Abstract
The plant growth-repressing DELLA proteins (DELLAs) are known to represent a convergence point in integration of multiple developmental and environmental signals in planta, one of which is hormone gibberellic acid (GA). Binding of the liganded GA receptor (GID1/GA) to the N-terminal domain of DELLAs is required for GA-induced degradation of DELLAs via the ubiquitin-proteasome pathway, thus derepressing plant growth. However, the conformational changes of DELLAs upon binding to GID1/GA, which are the key to understanding the precise mechanism of GID1/GA-mediated degradation of DELLAs, remain unclear. Using biophysical, biochemical, and bioinformatics approaches, we demonstrated for the first time that the unbound N-terminal domains of DELLAs are intrinsically unstructured proteins under physiological conditions. Within the intrinsically disordered N-terminal domain of DELLAs, we have identified several molecular recognition features, sequences known to undergo disorder-to-order transitions upon binding to interacting proteins in intrinsically unstructured proteins. In accordance with the molecular recognition feature analyses, we have observed the binding-induced folding of N-terminal domains of DELLAs upon interaction with AtGID1/GA. Our results also indicate that DELLA proteins can be divided into two subgroups in terms of their molecular compactness and their interactions with monoclonal antibodies.
Keywords: Organisms/Plant, Protein/Protein-Protein Interactions, Protein/Folding, Signal Transduction, Biophysics, DELLA Proteins, GA Signaling, GID1 Receptor, Binding-induced Folding, Intrinsically Unstructured Proteins
Introduction
DELLA proteins (DELLAs),3 named after a highly conserved DELLA amino acid motif in their N-terminal domains, function as negative regulators to repress gibberellin (GA)-responsive plant growth and are the key regulatory targets in the GA signaling pathway (1). The Arabidopsis thaliana genome encodes five DELLAs: AtGAI, AtRGA, AtRGL1, AtRGL2, and AtRGL3. Monocot plants, such as rice, wheat, and barley, have each a single DELLA protein, SLR1, RHT1, and SLN1, respectively. The functions of DELLAs in repressing GA responses are highly conserved between dicots and monocots (2). DELLAs have a unique N-terminal domain that is the receiver of GA signal and contains conserved DELLA and VHYNP motifs. DELLAs also have a highly conserved C-terminal domain that includes several conserved motifs. The features of these motifs as well as genetic evidence have led to the proposal that DELLAs are putative transcriptional repressors regulating GA responses (1–3). DELLAs have been demonstrated recently to act as co-repressors or co-activators by interacting with other transcription factors to regulate GA-responsive gene expression (4, 5).
Other important components of the GA signaling pathway are the rice GA receptor GID1 (6) and its three homologous genes in A. thaliana (AtGID1a, AtGID1b, and AtGID1c) (7). Crystal structure has revealed that the bioactive GA3- or GA4-bound AtGID1a provides a binding interface required for specific interactions with the DELLA and VHYNP motifs in the N-terminal domain of AtGAI (8). The GA signal modulates plant growth by causing degradation or deactivation of DELLAs, thereby derepressing GA-regulated genes (9). An F-box protein, referred to as GID2 (GA INSENSITIVE2) in rice and SLY1 (SLEEPY1) in A. thaliana, binds to the C-terminal domain of DELLAs once they have perceived the GA signal. This GA-dependent interaction confers specificity of the SCFGID2/SLY1 (Skp1/cullin/F-box) E3 ubiquitin ligase complex toward DELLAs, which promotes the addition of a polyubiquitin chain, thereby targeting DELLAs for their subsequent degradation by the 26 S proteasome complex in response to GAs (10). It was shown that the interaction between the C-terminal domain of AtRGA and SLY1 is enhanced by AtGID1/GA3 binding to the N-terminal domain of AtRGA. Hence, a hypothesized conformational change within the DELLA protein may occur during protein interactions (11).
DELLAs are not only the repressors of plant growth in response to GAs but also serve as integrators of signals from other growth-regulatory inputs. Multiple phytohormone and environmental signals regulate plant processes by affecting the GA-dependent degradation of DELLAs (12, 13). Global microarray analysis has shown that DELLAs control the expression of many genes during seed germination and floral development, including genes involved in stress and disease responses (14, 15).
In recent years, intrinsically unstructured proteins (IUPs) have been found to carry out a variety of important biological functions under physiological conditions, such as signaling and molecular recognition (16, 17, 63). Bioinformatics studies have predicted that about 23% of 28,564 sequences from the A. thaliana genome are mostly disordered, and more than 50% of eukaryotic proteins and 70% of signaling proteins contain long disordered regions (18–20). Most well studied IUPs are involved in molecular recognition during signaling events. Molecular recognition features (MoRFs) are short fragments within IUPs, which are responsible for molecular recognition. MoRFs undergo disorder-to-order transitions upon binding to their interacting partners (21–25). Although the crystal structure of the AtGID1a-AtGAIn complex has shown that the DELLA and VHYNP motifs are ordered in the bound form (8), no data are available for the DELLAs in the unbound (free) form. Thus, there remains a gap in understanding the putative conformational changes that occur during DELLA-GID1 interactions.
Using biophysical, biochemical, and bioinformatic analyses, we have observed, from experiments on DELLAs in both unbound and bound form, that unbound N-domains of DELLAs are IUPs under physiological conditions and that the conserved DELLA and VHYNP motifs in the N-domains of DELLAs act as MoRFs in seeding the DELLA-AtGID1 interactions. Our work provides the key insight into the structure of unbound N-domains of DELLAs and the mechanism by which DELLAs bind to liganded GA receptors.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Recombinant N-domains of DELLA Proteins, AtGID1a, and AtGID1a-AtRGL2n Complex
The N-domains of DELLA proteins were prepared as described previously (26). Recombinant AtGID1a with a C-terminal His tag was prepared using the same protocol. To make a AtGID1a-AtRGL2n complex, the purified maltose-binding protein-fused AtGID1a was incubated with equal molar AtRGL2n in Tris buffer containing 0.1 mm GA3 for 8 h at 4 °C before adding recombinant tobacco etch virus protease (1:100 w/w). The mixture was shaken overnight to cleave maltose-binding protein fusion tag and centrifuged for 15 min at 30,000 × g to remove aggregated uncomplexed AtGID1a. The supernatant was subjected to maltose-binding protein affinity (MBPTrap HP, GE Healthcare), His tag (HiTrap Chelating HP, GE Healthcare), anion exchange (HiTrap DEAE FF, GE Healthcare) and gel filtration (Superdex75 16/60, Amersham Biosciences) chromatography. The Tris buffer used for various separations contained 0.1 mm GA3. The purified proteins were analyzed on both 10% native and 12.5% SDS-polyacrylamide gels that were stained with Coomassie Brilliant Blue R to monitor protein bands. LAS MC3000 (Fuji) was used to digitize images of protein gels.
Plant Extracts from A. thaliana ga1–3 and Quadruple-DELLA Mutants
The inflorescence tissues of A. thaliana ga1–3 mutant (GA-deficient with increased DELLA accumulation) (27) and A. thaliana quadruple-DELLA mutant (QUAD) (lacking GAI, RGA, RGL1, and RGL2) (28) were extracted in buffer (1 g of tissue/3 ml; 50 mm phosphate, pH 7.5, containing 400 mm sodium chloride, 50 μm GA3, 1 mm EDTA, 20 μm proteasome inhibitor MG132, and Roche Complete protease inhibitors). Extracts were centrifuged for 30 min at 40,000 × g to sediment the particulate material, and the supernatant was stored on ice.
Production and Specificities of Monoclonal Antibodies
Recombinant N-domains of DELLAs (26) were used to immunize BALB/c PN mice. Splenocytes from immunized mice were hybridized to myeloma cell line NS1 using electroporation and cloned by limiting dilution to monoclonality. Monoclonal antibodies (mAbs) were prepared as ascitic tumors and purified by ammonium sulfate fractionation, followed by immunoaffinity chromatography on Protein A-Sepharose (Repligen Corp.). mAb BC9 reacted with all DELLA proteins, whereas mAb AD7 reacted with AtRGL1n, AtRGL2n, and AtRGL3n but not with AtRGAn and AtGAIn and the DELLAs from monocots, such as SLR1n. mAbs AB8, BB7, and AF2 react specifically with AtRGL1n, AtRGL2n, and AtGAIn, respectively, and none of these bind in the vicinity of either the DELLA motif or VHYNP motif. Therefore, mAbs AB8, BB7, and AF2 were used as capture antibodies, and biotin-labeled mAbs BC9 and AD7 were used as detection antibodies in double antibody sandwich immunoassays.
Western Blotting of A. thaliana ga1–3 Mutant Proteins, AtRGL2n, AtGID1a-AtRGL2n complex, and AtGAIn
Plant extracts (20 μl/lane), recombinant AtRGL2n, and AtGAIn (10 ng) were separated on a 7.5% native polyacrylamide gel, and AtGID1a-AtRGL2n complex (∼12 ng) was separated on both 10% native and 12.5% SDS-polyacrylamide gels. The protein gels were then transferred to polyvinylidene difluoride membrane (Pall) overnight. The membrane was blocked with 0.5% I-block (PerkinElmer Life Sciences) in phosphate-buffered saline, pH 7.5, containing 0.1% Tween 20 (Sigma). Western blotting of the native form of recombinant AtGAIn, AtRGL2n, and DELLA proteins in the A. thaliana ga1–3 mutants was developed by incubation (2 h) with mAb AF2, BB7, AD7, and BC9. Both the native and SDS-denatured forms of the AtGID1a-AtRGL2n complex were developed by incubation (2 h) with mAb BB7 and rabbit polyclonal anti-AtGID1 antibody. The DELLA proteins and AtGID1a-AtRGL2n complex were detected by further incubation (1 h) with peroxidase-conjugated anti-mouse IgG Fc-specific antibody (Sigma; 1:50,000 dilution) for mAbs or peroxidase-conjugated goat anti-rabbit IgG (Sigma; 1:50,000 dilution) for the polyclonal antibody. Proteins were visualized using chemiluminescence reagent ECL Advance (GE Healthcare) and imaged using a LAS MC3000 imager (Fuji). Protein concentration was determined using the Qubit fluorometer as per the instrument manual (Invitrogen).
