis, and growth. They are necessary for the organization of the actin cytoskeleton, polarized secretion, and the activation and location of the cell wall biosynthetic enzymes, which are essential for yeast cell viability (3). Rho GTPases act as molecular switches cycling between an active GTP-bound state and an inactive GDP-bound state (1). This activity is controlled by (i) guanine nucleotide exchange factors that catalyze exchange of GDP for GTP to activate the switch (4); (ii) GTPase-activating proteins (GAPs)5 that stimulate the intrinsic GTPase activity to inactivate the switch (5); and (iii) guanine nucleotide dissociation inhibitors, whose role appears to block spontaneous activation (1). The genome of fission yeast Schizosaccharomyces pombe contains six genes coding for putative Rho GTPases: rho1+ to rho5+ and Cdc42+ (6). Rho1 is essential and required to maintain cell wall integrity and organization of the actin cytoskeleton (6, 7). Cdc42 is also essential to establish polarized growth and the formation of actin cables (8–10). Rho2, although not essential, plays a role in the biosynthesis of the cell wall (1–3)β-d-glucan (11). Rho2 also stabilizes and activates kinase Pck2, one of the two S. pombe protein kinase C orthologs that is required for Mok1/Ags1 localization and biosynthesis of (1–3)β-d-glucan (11, 12). Additionally, Rho2 and Pck2 are essential both for the basal activity and the activation of the cell integrity MAPK pathway (13–15) that is related to the maintenance of cell integrity, cytokinesis, ion homeostasis, and vacuole fusion (16–20). The MAPK module of this pathway is composed by MAPK kinase kinase Mkh1 (21), MAPK kinase Pek1/Skh1 (19, 20), and MAPK Pmk1/Spm1 (16, 17), an extracellular signal-regulated kinase (ERK)-type kinase that becomes dually phosphorylated and activated in response to a variety of external stimuli (14). Rho2 and Pck2 act upstream of Mkh1 and regulate Pmk1 activation in response to hypertonic stress and hypotonic shock (15).
S. pombe genome contains nine genes coding for predicted Rho GAP proteins (22). Three of these proteins, Rga1, Rga5, and Rga8, are Rho1 GAPs. Rga1 is involved in the F-actin patch polarization and cell morphogenesis (22). Rga5 mainly regulates the Rho1-Pck1 interaction and cytokinesis (23), whereas Rga8 is modulated by Shk1 (24), a p21-actived kinase that in turn is regulated by Cdc42 (25). We have recently reported the characterization of Rga2 as a Rho2 GAP that plays a role in the regulation of cell morphogenesis and the cell integrity MAPK pathway (26). We also pointed out that, likely, Rga2 was not the only GAP regulating Rho2 and that other GAPs might inhibit this GTPase. In this context, Rga4 has been recently described as a Cdc42 GAP involved in the control of cell diameter and symmetry in fission yeast (27, 28). Rga4 would play a critical role in the polarized distribution of active GTP-loaded Cdc42 by defining the cortical region where the GTPase is kept in an inactive state (27, 28). Moreover, Pom1, a DYRK-type Ser/Thr-protein kinase, regulates the localization and phosphorylation state of Rga4, although it appears likely that this GAP is not a direct substrate for Pom1 (28). In this paper we present evidence to show that Rga4 is also a Rho2 GAP that negatively regulates the activity of the Pmk1 cell integrity pathway and that this role is independent of its function as a GAP for Cdc42. Finally, we show that Rga7, a Rho2 GAP, is also a negative regulator of the above pathway.
EXPERIMENTAL PROCEDURES
Strains, Growth Conditions, and Plasmids
The S. pombe strains (Table 1) were grown at 28 °C in rich medium (YES) or minimal medium (EMM2) supplemented with adenine, leucine, histidine, or uracil (100 mg/liter; Sigma) depending on their particular requirements (29). Transformation of yeast strains was performed by the lithium acetate method (29). Mutant strains were constructed by the random spore germination method after purification by glusulase treatment (30). Correct construction of strains was verified by PCR and Western blot analyses (see below). Escherichia coli DH5α was used as host for propagation of plasmids. Bacterial strains were grown in LB medium supplemented with 50 μg/ml ampicillin. Plasmids pREP3X-Rga4, pREP3X-Rga7, and pREP3X-Rga4ΔN were constructed to express either full-length Rga4 and Rga7 or the GAP domain of Rga4 under the control of the wild type thiamine-repressible promoter (nmt1+) (31), respectively. To construct pREP3X-Rga4, the rga4+ open reading frame was amplified by PCR using genomic S. pombe DNA as the template and the oligonucleotides PPG-83 (ATATAGTCGACCATGGCTGCTTTCAAAAAGAG, SalI site is underlined) and PPG-84 (ATATAAGATCTAAGCTTTTATAAAATCACGCAAGAC, BglII site is underlined). The resulting ∼ 3.1-kbp DNA fragment was digested with SalI and BglII and cloned into plasmid pREP3X. To obtain plasmid pREP3X-Rga4ΔN, a 480-bp DNA fragment of rga4+ open reading frame corresponding to the GAP domain in Rga4 (amino acids 645–933) was amplified with the oligonucleotides RGA4GAP-5 (TATATCTCGAGGTCCCGACGCTTATCGTTTC, XhoI site is underlined) and RGA4GAP-3 (TATATGGATCCTTAGGCAAAGACTTCATGTAC, BamHI site is underlined), and the resulting DNA fragment was digested and cloned as above. To construct pREP3X-Rga7, the rga7+ open reading frame was amplified by PCR using oligonucleotides PPG-83 (ATATAGTCGACCATGGCTGCTTTCAAAAAGAG, SalI site is underlined) and PPG-84 (ATATAAGATCTAAGCTTTTATAAAATCACGCAAGAC, BglII site is underlined). The resulting ∼ 3.1-kbp DNA fragment was digested with SalI and BglII and cloned into plasmid pREP3X. Plasmid pREP41X-Ha-Cdc42(G12V) expresses an hyperactive allele of Cdc42 under the control of the attenuated variant (41X) of the thiamine-repressible promoter nmt1 (14). In experiments employing either pREP3X-Rga4, pREP3X-Rga7, pREP3X-Rga4ΔN, or pREP41X-Ha-Cdc42(G12V), yeast transformants were grown in EMM2 with or without thiamine (5 mg/liter) for 16–24 h.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source | |
|---|---|---|---|
| PPG4546 | h+ | leu1-32 ura4-D18 HA-rho2:KanR | Ref. 26 |
| PPG1811 | h− | leu1-32 rga4-GFP:KanR | Pérez's lab |
| PPG7114 | h− | leu1-32 rga4-GFP:KanR HA-rho2:KanR | This work |
| PPG1848 | h− | leu1-32 ura4-D18 rga4::KanR | Pérez's lab |
| PPG4465 | h− | leu1-32 ura4-D18 rga4::kanR HA-rho2:KanR | This work |
| MI102 | h+ | ade6-M210 pmk1::KanR leu1-32 ura4D-18 | Ref. 14 |
| TS320 | h+ | ade6-M216 pmk1::KanR rga4::KanR leu1-32 ura4D-18 | This work |
| TS321 | h+ | ade6-M216 pmk1::KanR pom1::KanR leu1-32 ura4D-18 | This work |
