Abstract
Coagulation factor XII (FXII) is a liver-derived serine protease involved in fibrinolysis, coagulation, and inflammation. The regulation of FXII expression is largely unknown. Transforming growth factor-β1 (TGF-β1) is a multifunctional cytokine that has been linked to several pathological processes, including tissue fibrosis by modulating procoagulant and fibrinolytic activities. This study investigated whether TGF-β1 may regulate FXII expression in human lung fibroblasts. Treatment of human lung fibroblasts with TGF-β1 resulted in a time-dependent increase in FXII production, activation of p44/42, p38, JNK, and Akt, and phosphorylation and translocation into the nucleus of Smad3. However, TGF-β1-induced FXII expression was repressed only by the JNK inhibitor and JNK and Smad3 antisense oligonucleotides but not by MEK, p38, or phosphoinositide 3-kinase blockers. JNK inhibition had no effect on TGF-β1-induced Smad3 phosphorylation, association with Smad4, and its translocation into the nucleus but strongly suppressed Smad3-DNA complex formation. FXII promoter analysis revealed that the −299/+1 region was sufficient for TGF-β1 to induce FXII expression. Sequence analysis of this region detected a potential Smad-binding element at position −272/−269 (SBE-(−272/−269)). Chromatin immunoprecipitation and streptavidin pulldown assays demonstrated TGF-β1-dependent Smad3 binding to SBE-(−272/−269). Mutation or deletion of SBE-(−272/−269) substantially reduced TGF-β1-mediated activation of the FXII promoter. Clinical relevance was demonstrated by elevated FXII levels and its co-localization with fibroblasts in the lungs of patients with acute respiratory distress syndrome. Our results show that JNK/Smad3 pathway plays a critical role in TGF-β1-induced FXII expression in human lung fibroblasts and implicate its possible involvement in pathological conditions characterized by elevated TGF-β1 levels.
Keywords: Blood Coagulation/Coagulation Factors, Chromatin/Immunoprecipitation/ChIP, Cytokines/TGFbeta, Gene/Promoters, Signal Transduction/Protein Kinases/MAP, Transcription/Smad
Introduction
Coagulation factor XII (FXII,2 Hageman factor) is a serine protease that is capable of initiating blood coagulation as well as activating the fibrinolytic system. FXII is predominantly synthesized in the liver and secreted into the blood as a single-chain zymogen. Upon binding to negatively charged surfaces, FXII becomes converted into a two-chain active enzyme (FXIIa), which exhibits serine proteinase-like activity at its C terminus. Factor XIIa converts factor XI into activated factor XI and prekallikrein into kallikrein. Consequently, factor XI activation culminates in a series of proteolytic reactions resulting in thrombin generation and subsequent clot formation. Kallikrein cleaves high molecular weight kininogen releasing a potent proinflammatory and vasodilatory peptide, bradykinin. Under physiological conditions, the C1 inhibitor controls this cascade by inactivating FXIIa and kallikrein (1, 2).
The function of FXII in hemostasis is not clear because FXII deficiency is not associated with bleeding disorders. However, accumulating evidence suggests that FXII may play an important role in pathological thrombus formation (3) and inflammation, for example by kallikrein and bradykinin generation (4). In addition, FXII may down-regulate Fc receptors on monocytes (5) and can stimulate monocytes and macrophages to release interleukin (IL)-1 and IL-6 (6).
There is not much known about the regulation of FXII production in the liver and other organs. Previous reports demonstrated a strong induction of FXII expression by 17β-estradiol in transiently transfected mouse NIH3T3 fibroblasts and human HepG2 hepatoma cells (7, 8). These results were further supported by the identification of four putative binding sites for the hepatocyte nuclear factor-3 in the 5′-flanking region of the FXII gene (9). Furthermore, elevated FXII plasma levels and hepatic FXII mRNA steady states were noted in ovariectomized rats treated with 17β-estradiol (10, 11). In line with these data, increased FXII plasma levels were also detected in women during pregnancy (12), treatment with oral contraceptives (13), or women receiving estrogen replacement therapy (14). In addition, IL-6 was found to down-regulate FXII expression in HepG2 cells, whereas no change in FXII production was observed upon stimulation with IL-1β or tumor necrosis factor-α (15).
Although regulation of FXII expression by estrogen and IL-6 is well described, no data are available about the regulation of FXII synthesis by other molecules such as growth factors. Transforming growth factor-β1 (TGF-β1) is a highly pleiotropic cytokine that plays a fundamental role in wound healing, embryonic development, and diseases associated with inflammation and proliferation, such as tissue fibrosis. TGF-β1 participates in these processes by enhancing synthesis and inhibiting degradation of extracellular matrix components and by regulating fibroblast differentiation and proliferation (16, 17). Moreover, recent studies suggest that TGF-β1 may contribute to pathological conditions by modulating procoagulant and fibrinolytic activities. In particular, TGF-β1 has been shown to up-regulate the expression of tissue factor (18), the key initiator of the extrinsic coagulation pathway and of plasminogen activator inhibitor-1 in different cell populations, including fibroblasts (19–21). The cellular response to TGF-β1 involves ligand binding to TGF-β receptor type II, which phosphorylates the TGF-β receptor type I (TβRI). Activated TβRI phosphorylates receptor-associated Smad proteins (Smad1–3, -5, and -8), promoting their heterodimerization with the common mediator Smad4 and translocation from the cytoplasm into the nucleus. Within the nucleus, the Smad heterocomplex interacts with canonical Smad-binding elements (SBEs) of target genes to activate their transcription (22, 23). Human Smad3 and Smad4 have been shown to bind to an SBE comprising a CAGA box (19).
It is currently unknown whether TGF-β1 may modulate FXII expression, and whether this phenomenon might contribute to disease conditions that exhibit perturbed procoagulant and/or fibrinolytic activity. Therefore, the objective of this study was to determine the potential role of TGF-β1 in the regulation of FXII synthesis in human lung fibroblasts (HLF). We further explored the intracellular mechanism by which TGF-β1 may modulate FXII expression and identified the FXII promoter region necessary for TGF-β1-mediated FXII production. There are several reasons why we chose HLF to study FXII expression. Abnormalities of local coagulation favoring excessive deposition of extravascular fibrin occur in different forms of acute and chronic lung diseases (24–28). Moreover, several lines of evidence indicate that TGF-β1 modulates the function of lung fibroblasts during the course of various pulmonary diseases (29–32) and that the interaction of fibroblasts with the disordered coagulation system plays an important role in lung tissue injury and pathological lung tissue remodeling (33–41). In this respect, fibroblasts have already been identified as an important source of extrinsic coagulation cascade factors in experimental animal models of lung injury as well as in in vitro studies after stimulation with TGF-β1 and other cytokines that occur in the diseased lung (18, 20, 42).