Epitope Mapping of mAb BC9 and AD7
Epitope mapping for mAb BC9 and AD7 was carried out in an ELISA format. Biotinylated peptides covering part of the DELLA motif (DELLAVLGYKVR-biotinylated K) and the VHYNP motif (SVHYNPSDLSNWVESMLSE-biotinylated K) were synthesized with replacement of each of the amino acids of the sequence with a single alanine (Mimotopes). Each biotinylated peptide was incubated (1 h) with streptavidin (10 μl/ml)-coated 96-microwell plates. mAb BC9 and AD7 (10 μg/ml phosphate-buffered saline containing 0.1% Tween 20 (PBST) plus 2% bovine serum albumin; 100 μl/well) were incubated (2 h) at room temperature. Antibody binding was detected using peroxidase goat anti-mouse IgG (Sigma) and o-phenylene diamine substrate (see below). Amino acids that formed part of the epitope for BC9 and AD7 were identified by observing no reaction or reduced reaction with the alanine-substituted peptides.
ELISAs of QUAD Extract and the N-domain of DELLA Proteins
All ELISA assays were carried out at 37 °C. The immunoassay plates were washed six times with wash buffer (PBST, 50 μm GA3, 20 μm proteasome inhibitor MG132 (Sigma), and Roche Complete protease inhibitors) between the additions of reagents. Reagent buffer was 2% bovine serum albumin in wash buffer. The peroxidase substrate used was phosphate citrate buffer, pH 5.0, containing o-phenylene diamine (40 mg) and 30% hydrogen peroxide (40 μl) per 100 ml. Peroxidase activity was stopped by the addition of 50 μl of 4 m sulfuric acid. Wells of 96-well immunoplates (Nunc; Maxisorb) were incubated (3 h) with AtRGL1n-specific mAb AB8, AtRGL2n-specific mAb BB7, and AtGAIn-specific mAb AF2 (30 μg/ml in phosphate-buffered saline; 100 μl/well) and were further blocked by incubation with 2% bovine serum albumin (Sigma) in phosphate-buffered saline. Serial dilutions of the recombinant N-domain of DELLAs in either extraction buffer or QUAD extracts (containing AtGID1 receptors) were incubated (1 h) on ice before addition to the wells. The recombinant N-domains of DELLAs, captured by their specific mAbs, were further developed by the addition of either biotinylated mAb BC9 or AD7 and detected by the addition of peroxidase-labeled streptavidin (Amersham Biosciences) and the addition of the substrate. A rabbit polyclonal anti-AtGID1 antibody (1:5000 dilution) was used to verify that the AtGID1 receptors complexed with recombinant N-domains of DELLAs had been captured by their specific mAbs. The rabbit anti-AtGID1 antibody was detected by incubation with peroxidase-labeled goat anti-rabbit antibody (Sigma) followed by the addition of peroxidase substrate. Absorbance was measured at 492 nm using a Dynal 5000 microplate absorption reader.
Hydrodynamic Analyses of N-domains of DELLA Proteins and AtGID1a-AtRGL2n Complex
Six natively folded globular proteins (IgG1 (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), myoglobin (17 kDa), and cytochrome c (12.3 kDa)) were used as standards to calibrate the Superdex 75-16/60 column. Both calibration and elution of DELLAs and the AtGID1a-AtRGL2n complex were run under the same flow rate (0.2 ml/min) and column buffer (50 mm Na2HPO4/NaH2PO4, pH 7.4, 0.5 mm EDTA, containing 0.1 mm GA3 in the case of the AtGID1a-AtRGL2n complex). Theoretical Stokes radii for natively folded standard proteins (RSNF) were calculated from the equation (62), log(RSNF) = 0.369 log(MrTheo) − 0.254, where MrTheo represents theoretical molecular weight). The experimentally determined migration rate (1000/Velution) of each standard protein was plotted against theoretical Stokes radius and theoretical molecular weight. These plots were used to determine the experimental Stokes radius (RSD) and apparent molecular weight (MrApp) of DELLAs and the complex. The theoretical Stokes radii of DELLAs and the complex in various folding states were derived from their theoretical molecular weight using the appropriate equations (24).
Far-UV CD Spectroscopy
The CD spectra were obtained using a J-720 spectrometer (Jasco). The spectra of the protein samples were measured in a 0.5-mm cell. The samples in buffer (50 mm Na2HPO4/NaH2PO4, pH 7.4, 0.5 mm EDTA) were scanned with 1 nm bandwidth, and the average of 25 scans was recorded. The resultant machine unit θ values (m°) were converted to the mean residue ellipticity [θ] using the formula, [θ] = 0.1 × θ × MRW/P × C, where θ is the machine-assigned m°, MRW is the mean residue weight (protein molecular weight/number of residues), P is path length of the cell, and C is the protein concentration (mg/ml). 2,2,2-Trifluoroethanol (TFE; >99.5% NMR grade; Sigma) was used to measure the CD spectra of TFE-induced folding. TFE was added to protein samples to give a final concentration of 10–50% (v/v). CD spectra were recorded over the temperature range 20–80 °C. The α-helical content was estimated using the equation (29), percentage of α-helix = (−[θ]222 nm + 3000)/39,000, where [θ]222 nm is the mean residue ellipticity at 222 nm.
NMR Spectroscopy
260-μl samples of 0.2 mm AtRGL2n and 0.5 mm AtRGAn in buffer (50 mm sodium phosphate, pH 6.6, 0.5 mm EDTA, and 5% D2O) were used for the NMR data collection. Protein samples were placed into the susceptibility-matched Shigemi NMR tubes (5 mm; Shigemi Inc. (Allison Park, PA)). Both samples were labeled with 13C and 15N. A 15N HSQC and a 1H-13C plane of a CBCA(CO)NH experiment were recorded at 298 K for both proteins on a Bruker Avance 700-MHz spectrometer (Fällanden, Switzerland) equipped with a cryoprobe. For AtRGAn 15N HSQC, 1024 complex points with a sweep width of 11.3 kHz and 200 complex points with a sweep width of 2.8 kHz were recorded in F2 and F1, respectively. For AtRGL2n, the corresponding parameters were 1024 complex points and a sweep width of 11.3 kHz in F1 and 128 complex points and a sweep width of 2.3 kHz in F2. Broadband 15N decoupling was applied during acquisition. The 1H-13C plane of a CBCA(CO)NH experiment was acquired for each sample using 1024 complex points with a sweep width of 8.1 kHz in F2 and 64 complex points with a sweep width of 10.2 kHz in F1. Broadband 13C decoupling was applied during acquisition. Spectra were processed with standard parameters using the Topspin software package (Bruker BioSpin). All chemical shifts were referenced to the residual water peak at 4.765 ppm.
Primary Sequence Analysis and Disorder Prediction
DELLA protein sequences were aligned using ClustalW2 with default parameter settings. In addition, manual alignment was applied to the N terminus and poly(S/T) regions. Amino acid compositional analysis was carried out by comparing the average amino acid frequencies between N-domains of DELLAs, the DisPro data base, and an ordered protein data base. The mean net charge 〈R〉 of each DELLA protein was calculated as the absolute value of the difference between the numbers of positively charged and negatively charged amino acid residues divided by the total number of amino acids. The hydrophobicity of each amino acid residue was computed by the Kyte and Doolittle approximation using tools from the EXPASY server, with a window size of 5 and normalization on a scale from 0 to 1. The mean hydrophobicity 〈H〉 is defined as the sum of the normalized hydrophobicities of all residues divided by the total number of residues minus 4. The order/disorder of DELLAs was predicted by the program PONDR (predictor of natural disordered regions) VL-XT (available on the World Wide Web). Access to PONDR® was provided by Molecular Kinetics (Indianapolis, IN). A PONDR-based cumulative distribution function (CDF) curve was plotted from the PONDR scores (20), and the CDF charge-hydrophobicity (CH) plot was presented as the distance of a DELLA protein from the order/disorder boundary of CDF plot (x coordinate; negative for disorder) and CH plot (y coordinate; positive for disorder). An α-helix-forming molecular recognition feature predictor (α-MoRFs-II) based on PONDR prediction and a large positive data set (30, 31) was used to identify potential α-MoRFs. The non-regular secondary structure was predicted using the ROSTLAB server (NORSp) at Columbia University (available on the World Wide Web) with window size of 60 and structure content cut-off of 25%. Secondary structure and solvent accessibility were predicted using the Jpred method (32) (available on the World Wide Web).
RESULTS
The N-terminal Domains of DELLA Proteins Are Intrinsically Unstructured
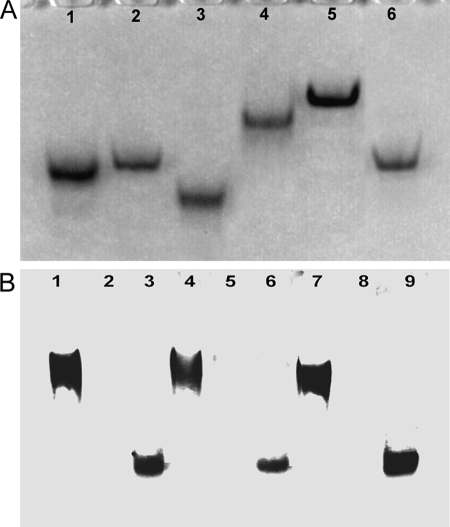
To gain insight into the biophysical and biochemical properties of unbound N-domains of DELLAs, we have expressed and purified the N-domains (n) of all five DELLAs of A. thaliana (AtRGL1n, AtRGL2n, AtRGL3n, AtGAIn, and AtRGAn), the N-domain of wheat DELLA protein (RHT1n). The domain structure and sequence alignment of DELLAs together with the conserved motifs and other sequence features are shown in Fig. 1. All of recombinant DELLA proteins expressed in E. coli are soluble, entering a native polyacrylamide gel (Fig. 2A). mAbs BC9 and AD7, interacting with the DELLA and VHYNP motifs (the GID1/GA3 binding sites of DELLAs (8)) respectively, and mAb BB7 have been used for native Western blotting (Fig. 2B). All three different mAbs recognize both DELLAs extracted from the ga1–3 and recombinant AtRGL2n, but no signal was detected with QUAD extract (a negative control), indicating that the conformations of these three epitopes in recombinant N-domains are similar to those of DELLAs extracted from plant (see also Fig. 3 and Fig. 8 for further evidence).