| JB151 | h− | 3nmt1-pom1+:KanR | Ref. 21 |
| TS322 | h− | 3nmt1-pom1+:KanR pmk1-HA6H::ura4+ | This work |
| MI200 | h+ | ade6-M216 pmk1-HA6H::ura4+leu1-32 ura4D-18 | Ref. 14 |
| MI201 | h− | ade6-M210 pmk1-HA6H::ura4+leu1-32 ura4D-18 | Ref. 14 |
| TS309 | h− | ade6-M210 rga4::KanMX6 pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| PPG4460 | h− | ade6-M216 rga2::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | Ref. 26 |
| TS307 | h− | ade6-M210 rga6::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | This work |
| TS308 | h− | ade6-M210 rga7::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | This work |
| TS326 | h+ | ade6-M216 rga2::KanMX6 rga4::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | This work |
| TS327 | h+ | ade6-M216 rga2::KanMX6 rga7::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | This work |
| TS328 | h+ | ade6-M216 rga4::KanMX6 rga7::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | This work |
| TS325 | h+ | ade6-M216 rga7::KanMX6 pmk1::KanMX6 leu1-32 ura4-D18 | This work |
| MI700 | h+ | ade6-M216 rho2::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | Ref. 15 |
| TS323 | h+ | ade6-M216 rga6::KanMX6 rho2::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | |
| TS324 | h− | ade6-M210 rga7::KanMX6 rho2::KanMX6 pmk1-HA6H:ura4+leu1-32 ura4-D18 | |
| TS310 | h+ | ade6-M216 pck2::HygR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS311 | h+ | ade6-M216 rga4::KanR rho2::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS312 | h+ | ade6-M216 rga4::KanR pck2::HygR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS313 | h− | ade6-M216 pom1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS314 | h+ | ade6-M216 pom1::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS315 | h+ | ade6-M216 pom1::KanR rga4::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS316 | h+ | ade6-M216 pom1::KanR rho2::KanR pmk1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS317 | h+ | ade6-M216 pom1::KanR pck2::HygR pmk1-HA6H::ura4+leu1-32ura4D-18 | This work |
| JM1521 | h+ | ade6-M216 his7-366 sty1-HA6H:: ura4+leu1-32 ura4D-18 | Ref. 43 |
| TS318 | h− | ade6-M210 rga4::KanMX6 sty1-HA6H::ura4+leu1-32 ura4D-18 | This work |
| TS319 | h+ | ade6-M216 pom1::KanMX6 sty1-HA6H::ura4+leu1-32 ura4D-18 | This work |
Two-hybrid Assay
Protein interactions were analyzed using the two-hybrid system (32). All of the constitutively active rho1G15VC199RΔILL, rho2G17VC198SΔIIS, cdc42G12VC198SΔLVL, and rho4G23VC199SΔVIL alleles, with the corresponding C-terminal cysteine mutated and lacking the last three amino acid residues to avoid prenylation, were cloned into the NcoI-BamHI sites of pAS2 as described (33) and used as bait against rga4+ cloned in the pACT2 plasmid. The Saccharomyces cerevisiae Y190 (MATa gal4 gal80 his3 trp1-901 ura3-52 leu2-3,-112 URA3::GAL–Δ lacZ, lys2::GAL (UAS)–ΔHIS3 cyhr) cells were transformed, and β-galactosidase activity was analyzed in the transformants as described (34).
In Vivo Analysis of Rho GAP Activity
The amount of GTP-bound Rho2 was analyzed using a Rho-GTP pull-down assay as previously described (23). Briefly, the assay was performed on cells containing HA-rho2+ expressed from their own promoter and lacking or overexpressing rga4+. The cell extracts were obtained using 500 μl of lysis buffer (50 mm Tris-HCl, pH 7.5, 20 mm NaCl, 0.5% Nonidet P-40, 10% glycerol, 0.1 mm dithiothreitol, 1 mm NaF, and 2 mm MgCl2 containing 100 μm p-aminophenyl methanesulfonyl fluoride, leupeptin, and aprotinin. Some 10 μg of GST-RBD (Rhotekin Rho-binding domain), previously obtained from E. coli DNA expression, purified, and coupled to glutathione-Sepharose beads, was used to precipitate the GTP-bound Rho2 from 2 mg of the total cell lysates. The extracts were incubated with the beads for 2 h at 4 °C, washed four times, and blotted against anti-HA monoclonal antibody (12CA5; Roche Applied Science) to detect the corresponding GTP-bound HA-Rho2. The total amounts of HA-Rho2 from extracts were determined by Western blot using the anti-HA monoclonal antibody.
In Vitro Analysis of Rho GAP Activity
The entire coding sequences for Rga4, Rho1, Rho2, and Cdc42 were fused in-frame to the C terminus of the GST sequence using the NdeI-NotI sites in the pREP-KZ vector (35). These constructs were used to transform S. pombe, and expression of the proteins was induced by growing the cells in the absence of thiamine for 18 h. GST-tagged proteins were purified by affinity chromatography. Briefly, the cell extracts from 100-ml cultures (A600 of 0.8) were obtained as described above using as lysis buffer (50 mm Tris-HCl, pH 7.6, 0.1% Nonidet P-40, 20 mm NaCl, 0,1 mm dithiothreitol, 1 mm EDTA, 100 μm p-aminophenylmethanesulfonyl fluoride, and protease inhibitors). The cell extracts were incubated with glutathione-Sepharose beads at 4 °C for 2 h. After centrifugation for 10 min at 5000 × g, the beads were washed three times with lysis buffer and resuspended in 100 ml of the lysis buffer. The GTP hydrolysis by Rho proteins in response to Rga4 activation was performed as described (36). Briefly, 30 μl of purified GST-Rho proteins on glutathione-Sepharose beads were first incubated for 10 min at 30 °C with [γ-32P]GTP (5 μCi) in 30 μl of 50 mm Tris-HCl, pH 7.6, 0.1% Nonidet P-40, 20 mm NaCl, 0.1 mm dithiothreitol, 1 mm EDTA to allow loading of the GTPase. The mixture was then placed on ice, and 10 μl of 25 mm MgCl2 were added. Aliquots of 10 μl were mixed with 40 μl of the reaction buffer (20 mm Tris-HCl, pH 7.6, 20 mm NaCl, 0,1 mm dithiothreitol, 2 mm GTP, 1 mg ml−1 bovine serum albumin, 5 mm MgCl2) and 30 μl of either purified GST or GST-Rga4. The reactions were placed at 20 °C, and 10-μl aliquots were removed at 2-min intervals and diluted with cold reaction buffer (1 ml). After filtration through nitrocellulose filters (Protran BA85; Whatman GmbH), the amount of radioactivity bound to the protein was determined by scintillation counting.
Gene Disruption and Epitope Tagging
The rga4+, rga6+, rga7+, pom1+, rho2+, and pck2+ null mutants were obtained by entire deletion of the corresponding coding sequence and its replacement with the KanMX6 or hygromycin cassettes by a PCR-mediated strategy (37). To construct strains expressing C-terminal GFP-tagged versions of rga4+, we employed plasmid pFA6a-GFP-kanMX6 (37). Strains expressing genomic versions of either Pmk1 or Sty1 fused to HA6H epitope at its C terminus in different genetic backgrounds were obtained after random spore analysis of appropriate crosses.