EXPERIMENTAL PROCEDURES
Isolation of HLF and Cell Culture
Lung specimens of the pulmonary parenchyma were chopped into <1-mm3 pieces. The minced pieces were washed twice with PBS (137 mm NaCl, 1.7 mm KCl, 10 mm Na2HPO4, 1.7 mm KH2PO4, pH 7.4) and then plated in 100-mm dishes (Greiner Bio-One, Frickenhausen, Germany). The specimens were cultured with Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (HyClone, South Logan, UT) and 1% penicillin/streptomycin (Invitrogen) in humidified atmosphere of 5% CO2 at 37 °C. The purity of isolated fibroblasts was verified by positive staining for vimentin, fibronectin, and collagen IV and by negative staining for α-smooth muscle actin, pro-surfactant protein C, and von Willebrand factor. All experiments were carried out with HLF between passages 3 and 4. The mouse NIH3T3 fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1% penicillin/streptomycin in humidified atmosphere of 5% CO2 at 37 °C.
Bronchoalveolar Lavage
Bronchoalveolar lavage (BAL) fluids were obtained by flexible fiberoptic bronchoscopy from healthy volunteers (n = 9) and from patients with acute respiratory distress syndrome (ARDS; n = 9) as described previously (43). In addition to BAL fluids, lung specimens from seven patients with ARDS were obtained by autopsy. All patients met the ARDS American-European Consensus criteria (44). All investigations were approved by the local ethics committee, and written informed consent was obtained from all participants or their next-of-kin.
Cell Stimulation
HLF and NIH3T3 were seeded in 6-well plates and were either not stimulated or were stimulated for various times with 10 ng/ml TGF-β1 (R & D Systems, Wiesbaden, Germany). In some experiments, the cells were preincubated with various concentrations of PD98059, SP600125, wortmannin, SB203580, or SB431542 inhibitors (all from Calbiochem) 1 h prior to the addition of TGF-β1.
FXII Immunoassay
FXII levels were quantified in BAL fluid samples using a matched pair antibody set for enzyme-linked immunosorbent assay of human FXII antigen according to the manufacturer's instructions (Affinity Biologicals, Ancaster, Canada).
Western Blotting
Cell lysates were separated on SDS-polyacrylamide gels under reducing conditions, followed by electrotransfer to a polyvinylidene difluoride membrane. After blocking with 5% nonfat dry milk in Tris-buffered saline (TBS: 25 mm Tris-Cl, 150 mm NaCl, pH 7.5) containing 0.1% (v/v) Tween 20 (TBS-T), the membranes were incubated at 4 °C overnight with one of the following antibodies: mouse anti-FXII (Abcam, Cambridge, UK), mouse anti-phospho-p44/42, rabbit anti-phospho-Akt, rabbit anti-phospho-p38, rabbit anti-Smad4, rabbit anti-phospho-Smad3 (Ser-423/Ser-425) (all from Cell Signaling, Frankfurt am Main, Germany), rabbit anti-phospho-Smad3 (Ser-208), rabbit anti-phospho-Smad3 (Ser-213; Abnova, Heidelberg, Germany), or rabbit anti-phospho-JNK (R & D Systems). After 1 h of incubation with peroxidase-labeled secondary antibody (Dako, Gostrup, Denmark), proteins were detected using an ECL Plus kit (Amersham Biosciences). To determine the amounts of protein loaded on the gel, blots were stripped and reprobed using rabbit anti-p44/42, rabbit anti-Akt, rabbit anti-p38, rabbit anti-Smad3 (all from Cell Signaling), rabbit anti-JNK (R & D Systems), rabbit anti-lamin B, rat anti-tubulin (all from Abcam, Cambridge, MA), and mouse anti-β-actin (Sigma) antibodies.
Co-immunoprecipitation
HLF lysates were incubated overnight at 4 °C with 1 μg of rabbit anti-Smad3 antibody (Cell Signaling) or IgG as an isotype control (R & D Systems). Samples were transferred to tubes containing 50 μl of protein A-SepharoseTM CL-4B beads (Amersham Biosciences). After 1 h of incubation at 4 °C, the immunoprecipitates were washed several times with lysis buffer, boiled in SDS sample buffer, separated by SDS-PAGE under reducing conditions, and transferred to a polyvinylidene difluoride membrane. Immunoblots were analyzed using mouse anti-Smad4 (Abcam) and rabbit anti-Smad3 antibodies (Cell Signaling).
RNA Isolation and Real Time Reverse Transcription-PCR
Total RNA was extracted using a PeqGOLD total RNA kit (PeqLab, Erlangen, Germany) according to the manufacturer's instructions. One μg of RNA was reverse-transcribed as described previously (45). Real time PCR was performed by Sequence Detection System 7700 (PerkinElmer Life Sciences). Reactions were set up with Platinum SYBR Green quantitative PCR SuperMix uracil-DNA glycosylase (Invitrogen) using 2 μl of cDNA. The following pairs of oligonucleotide primers were used: human FXII forward, 5′-ACGACCTGGCTCTGTTGC-3′, and human FXII reverse, 5′-CTTGGCAGGCACACCGG-3′; human β-actin forward, 5′-ATTGCCGACAGGATGCAGGAA-3′, and human β-actin reverse, 5′-GCTGATCCACATCTGCTGGAA-3′. The β-actin gene was used as a reference gene. Cycling conditions were 95 °C for 6 min, followed by 45 cycles of 95 °C for 20 s, 55 °C for 30 s, and 73 °C for 30 s. Melting curve analysis and gel electrophoresis were performed to confirm the exclusive amplification of the expected PCR product. Gene expression was assessed using the 2ΔΔCT method for the calculation of fold change as described previously (46).
Immunocytochemistry
For immunocytochemical analysis, HLF were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked with 3% bovine serum albumin in PBS for 1 h at room temperature, and incubated overnight at 4 °C with one of the following antibodies: mouse anti-FXII, rabbit anti-collagen IV, mouse anti-fibronectin (all from Abcam), goat anti-vimentin (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-α-smooth muscle actin (Sigma), rabbit anti-pro-surfactant protein C (Millipore; Schwalbach, Germany), rabbit anti-von Willebrand factor (Dako), and rabbit anti-phospho-Smad3 (Ser-423/Ser-425; Cell Signaling). Slides were incubated with rhodamine- or fluorescein-conjugated secondary antibody (Dianova, Hamburg, Germany) and mounted with Vectashield mounting medium (Vector, Burlingame, CA). Nuclei were visualized by 4′,6-diamidino-2-phenylindole (Sigma) staining. Controls were performed by substituting the primary antibody by a nonspecific antibody. Images were captured by a Leica DMR microscope (Leica, Heidelberg, Germany) with 40×/1.25–0.75 oil objective at room temperature and photographed using MetaMorph 7.0 (Molecular Devices, Downingtown, PA). All images illustrated are representative of at least four other areas per section, seen on at least three independent sections.
Immunohistochemistry
Immunohistochemistry for the detection of FXII in formalin-fixed paraffin-embedded lung tissue was performed using a Histostain-SP kit according to the manufacturer's instructions (Zymed Laboratories Inc.). The sections were incubated with a rabbit anti-FXII antibody (Abcam) or a mouse anti-vimentin antibody (Zymed Laboratories Inc.). The slides were scanned by a Mirax Desk Digital Slide Scanner (Zeiss, Göttingen, Germany) and analyzed by Mirax Viewer (Zeiss).