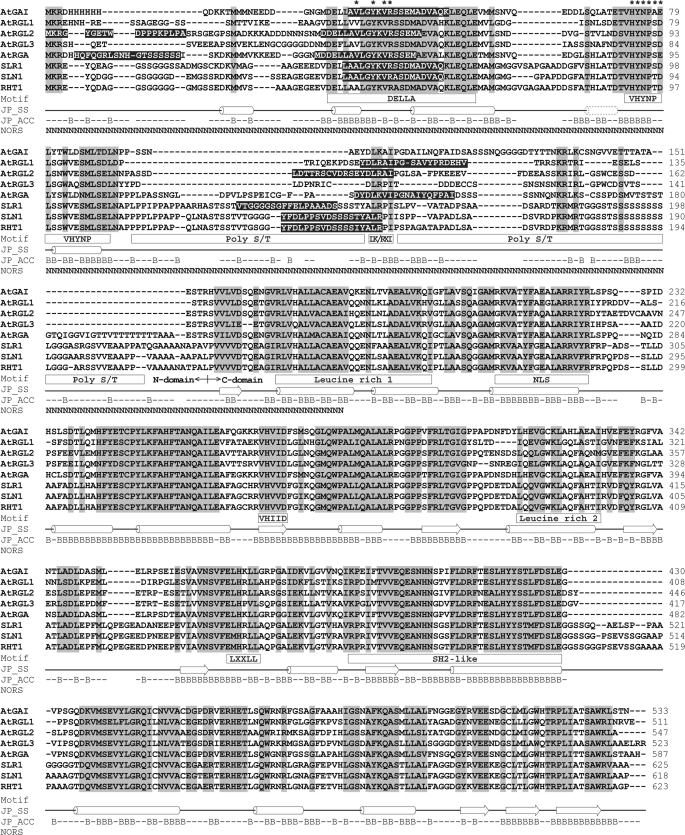
FIGURE 1.
Sequence alignment and domain structures of DELLA proteins. The identical and similar amino acids are shaded in light gray. Conserved motifs are marked in the square under the sequences. Predicted common secondary structures (JP_SS) are presented as a solid line (loop), cylinder (α-helix), and block arrow (β-strand). The dotted cylinder indicates an extra α-helix predicted for AtGAI, AtRGA, SLR1, SLN1, and RHT1. Solvent accessibility (JP_ACC) of AtRGA is shown with a B for solvent accessibility less than 25% (buried) or a short line for solvent accessibility more than 25% (exposed). Predicted non-regular secondary structure (NORS) is marked with an N. The amino acids predicted as α-MoRFs are shaded in black. The epitopes of mAbs BC9 and AD7 are marked with asterisks.
FIGURE 2.
Native PAGE and Western blotting of DELLA proteins. A, 10% native polyacrylamide gel electrophoresis. Lane 1, AtRGL1n; lane 2, AtRGL2n; lane 3, AtRGL3n; lane 4, AtRGAn; lane 5, RHT1n; lane 6, AtGAIn. B, native Western blotting of proteins extracted from A. thaliana mutants ga1–3 and QUAD and of recombinant AtRGL2n. Lanes 1, 4, and 7, proteins extracted from ga1–3; lanes 2, 5, and 8, proteins from QUAD extract; lanes 3, 6, and 9, proteins from QUAD extract spiked with 10 ng of recombinant AtRGL2n. Lanes 1–3 were developed with mAb AD7 (reactive to the VHYNP motif of AtRGL1n, AtRGL2n, and AtRGL3n), lanes 4–6 were developed with mAb BB7 (specific for AtRGL2n only), and lanes 7–9 were developed with mAb BC9 (reactive to the DELLA motif of all DELLAs).
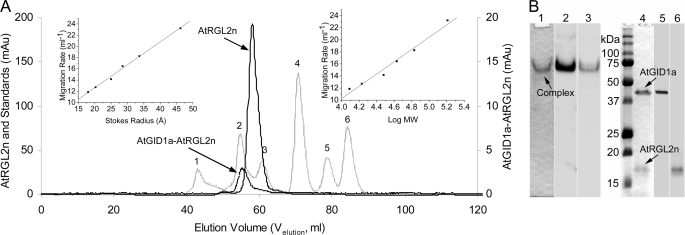
FIGURE 3.
Hydrodynamic properties of N-domains of DELLA proteins and AtGID1a-AtRGL2n complex. A, size exclusion chromatography. Six natively folded globular proteins (IgG1 (158 kDa) (1), bovine serum albumin (67 kDa) (2), ovalbumin (43 kDa) (3), carbonic anhydrase (30 kDa) (4), myoglobin (17 kDa) (5), and cytochrome c (12.3 kDa) (6)), corresponding to numbered peaks of the hatched line, were used as standards to calibrate the Superdex 75–16/60 column. The resultant migration rate (1000/Velution) plotted against the Stokes radius (left inset) and molecular weight (right inset) were used to determine the RSD and MrApp of DELLAs and the complex. As examples, the peaks of AtRGL2n and AtGID1a-AtRGL2n complex are shown in solid and dotted lines, respectively. B, PAGE and Western blotting of the AtGID1a-AtRGL2n complex. Lane 1, native polyacrylamide gel stained with Coomassie Blue dye; lane 2, Western blotting of the native gel developed with rabbit polyclonal anti-AtGID1 antibody; lane 3, as for lane 2 but developed with AtRGL2n-specific mAb BB7; lane 4, SDS-polyacrylamide gel stained with Coomassie blue dye; lane 5, Western blotting of the SDS gel developed with rabbit polyclonal anti-AtGID1 antibody; lane 6, Western blotting of the SDS gel developed with AtRGL2n-specific mAb BB7.
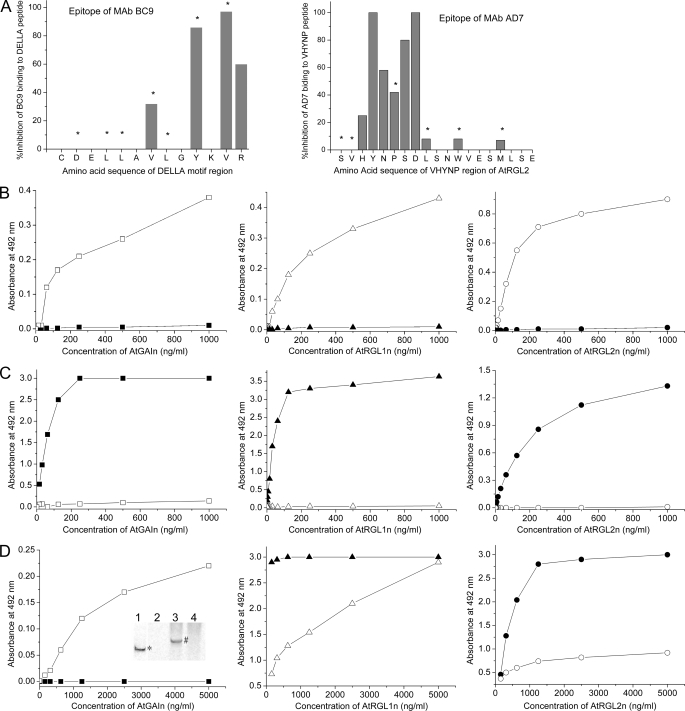
FIGURE 8.
Epitope mapping for mAbs and interaction of AtGID1 receptors from A. thaliana QUAD with recombinant DELLA proteins using antibody sandwich immunoassays. AtGAIn, AtRGL1n, and AtRGL2n were incubated with either extraction buffer (solid symbols) or QUAD extract containing AtGID1 receptors (open symbols) for 1 h. DELLA proteins or their complexes with AtGID1 receptors were captured by specific mAb AF2, AB8, and BB7 for AtGAIn, AtRGL1n, and AtRGL2n, respectively. All assays were carried out for AtGAIn (□), AtRGL1n (▵), and AtRGL2n (○) mixed with QUAD extract and AtGAIn (■), AtRGL1n (▴), and AtRGL2n (●) mixed with extraction buffer. A, the epitopes of mAb BC9 (reactive to the DELLA motif of all DELLAs) and AD7 (reactive to the VHYNP motif of AtRGL1n, AtRGL2n, and AtRGL3n) were identified by synthesizing the respective peptides with replacement of each individual amino acid with alanine. The amino acids marked with asterisks are involved in interactions with AtGID1a. B, the AtGID1 receptors in QUAD extracts bind to recombinant DELLA proteins. The AtGID1 in resultant complexes was detected with rabbit polyclonal anti-AtGID1c antibody and peroxidase-labeled goat anti-rabbit secondary antibody. C, the interaction of AtGID1 receptors with DELLA proteins inhibits the binding of biotin-labeled mAb BC9, which recognizes the DELLA motif of all DELLAs examined. mAb BC9 was detected by peroxidase-labeled streptavidin. D, the interaction of AtGID1 receptors with AtRGL1n and AtRGL2n reduces the binding of biotin-labeled mAb AD7, which recognizes the VHYNP motif of AtRGL1n and AtRGL2n. mAb AD7 does not recognize the VHYNP motif of AtGAIn except after binding of AtGID1 receptors to AtGAIn. mAb AD7 was detected by peroxidase-labeled streptavidin. The native Western blotting confirmed the ELISA result (inset in the left panel); AtGAIn incubated with extraction buffer (lanes 1 and 2), AtGAIn incubated with QUAD extract (lane 3), and QUAD extract control (lane 4) were separated on a 7.5% native polyacrylamide gel and then transferred to polyvinylidene difluoride membrane. An AtGAIn (band marked with asterisk in lane 1) was detected with AtGAIn-specific mAb AF2; the AtGID1-AtGAIn complex (band marked with # in lane 3) was detected with mAb AD7; and mAb AD7 did not detect AtGAIn incubated with extraction buffer (lane 2) or any corresponding components in QUAD extract control (lane 4).