Purification and Detection of Activated Pmk1-HA6H and Sty1-HA6H Fusions
Log phase cell cultures (A600 of 0.5) growing at 28 °C in either YES medium or EMM2 medium, with or without thiamine, were subjected to a salt stress with 0.6 m KCl. In all cases the cells from 40 ml of culture were harvested at different times by centrifugation and washed with cold phosphate-buffered saline, and the yeast pellets were immediately frozen in liquid nitrogen and stored for subsequent analysis. To analyze dual phosphorylation of either Pmk1 or Sty1, total cell extracts were prepared under native conditions employing chilled acid-washed glass beads and lysis buffer (10% glycerol, 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40, plus specific protease and phosphatase inhibitor cocktails for fungal and yeast extracts, obtained from Sigma). After clarifying cell extracts by centrifugation at 20,000 × g for 20 min, HA6H-tagged Pmk1 or Sty1 were purified with Ni2+-nitrilotriacetic acid-agarose beads (Qiagen), as described previously (38). The purified proteins were resolved in 10% SDS-PAGE gels and transferred to Hybond-ECL membranes (GE Healthcare). Dual phosphorylation in either Pmk1 or Sty1 was detected after incubation with mouse polyclonal anti-phospho-p44/42 (Cell Signaling) or mouse monoclonal anti-phospho-p38 (Cell Signaling) antibody, respectively. Total Pmk1 and Sty1 proteins were detected after incubation with anti-HA antibody (loading control). The immunoreactive bands were revealed with an anti-mouse horseradish peroxidase-conjugated secondary antibody (Sigma) and the ECL system (Amersham Biosciences). Densitometric quantification of Western blot signals was performed using Molecular Analyst Software (Bio-Rad).
Immunoprecipitation
Extracts from 5 × 108 cells expressing the different tagged proteins were obtained using 200 ml of lysis buffer (20 mm Tris-HCl, pH 8.0, 2 mm EDTA, 100 mm NaCl, 0.5% Nonidet P-40, and 10% glycerol containing 100 mm p-aminophenyl methanesulfonyl fluoride, 2 mg ml−1 leupeptin, and 2 mg ml−1 aprotinin) (33). Cell extracts (4 mg of total protein) were incubated with the anti-GFP polyclonal antibody and protein A-Sepharose beads for 2–4 h at 4 °C. The beads were washed four times with lysis buffer and resuspended in sample buffer. The proteins were separated by SDS-PAGE, transferred to Immobilon-P membranes (Millipore), and blotted to detect the GFP- and HA-fused epitopes with the corresponding antibodies and the ECL detection kit (GE Healthcare). Some 40 μg of total protein was used for Western blot as control of the total amount of tagged protein.
Plate Assay of Stress Sensitivity for Growth
Wild type and mutant strains of S. pombe were grown in YES liquid medium to an A600 of 0.6. Appropriate dilutions were spotted per duplicate on either YES or EMM2 solid medium or in the same medium supplemented with different concentrations of MgCl2. The plates were incubated at 28 °C for 3–4 days and then photographed.
Cell Wall Resistance to β-1,3-Glucanase Treatment
Strains were grown in YES medium to an A600 of 0.6; washed with 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm β-mercaptoethanol; and incubated with vigorous shaking at 30 °C in the same buffer supplemented with 20 μg/ml of Zymolyase 100T (Seikagaku Corporation). Samples were taken every 15 min, and cell lysis was monitored by measuring A600 decay.
Microscopy
To measure cell length and width at division, the yeast strains were grown in EMM2 medium to an A600 of 0.5, stained with Calcofluor white (39) and observed under fluorescence microscopy. A minimum of 100 septated cells was scored for each mutant. To determine the percentage of multiseptated cells, the number of septated cells scored was ≥400 in each case. The images were captured with a Leica DM 4000B fluorescence microscope coupled to a cooled Leica DC 300F camera and IM50 software.
Reproducibility of Results
All of the experiments were repeated at least three times with similar results. Representative results are shown.
RESULTS
Rga4 Is a Rho2 GAP
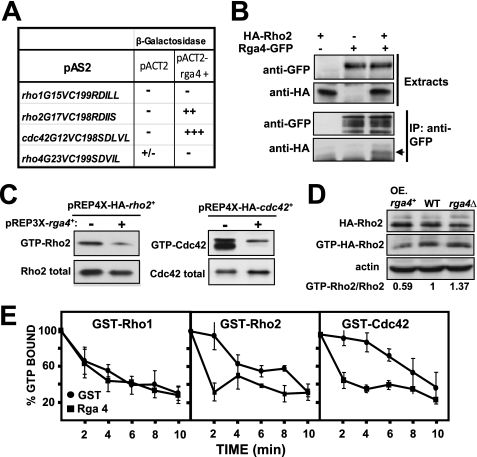
It has been recently described that Rga4 is a GAP for Cdc42 GTPase without apparent activity on other GTPases like Rho1 and Rho3 (28). Indeed, in a yeast two-hybrid assay, we observed a strong interaction between Rga4 and Cdc42G12V, a constitutively active GTP-locked version of Cdc42, as estimated by a colony filter assay for β-galactosidase activity (Fig. 1A). In the same assay neither Rho1 nor Rho4 GTPases interacted with Rga4 but, notably, a GTP-locked version of Rho2 GTPase consistently showed a reproducible interaction with Rga4 (Fig. 1A). To further characterize the Rga4-Rho2 association, we next performed coimmunoprecipitation analysis by constructing S. pombe strains in which the chromosomal rho2+ and rga4+ genes were fused at their 5′ (rho2+) and 3′ (rga4+) ends with HA or GFP epitopes, respectively. The phenotypes of the resulting strains were indistinguishable from those expressing untagged Rho2 or Rga4 (not shown). As illustrated in Fig. 1B, Rga4-GFP immunoprecipitation resulted in the detection of HA-Rho2 in cell extracts of strain PPG7114 (Rga4-GFP and HA-Rho2) but not in strain PPG4546 expressing HA-Rho2 (negative control), thus supporting the existence of a Rga4-Rho2 interaction in vivo. The levels of GTP-bound Rho2 and Cdc42 were similarly reduced by overexpression of Rga4 (Fig. 1C). Thus, we next performed pull-down experiments to determine the amount of endogenous GTP-bound Rho2 in wild type, rga4Δ, and rga4+-overexpressing cells harboring a genomic version of HA-Rho2. Fig. 1D shows that the amount of GTP-bound Rho2 was significantly reduced in rga4+-overexpressing cells and that the binding increased in the rga4Δ background. Finally, we performed an in vitro assay to corroborate the in vivo Rho2 GAP activity of Rga4. To this end, the GTPase activity of affinity-purified GST-Rho2, GST-Cdc42 (positive control), and GST-Rho1 (negative control) was measured in the presence of GST-Rga4 or GST alone. As can be seen in Fig. 1E, Rga4 increased significantly the rate of hydrolysis of GTP by Cdc42 and Rho2, but not by Rho1. Taken together, these results indicate that Rga4 is a Rho2 GAP that negatively regulates Rho2 activity in fission yeast.
FIGURE 1.