Clotting Assay
A clotting assay was performed with human FXII-deficient plasma (CSL-Behring, Marburg, Germany) as described previously (47). In some experiments, ARDS BAL fluids were supplemented with 100 nm corn trypsin inhibitor (CTI, Calbiochem), 10 μg/ml anti-TF blocking antibody (kindly provided by W. Ruf, Scripps Research Institute, La Jolla, CA), or 10 μg/ml IgG isotype control (R & D Systems).
Generation of FXII Promoter Constructs and Site-directed Mutagenesis
The human FXII promoter fragments were amplified by PCR from human lung DNA using Long PCR Enzyme Mix (Fermentas, St. Leon-Rot, Germany) according to the manufacturer's instructions. The following primers were used: human FXII-1630 forward, 5′-CCGCTCGAGTGCTCTGTGCTTAGTAACC-3′; human FXII-907 forward, 5′-CCGCTCGAGCAGCTACCCAGGAGGCT-3′; human FXII-577 forward, 5′-CCGCTCGAGGCGTGGTGGTGGGCTCCT-3′; human FXII-541 forward, 5′-CCGCTCGAGGAGGCTGAGGCAGGAG-3′; human FXII-504 forward, 5′-CCGCTCGAGGGAGCTTGCAGTGAGC-3′; human FXII-455 forward, 5′-CCGCTCGAGCAGAGCGAGACTCCGTC-3′; human FXII-404 forward, 5′-CCGCTCGAGGTGGGTATTGTTGTAAG-3′; human FXII-346 forward, 5′-CCGCTCGAGGAACACACTTCACAGTG-3′; human FXII-299 forward, 5′-CCGCTCGAGCTTAACCTCCTGATCTCC-3′; human FXII-183ΔSBE-(−272/−269) forward, 5′-CCGCTCGAGAAACTCCCAAACTTTCC-3′; and human FXII reverse, 5′-CCCAAGCTTCGTTGGTCCAGCTGCCTATC-3′. The PCR fragments were cloned into the pGL3 Enhancer Vector (Promega, Mannheim, Germany) using XhoI and HindIII restriction sites (shown in boldface). A point mutation introduced into the CAGA box and a deletion of the repressor element were performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Successful cloning, insertion of the mutation into the CAGA box, and deletion of the repressor element were confirmed by sequencing.
Transient Transfection and Luciferase Assay
NIH3T3 cells were transfected with the indicated plasmids using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. After 48 h, cells were either not stimulated or were stimulated with 10 ng/ml TGF-β1 (R & D Systems) for 16 h. Subsequently, the cells were harvested and assayed for luciferase reporter activity using Promega luciferase assay kit according to manufacturer's instructions. Co-transfection with pEGF-N1 (Clontech) control vector was used to normalize for transfection efficiency.
Antisense Oligonucleotides
Pre-designed, commercially available siRNA sequences directed against human Smad3 (Dharmacon, Chicago, IL), human JNK (Abnova, Heidelberg, Germany), and a universal negative control siRNA (Ambion, Austin, TX) were employed. Cells were treated with siRNA (250 nm each) using the X-treme Gene siRNA transfection reagent (Roche Applied Science). The siRNA-mediated down-regulation of the target genes was assessed 48 h after transfection by Western blotting. At this time point, cells were either not stimulated or were stimulated with 10 ng/ml TGF-β1 for 24 h, and the Western blots for FXII or JNK were prepared as described above.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using the chromatin immunoprecipitation assay kit from Millipore (Schwalbach, Germany) according to the manufacturer's instructions. Briefly, HLF either not stimulated or stimulated with 10 ng/ml TGF-β1 (R & D Systems) were treated with 1% formaldehyde for 10 min. The cross-linked chromatin was then sonicated to an average size of 300–400 bp. The DNA fragments were immunoprecipitated with rabbit anti-Smad3 antibody (Cell Signaling) or IgG isotype control antibody, overnight at 4 °C. After reversal of cross-linking, the immunoprecipitated chromatin was amplified by PCR using the following primers: human FXII-299 forward, 5′-CTTAACCTCCTGATCTCC-3′; human FXII-299 reverse, 5′-CGTTGGTCCAGCTGCCTATC-3′. PCR products were separated on a 2% agarose gel and visualized by ethidium bromide staining. Amplicons comprise the FXII promoter region between −299 and +1 bp.
Streptavidin Pulldown Assay
The double-stranded biotinylated DNA fragment (T1-(−282/−258), 5′-biotin, CTTAACCTCCTGATCTCCACAGGACCCAGAGCATAAGAATGTCCC-3′, or T2-(−282/−258 C/T) 5′-biotin, CTTAACCTCCTGATCTCCACAGGACCTAGAGCATAAGAATGTCCC-3′) spanning the SBE was assayed for protein interaction in 100 μl of binding reaction containing 20 μg of nuclear extract, 20 pmol/μl biotinylated template, and 1 μg of poly(dI-dC). After incubation for 1 h at 30 °C, streptavidin MagneSphere paramagnetic particles (Promega) pre-equilibrated in binding buffer (20 mm HEPES, pH 7.9, 80 mm KCl, 10 mm MgCl2, 10% (v/v) glycerol, 2 mm dithiothreitol, 500 μg/ml of bovine serum albumin and 0.05% (v/v) Nonidet P-40) were added to the reaction and incubated for another 1 h at 30 °C. The DNA-protein complexes were washed three times with wash buffer (20 mm HEPES, pH 7.9, 50 mm KCl, 6.25 mm MgCl2, 0.5 mm EDTA, 2 mm dithiothreitol, and 8.5% (v/v) glycerol) using a magnetic device (Dynal MPC®-E, Magnetic Particle Concentrator). After boiling, the DNA-bound proteins were analyzed by Western blot using rabbit anti-phospho-Smad3 antibody (Cell Signaling). Nuclear extracts were prepared using NE-PER® nuclear and cytoplasmic extraction reagent (Pierce) according to the manufacturer's instructions.
Statistics
Data are given as means ± S.D. Statistical significance was assessed using one-way analysis of variance followed by the Tukey-HSD post hoc test, and paired samples were analyzed using the two-tailed Student's t test. For evaluation of differences between healthy volunteers and patients, the two-tailed Mann-Whitney U test was used. A level of p < 0.05 was considered statistically significant.