Experimental Evidence
Recombinant N-domains of DELLA Proteins Possess Hydrodynamic Dimensions Typical of Intrinsically Unstructured Proteins
The hydrodynamic properties of the N-domains of DELLAs were determined by size exclusion chromatography (Fig. 3A). The resultant apparent molecular weight (MrApp) and Stokes radii (RSD) of N-domains of the analyzed DELLAs are listed in Table 1. The MrApp of each N-domain of DELLAs is approximately 2–3 times larger than its MrTheo, indicating that these proteins are not compatible with a monomeric, globular protein structure. Rather, such a large value of MrApp can be attributed to an extended conformation with low compactness of the polypeptide chain typical of IUPs (24).
TABLE 1.
Determined apparent molecular weight and Stokes radius of DELLA proteins
Based on Uversky (24, 62), a protein molecule can exist in NF, MG, PMG, native coil, and completely unfolded states and is characterized by the following relationships of its RS and MrTheo: natively folded state (log RSNF = 0.369 × log(MrTheo) − 0.254); molten globule state (log RSMG = 0.334 × log(MrTheo) − 0.053); native premolten globule (log RSPMG = 0.403 × log(MrTheo) − 0.239); native coil (log RSCoil = 0.493 × log(MrTheo) − 0.551); complete unfolded random coil state in urea (log RSUurea = 0.521 × log(MrTheo) − 0.649). MrApp and RSD are apparent molecular weight and Stokes radius determined using the migration rate values obtained from size exclusion chromatography (Fig. 3A).
| Proteins | MrTheo | MrApp | RSD | RSNF | RSMG | RSPMG | RSCoil | RSUurea | RSD/RSPMG |
|---|---|---|---|---|---|---|---|---|---|
| kDa | kDa | Å | Å | Å | Å | Å | Å | Å | |
| AtRGL2n | 18.2 | 48.4 | 31.3 | 20.8 | 23.5 | 30.1 | 35.5 | 37.2 | 1.04 |
| AtRGL1n | 15.3 | 39.0 | 28.9 | 19.5 | 22.1 | 28.0 | 32.5 | 34.0 | 1.03 |
| AtRGL3n | 15.7 | 38.3 | 28.7 | 19.7 | 22.3 | 28.3 | 32.9 | 34.4 | 1.02 |
| RHT1n | 25.0 | 60.3 | 33.7 | 23.4 | 26.1 | 34.2 | 41.4 | 43.9 | 0.98 |
| AtRGAn | 22.0 | 52.1 | 32.1 | 22.3 | 25.0 | 32.5 | 38.9 | 41.1 | 0.99 |
| AtGAIn | 15.8 | 34.8 | 27.7 | 19.7 | 22.4 | 28.4 | 33.0 | 34.5 | 0.98 |
| AtGID1a-AtRGL2n | 59.4 | 58.9 | 33.4 | 32.2 | 34.8 | 48.4 | 63.5 | 68.9 | 0.69 |
By analogy to a denatured globular protein existing in different unfolded states, intrinsically disordered proteins can be divided into three subclasses: molten globule (MG)-like, premolten globule (PMG)-like, or random coil-like, depending on their hydrodynamic characteristics. Whereas random coils have extended hydrodynamic dimension and no or little ordered secondary structure, premolten globules are relatively more compact (but still less compact than native globules) and possess some residual ordered secondary structure. Molten globules possess a high degree of compactness and a very well developed secondary structure (25). By comparing each experimentally determined Stokes radius (RSD) of a particular N-domain with various theoretical Stokes radii (RSNF, RSMG, RSPMG, RSCoil, and RSUurea in Table 1) of the same protein, all of the N-domains of the DELLAs analyzed have Stokes radii similar to the expected values of PMG-like IUPs. In other words, the RSD values for all of N-domains are much larger than the RSNF and RSMG values but are within 1–4% of the values estimated for each RSPMG (Table 1). RHT1n, AtGAIn, and AtRGAn exhibit RSD values that are 1–2% smaller than their respective RSPMG values, whereas AtRGL1n, AtRGL2n, and AtRGL3n exhibit RSD that are 2–4% larger than their respective RSPMG values, suggesting that the latter have a tendency to be relatively more flexible with less residual structures. Overall, the very small deviations from the expected RSPMG values indicate that the N-domains of DELLAs can be structurally classified as the premolten globule (24).
To investigate the potential disorder-to-order transition of N-domains of DELLAs upon binding to AtGID1, the hydrodynamic properties of the AtGID1a-AtRGL2n complex were determined under conditions identical to those used for calibration of the Superdex 75 column and for unbound N-domains of DELLAs (Fig. 3A and Table 1). The MrApp of the complex is almost equal to the MrTheo, with its RSD value approximating the theoretical RSNF value, indicating that both proteins, AtRGL2n and AtGID1a/GA3, in the complex are in natively folded states.
The AtGID1a-AtRGL2n complex, isolated from the gel filtration, was analyzed using native PAGE and SDS-PAGE. The complex appeared as a single band under native electrophoresis conditions (Fig. 3B, lane 1) and split into two bands on SDS-PAGE (Fig. 3B, lane 4) corresponding in size to recombinant AtGID1a (41.3 kDa) and AtRGL2n (18.2 kDa). Western blotting using polyclonal anti-AtGID1 antibody and AtRGL2n-specific mAb BB7 as probes, confirmed that the complex contains AtGID1a and AtRGL2n (Fig. 3B, lanes 2, 3, 5, and 6). The formation of the AtGID1a-AtRGL2n complex indicates that recombinant AtRGL2n is functional in binding to AtGID1.
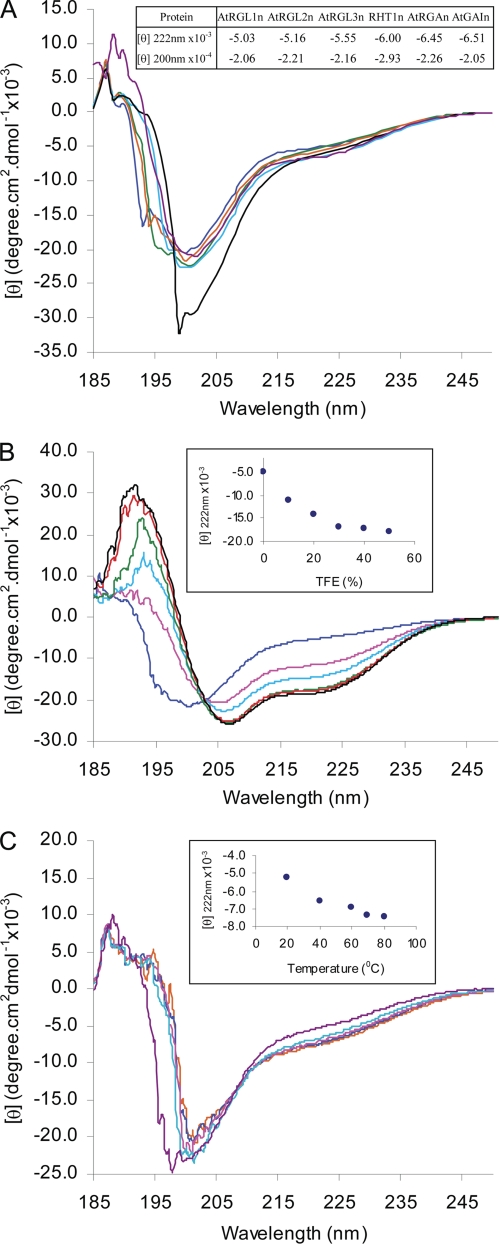
CD Spectra of N-domains of DELLA Proteins
The far-UV CD spectra of the N-domains of DELLAs display a large negative ellipticity at ∼200 nm and low ellipticity at 190 nm (Fig. 4A), characteristic of proteins in a largely disordered conformation (33). The negative ellipticity at 222 nm is typical of PMG-like IUPs, whereas negative ellipticity at 200 nm is typical of random coil-like IUPs (25) (Fig. 4A, inset table). Furthermore, the ellipticity at 222 nm shows that AtGAIn, AtRGAn, and RHT1n may contain slightly more residual α-helicity than AtRGL1n, AtRGL2n, and AtRGL3n. Although this difference is marginal, it is consistent with differences in the Stokes radii derived from the size exclusion chromatography.
FIGURE 4.
Far-UV CD spectra of N-domains of DELLA proteins. A, CD spectra of AtRGL1n (blue), AtRGL2n (green), AtRGL3n (orange), AtRGAn (cyan), RHT1n (black), and AtGAIn (purple). The ellipticity [θ] at both 200 and 222 nm are shown in the inset. B, 2,2,2-trifluoroethanol (TFE)-induced folding of AtRGL2n with 2,2,2-trifluoroethanol percentage: 0% (blue), 10% (pink), 20% (cyan), 30% (green), 40% (red), and 50% (black). The ellipticity [θ] at 222 nm versus TFE percentage is shown in the inset. C, temperature effect on folding of AtRGL2n at room temperature (purple), 40 °C (cyan), 60 °C (pink), 70 °C (blue), and 80 °C (orange). The ellipticity [θ] at 222 nm versus temperature is shown in the inset.
TFE-induced Folding of the N-domain of AtRGL2
The solvent TFE mimics the hydrophobic environment experienced by protein-protein interactions and has therefore been widely used as a probe for the propensity of IUPs to undergo an induced folding upon target binding (23). To test the potential binding-induced folding of N-domains of DELLAs, far-UV CD spectra of AtRGL2n were recorded in the presence of increasing concentrations of TFE (Fig. 4B). AtRGL2n showed an increased α-helicity upon the addition of TFE, as indicated by the characteristic maximum at 192 nm and double minima at 208 and 222 nm. Most of the unfolding-to-α-helix transition takes place in the presence of 30% TFE, at which point the α-helical content estimated from the ellipticity at 222 nm reached 51.5%. The transition slows down dramatically after 30% TFE (Fig. 4B, inset). The TFE results reveal a potential of AtRGL2n to form α-helices upon binding to its interacting partners.