Rga4 is Rho2 GAP. A, two-hybrid analysis of the interactions between different Rho GTPases (pAS2), and Rga4 (pACT2) used as bait. +++, very strong interaction; ++, strong interaction; +/−, weak interaction; −, no interaction. B, coimmunoprecipitation analysis of Rga4 and Rho2. Extracts from cells expressing either HA-Rho2 (strain PPG4546), Rga4-GFP (strain PPG1811), or both HA-Rho2 and Rga4-GFP (strain PPG7114) were immunoprecipitated with anti-GFP and blotted against anti-GFP or anti-HA antibodies (lower panels). C, extracts from cells transformed with pREP3X-rga4+ and pREP4X-HA-rho2+ or pREP4X-HA-cdc42+ were precipitated with GST-C21RBD (HA-Rho2) or GST-CRIB (HA-Cdc42) and blotted against anti-HA antibody (upper panel). Total HA-Rho proteins in the extracts were visualized by Western blot using anti-HA antibody (lower panel). D, effect of Rga4 on Rho2 GTPase. Cell extracts from wild type (WT) or from cells expressing HA-Rho2 and lacking or overexpressing (OE) rga4+ were precipitated with GST-C21RBD and blotted against anti-HA antibody (middle panel). Total HA-Rho2 was estimated by Western blot employing anti-HA antibody (top panel), and actin was detected in the same extracts as loading control (bottom panel). Quantification of the ratio GTP-bound versus total Rho2 is shown. E, Rga4 stimulates Rho2 and Cdc42 GTPase activity in vitro. Purified GST-Rho1, GST-Rho2, and GST-Cdc42 were preloaded with [γ-32P]GTP, as described under “Experimental Procedures,” followed by addition of GST (closed circles) or GST-Rga4 (closed squares). The aliquots were removed at different times, and the amount of radioactivity bound to the protein was determined. The values are the means of three independent experiments, and the error bars represent standard deviations.
Rga4 Negatively Regulates Pmk1 Activity Acting as a Rho2 GAP
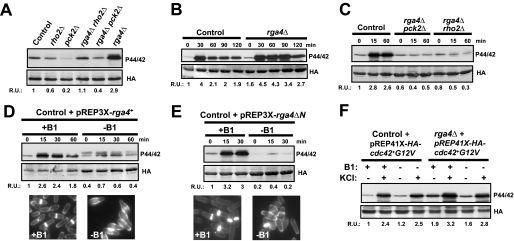
Rho2 and its downstream target Pck2 (protein kinase C ortholog) are members of the fission yeast MAPK cell integrity pathway (13, 15), with a positive role in both the basal and the induced activation of MAPK Pmk1 under specific triggering stimuli such as osmostress and hypotonic shock (15). After identifying Rga4 as a Rho2 GAP, we next investigated its putative function as negative regulator of the above pathway by measuring with a p44/42 antibody (14) the basal Pmk1 activity in control, rho2Δ, pck2Δ, rga4Δ, rga4Δ rho2Δ, and rga4Δ pck2Δ cells that express a chromosomal HA6H-tagged version of Pmk1. As shown in Fig. 2A and congruent with our earlier report (15), Pmk1 phosphorylation was partially compromised in rho2Δ cells and even lower in the pck2Δ mutant. Importantly, rga4+ deletion elicited a significant increase in basal Pmk1 phosphorylation, which was almost completely abolished in rga4Δ rho2Δ and rga4Δ pck2Δ cells (Fig. 2A). Moreover, we have previously shown that Rho2p and/or Pck2p are critical for the quick and transient increase in Pmk1 phosphorylation observed when S. pombe cells are under osmostress (15). As shown in Fig. 2B, rga4Δ cells displayed a marked increase in Pmk1 activity under saline osmostress, which was maintained as compared with control cells, and the rise was completely dependent on the presence of Rho2 and/or Pck2 (Fig. 2C). We considered that if Rga4 was a negative regulator of Pmk1 activity, its overexpression should then decrease Pmk1 phosphorylation. This prediction turned out to be correct because ectopic expression of the rga4+ gene caused a clear reduction in the level of Pmk1 phosphorylation both in growing cells and in response to hyperosmotic stress (Fig. 2D). Furthermore, we found that overexpression of the C-terminal GAP domain of Rga4 (Rga4ΔN, amino acids 645–993) was enough to strongly decrease Pmk1 activity to an extent similar to that caused by full-length rga4+ under the above conditions (Fig. 2E). In both cases, the previously described “dumpy” phenotype and reduced diameter of the growing cell tip in cells overexpressing either Rga4 or the Rga4 GAP domain (27) were evident (Figs 2, D and E, lower panels), thus confirming the full functionality of both constructs.
FIGURE 2.
Rga4 negatively regulates Pmk1 phosphorylation by acting as a Rho2 GAP. A, cells from strains MI200 (control), MI700 (rho2Δ), TS310 (pck2Δ), TS309 (rga4Δ), TS311 (rga4Δ rho2Δ), and TS312 (rga4Δ pck2Δ), carrying a HA6H-tagged chromosomal version of pmk1+, were grown in YES medium to mid-log phase and Pmk1-HA6H was purified by affinity chromatography. Activated Pmk1 was detected by immunoblotting with anti-(phosho-p44/42) antibody, and total Pmk1 was detected with anti-HA antibody as loading control. B, strains MI200 (control) and TS309 (rga4Δ) were grown as above and treated with 0.6 m KCl. The aliquots were harvested at the times indicated, Pmk1 was purified by affinity chromatography, and activated and total Pmk1 was detected as indicated before. C, strains MI200 (control), TS312 (rga4Δ pck2Δ), and TS311 (rga4Δ rho2Δ) were grown in YES medium to mid-log phase and treated with 0.6 m KCl. At different times Pmk1-HA6H was purified, and the activated and total Pmk1 was detected as described above. D, strain MI200 transformed with pREP3X-Rga4 plasmid was grown for 18 h in the presence (+B1) or absence (−B1) of thiamine and treated with 0.6 m KCl. At timed intervals Pmk1-HA6H was purified, and the activated and total Pmk1 was detected as already indicated. Cell morphology was analyzed by fluorescence microscopy after staining cells with Calcofluor white. E, strain MI200 transformed with pREP3X-Rga4ΔN (expressing the GAP domain of Rga4) was grown for 18 h in the presence (+B1) or absence (−B1) of thiamine and treated with 0.6 m KCl. At different times Pmk1-HA6H was purified, and the activated and total Pmk1 was detected as described above. Cell morphology was analyzed by staining with Calcofluor white. F, strains MI200 and TS309 were transformed with plasmid pREP41-Cdc42(G12V)-Ha6H and grown in EMM2 medium with or without thiamine in the presence or absence of 0.6 m KCl for 15 min. The purification and detection of active or total Pmk1-HA6H was performed as described above. R.U., relative units.
We have previously described that the Cdc42-Pak1/Pak2 cascade does not regulate Pmk1 activity in fission yeast (14). Interestingly, the above results indicate that Rga4 is a GAP for both Cdc42 and Rho2, suggesting that some type of cross-talk might exist between Cdc42 and the Pmk1 pathway. However, the basal and salt-induced levels of phosphorylated Pmk1 in rga4Δ cells were still higher than in control cells, when in both cases the cells expressed a constitutively active allele of cdc42+ (Cdc42-G12V), which is insensitive to regulation by Rga4 (Fig. 2F). Altogether, these results strongly support that Rho2 is a specific target for Rga4 to negatively regulate Pmk1 activity in fission yeast and that the GAP domain of Rga4 is responsible for this control.