RESULTS
TGF-β1 Up-regulates FXII mRNA and Protein Levels in HLF
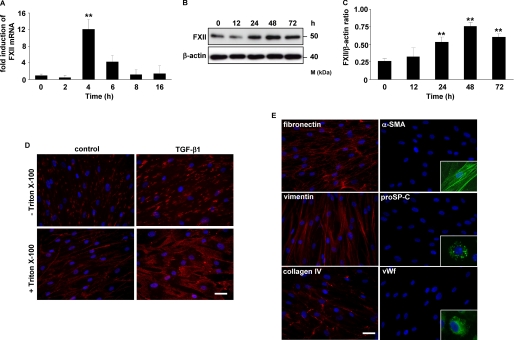
Exposure of HLF to 10 ng/ml TGF-β1 stimulated the synthesis of FXII in a time-dependent manner. Real time reverse transcription-PCR analysis demonstrated the strongest induction of FXII mRNA expression at 4 h of treatment (Fig. 1A). The maximal FXII protein level was achieved within a 48-h stimulation period and slightly declined at 72 h (Fig. 1, B and C). Immunofluorescence staining revealed pronounced expression of FXII in HLF in response to TGF-β1. FXII was detected on the cell surface (Fig. 1D, top panel) as well as in the cytoplasmic compartment of HLF (Fig. 1D, bottom panel). The purity of isolated HLF was verified by positive staining for fibronectin, vimentin, and collagen IV and by negative staining for α-smooth muscle actin, pro-surfactant protein C, and von Willebrand factor (vWF, Fig. 1E).
FIGURE 1.
TGF-β1 up-regulates FXII expression in HLF. A and B, time course of FXII expression in HLF following TGF-β1 stimulation as assessed by real time PCR (A) and Western blotting (B). Real time PCR results are expressed as the fold increase in FXII expression (normalized for β-actin expression) versus values obtained for unstimulated cells and are means ± S.D.; n = 3; **, p < 0.01. The Western blot illustrated is from one representative experiment out of four. C, densitometric analysis of B. Data are presented as means ± S.D.; n = 4; **, p < 0.01, all versus unstimulated cells. D, immunofluorescence for the detection of FXII in unstimulated (control) or TGF-β1-treated HLF. Original magnification was ×40/1.25–0.75 oil objective. Bar size, 10 μm. E, immunofluorescence staining of HLF for fibronectin, vimentin, collagen IV, α-smooth muscle actin, pro-surfactant protein C, and von Willebrand factor. Original magnification was 40×/1.25–0.75 oil objective. Bar size, 10 μm. The insets are controls and show the positive staining of pulmonary artery smooth muscle cells for α-smooth muscle actin, of alveolar type II cells for pro-surfactant protein C, and of pulmonary artery endothelial cells for von Willebrand factor (vWf). α-SMA, α-smooth muscle actin; proSP-C, pro-surfactant protein C.
TGF-β1 Induces Phosphorylation of MAPK, Akt, and Smad3 in HLF
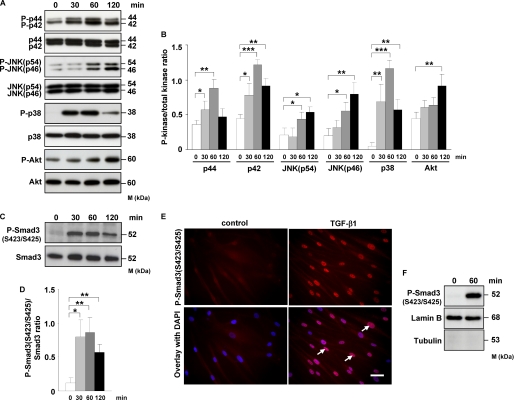
To dissect the contribution of different signal transduction pathways to TGF-β1-induced FXII production in HLF, we first analyzed phosphorylation kinetics of different MAPKs, Akt, and Smad3 in response to TGF-β1 stimulation. Phosphorylation of p42/44 and p38 kinases peaked at 60 min and then decreased. A marked increase in JNK activity was visible after 60 min, whereas enhanced phosphorylation of Akt was noted after 120 min (Fig. 2, A and B). No differences in activation of c-Jun were detected (data not shown). As expected, TGF-β1 induced rapid C-terminal phosphorylation of Smad3 (Ser-423/Ser-425) with maximal response within 30–60 min (Fig. 2, C and D). Immunofluorescence analysis demonstrated TGF-β1-induced translocation of phospho-Smad3 (Ser-423/Ser-425) into the nucleus of HLF (Fig. 2E). Accordingly, increased levels of phospho-Smad3 (Ser-423/Ser-425) were observed in the nuclear extracts after TGF-β1 treatment as assessed by Western blotting (Fig. 2F).
FIGURE 2.
TGF-β1 induces phosphorylation of MAPK, Akt, and Smad3, as well as translocation of phospho-Smad3 into the nucleus of HLF. A, HLF were treated for the indicated time points with TGF-β1, and the activation of p44/42, JNK, p38, and Akt kinases as assessed by phosphorylation was analyzed by Western blotting. Phosphoproteins were detected with phospho-specific antibodies. Equal loading was confirmed with pan-specific antibodies. Data are representative of four independent experiments. B, densitometric analysis of A. Data are presented as means ± S.D.; n = 4; *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, Western blot analysis of Smad3 phosphorylation in TGF-β1-stimulated HLF. Data are representative of four independent experiments. D, densitometric analysis of C. Data are presented as means ± S.D.; n = 4; *, p < 0.05; **, p < 0.01. E, TGF-β1-dependent translocation of phospho-Smad3 (Ser-423/Ser-425) into the nucleus as assessed by immunofluorescence. HLF were incubated with TGF-β1 for 1 h and then washed, fixed, and stained with phospho-Smad3 (Ser-423/Ser-425) antibody. Arrows indicate nuclear localization of Smad3. Original magnification was ×40/1.25–0.75 oil objective. Bar size, 10 μm. Data are representative of three independent experiments. F, Western blot analysis of TGF-β1-driven translocation of phospho-Smad3 (Ser-423/Ser-425) into the nucleus. HLF were unstimulated or treated for 1 h with TGF-β1; nuclear extracts were prepared and immunoblotted with antibodies against phospho-Smad3, lamin B, and tubulin. Lamin B was used as a loading control, and tubulin was used to assess the purity of the nuclear fraction. Data are representative of three independent experiments.