Temperature Effects on the N-domain of AtRGL2
The effect of temperature on CD spectra of AtRGL2n is shown in Fig. 4C. With increasing temperature, the CD spectra showed a modest but discernible heat-induced folding. The significant spectral changes occurred during temperature increase in the range 20–40 °C; further heating up to 80 °C resulted in continuous but much less pronounced heat-induced folding (Fig. 4C, inset). At 80 °C, the α-helical content reached 26.9%. These changes of CD spectra with temperature are completely reversible. The effect of heat-induced folding may be attributed to the increased strength of hydrophobic interactions at higher temperature and therefore a stronger hydrophobic driving force for folding (24). Contrary to globular proteins that become more unfolded during thermal denaturation, the heat-induced folding has also been observed in other IUPs (34).
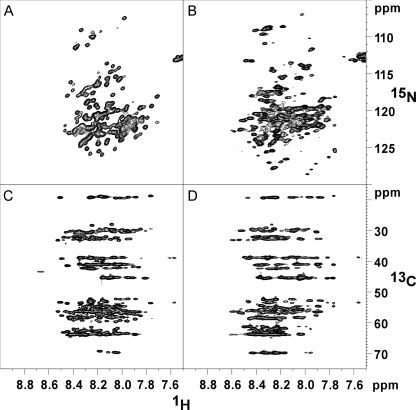
Two-dimensional 15N,1H HSQC and 13C CBCA(CON)H Spectra of AtRGL2n and AtRGAn
To obtain further structural insights, AtRGL2n and AtRGAn were compared by NMR spectroscopy. The recombinant N-domains were double-labeled (15N and 13C) for collecting two-dimensional 15N,1H HSQC spectra and 1H-13C planes from a CBCA(CO)NH experiment. The narrow ranges of chemical shifts in the HSQC spectra for AtRGL2n and AtRGAn are characteristic of unstructured proteins (Fig. 5, A and B). The CBCA(CO)NH planes (Fig. 5, C and D) correlate the amide proton chemical shift with the Cα and Cβ shifts of the previous residue. Horizontal rows of peaks at chemical shifts typical of random coil structures can be seen. For example, the row of peaks near 19 ppm arises almost certainly from alanine Cβ resonances, whereas the row near 70 ppm is consistent with threonine Cβ protons (Biological Magnetic Resonance Data Bank). Neither of these rows displays a significant spread in 13C shift values, suggesting a nearly uniform and therefore disordered environment. Structured proteins can display similar average chemical shifts as are observed here, but the chemical shift dispersion as represented by the S.E. value (approximately at ±1.8 ppm) increases by nearly an order of magnitude relative to disordered proteins (Biological Magnetic Resonance Data Bank). Although the narrow range of 13C shifts seen here is consistent with disorder, the chemical dispersion of AtRGAn (Fig. 5B) does appear to be slightly higher than that of AtRGL2n (Fig. 5A). Again this is a marginal effect, but it is consistent with differences observed in the Stokes radii and CD spectra.
FIGURE 5.
NMR spectra showing chemical shift dispersion of AtRGL2n and AtRGAn. A and B, 1H-15N HSQC of AtRGL2n and AtRGAn, respectively. C and D, 1H-13C plane of a CBCA(CO)NH experiment for AtRGL2n and AtRGAn, respectively.
Bioinformatic Analyses
Amino Acid Composition of the N-domains of DELLA Proteins
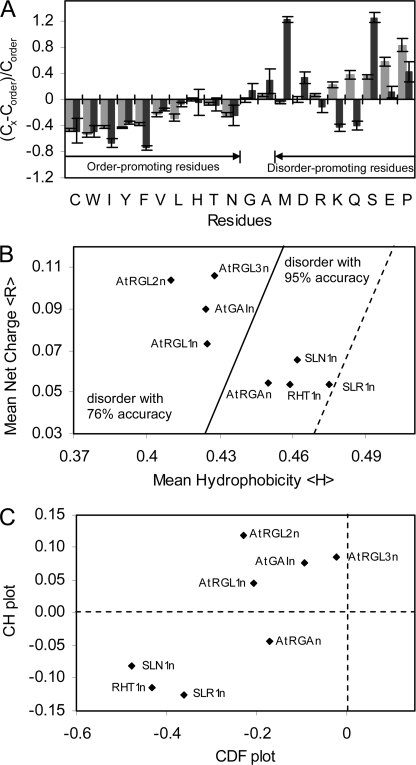
It has been shown that most IUPs lack amino acids Trp, Cys, Phe, Ile, Tyr, Val, Leu, His, Thr, and Asn (order-promoting residues) and are enriched in Lys, Glu, Pro, Ser, Gln, Arg, Asp, and Met (disorder-promoting residues). Amino acids Ala and Gly are neutral in regard to the order and disorder (35). Fig. 6A illustrates the deviation of average amino acid frequencies in N-domains of eight DELLAs (as in Fig. 1) in comparison with the average frequencies found in the ordered globular proteins from the Protein Data Bank. Similar to disordered proteins from the DisProt data base (36), the N-domains of DELLAs showed an overall lack of order-promoting residues and enrichment in disorder-promoting residues, remarkably Ser, Met, and Asp, a characteristic of IUPs. This feature of amino acid composition also accounts for the propensity of an overall low hydrophobicity and relatively high net charge of the N-domains of DELLAs, as found in other IUPs. A special feature of the N-domains of DELLAs is depletion of Lys, Gln, and Arg residues. This may indicate that N-domains of DELLAs are a special group of IUPs.
FIGURE 6.
Primary sequence and disorder analyses. A, deviations of average amino acid frequencies of N-domains of DELLAs (black bars) and disordered proteins of DisPro data base (gray bars) from average values of ordered protein data base. The fractional difference was calculated as (CX − Corder)/Corder, where Cx is the content of a given amino acid in eight N-domains of DELLAs studied or a set of well characterized IUPs from DisProt, and Corder is the corresponding content in a set of ordered proteins. B, mean net charge versus mean hydrophobicity plot of N-domains of DELLAs. The major boundary line (solid) (〈R〉 = 2.785〈H〉 − 1.151) distinguishes natively disordered (left side of the line) from ordered proteins (right side of the line). A boundary margin of +0.045 (dotted) extends the disorder estimation accuracy to 95%. C, combined CDF CH plot of N-domains of DELLAs. y axis, distance of each DELLA protein from major boundary line in CH plot (positive for disordered, negative for ordered or small number of disordered); x axis, distance of each DELLA protein from boundary line in CDF plot (positive for ordered, negative for disordered).
CH and CDF Analysis
Statistical analyses of both natively ordered and disordered proteins utilize a plot of the net charge of a protein against its mean hydropathy and show that ordered and disordered proteins tend to occupy two different areas within the charge-hydrophobicity phase space, separated by an estimated boundary line (37). As shown in the charge-hydropathy plot (Fig. 6B), AtGAIn, AtRGL1n, AtRGL2n, and AtRGL3n fit well into the disordered area, whereas AtRGAn and three non-redundant DELLAs of monocot plants (SLR1n, SLN1n, and RHT1n) fall onto the right side of the boundary, which hosts natively ordered proteins as well. According to statistics (20), mean net charge and mean hydrophobicity of natively disordered proteins are scattered over the charge-hydrophobicity phase space, sometimes crossing into the area of ordered proteins. All of the N-domains of DELLAs fit into the disordered area with a boundary margin for accuracy of the estimation reaching up to 95% (Fig. 6B).
The CH plot is a linear disorder classifier that takes into account only two parameters of the particular sequence, charge and hydrophobicity, and is predisposed to discriminate proteins with substantial amounts of extended disorder (random coils and premolten globules) from proteins with globular conformations (molten globule-like and rigid well structured proteins) (20). Alternatively, another binary disorder classifier, CDF analysis, which is based on the output of PONDR, may discriminate all disordered conformations, including random coils, premolten globules, and molten globules from rigid well folded proteins (20). Simultaneous CDF CH plot analysis of the N-domains of DELLAs (Fig. 6C) shows that all N-domains of DELLAs analyzed are predicted to be intrinsically disordered by CDF.
Disorder Prediction Analysis
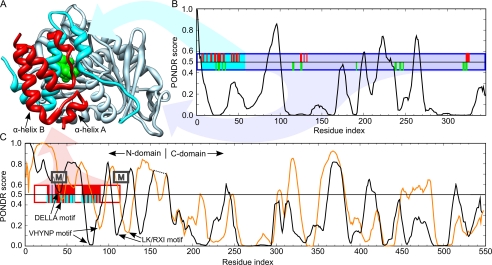
To investigate the degree of disorder along the primary sequence of DELLAs, sequences of the eight DELLAs studied above in the CDF CH plot, together with AtGID1a, were further analyzed using PONDR (38) to identify the intrinsically unstructured regions. The PONDR score pattern of AtGID1a shows basically a folded structure (Fig. 7, A and B). The resultant PONDR score patterns of all DELLAs are similar to each other. As shown in Fig. 7C, the C-domains of both AtGAI and AtRGL2 are dominated by predicted ordered structures, with most residues having PONDR score below 0.5, the threshold for order/disorder states. In contrast, the N-domains of AtGAI and AtRGL2 are clearly natively disordered except for some short segments corresponding to the DELLA, VHYNP, and L(K/R)XI motifs (Fig. 7C, indicated by arrows).
FIGURE 7.
Comparison of the PONDR disorder predictions for AtGID1a, AtGAI, and AtRGL2 with the x-ray crystal structure of AtGAIn-AtGID1a/GA3 complex from A. thaliana (Protein Data Bank code 1ZSH). A, ribbon representation of the complex showing the AtGAIn (red ribbon), the N-terminal binding pocket cap of AtGID1a (cyan ribbon), gibberellin receptor domain of AtGID1a (light blue ribbon), and gibberellin (green van der Waals surface). B, disorder prediction for AtGID1a. C, disorder predictions for AtGAI (black line; its C-domain is shifted right to align with that of AtRGL2) and AtRGL2 (orange line). Disorder predictions were made with PONDR VLXT, where a prediction of ≥0.5 indicates a propensity for disorder, and <0.5 indicates a propensity for order. The boxes indicate the fragments of AtGAIn and AtGID1a crystallized in the complex, with the filled positions indicating the region of defined density in the crystal structure. The ticks indicate residues that interact with the portion of the complex with the corresponding colors. Interacting residues were assessed as those having a ΔASA of >10 Å2, where ΔASA is the surface area buried in the interaction calculated as the difference in surface areas of the residue in the isolated monomer and the interaction complex. Approximate positions of α-MoRFs predicted for most DELLA proteins are indicated by boxes labeled M (α-MoRF predictions shown in Fig. 1).