Pom1 Kinase Regulates Pmk1 Activity Independently of Rga4-Rho2 Function
Pom1 is a member of the DYRK family of Ser/Thr protein kinases that physically interact with Rga4 and regulate its phosphorylation status (28). Following the demonstration that Rga4 negatively regulates Pmk1 activity by acting as a Rho2 GAP, we next analyzed the possible role for Pom1 as a regulator of the cell integrity pathway through modulation of Rga4 function. Fig. 3A indicates that, in comparison with control cells, Pmk1 basal phosphorylation increased in pom1Δ cells to an extent similar to that of rga4Δ cells, suggesting that Pom1 might act as a positive regulator for Rga4 activity via Rho2 GTPase. However, and contrary to this rationale, we found that basal Pmk1 phosphorylation was higher in pom1Δ rga4Δ cells than in single pom1Δ or rga4Δ cells (Fig. 3A), supporting the idea that Pom1 negatively regulates Pmk1 activity by an independent pathway not involving Rga4. Congruent with this model, pom1+deletion promoted a clear increase in Pmk1 phosphorylation in both rho2Δ and pck2Δ cells (Fig. 3A). In this context, the phosphorylation state of Pmk1 is critical for chloride homeostasis in fission yeast, and Pmk1 hyperactivation leads to strong sensitivity to this anion (40). As shown in Fig. 3B, cells lacking pom1+ did not grow in the presence of 0.2 m MgCl2, but the chloride sensitivity phenotype of rga4Δ cells was not as strong as in pom1Δ or in rga4Δ pom1Δ cells. Importantly, the phenotype of these mutants was rescued by additional deletion of any member of the MAPK cascade (Fig. 3B), suggesting that the Pmk1 hyperactivation observed in either pom1Δ or rga4Δ cells is physiologically relevant. Because of the putative role of Pom1 as a negative regulator of Pmk1, we further tested whether pom1+ overexpression might decrease Pmk1 phosphorylation. To this end we employed strain TS322, which expresses a genomic version of Pom1 under the regulation of the full strength thiamine-repressible promoter (41) in a Pmk1-HA6H background. After 18 h of growth in the absence of thiamine, many cells adopted a clearly branched phenotype because of Pom1 overexpression (Fig. 3C) (41). Under these conditions, pom1+ overexpression did not reduce Pmk1 phosphorylation levels in response to hyperosmotic stress, and a very low increase in basal Pmk1 phosphorylation was observed (Fig. 3C). Hence, these results further support that Pom1 modulates Pmk1 activity through a distinct pathway that is Rga4-independent.
FIGURE 3.
Pom1 kinase negatively regulates Pmk1 independently of Rga4-Rho2 function. A, cells from strains MI200 (control), MI700 (rho2Δ), TS310 (pck2Δ), TS313 (pom1Δ), TS309 (rga4Δ), TS315 (pom1Δ rga4Δ), TS316 (pom1Δ rho2Δ), TS317 (pom1Δ pck2Δ), TS311 (rga4Δ rho2Δ), and TS312 (rga4Δ pck2Δ), carrying a HA6H-tagged chromosomal version of pmk1+, were grown in YES medium to mid-log phase, and Pmk1-HA6H was purified by affinity chromatography. Activated Pmk1 was detected by immunoblotting with anti-(phosho-p44/42) antibody, and total Pmk1 was detected with anti-HA antibody as loading control. B, chloride sensitivity assays for strains MI200 (control), MI102 (pmk1Δ), TS309 (rga4Δ), TS313 (pom1Δ), TS315 (pom1Δ rga4Δ), TS320 (pmk1Δ rga4Δ), and TS321 (pmk1Δ pom1Δ). After growth in YES medium, 104, 103, or 102 cells were spotted onto YES plates supplemented with either 0.15 or 0.2 m MgCl2 and incubated for 3 days at 28 °C before being photographed. C, strain TS322 (nmt1-pom1+) was grown for 24 h in the presence (+B1) or absence (−B1) of thiamine and treated with 0.6 m KCl. At different times Pmk1-HA6H was purified, and the activated and total Pmk1 was detected as described above. Cell morphology was analyzed by staining with Calcofluor white. D, strains JM1521 (sty1-HA6H, control), TS318 (sty1-HA6H, rga4Δ), and TS319 (sty1-HA6H, pom1Δ) were grown in YES medium to mid-log phase and treated with 0.6 m KCl. At timed intervals Sty1-HA6H was purified by affinity chromatography, and the activated and total Sty1 was detected by immunoblotting with anti-p38 or anti-HA antibodies, respectively. R.U., relative units.
Finally, we examined the involvement of Rga4 and/or Pom1 in the regulation of the activation of the stress-activated protein kinase pathway by analyzing the phosphorylation status of Sty1 MAPK, which is the key element of the pathway (42, 43). This study included the analysis of control, rga4Δ, and pom1Δ cells expressing a genomic copy of Sty1 tagged with the HA6H epitope, during exponential growth, and under a salt stress with KCl. The Sty1-HA6H fusion was purified by affinity chromatography. and its activation was assessed by Western immunoblotting with anti-phospho-p38 antibodies (38). As shown in Fig. 3D, neither the basal level nor the salt induced activation of Sty1 were significantly affected by rga4+ or pom1+deletion, suggesting that Rga4 and Pom1 are specific regulators of the cell integrity MAPK pathway.
Pmk1 Activity Is Not Involved in the Rga4-dependent Control of Cell Shape
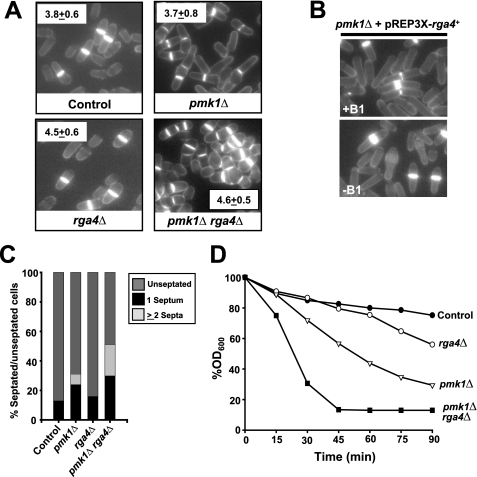
Recently, it has been proposed that Rga4 is involved in the control of cell diameter and symmetry in fission yeast by acting as a GAP for Cdc42 (28). The less polarized distribution of active Cdc42 in rga4Δ cells might be the cause whereby these cells are shorter and larger in diameter at division as compared with control cells (27, 28). However, our finding that Rga4 also functions as a Rho2 GAP in vivo raised the question of whether any of the morphological defects associated with Rga4 deletion might be due, at least in part, to Pmk1 hyperactivation. To tackle this possibility we performed a comparative study of the cell length and diameter at the time of division in control, pmk1Δ, rga4Δ, and pmk1Δ rga4Δ cells. As described earlier (22, 27, 28), rga4Δ cells were 15% shorter and 20% wider than control cells (Fig. 4, A and B). Importantly, the cell width of rga4Δ cells was not significantly altered in pmk1Δ rga4Δ double mutants (Fig. 4A). Because, as indicated above, Pmk1 is hyperphosphorylated in rga4Δ cells (Fig. 2A), these results strongly suggest that the role of Rga4 in the regulation of cell diameter and symmetry in fission yeast is unrelated to its function as a Rho2 GAP regulating the activity of Pmk1. Consistent with this observation, rga4+ overexpression in pmk1Δ cells induced a reduction in the diameter of the growing cell tip similar to that described in control cells (Fig. 4B).