Smad3 and JNK Kinase Regulate TGF-β1-induced FXII Expression in HLF
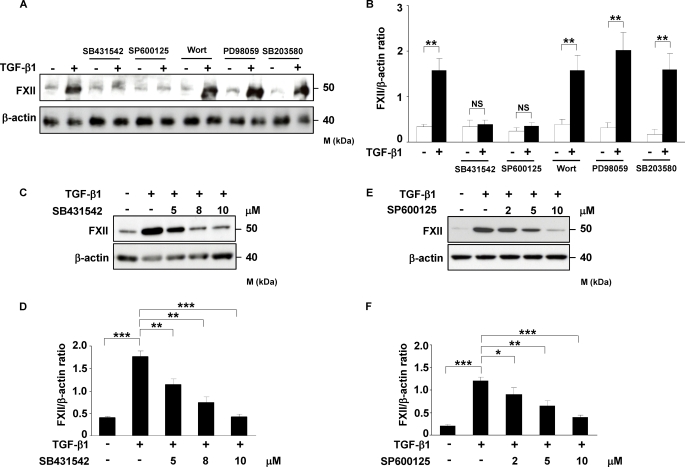
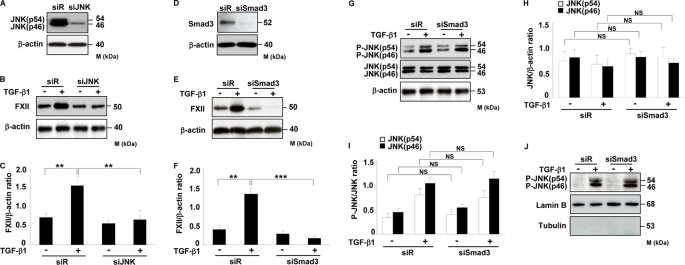
After determination of the phosphorylation kinetics, we next sought to analyze whether interference with these pathways would affect TGF-β1-induced FXII expression in HLF. To address this, we used specific inhibitors of TβRI, JNK, PI3K, MEK, and p38 kinase (SB431542, SP600125, wortmannin, PD98059, and SB203580, respectively) and evaluated their effect on TGF-β1-stimulated expression of FXII. As depicted in Fig. 3, A–F, inhibition of TβRI and JNK activity by SB431542 and SP600125, respectively, led to strong and dose-dependent reduction in FXII expression in response to TGF-β1. No changes in FXII expression were visible when HLF were pretreated with inhibitors of PI3K, MEK, and p38 kinase (Fig. 3, A and B, and supplemental Fig. 1S). To further confirm these results, HLF were transfected with JNK- or Smad3-specific siRNAs, which caused significant knockdown of these proteins as demonstrated by Western blotting (Fig. 4, A and D). As shown in Fig. 4, B and C, knockdown of JNK resulted in significant inhibition of FXII expression after TGF-β1 stimulation. Similar results were obtained when Smad3 was depleted (Fig. 4, E and F). Of note, knockdown of Smad3 did not have any influence on JNK expression (Fig. 4, G and H), phosphorylation (Fig. 4, G and I), and translocation into the nucleus (Fig. 4J) indicating that JNK production, activation, and nuclear import are not affected by the Smad signaling pathway.
FIGURE 3.
TβRI and JNK activities are required to regulate TGF-β1-induced FXII expression in HLF. A, Western blot analysis of TGF-β1-induced FXII expression in HLF. HLF were treated with TβRI (SB431542; 8 μm), JNK (SP600125; 5 μm), PI3K (wortmannin; 80 nm), MEK (PD98059; 30 μm), or p38 (SB203580; 8 μm) inhibitors for 1 h prior to incubation with TGF-β1 for 48 h. Cell lysates were prepared, and FXII expression was examined. β-Actin was used as a loading control. The Western blot illustrated is from one representative experiment out of four. B, densitometric analysis of A. Data are presented as means ± S.D.; n = 4; **, p < 0.01. C and E, Western blot analysis of TGF-β1-induced FXII expression in HLF pretreated for 1 h with various concentrations of SB431542 (C) or SP600125 (E). β-Actin was used as a loading control. The Western blot illustrated is from one representative experiment out of three. D and F, densitometric analysis of C and E, respectively. Data are presented as mean ± S.D.; n = 3; *, p < 0.05; **, p < 0.01; ***,p < 0.001. wort, wortmannin; NS, not significant.
FIGURE 4.
Smad3 and JNK regulate TGF-β1-induced FXII expression in HLF. A and D, determination of knockdown efficiency in HLF after transfection with siRNA against (A) JNK or (D) Smad3 as assessed by Western blotting. Data are representative of three independent experiments. B and E, effect of JNK (B) or Smad3 (E) knockdown on TGF-β1-induced FXII expression in HLF. Data are representative of four independent experiments. C and F, densitometric analysis of B and E, respectively. Data are presented as means ± S.D.; n = 4; **, p < 0.01; ***, p < 0.001. G, impact of Smad3 knockdown on TGF-β1-induced JNK expression and phosphorylation. Data are representative of three independent experiments. H and I, densitometric analysis of G. Data are presented as means ± S.D.; n = 3. J, effect of Smad3 depletion on JNK translocation into the nucleus. Nuclear extracts were prepared and immunoblotted with antibodies against phospho-JNK, lamin B, and tubulin. Lamin B was used as a loading control, and tubulin was used to assess the purity of the nuclear fraction. Data are representative of three independent experiments. siR, scramble siRNA; NS, not significant.
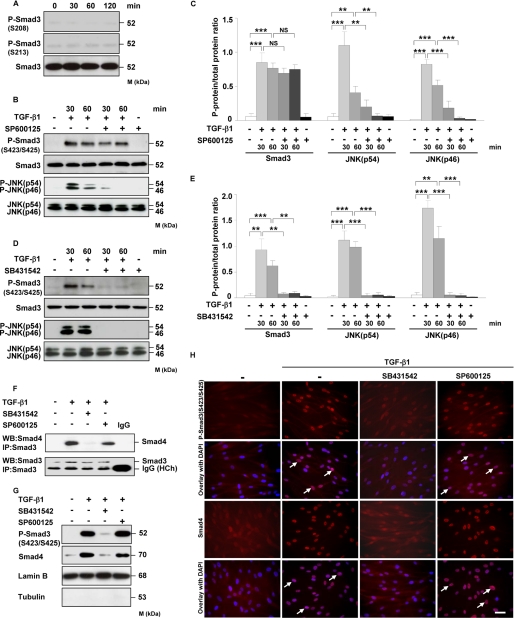
JNK Kinase Does Not Regulate Smad3 Phosphorylation, Smad3-Smad4 Complex Formation, and Its Translocation into the Nucleus
As putative JNK phosphorylation sites located within Smad3 linker region (Ser-208/Ser-213) have been identified (48), we examined their phosphorylation in HLF in response to TGF-β1. However, none of the aforementioned serine residues was phosphorylated in the presence of TGF-β1 (Fig. 5A). We next tested the role of JNK in Smad3 C-terminal phosphorylation, complex formation with Smad4, and its translocation into the nucleus. Incubation of TGF-β1-treated HLF with JNK inhibitor (SP600125) did not interfere with the C-terminal phosphorylation of Smad3 but abolished JNK activity (Fig. 5, B and C). These results suggest that the JNK pathway does not affect the upstream interaction between Smad3 and TβRI kinase. A TβRI inhibitor (SB431542) attenuated phosphorylation of Smad3 and JNK in response to TGF-β1 (Fig. 5, D and E) indicating that phosphorylation of Smad3 and JNK originates from TβRI, the most proximal molecule in TGF-β1 signal transduction pathway.
FIGURE 5.