An independent predictor of non-regular secondary structure (NORSp) that adopts different definitions of disorder to distinguish unstructured loop regions from regular structured loops was also used to predict disorder (39). The entire N-domain of each DELLA was predicted as an unstructured loop region (Fig. 1, NORS marks), giving results that are consistent with the predictions made by PONDR.
α-Helix-forming Molecular Recognition Feature (α-MoRF) Prediction
Many long disordered regions contain a short region or regions responsible for molecular recognition via a disorder-to-order transition. The short regions, termed MoRFs (40), are often detected in PONDR predictions as short downward spikes of predicted order within longer regions of predicted disorder. Such downward spikes appear in the N-domains of DELLAs. Using the α-MoRFs-II predictor, which defines a heuristic for binding-associated downward spikes and removes false positive predictions (30, 31), several α-MoRFs have been predicted in three regions of the DELLA sequences (Fig. 1), at or near the N terminus, the DELLA motif, and the L(K/R)XI motif. The latter two have corresponding α-MoRFs predicted in the majority of DELLA sequences, which suggests that, allowing for false negative predictions, α-MoRFs are a conserved feature of these motifs. In contrast, the α-MoRFs at the N terminus are only predicted in two of DELLA sequences, indicating that these are likely to be false positive predictions. There were no α-MoRFs predicted to coincide with the VHYNP motif for any of the DELLAs.
Secondary Structure Element Prediction
Predicted secondary structures are shown in Fig. 1. The C-domains of DELLAs are predicted to contain highly conserved α-helix/β-sheet conformation with limited solvent accessibility, indicating their natively folded state. In contrast, the N-domains of DELLAs are largely dominated by loops with high solvent accessibility. Four common residual α-helices (solid cylinders) are predicted in the vicinity of DELLA and VHYNP motifs, and an extra short α-helix (dotted cylinder) is predicted only for AtGAI, AtRGA, SLR1, SLN1, and RHT1. All of these structural features are consistent with PONDR prediction and commensurate with an open, unstructured N-domain and a well ordered C-domain.
Interaction-triggered Conformational Change in the VHYNP Motif of AtGAI
To gain insight into conformations of the unbound form of DELLAs and their folding upon binding to the AtGID1 extracted from plant tissue, mAbs BC9 and AD7, binding to DELLA and VHYNP motif, respectively, were used as probes in the ELISAs. The epitopes of both mAbs have been determined (Fig. 8A); the conserved V—Y-VR residues within the DELLAVLGYKVR peptide comprise the epitope for mAb BC9, and the conserved HYNPSD residues within the SVHYNPSDLSNWVESMLSE peptide are primary amino acids comprising the epitope for mAb AD7 (Fig. 1, amino acids marked with asterisks).
To verify that AtGID1 extracted from plant tissue binds to immobilized N-domains of DELLAs, a rabbit polyclonal antibody raised against recombinant AtGID1c was first used in the assays. The results showed that AtGID1 extracted from plant tissue binds in vitro to recombinant AtGAIn, AtRGL1n, and AtRGL2n (Fig. 8B). Detection of the AtGID1 in the ELISAs was dependent on the existence of both recombinant DELLAs and QUAD extract, indicating that the AtGID1 was specifically interacting with the recombinant DELLAs.
The AtGID1/GA3 in QUAD extract competes with mAb BC9 and AD7 for binding to immobilized recombinant N-domains of DELLAs. The competitive ELISAs were further carried out to determine essentiality of conserved DELLA and VHYNP motifs for interaction with plant AtGID1. The ELISAs showed that AtGID1 blocks mAb BC9 from binding to AtGAIn, AtRGL1n, and AtRGL2n (Fig. 8C) and vice versa (data not shown). Given that mAb BC9 is a DELLA motif-specific, universal antibody to the whole DELLA family, this mutual blocking between mAb BC9 and AtGID1 is probably applicable to all DELLAs. AtGID1 only partially blocks mAb AD7 from binding to both AtRGL1n and AtRGL2n (Fig. 8D, middle and right). On the other hand, in a reverse experiment, mAb AD7 completely blocks AtGID1 from binding to both AtRGL1n and AtRGL2n (data not shown).
In contrast to mAb BC9, mAb AD7 does not bind unbound AtGAIn. Interestingly, binding of AtGID1 to AtGAIn renders mAb AD7 able to bind the AtGID1-AtGAIn complex (Fig. 8D, left). This suggests that a conformational change in the VHYNP motif of AtGAIn occurs upon formation of the AtGID1-AtGAIn complex, resulting in at least the partial reconstitution of AD7 epitope. As additional evidence of this conformational change, the interactions between AtGID1, AtGAIn, and mAb AD7 shown in the ELISA were reproduced by using the native Western blotting approach (Fig. 8D, left, inset). It shows that mAb AD7 does not bind AtGAIn in extraction buffer (lane 2) but does in QUAD extract, appearing at a higher molecular weight in the form of the AtGID1 complex (lane 3).
In summary, these results show that AtGID1 extracted from plant tissue not only binds to recombinant DELLAs, but it does so in a site-specific manner, which is in agreement with previous studies (8, 11). Furthermore, binding of AtGID1 to AtGAIn stimulates binding of mAb AD7, suggesting that the VHYNP motif undergoes conformational change upon complex formation.
DISCUSSION
Recombinant N-domains of DELLA Proteins Are Functionally Active in Binding to Native and Recombinant AtGID1 Receptors
Due to the low abundance of DELLAs in plants tissues and hence the impracticality of purifying plant DELLAs in sufficient quantities for protein characterization, recombinant DELLAs from either E. coli or yeast (yeast two-hybrid system) have been commonly used in previous studies of DELLAs without validating the recombinant DELLAs as a substitute of plant DELLAs. We have now compared E. coli expressed and plant DELLAs using a set of monoclonal antibodies that recognize amino acid conformation (epitope) of DELLAs. The native Western blotting (Fig. 2B) has shown that three different mAbs, binding to independent epitopes, recognize both recombinant AtRGL2n and plant DELLAs (including AtRGL2) from A. thaliana ga1–3 extract under the same experimental conditions. This implies that the conformation of three different epitopes of recombinant AtRGL2n are similar to those of plant DELLAs extracted from A. thaliana. AtGID1 from QUAD extract has been shown to bind in vitro to recombinant AtGAIn, AtRGL1n, and AtRGL2n (Fig. 8B). These results further reveal that recombinant N-domains of DELLAs are functionally active with regard to interactions with native AtGID1. Moreover, recombinant AtRGL2n has also been shown to bind in vitro to recombinant AtGID1a to form a folded complex (Fig. 3, A and B), indicating again that the recombinant N-domains of DELLAs are functionally active.
In addition to the yeast two-hybrid system, glutathione S-transferase- or thioredoxin-fused DELLA proteins have often been used in previous studies to examine in vitro binding between GID1 receptors and DELLAs. The use of a yeast two-hybrid system to elucidate protein-protein interactions frequently produces false positive results (41). The potential of fusion tags to interfere with the protein interactions in the in vitro binding studies has also been noticed (42). The use of mAbs of known epitope sites has enabled us to approach these studies in a different way by investigating the effect of the interactions between the recombinant N-domains of DELLAs and native GID1 receptors extracted from plant tissue. These types of methods would increase the reliability of the deductions that have been made based on protein-protein interactions.
Functional Implications of IUP Characteristics of the Unbound DELLA Proteins
Functional, nonfibrous proteins have been proposed to exist in one of four states: namely ordered globule, molten globule-like, premolten globule-like, and random coil-like forms (21, 25). Any of these four distinct forms or transitions between them can be the “native state” of a protein and specify the protein's functions. It was proposed (24) that the premolten globule state arises as a result of water being a poor solvent for certain proteins. Recent studies on polar but uncharged polypeptides have given further support to the idea that these molecules exist as collapsed random coils due to water being a poor solvent for the peptide backbone and for polar but uncharged side chains (43, 44).
Using multiple lines of evidence, we have demonstrated that under physiological conditions, the unbound N-domains of DELLAs are essentially premolten-like IUPs with a few regions likely to form transient α-helices. In contrast, the C-domains are well ordered with a regular α-helix/β-sheet structure. We believe that this is a common structural feature shared by the whole family of DELLAs, given their similar domain structure and highly conserved sequences in both the entire C-domain and the essential motifs in the N-domain. In an independent study (45), the N-domain of rice DELLA (SLR1) has also been found to have large hydrodynamic dimensions with its apparent molecular weight 2.1 times (in Trx-His-tagged form) and 3.1 times (without tag) larger than its theoretical molecular weight. This matches our results obtained from six other DELLAs (Table 1). All of the N-domains of DELLAs we have analyzed migrate into the native gel as a single band (Fig. 2A), with no indication of multiple bands that would be typical for the multimeric proteins. Thus, the large apparent molecular weight of SLR1 is probably attributable to its being an extended monomer rather than a globular dimer or trimer.
Both experimental and bioinformatics analyses indicate that IUPs are overrepresented in the cell nucleus and overwhelmingly prevalent in cell signaling and transcription regulation. For example, over 90% of transcription factors were predicted to contain extended intrinsic disordered regions (46, 47). An intrinsically disordered region enables an IUP to be folded differently to recognize and bind different partners at various binding interfaces (48, 49). Such multiple binding activities are expected for DELLAs, given that they not only regulate plant growth in response to GA signal but also cross-talk with many other signal pathways to maintain GA homeostasis and adapt to environmental changes (12, 13). In order to perceive multiple signals, the N-domains of DELLAs are functionally required to accommodate different partners involved in different signal transduction events. Thus, the intrinsically unstructured feature of the N-domain of DELLAs fits very well with their potential role of a convergence point in integration of multiple developmental and environmental signals, acting like hub proteins (49).