FIGURE 4.
Rga4-dependent control of cell shape is independent on Pmk1 activity. A, strains MI200 (control), TS309 (rga4Δ), MI102 (pmk1Δ), and TS320 (pmk1Δ rga4Δ) were grown in YES medium to mid-log phase, and cell width at division was determined by fluorescence microscopy after staining the cells (n > 200) with Calcofluor white. B, strain MI102 (pmk1Δ) transformed with pREP3X-Rga4 was grown for 18 h in the presence (+B1) or absence (−B1) of thiamine, and cell morphology was analyzed by after staining with Calcofluor white. C, percentage of septated/unseptated cells in cultures of strains described in A. (n > 400). D, the same strains were grown in YES medium (A600 = 0.5) and assayed for β-glucanase sensitivity. Cell lysis was monitored by measuring decay in A600 at different incubation periods. The results shown are the mean values of three independent experiments.
Rga4 Positively Regulates Cell Wall Integrity and Cell Separation
Cell separation is partially defective in pmk1Δ cells and causes a modest multiseptate phenotype with ∼7% of septated cells containing two or three septa (Fig. 4, A and C;) (14, 16, 17), whereas rga4Δ cells do not show any evident cell separation defect. Interestingly, pmk1Δ rga4Δ double mutant cells displayed an exacerbated multiseptate phenotype (∼20% multiseptated cells; Fig. 4, A and C) as compared with pmk1Δ cells. Another feature of pmk1+-deleted cells is their sensitivity to digestion with β-glucanase, which is indicative of structural cell wall defects (16, 17). The sensitivity to β-glucanase in rga4Δ pmk1Δ double mutant cells was higher than in rga4Δ or pmk1Δ cells (Fig. 4D). Likely, the increased defect in cell wall integrity in pmk1Δ rga4Δ cells is responsible for their different overall shape as compared with rga4Δ cells, although both mutants display a similar cell width. As a whole, these results suggest that Rga4 positively regulates cell wall integrity and cell separation independently of the Pmk1 pathway, probably by acting as a Cdc42 GAP.
Rga7 Is a Rho2 GAP That Negatively Regulates Pmk1 Activity in Addition to Rga4
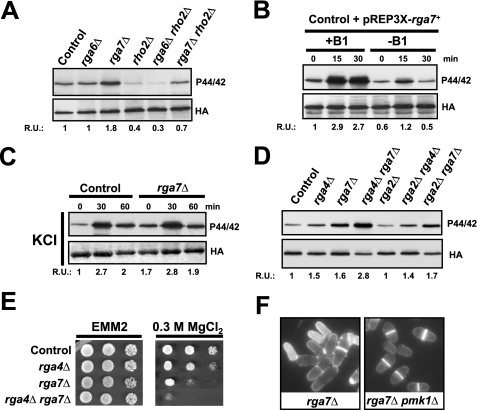
Based on in vivo measurements of Rho activity, we have previously described that Rga6 and Rga7 are Rho2 GAPs (26). We thus explored their putative roles as negative regulators of the Pmk1 MAPK pathway. Whereas the phosphorylation status of Pmk1 was unaffected by rga6+ deletion, a clear increase in MAPK phosphorylation was detected in extracts from rga7Δ cells as compared with control cells (Fig. 5A). Notably, this increase was mainly dependent on the presence of Rho2 (Fig. 5A). Similar to rga4+, rga7+ overexpression decreased both the basal and the salt stress-induced phosphorylation of Pmk1 (Fig. 5B). However, the kinetics of Pmk1 deactivation in rga7+-deleted cells during saline stress was very similar to that of control cells (Fig. 5C), supporting the possibility that Rga7 negatively regulates the Rho2-Pmk1 cascade during vegetative growth but not under stress. We performed a comparative analysis of basal Pmk1 phosphorylation in growing cultures of wild type cells versus rga2Δ, rga4Δ, rga7Δ, rga2Δ rga4Δ, rga2Δ rga7Δ, and rga4Δ rga7Δ mutants. Rga2 null mutants do not display an obvious increase in basal Pmk1 phosphorylation (26) (Fig. 5D). As a result, MAPK hyperphosphorylation in either rga4Δ or rga7Δ cells was not further enhanced by simultaneous deletion of rga2+ (Fig. 5D). Nevertheless, Pmk1 phosphorylation was higher in rga4Δ rga7Δ cells than in rga4Δ or rga7Δ single mutants, strongly suggesting that both Rga4 and Rga7 negatively regulate the Rho2-Pmk1 pathway in an additive, independent fashion. Further support for this hypothesis came from the fact that the chloride sensitivity phenotype of rga4Δ rga7Δ cells was higher than that of rga4Δ or rga7Δ single mutants (Fig. 5E). The presence of shrunken, lysed cells is a typical feature of rga7Δ cells (22) (Fig. 5F). However, this phenotype was rescued by simultaneous deletion of the pmk1+ gene (Fig. 5F), indicating that Pmk1 hyperactivation promotes cell lysis in rga7Δ cells.
FIGURE 5.
Rga7 is a Rho2 GAP that negatively regulates Pmk1 activity. A, cells from strains MI200 (control), MI700 (rho2Δ), TS307 (rga6Δ), TS308 (rga7Δ), TS323 (rga6Δ rho2Δ), and TS324 (rga7Δ rho2Δ), were grown in YES medium to mid-log phase, and Pmk1-HA6H was purified by affinity chromatography. Activated Pmk1 was detected by immunoblotting with anti-(phosho-p44/42) antibody, and total Pmk1 was detected with anti-HA antibody as loading control. B, strain MI200 transformed with pREP3X-Rga7 plasmid was grown for 18 h in the presence (+B1) or absence (−B1) of thiamine and treated with 0.6 m KCl. At timed intervals Pmk1-HA6H was purified, and the activated and total Pmk1 was detected as above. C, growing cultures of strains MI200 and TS308 (rga7Δ) were treated with 0.6 m KCl. The aliquots were harvested at the times indicated, Pmk1 was purified by affinity chromatography, and activated and total Pmk1 was detected as indicated before. D, cells from strains MI200 (control), TS309 (rga4Δ), TS308 (rga7Δ), TS328 (rga4Δ rga7Δ), PPG4460 (rga2Δ), TS326 (rga2Δ rga4Δ), and TS327 (rga2Δ rga7Δ), carrying a HA6H-tagged chromosomal version of pmk1+, were grown to mid-log phase, and Pmk1-HA6H was purified by affinity chromatography. Activated and total Pmk1 was detected as above. E, chloride sensitivity assays for strains MI200 (control), TS309 (rga4Δ), TS308 (rga7Δ), and TS321 (rga4Δ rga7Δ). 104, 103, 102, or 10 cells were spotted onto EMM2 plates supplemented with 0.3 m MgCl2 and incubated for 4 days at 28 °C before being photographed. F, cell morphology was analyzed in strains TS308 (rga7Δ) and TS325 (rga7Δ pmk1Δ) after staining with Calcofluor white. R.U., relative units.