JNK does not regulate Smad3 phosphorylation, association with Smad4, and its translocation into the nucleus. A, HLF were treated for the indicated time points with TGF-β1, and the phosphorylation of Smad3 at Ser-208 and Ser-213 was examined by Western blotting. Equal loading was confirmed with pan-specific antibody. Data are representative of four independent experiments. B and D, HLF were treated with TGF-β1 in the absence or presence of JNK inhibitor (SP600125) (B) or TβRI inhibitor (SB431542) (D) as indicated, and the activation of Smad3 and JNK as assessed by phosphorylation was analyzed by Western blotting. Data are representative of four independent experiments. C and E, densitometric analysis of B and D, respectively. Data are presented as mean ± S.D.; n = 4; **, p < 0.01; ***, p < 0.001. F, HLF were preincubated with SB431542 or SP600125 1 h prior to addition of TGF-β1. After 1 h, the cell lysates were prepared and immunoprecipitated (IP) with anti-Smad3 antibody. Coprecipitating Smad4 was detected by Western blotting (WB). The lower band present in the bottom panel represents a heavy chain (HCh) of anti-Smad3 antibody and isotype control. Data are representative of three independent experiments. G, Western blot analysis of TGF-β1-driven translocation of phospho-Smad3 (Ser-423/Ser-425) and Smad4 into the nucleus. HLF were pretreated with SB431542 or SP600125 and then either not stimulated or stimulated for 1 h with TGF-β1. Nuclear extracts were prepared and immunoblotted with antibodies against phospho-Smad3 (SER-423/Ser-425), Smad4, lamin B, and tubulin. Lamin B was used as a loading control, and tubulin was used to assess the purity of the nuclear fraction. Data are representative of three independent experiments. H, TGF-β1-driven translocation of phospho-Smad3 (Ser-423/Ser-425) and Smad4 into the nucleus as assessed by immunofluorescence. HLF were preincubated with SB431542 or SP600125 1 h prior to addition of TGF-β1. After 1 h, the cells were washed, fixed, and stained with phospho-Smad3 (Ser-423/Ser-425) and Smad4 antibodies. Arrows indicate nuclear localization of Smad3 and Smad4. Original magnification was ×40/1.25–0.75 oil objective. Bar size, 10 μm. Data are representative of three independent experiments.
To determine the impact of JNK on Smad3-Smad4 interaction, we performed a series of co-immunoprecipitations. As indicated in Fig. 5F, treatment of HLF with TGF-β1 stimulated the interaction between Smad3 and Smad4. Moreover, Smad3-Smad4 association was visible when HLF were pretreated with SP600125 but completely abolished in the presence of SB431542. As Smad3-Smad4 interaction leads to changes in the subcellular localization of these proteins, we next investigated Smad3 and Smad4 movement into the nucleus. As shown in Fig. 5G, exposure of HLF to SP600125 had no effect on TGF-β1-induced Smad3 and Smad4 translocation into the nucleus, whereas SB431542 completely blocked this process. Similar results were obtained by immunofluorescence analysis (Fig. 5H). SB431542 and SP600125 alone did not impact Smad3-Smad4 complex formation and its accumulation in the nucleus (data not shown). These results indicate that JNK kinase can regulate TGF-β1-induced FXII expression in HLF in the absence of any effects on Smad3 phosphorylation, association with Smad4, and its translocation into the nucleus.
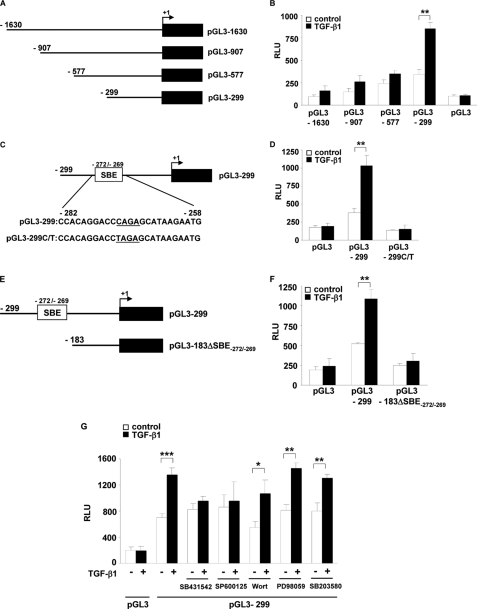
TGF-β1 Induces FXII Promoter Activity via a Smad-binding Element Located at the Position −272 to −269 (SBE-(−272/−269)) in a JNK-dependent Fashion
To identify DNA elements required for TGF-β1-induced FXII production, we transiently transfected NIH3T3 cells with a series of human FXII promoter deletion constructs (pGL3-1630, pGL3-907, pGL3-577, and pGL3-299; Fig. 6A) and then measured luciferase activity in untreated and TGF-β1-treated cells. We used the mouse embryonic fibroblast cell line NIH3T3 in these studies because of their high transfection efficiency. Cells transfected with pGL3-1630, pGL3-907, and pGL3-577 constructs displayed a moderately increased luciferase activity in response to TGF-β1, whereas strong induction of FXII promoter activity was observed in pGL3-299 transfected cells (Fig. 6B). These results indicate the presence of a TGF-β1-responsive element within the −299/+1-bp region in the human FXII promoter. In addition, our data suggest that repressor element(s) located in the region upstream of −299 bp may dampen the stimulatory effects of TGF-β1.
FIGURE 6.
TGF-β1 induces FXII promoter activity via SBE at position −272 to −269 (SBE-(−272/−269)) in a JNK-dependent fashion. A, schematic representation of FXII promoter luciferase reporter constructs. The angled arrow indicates the transcription start site. B, NIH3T3 cells were transfected with the indicated FXII promoter deletion constructs and then were either not stimulated (open columns) or were stimulated with TGF-β1 (closed columns). Luciferase activity is expressed in relative luminescence units (RLU). Data represent mean values ± S.D. from four independent experiments, each performed in triplicate; **, p < 0.01. C, schematic representation of the FXII promoter region containing a putative SBE at position −272 to −269. pGL3-299 C/T represents a construct in which the SBE-(−272/−269) was mutated by the replacement of the C residue at position −272 by T. D, NIH3T3 cells were transfected with the pGL3-299 or pGL3-299 C/T constructs. Luciferase activity was determined in untreated (open columns) and TGF-β1 treated cells (closed columns). Data represent mean values ± S.D. from four independent experiments, each performed in triplicate; **, p < 0.01. E, schematic representation of the pGL3-299 construct and the construct lacking SBE-(−272/−269) (pGL3-183ΔSBE-(−272/−269)). F, NIH3T3 cells were transfected with pGL3-299 or pGL3-183ΔSBE-(−272/−269), and the luciferase activity was measured in unstimulated (open columns) and TGF-β1-stimulated cells (closed columns). Data represent mean values ± S.D. from four independent experiments, each performed in triplicate; **, p < 0.01. G, NIH3T3 cells, transfected with the pGL3-299 construct, were pretreated with SB431542, SP600125, wortmannin (Wort), PD98059, or SB203580 for 1 h prior to incubation with TGF-β1. Luciferase activity was determined in untreated (open columns) and TGF-β1 treated cells (closed columns). Data represent mean values ± S.D. from four independent experiments, each performed in triplicate; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To identify the TGF-β1-responsive element of the FXII promoter lying between −299/+1 bp, we examined this DNA region for consensus SBEs. This analysis identified a putative SBE at the position −272 to −269 (SBE-(−272/−269), Fig. 6C). A point mutation of SBE-(−272/−269) (C→T mutation at the position −272) completely abolished FXII promoter activity in response to TGF-β1 (Fig. 6D). To confirm these results, we generated a construct lacking SBE-(−272/−269) (Fig. 6E). Deletion of the sequence between −299 and −183 abrogated the ability of the FXII promoter to confer responsiveness to TGF-β1 (Fig. 6F). These experiments were also performed with HLF and demonstrated similar results (data not shown).