A recent study (50, 51) revealed that most IUPs are tightly regulated in cells to increase fidelity in signaling and recognition and are often reused in multiple signaling pathways to produce different physiological outcomes. This study further revealed that IUPs were highly targeted by many different kinases, the levels of which were themselves modulated in response to cell division cycle or were activated upon exposure to specific stimuli or stress. The function and availability of IUPs can be finely tuned by post-translational modifications through phosphorylation and interactions with other proteins under different conditions. In a similar situation, DELLAs not only respond to the GA signal but also to ethylene, abscisic acid, auxin, and environmental signals, such as light and salt stress (12, 13). In addition to GA-induced degradation, other mechanisms probably exist that regulate GA signaling through interactions between DELLAs and regulatory proteins from other pathways (52), a characteristic of the tight regulation of IUPs. It has been shown that the O-linked N-acetylglucosamine transferase SPY (SPINDLY) activated the repressive function of DELLAs possibly by GlcNAc modification (53). The phosphorylation and dephosphorylation of DELLAs were also correlated to their stability, plant growth-repressive activity, and GA-induced degradation (54, 55). Although these studies have not been able to define the exact interacting sites between DELLAs and SPY or kinases, it seems very likely that GlcNAc modification and phosphorylation can be used in the GA signaling for fine-tuning the availability and function of DELLAs. These interactions may also reflect how DELLAs perceive environmental stress and multiple phytohormone signals.
Binding-induced Folding Is Involved in DELLA-AtGID1 Interactions
It is known that a large decrease in conformational entropy due to folding of disordered regions in the disorder-to-order transition can uncouple specificity from binding strength. With such a high specificity/low affinity, the regulatory interaction between an IUP and its partner is both highly specific and easily dispersed; activating and terminating a signal are equally important (22). An IUP has been suggested to contain a “conformational preference” for the structure it will take upon binding (56). This preferred conformation could be α-helix (α-MoRFs) (30, 31), β-strand (β-MoRFs), or an irregular structure (ι-MoRFs) (40, 57). TFE is a powerful tool to define a correspondence between induced structure and the nascent structure of proteins (58). AtRGL2n exhibits TFE- and temperature-induced folding (Fig. 4, B and C), forming transient α-helices. As shown in Fig. 3 and Table 1, unbound AtRGL2n is an IUP with a premolten globule-like conformation, and the AtGID1a-AtRGL2n complex becomes natively folded. Given that AtGID1a/GA3 possesses a folded structure (Fig. 7, A and B), AtRGL2n must have a folding transition during the complexing of AtGID1a/GA3 and AtRGL2n. These lines of evidence suggest that DELLA proteins, in view of the whole N-domain, undergo disorder-to-order transitions upon binding to their GID1 receptors.
The ELISAs with mAbs of known epitope enabled us to gain insight into interactions between AtGID1 and DELLAs and therefore any potential conformational changes in view of local motifs. The DELLA motif was proposed as one of the GID1 binding sites of AtGAI, AtRGA, and SLR1 (11, 45, 59), which was recently confirmed by the crystal structure of AtGID1a-AtGAIn complex (8). We have shown that the DELLA motif-specific mAb BC9 and AtGID1 from QUAD extract completely blocked each other from binding to AtGAIn, AtRGL1n, and AtRGL2n (Fig. 8C), suggesting that the DELLA motif of these three DELLA proteins is a binding site for AtGID1. The amino acids comprising the epitope of mAb BC9 overlap with those shown by the crystal structure of the AtGID1a-AtGAIn complex (8) to interact with AtGID1. The DELLA motifs are predicted to be α-MoRFs for most DELLAs analyzed (black-shaded amino acids in Fig. 1). In agreement with these experiments, α-MoRF predictions assign the DELLA motif to a binding site in all DELLAs and propose that the DELLA motif undergoes disorder-to-α-helix transition upon binding to GID1 receptors. The formation of this α-helix has been confirmed and shown as α-helix A and α-helix B (Fig. 7A) in the crystal structure of AtGID1a-AtGAIn complex (8).
mAb AD7, specific to VHYNP motif, does not bind AtGAIn, AtRGAn, and SLR1n but does bind AtRGL1n, AtRGL2n, and AtRGL3n. Given that conformations of epitopes are recognized by antibodies, this implies the different conformations of the VHYNP motif between these two groups in their unbound form. Based on the AD7 epitope and the crystal structure of the AtGID1a-AtGAIn complex, mAb AD7 epitope sits on the VHYNPSD loop in which the side chains of all of the residues point outward from the AtGID1a-AtGAIn interacting interface and therefore are accessible to AD7, except for the proline and valine that point inward to sandwich the aromatic ring of a tyrosine from AtGID1a. This proline plays a key role in the interaction with AtGID1 but is less important in the interaction with AD7 due to the availability of five other stronger epitope residues (HYNSD). This is compatible with our ELISA findings (Fig. 8D, middle and right) that AD7 completely blocks AtGID1 binding but AtGID1 only partially blocks AD7 from binding to AtRGL1n and AtRGL2n. This result also suggests that the VHYNPSD loop in AtRGL1n and AtRGL2n undergoes no significant conformational changes during binding to AtGID1 receptors. In contrast, AtGID1 binding to AtGAIn allows the complex to interact with AD7 (Fig. 8D, left), indicating that the VHYNPSD loop in the unbound AtGAIn undergoes conformational changes induced by AtGID1 binding. This result was further confirmed by the native Western blotting (Fig. 8D, inset) in which AtGAIn, in the form of a complex with AtGID1, is recognized by mAb AD7. Based on the reactivity of mAb AD7 to DELLAs, we believe that a similar conformational change would occur in AtRGAn and non-redundant DELLAs of monocot plants, such as SLR1n. Although the VHYNP motif appears as a large downward spike in the PONDR score pattern (Fig. 7C), which is consistent with its binding function, it was not identified in our α-MoRFs prediction for any of the DELLAs. We now understand that the irregular VHYNPSD loop (ι-MoRFs rather than α-MoRFs) is involved in the binding-induced folding, at least for AtGAIn. The prediction of ι-MoRFs is under development. Both mAb BC9 and AD7 alone blocked AtGID1 from binding to both AtRGL1n and AtRGL2n, indicating that DELLAs need a synergistic action between the DELLA and VHYNP motif during folding and binding to AtGID1 receptors. This is consistent with the results from yeast two-hybrid assays (11, 45).
Interestingly, the L(K/R)XI motif was also identified in our α-MoRF prediction for most DELLAs analyzed (Figs. 1 and 7C). It is possibly a potential binding site of DELLAs for an unknown component in the signaling pathway because this motif was not involved in interactions with GID1 receptors. A three-dimensional structural data base search showed that fragments containing the L(K/R)XI motif mostly exist in the α-helix conformation (data not shown), which is in agreement with α-MoRFs forming a transient α-helix that is stabilized upon binding to its partner. Verification of the L(K/R)XI motif as a potential new binding site of DELLAs will require further investigations, such as pull-down experiments.
DELLA Proteins May Be Divided into Two Subgroups Based on Their Biophysical Properties and Interactions with Monoclonal Antibodies
Although the individual lines of biophysical evidence show only marginal differences between two groups of DELLAs, the experimentally determined Stokes radii (RSD) and CD and NMR spectra collectively suggest that DELLAs, in terms of their compactness and residual structures, can be divided into two subgroups; AtGAIn, AtRGAn, and non-redundant DELLAs of monocot plants, such as SLR1n and RHT1n (AtRGA group), may contain more residual structure and are less flexible than AtRGL1n, AtRGL2n, and AtRGL3n (AtRGL group). Consistent with the suggested two grouping of DELLAs, an extra short α-helix immediately before the VHYNP motif (Fig. 1, dotted cylinder) was predicted for the AtRGA group but not for the AtRGL group. Correspondingly, the CH-plot (Fig. 6B) also supports this two grouping of DELLAs in terms of average hydrophobicity and net charge, with an exception for AtGAIn. Over the entire sequence, AtRGA and AtGAI are 82% identical, but both share about 60% identity with members of the AtRGL group. Sequence alignments among the N-domains give the same trend that AtRGAn and AtGAIn are 61% identical, but both share lower than 42% identity with members of the AtRGL group. Biologically, AtRGA and AtGAI have partially redundant functions, and both transcripts were detected in all plant tissues, and AtRGA functions in a manner similar to SLR1 (60, 61).
Furthermore, mAb AD7 distinguishes AtRGL group from the AtRGA group with respect to the conformation of the VHYNP motif of the unbound DELLAs. Upon binding to AtGID1, AtGAIn and possibly the other members of the AtRGA group undergo conformational changes in the region of the HYNPSD loop to partially release the epitope of mAb AD7. In contrast, the mAb AD7 epitope is available for binding to the AtRGL group, and these DELLAs all display a similar dose-dependent decrease in mAb avidity during AtGID1 interactions. Both biophysical properties measured for the whole N-domain of DELLAs and monoclonal antibody reactivity in regard to the local HYNPSD region show differences between these two subgroups.
Acknowledgments
We thank Nicholas Harberd for providing A. thaliana quadruple-DELLA mutant and the Arabidopsis Information Resource for A. thaliana ga1–3 mutant seeds.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 LM007688-01A1 and GM071714-01A2. This work was also supported by New Zealand Foundation for Research, Science, and Technology Grant C06X0207 and the Program of the Russian Academy of Sciences for “Molecular and Cellular Biology.” This work was also supported by the Indiana University Purdue University Indianapolis Signature Centers Initiative.
- DELLA
- DELLA protein
- IUP
- intrinsically unstructured protein
- GA
- gibberellic acid
- mAb
- monoclonal antibody
- QUAD
- A. thaliana quadruple-DELLA mutant
- TFE
- 2,2,2-trifluoroethanol
- HSQC
- heteronuclear single quantum coherence
- MoRF
- molecular recognition feature
- ELISA
- enzyme-linked immunosorbent assay
- CDF
- cumulative distribution function
- MG
- molten globule
- PMG
- premolten globule
- α-MoRF
- α-helix-forming MoRF
- NF
- natively folded
- N-domain
- N-terminal domain
- C-domain
- C-terminal domain
- MrTheo and MrApp
- theoretical and apparent molecular weight, respectively.