DISCUSSION
In eukaryote organisms, and fission yeast is not an exception, the number of Rho GAPs is higher than that of Rho GTPases (2). However, it is widely accepted that most GAPs display activity toward various Rho proteins (2). Rga4 is a Rho GAP that has been recently demonstrated to negatively regulate Cdc42, an essential Rho-type GTPase involved in the control of cell diameter and symmetry of fission yeast (28). In this work we provide strong genetic and biochemical evidence to demonstrate that Rga4 also regulates negatively the activity of the Pmk1 MAPK cell integrity pathway by acting as a GAP for Rho2 GTPase, which acts upstream of the protein kinase C ortholog Pck2 and modulates Pmk1 activation in response to several environmental stimuli (13, 15). Two-hybrid and coimmunoprecipitation studies indicated that Rga4 interacts in vivo with Rho2, and biochemical data showed that Rga4 activity negatively modulates GTP-Rho2 levels both in vivo and in vitro. Also, confirming previous results, we found that Rga4 does not display significant interaction with other GTPases such as Rho1, Rho3, or Rho4. Hence, Rga4 appears to be a Rho GAP specific for both Cdc42 and Rho2 in fission yeast.
In a previous work, we described that another Rho2 GAP, Rga2, is involved in the activation of Pmk1 in fission yeast (26). Although rga2Δ cells did not increase basal Pmk1 phosphorylation as compared with control cells, rga2+ overexpression significantly reduced both basal and MAPK activation in response to saline osmostress (26). These and other indications led us to propose that Rga2 was not the only GAP regulating the subset of Rho2 that is part of the Pmk1 signaling pathway (26). The results obtained in this work favor this suggestion and show that Rga4 is an integral component of the above pathway. Moreover, we analyzed the putative role of Rga6 and Rga7, two Rho2 GAPs (26), on Pmk1 activity and found that Rga7 is also a negative regulator of the MAPK cascade. Importantly, and contrary to rga2+ null cells, both rga4Δ and rga7Δ cells displayed a clear increase in Pmk1 phosphorylation during growth that was largely abolished in rho2Δ or pck2Δ cells and also in cells overexpressing either Rga4, Rga7, or its GAP domains. Taken together, these data indicate that the role of Rga4 and Rga7 in the regulation of Pmk1 activity is physiologically more relevant than that of Rga2, which appears involved in the regulation of cell wall synthesis independently of the Pmk1 pathway (26) (Fig. 6). However, Rga4 and Rga7 appear to negatively regulate Pmk1 independently, because both Pmk1 phosphorylation and the chloride sensitivity phenotype were higher in rga4Δ rga7Δ cells than in rga4Δ or rga7Δ single mutants. Furthermore, and contrary to Rga7, Rga4 modulates Rho2-dependent Pmk1 activation also during stress.
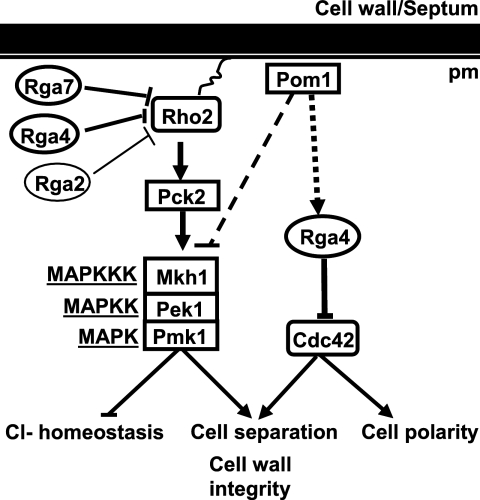
FIGURE 6.
Proposed model showing the differential roles of Rho-GAPs as negative regulators of the Pmk1 MAPK pathway.
Rho2 is a farnesylated protein localized to the plasma membrane and the cell division site (44). Rga4 is excluded from the growing areas and localizes to the cell sides and at the septum (27, 28), Rga7 localizes at both at the cytosol and septum (45), whereas Rga2 is mainly found at the cell tips and the septum (26). It seems reasonable that differential subcellular localization of both GAPs might determine which subset of membrane-bound Rho2 becomes down-regulated during S. pombe cell cycle, thus providing specificity in the signaling through the Pmk1 MAPK cascade. Alternatively, Rga4 or Rga7 might functionally replace Rga2 for its activity on the Pmk1 pathway.
Pom1 is recruited to cell ends of the fission yeast by the Tea1–Tea4·Wsh3 complex and becomes essential for proper localization of Rga4, which ensures bipolar localization of GTP-bound, active Cdc42 (28). Recently, it has also been described that Pom1 gradients from the cell ends play a key role in the control of the progression of the cell cycle by coupling cell length to G2/M transition (46, 47). The results obtained in this work show that Pom1 kinase is also a negative regulator of the Pmk1 pathway, as demonstrated by the increased basal Pmk1 phosphorylation in pom1Δ growing cells. However, this control is not dependent on the Rga4-Rho2-Pck2 branch, because Pmk1 hyperactivation in pom1Δ cells was not abolished by simultaneous deletion of rho2+ or pck2+. Importantly, both Pmk1 basal phosphorylation and the growth sensitivity in the presence of chloride were higher in rga4Δ pom1Δ cells than in single rga4Δ and pom1Δ mutants, indicating that Rga4 and Pom1 negatively regulate Pmk1 activity through independent pathways (Fig. 6). Supporting this hypothesis, overexpression of the GAP domain of Rga4, which does not interact with Pom1 (28), decreased Pmk1 activity to an extent similar to full-length Rga4. Nevertheless, the identity of the putative targets of Pom1 in this control remains unknown.
Rga4, acting as a GAP for Cdc42, plays a critical role in the establishment of growth polarity in fission yeast (28). However, Rho2 and the Pmk1 cell integrity pathway are not involved in this control, despite our findings showing that Rga4 is also a GAP for Rho2. Remarkably, our results also reveal a positive role for Rga4, independent of Pmk1, in the control of cell wall integrity and cell separation, because both cell wall sensitivity against β-glucanase and multiseptation were exacerbated in rga4Δ pmk1Δ cells as compared with pmk1Δ cells. Although we have not demonstrated that this role of Rga4 is mediated through Cdc42 regulation, it appears that in fission yeast there are two subsets of Rga4 that negatively regulate Cdc42 and Rho2 GTPases in an independent way (Fig. 6). The fact that Pom1 negatively regulates Pmk1 phosphorylation in the absence of Rho2 would favor this interpretation.
In conclusion, the results presented in this work draw a complex scenario, similar to that of S. cerevisiae and mammalian cells, in which a given Rho GAP might act on different Rho GTPases and different GAPs might regulate the same Rho GTPase (1, 2, 4). The overabundance and promiscuity of Rho GAPs suggest that they may selectively regulate specific Rho GTPases functions. Unfortunately, little is known about their spatial and temporal regulation and on the factors that control their specificity toward Rho, an issue of paramount importance that should guide future studies.
Acknowledgments
We thank J. B. Millar (University of Warwick, Warwick, United Kingdom) and P. Nurse (Rockefeller University) for kind supply of yeast strains and F. Garro for technical assistance.
This work was supported by Grants BFU2007-60675 from Ministerio de Cienicia e Innovación and GR231 from Junta de Castilla y León, Spain (to P. P.) and Grants BFU2008-01653 from Ministerio de Cienicia e Innovación and 08725/PI/08 from Fundación Séneca (Región de Murcia), Spain (to J. C.).
- GAP
- GTPase-activating protein
- EMM2
- Edinburgh minimal medium
- HA6H
- epitope comprising hemagglutinin antigen plus six histidine residues
- MAPK
- mitogen-activated protein kinase
- YES
- yeast extract plus supplements
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein.