We next tested the role of the TβRI/JNK signaling pathway in the regulation of FXII promoter activity by gene luciferase assay. The cells were transfected with the pGL3-299 construct, pretreated with the indicated inhibitors, and then either unstimulated or stimulated with 10 ng/ml TGF-β1. As depicted in Fig. 6G, TGF-β1-driven luciferase activity was strongly reduced only in the presence of TβRI and JNK (SB431542 and SP600125, respectively) inhibitors but not by PI3K, MEK, and p38 (wortmannin, PD98059, and SB203580, respectively) blockers.
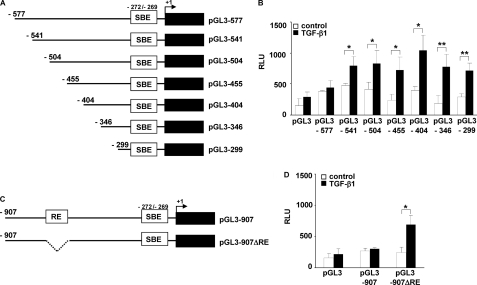
FXII Promoter Contains a Repressor Element Located at Position −577/−541
Our previous experiments demonstrated low luciferase activity in NIH3T3 cells transfected with the reporter constructs containing longer portions of FXII promoter (pGL3-1630, pGL3-907, and pGL3-577) and stimulated with TGF-β1. This observation led us to speculate that a repressor element might be located in the promoter region between −577 and −299 bp. To pinpoint further the repressor element itself, we generated a number of reporter constructs carrying luciferase gene together with upstream sequences of FXII promoter (−541/+1, −504/+1, −455/+1, −404/+1, and −346/+1 bp; Fig. 7A). All aforementioned FXII promoter constructs demonstrated high luciferase activity upon treatment of NIH3T3 cells with TGF-β1 indicating that a repressor element must be located in the promoter region between −577 and −541 bp (Fig. 7B). Accordingly, deletion of this region in the pGL3-907 construct (Fig. 7C) substantially enhanced luciferase activity upon stimulation of NIH3T3 cells with TGF-β1 (Fig. 7D).
FIGURE 7.
FXII promoter contains a repressor element located at the position −577/−541. A, schematic representation of FXII promoter luciferase reporter constructs. The angled arrow indicates the transcription start site. B, NIH3T3 cells were transfected with the indicated FXII promoter deletion constructs and then were either not stimulated (open columns) or were stimulated with TGF-β1 (closed columns). Luciferase activity is expressed in relative luminescence units (RLU). Data represent mean values ± S.D. from four independent experiments, each performed in triplicate; *, p < 0.05; **, p < 0.01. C, schematic representation of the pGL3-907 construct and the construct lacking repressor element (RE) located at the position −577/−541 (pGL3-907ΔRE). D, NIH3T3 cells were transfected with pGL3-907 or pGL3-907ΔRE, and the luciferase activity was measured in unstimulated (open columns) and TGF-β1-stimulated cells (closed columns). Data represent mean values ± S.D. from four independent experiments, each performed in triplicate; *, p < 0.05.
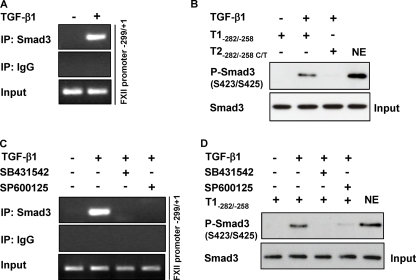
Smad3 Interacts with SBE-(−272/−269) within the FXII Promoter
To examine the interaction of Smad3 with SBE-(−272/−269), we performed ChIP and streptavidin pulldown assays. The ChIP assay clearly demonstrated TGF-β1-induced interaction of Smad3 with the FXII promoter region (−299/+1 bp) flanking SBE-(−272/−269) (Fig. 8A). To further analyze binding of Smad3 to SBE-(−272/−269), we performed a streptavidin pulldown assay using a biotinylated template spanning the region −282 and −258 bp. As expected, Smad3 was eluted from this template, whereas no interaction occurred when SBE-(−272/−269) was mutated (Fig. 8B). These findings indicate that Smad3-SBE-(−272/−269) forms a complex after stimulation of HLF with TGF-β1. No Smad3-SBE-(−272/−269) complex generation was noted when shorter biotinylated templates were used indicating the importance of neighboring DNA sequences for binding of Smad3 to SBE-(−272/−269) (data not shown).
FIGURE 8.
Smad3-SBE-(−272/−269) interaction is suppressed in the presence of a JNK inhibitor. A, HLF were either not stimulated or were stimulated with TGF-β1, and ChIP analysis was performed using a Smad3 antibody or IgG isotype control. PCR was performed with immunoprecipitated (IP) DNA as described under “Experimental Procedures,” thereby producing an amplicon comprising the −299/+1 region of the FXII promoter. PCR products were separated by agarose gel electrophoresis and detected by staining with ethidium bromide. Data are representative of three independent experiments. B, nuclear extracts (NE) from untreated or TGF-β1-treated HLF were incubated with biotinylated templates (T1−282/−258 or T2−282/−258 C/T), and bound Smad3 was detected by Western blotting. Data are representative of three independent experiments. Nuclear extract was used as positive control. C, HLF were pretreated with SB431542 or SP600125 for 1 h prior to incubation with TGF-β1. ChIP analysis was performed using Smad3 antibody or isotype IgG control. PCR was performed with immunoprecipitated chromatin as described under “Experimental Procedures,” thereby producing an amplicon comprising the −299/+1 region of the FXII promoter. Data are representative of three independent experiments. D, HLF were preincubated with SB431542 or SP600125 for 1 h prior to addition of TGF-β1. Nuclear extracts were prepared and then incubated with the biotinylated template T1-(−282/−258). Smad3 was detected by Western blotting. Data are representative of three independent experiments. Nuclear extract was used as positive control.
JNK Activity Is Required for Binding of Smad3 to SBE-(−272/−269)
Because inhibition of JNK did not impact the phosphorylation, complex formation with Smad4, and translocation of Smad3 into the nucleus, we investigated the involvement of this kinase in the formation of Smad3-SBE-(−272/−269) complex by ChIP and streptavidin pulldown assays. HLF, pretreated with TβRI or JNK inhibitors (SB431542 or SP600125, respectively), were either not stimulated or were stimulated with TGF-β1 and lysed, and a ChIP assay was performed using an anti-Smad3 antibody and an IgG isotype control. TGF-β1-induced binding of Smad3 to the FXII promoter region (−299/+1) flanking SBE-(−272/−269) was completely abolished when either SB431542 or SP600125 was used (Fig. 8C). SB431542 and SP600125 alone did not affect interaction of Smad3 with DNA (data not shown). Similar results were obtained when the streptavidin pulldown assay was performed (Fig. 8D).