REFERENCES
- 1.Peng J., Carol P., Richards D. E., King K. E., Cowling R. J., Murphy G. P., Harberd N. P. (1997) Genes Dev. 11, 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun T. P., Gubler F. (2004) Annu. Rev. Plant Biol. 55, 197–223 [DOI] [PubMed] [Google Scholar]
- 3.Peng J., Richards D. E., Hartley N. M., Murphy G. P., Devos K. M., Flintham J. E., Beales J., Fish L. J., Worland A. J., Pelica F., Sudhakar D., Christou P., Snape J. W., Gale M. D., Harberd N. P. (1999) Nature 400, 256–261 [DOI] [PubMed] [Google Scholar]
- 4.Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., Chen L., Yu L., Iglesias-Pedraz J. M., Kircher S., Schäfer E., Fu X., Fan L. M., Deng X. W. (2008) Nature 451, 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lucas M., Davière J. M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., Fankhauser C., Blázquez M. A., Titarenko E., Prat S. (2008) Nature 451, 480–484 [DOI] [PubMed] [Google Scholar]
- 6.Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T. Y., Hsing Y. I., Kitano H., Yamaguchi I., Matsuoka M. (2005) Nature 437, 693–698 [DOI] [PubMed] [Google Scholar]
- 7.Nakajima M., Shimada A., Takashi Y., Kim Y. C., Park S. H., Ueguchi-Tanaka M., Suzuki H., Katoh E., Iuchi S., Kobayashi M., Maeda T., Matsuoka M., Yamaguchi I. (2006) Plant J. 46, 880–889 [DOI] [PubMed] [Google Scholar]
- 8.Murase K., Hirano Y., Sun T. P., Hakoshima T. (2008) Nature 456, 459–463 [DOI] [PubMed] [Google Scholar]
- 9.Silverstone A. L., Jung H. S., Dill A., Kawaide H., Kamiya Y., Sun T. P. (2001) Plant Cell 13, 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGinnis K. M., Thomas S. G., Soule J. D., Strader L. C., Zale J. M., Sun T. P., Steber C. M. (2003) Plant Cell 15, 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z. L., Powers S. J., Gong F., Phillips A. L., Hedden P., Sun T. P., Thomas S. G. (2006) Plant Cell 18, 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N. P. (2006) Science 311, 91–94 [DOI] [PubMed] [Google Scholar]
- 13.Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N. P., Jones J. D. (2008) Curr. Biol. 18, 650–655 [DOI] [PubMed] [Google Scholar]
- 14.Cao D., Cheng H., Wu W., Soo H. M., Peng J. (2006) Plant Physiol. 142, 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou X., Hu W. W., Shen L., Lee L. Y., Tao Z., Han J. H., Yu H. (2008) Plant Physiol. 147, 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie H., Vucetic S., Iakoucheva L. M., Oldfield C. J., Dunker A. K., Uversky V. N., Obradovic Z. (2007) J. Proteome Res. 6, 1882–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vucetic S., Xie H., Iakoucheva L. M., Oldfield C. J., Dunker A. K., Obradovic Z., Uversky V. N. (2007) J. Proteome Res. 6, 1899–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunker A. K., Obradovic Z., Romero P., Garner E. C., Brown C. J. (2000) Genome Inform. Ser. Workshop Genome Inform. 11, 161–171 [PubMed] [Google Scholar]
- 19.Iakoucheva L. M., Brown C. J., Lawson J. D., Obradović Z., Dunker A. K. (2002) J. Mol. Biol. 323, 573–584 [DOI] [PubMed] [Google Scholar]
- 20.Oldfield C. J., Cheng Y., Cortese M. S., Brown C. J., Uversky V. N., Dunker A. K. (2005) Biochemistry 44, 1989–2000 [DOI] [PubMed] [Google Scholar]
- 21.Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., Obradovic Z. (2001) J. Mol. Graph. Model. 19, 26–59 [DOI] [PubMed] [Google Scholar]
- 22.Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradović Z. (2002) Biochemistry 41, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 23.Dyson H. J., Wright P. E. (2002) Curr. Opin. Struct. Biol. 12, 54–60 [DOI] [PubMed] [Google Scholar]
- 24.Uversky V. N. (2002) Eur. J. Biochem. 269, 2–12 [DOI] [PubMed] [Google Scholar]
- 25.Uversky V. N. (2002) Protein Sci. 11, 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X., Frearson N., Kirk C., Jones W. T., Harvey D., Rakonjac J., Foster T., Al-Samarrai T. (2008) Protein Expr. Purif. 58, 168–174 [DOI] [PubMed] [Google Scholar]
- 27.Koornneef M., Veen J. H. (1980) Theor. Appl. Genet. 58, 257–263 [DOI] [PubMed] [Google Scholar]
- 28.Cheng H., Qin L., Lee S., Fu X., Richards D. E., Cao D., Luo D., Harberd N. P., Peng J. (2004) Development 131, 1055–1064 [DOI] [PubMed] [Google Scholar]
- 29.Barrow C. J., Yasuda A., Kenny P. T., Zagorski M. G. (1992) J. Mol. Biol. 225, 1075–1093 [DOI] [PubMed] [Google Scholar]
- 30.Oldfield C. J., Cheng Y., Cortese M. S., Romero P., Uversky V. N., Dunker A. K. (2005) Biochemistry 44, 12454–12470 [DOI] [PubMed] [Google Scholar]
- 31.Cheng Y., Oldfield C. J., Meng J., Romero P., Uversky V. N., Dunker A. K. (2007) Biochemistry 46, 13468–13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson W. C., Jr. (1988) Annu. Rev. Biophys. Biophys. Chem. 17, 145–166 [DOI] [PubMed] [Google Scholar]
- 34.Soulages J. L., Kim K., Walters C., Cushman J. C. (2002) Plant Physiol. 128, 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radivojac P., Iakoucheva L. M., Oldfield C. J., Obradovic Z., Uversky V. N., Dunker A. K. (2007) Biophys. J. 92, 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sickmeier M., Hamilton J. A., LeGall T., Vacic V., Cortese M. S., Tantos A., Szabo B., Tompa P., Chen J., Uversky V. N., Obradovic Z., Dunker A. K. (2007) Nucleic Acids Res. 35, D786–D793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uversky V. N., Gillespie J. R., Fink A. L. (2000) Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 38.Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., Dunker A. K. (2001) Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 39.Schlessinger A., Liu J., Rost B. (2007) PLoS Comput. Biol. 3, 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohan A., Oldfield C. J., Radivojac P., Vacic V., Cortese M. S., Dunker A. K., Uversky V. N. (2006) J. Mol. Biol. 362, 1043–1059 [DOI] [PubMed] [Google Scholar]
- 41.Deane C. M., Salwiński Ł., Xenarios I., Eisenberg D. (2002) Mol. Cell Proteomics 1, 349–356 [DOI] [PubMed] [Google Scholar]
- 42.Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. (2002) Mol. Cell Proteomics 1, 930–946 [DOI] [PubMed] [Google Scholar]
- 43.Crick S. L., Jayaraman M., Frieden C., Wetzel R., Pappu R. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16764–16769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen A. A., Pappu R. V. (2007) J. Phys. Chem. B 111, 11884–11887 [DOI] [PubMed] [Google Scholar]
- 45.Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., Hongyu X., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. (2007) Plant Cell 19, 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 47.Liu J., Perumal N. B., Oldfield C. J., Su E. W., Uversky V. N., Dunker A. K. (2006) Biochemistry 45, 6873–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uversky V. N., Oldfield C. J., Dunker A. K. (2005) J. Mol. Recognit. 18, 343–384 [DOI] [PubMed] [Google Scholar]
- 49.Dunker A. K., Cortese M. S., Romero P., Iakoucheva L. M., Uversky V. N. (2005) FEBS J. 272, 5129–5148 [DOI] [PubMed] [Google Scholar]
- 50.Gsponer J., Futschik M. E., Teichmann S. A., Babu M. M. (2008) Science 322, 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uversky V. N., Dunker A. K. (2008) Science 322, 1340–1341 [DOI] [PubMed] [Google Scholar]
- 52.Schwechheimer C. (2008) Curr. Opin. Plant Biol. 11, 9–15 [DOI] [PubMed] [Google Scholar]
- 53.Silverstone A. L., Tseng T. S., Swain S. M., Dill A., Jeong S. Y., Olszewski N. E., Sun T. P. (2007) Plant Physiol. 143, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu X., Richards D. E., Ait-Ali T., Hynes L. W., Ougham H., Peng J., Harberd N. P. (2002) Plant Cell 14, 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussain A., Cao D., Cheng H., Wen Z., Peng J. (2005) Plant J. 44, 88–99 [DOI] [PubMed] [Google Scholar]
- 56.Fuxreiter M., Simon I., Friedrich P., Tompa P. (2004) J. Mol. Biol. 338, 1015–1026 [DOI] [PubMed] [Google Scholar]
- 57.Vacic V., Oldfield C. J., Mohan A., Radivojac P., Cortese M. S., Uversky V. N., Dunker A. K. (2007) J. Proteome Res. 6, 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hua Q. X., Jia W. H., Bullock B. P., Habener J. F., Weiss M. A. (1998) Biochemistry 37, 5858–5866 [DOI] [PubMed] [Google Scholar]
- 59.Willige B. C., Ghosh S., Nill C., Zourelidou M., Dohmann E. M., Maier A., Schwechheimer C. (2007) Plant Cell 19, 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen C. K., Chang C. (2002) Plant Cell 14, 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyler L., Thomas S. G., Hu J., Dill A., Alonso J. M., Ecker J. R., Sun T. P. (2004) Plant Physiol. 135, 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uversky V. N. (1993) Biochemistry 32, 13288–13298 [DOI] [PubMed] [Google Scholar]
- 63.Xie H., Vucetic S., Iakoucheva L. M., Oldfield C. J., Dunker A. K., Obradovic Z., Uversky V. N. (2007) J. Proteome Res. 6, 1917–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]