REFERENCES
- 1.Moon S. Y., Zheng Y. (2003) Trends Cell Biol. 13, 13–22 [DOI] [PubMed] [Google Scholar]
- 2.Tcherkezian J., Lamarche-Vane N. (2007) Biol. Cell 99, 67–86 [DOI] [PubMed] [Google Scholar]
- 3.Park H. O., Bi E. (2007) Microbiol. Mol. Biol. Rev. 71, 48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt A., Schmelzle T., Hall M. N. (2002) Mol. Microbiol. 45, 1433–1441 [DOI] [PubMed] [Google Scholar]
- 5.Bernards A. (2003) Biochim. Biophys. Acta 1603, 47–82 [DOI] [PubMed] [Google Scholar]
- 6.Arellano M., Durán A., Pérez P. (1996) EMBO J. 15, 4584–4591 [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano K., Arai R., Mabuchi I. (1997) Genes Cells 2, 679–694 [DOI] [PubMed] [Google Scholar]
- 8.Miller P. J., Johnson D. I. (1994) Mol. Cell. Biol. 14, 1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin S. G., Rincón S. A., Basu R., Pérez P., Chang F. (2007) Mol. Biol. Cell 18, 4155–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rincón S. A., Ye Y., Villar-Tajadura M. A., Santos B., Martin S. G., Pérez P. (2009) Mol. Biol. Cell 20, 4390–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calonge T. M., Nakano K., Arellano M., Arai R., Katayama S., Toda T., Mabuchi I., Perez P. (2000) Mol. Biol. Cell 11, 4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama S., Hirata D., Arellano M., Pérez P., Toda T. (1999) J. Cell Biol. 144, 1173–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y., Kuno T., Kita A., Asayama Y., Sugiura R. (2006) Mol. Biol. Cell 17, 5028–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madrid M., Soto T., Khong H. K., Franco A., Vicente J., Pérez P., Gacto M., Cansado J. (2006) J. Biol. Chem. 281, 2033–2043 [DOI] [PubMed] [Google Scholar]
- 15.Barba G., Soto T., Madrid M., Núñez A., Vicente J., Gacto M., Cansado J. (2008) Cell Signal. 20, 748–757 [DOI] [PubMed] [Google Scholar]
- 16.Toda T., Dhut S., Superti-Furga G., Gotoh Y., Nishida E., Sugiura R., Kuno T. (1996) Mol. Cell. Biol. 16, 6752–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaitsevskaya-Carter T., Cooper J. A. (1997) EMBO J. 16, 1318–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bone N., Millar J. B., Toda T., Armstrong J. (1998) Curr. Biol. 8, 135–144 [DOI] [PubMed] [Google Scholar]
- 19.Sugiura R., Toda T., Dhut S., Shuntoh H., Kuno T. (1999) Nature 399, 479–483 [DOI] [PubMed] [Google Scholar]
- 20.Loewith R., Hubberstey A., Young D. (2000) J. Cell Sci. 113, 153–160 [DOI] [PubMed] [Google Scholar]
- 21.Sengar A. S., Markley N. A., Marini N. J., Young D. (1997) Mol. Cell Biol. 17, 3508–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano K., Mutoh T., Mabuchi I. (2001) Genes Cells 6, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 23.Calonge T. M., Arellano M., Coll P. M., Perez P. (2003) Mol. Microbiol. 47, 507–518 [DOI] [PubMed] [Google Scholar]
- 24.Yang P., Qyang Y., Bartholomeusz G., Zhou X., Marcus S. (2003) J. Biol. Chem. 278, 48821–48830 [DOI] [PubMed] [Google Scholar]
- 25.Ottilie S., Miller P. J., Johnson D. I., Creasy C. L., Sells M. A., Bagrodia S., Forsburg S. L., Chernoff J. (1995) EMBO J. 14, 5908–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villar-Tajadura M. A., Coll P. M., Madrid M., Cansado J., Santos B., Pérez P. (2008) Mol. Microbiol. 70, 867–881 [DOI] [PubMed] [Google Scholar]
- 27.Das M., Wiley D. J., Medina S., Vincent H. A., Larrea M., Oriolo A., Verde F. (2007) Mol. Biol. Cell 18, 2090–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatebe H., Nakano K., Maximo R., Shiozaki K. (2008) Curr. Biol. 18, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno S., Klar A., Nurse P. (1991) Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 30.Soto T., Beltrán F. F., Paredes V., Madrid M., Millar J. B., Vicente-Soler J., Cansado J., Gacto M. (2002) Eur. J. Biochem. 269, 5056–5065 [DOI] [PubMed] [Google Scholar]
- 31.Forsburg S. L., Sherman D. A. (1997) Gene 191, 191–195 [DOI] [PubMed] [Google Scholar]
- 32.Durfee T., Becherer K., Chen P. L., Yeh S. H., Yang Y., Kilburn A. E., Lee W. H., Elledge S. J. (1993) Genes Dev. 7, 555–569 [DOI] [PubMed] [Google Scholar]
- 33.Arellano M., Valdivieso M. H., Calonge T. M., Coll P. M., Duran A., Perez P. (1999) J. Cell Sci. 112, 3569–3578 [DOI] [PubMed] [Google Scholar]
- 34.Coll P. M., Trillo Y., Ametzazurra A., Perez P. (2003) Mol. Biol. Cell 14, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craven R. A., Griffiths D. J., Sheldrick K. S., Randall R. E., Hagan I. M., Carr A. M. (1998) Gene 221, 59–68 [DOI] [PubMed] [Google Scholar]
- 36.Self A. J., Hall A. (1995) Methods Enzymol. 256, 67–76 [DOI] [PubMed] [Google Scholar]
- 37.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 38.Madrid M., Núñez A., Soto T., Vicente-Soler J., Gacto M., Cansado J. (2007) Mol. Biol. Cell 18, 4405–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfa C., Fantes P., Hyams J., Mcleod M., Warbrick E. (1993) Experiments with Fission Yeast, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 40.Sugiura R., Toda T., Shuntoh H., Yanagida M., Kuno T. (1998) EMBO J. 17, 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bähler J., Nurse P. (2001) EMBO J. 20, 1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiozaki K., Russell P. (1995) Nature 378, 739–743 [DOI] [PubMed] [Google Scholar]
- 43.Millar J. B., Buck V., Wilkinson M. G. (1995) Genes Dev. 9, 2117–21130 [DOI] [PubMed] [Google Scholar]
- 44.Hirata D., Nakano K., Fukui M., Takenaka H., Miyakawa T., Mabuchi I. (1998) J. Cell Sci. 111, 149–159 [DOI] [PubMed] [Google Scholar]
- 45.Matsuyama A., Arai R., Yashiroda Y., Shirai A., Kamata A., Sekido S., Kobayashi Y., Hashimoto A., Hamamoto M., Hiraoka Y., Horinouchi S., Yoshida M. (2006) Nat. Biotechnol. 24, 841–847 [DOI] [PubMed] [Google Scholar]
- 46.Martin S. G., Berthelot-Grosjean M. (2009) Nature 459, 852–856 [DOI] [PubMed] [Google Scholar]
- 47.Moseley J. B., Mayeux A., Paoletti A., Nurse P. (2009) Nature 459, 857–860 [DOI] [PubMed] [Google Scholar]