FXII Levels Are Elevated in the Lungs of ARDS Patients
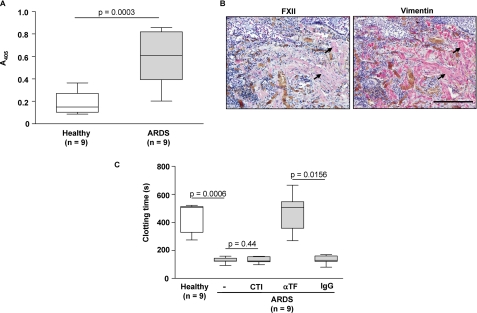
To provide the evidence that our observations may be clinically relevant, we investigated FXII levels in the lungs of patients with ARDS. ARDS is a prototype of an acute inflammatory lung disease in which severe abnormalities in the local coagulation system resulting in the formation of fibrin-rich hyaline membranes occur (24, 25). Furthermore, several lines of evidence indicate that fibroblasts as well as TGF-β1 play critical roles in the pathogenesis of this disorder (30–32). Significantly elevated FXII levels were noted in the ARDS BAL fluids compared with healthy controls (Fig. 9A). Moreover, immunohistochemical studies revealed a strong FXII co-localization with fibroblasts in the lung tissue of ARDS patients (Fig. 9B). Interestingly, we did not observe a significant contribution of FXII to the increased procoagulant activity in the BAL fluids of ARDS patients because the clotting time of these samples was not altered by FXII inhibitor (CTI). In contrast and in line with previous studies (24, 25, 49), a considerably prolonged clotting time of the ARDS BAL fluid samples was observed in the presence of a TF-neutralizing antibody (Fig. 9C). This underscores the importance of the extrinsic coagulation cascade for the increased alveolar procoagulant activity promoting extravascular fibrin deposition under these pathological conditions.
FIGURE 9.
Expression of FXII is elevated in the lungs of ARDS patients. A, quantification of FXII in BAL fluids from ARDS patients and healthy controls as assessed by immunoassay. The box-and-whiskers plots indicate the median, 1st, and 3rd quartile; the whiskers are extended to the most extreme value inside the 1.5-fold interquartile range. Significance levels are indicated. B, representative lung tissue sections from ARDS patient stained for FXII and vimentin. Arrows indicate fibroblast staining. Original magnification was ×20. Bar size, 200 μm. C, procoagulant activity of BAL fluids from ARDS patients was assessed by clotting assay in the absence or presence of FXII inhibitor (CTI), anti-TF blocking antibody (αTF), or an IgG isotype control (IgG). The box-and-whiskers plots indicate the median, 1st, and 3rd quartile; the whiskers are extended to the most extreme value inside the 1.5-fold interquartile range. Significance levels are indicated.
DISCUSSION
Factor XII is believed to participate in a number of pathophysiological processes, including inflammation, coagulation, and fibrinolysis (4). Previous reports demonstrated that FXII is mainly produced by the liver and that its expression is regulated by estrogen (50). The FXII promoter contains an estrogen-response element that mediates 17β-estradiol-stimulated induction of FXII gene expression (7, 8, 51). Moreover, hereditary angioedema, type III, characterized by increased activity of FXII, appears to be correlated with high estrogen levels (52).
Because the expression of FXII was also documented in other organs such as the lung and placenta (53), this study explores FXII production in the lung, and the molecular mechanisms underlying the regulation of FXII synthesis in primary human lung fibroblasts (HLF). We demonstrate, to the best of our knowledge and for the first time, that FXII is produced by HLF and that its expression is controlled by TGF-β1. Moreover, we show the requirement for JNK and Smad3 signaling pathways for TGF-β1-driven FXII synthesis in HLF. Our results are supported by a recent report that highlighted the importance of JNK in TGF-β1-induced expression of connective tissue growth factor in HLF (54).
The involvement of different signaling pathways, including p44/42, Akt, p38, and JNK, in the transcriptional induction of TGF-β1 target genes has already been demonstrated. TGF-β1-mediated activation of p38 kinase was found to be essential for mammary epithelial cell apoptosis, although it was not sufficient for epithelial-to-mesenchymal transdifferentiation (55). The p44/42 and Akt signaling pathways have been implicated in TGF-β1-driven expression of plasminogen activator inhibitor-1 in the endothelium and in mesangial cells, respectively (56, 57). In this study, we demonstrated that the blockade of MEK, PI3K, and p38 kinases did not affect the induction of FXII synthesis by TGF-β1, indicating that activation of these kinases is not essential for the enhancement of FXII expression in HLF. In contrast, blockage of JNK activity by the pharmacological inhibitor or JNK knockdown by siRNA significantly repressed TGF-β1-induced FXII expression in HLF. The crucial role of JNK kinase in TGF-β1-induced FXII expression is further confirmed by the luciferase promoter reporter assay, where preincubation of HLF with a JNK-specific inhibitor completely abolished TGF-β1-stimulated FXII promoter activity. Interestingly, JNK blockage did not interfere with the C-terminal phosphorylation of Smad3, indicating that inhibition of this pathway does not affect the upstream interaction between Smad3 and TβRI kinase. Furthermore, no phosphorylation was noted at the putative JNK phosphorylation sites (Ser-208/Ser-213) located within Smad3 linker region. This observation contrasts with data from other groups demonstrating TGF-β-induced JNK-dependent phosphorylation of Smad3 (58–61). The reason for this discrepancy is not clear; however, some differences in the experimental procedure, such as a choice of cell type and experimental conditions, may be of importance. In addition, other JNK-dependent phosphorylation sites may exist in the Smad3 linker region, and thus, a continuous effort has to be made to decipher JNK-dependent Smad3 phosphorylation sites to understand the cross-talk between Smad and JNK signaling pathways.
We also present evidence that JNK does not impact Smad3-Smad4 complex formation and its translocation into the nucleus. Therefore, we conclude that the Smad and JNK pathways may merge their signals within the nucleus, and we assume that JNK may target other transcription factors/coactivators that together with Smad3 can regulate FXII transcription by altering the binding of Smad3 to the FXII promoter region. To support this hypothesis, a strong reduction in Smad3-DNA complex formation was noted when HLF were preincubated with the JNK inhibitor prior to the addition of TGF-β1. Several transcription factors/coactivators, such as AP-1 (62), Sp-1 (63), IRF-7 (64), or CBP/p300 (65), were found to be able to interact with Smad molecules. Therefore, it is conceivable that JNK kinase may control the interaction of Smad3 with other proteins and, in this way, enhance TGF-β1-mediated FXII production. In line with these considerations, the role of other MAPKs such as p44/42 and of Akt in the modulation of DNA binding activities of various transcription factors, including Smads, has already been demonstrated in other systems (57, 66, 67). Further efforts are needed to clarify the detailed composition of transcriptional machinery that is responsible for TGF-β1-induced FXII expression in HLF. In this context, it is also important to note that the Smad signaling pathway did not influence JNK expression, activation, and translocation into the nucleus.
TGF-β1 induction of FXII gene transcription was further investigated by the generation of a series of FXII promoter luciferase reporter constructs. Transient transfection of NIH3T3 cells with these constructs revealed the importance of the sequence spanning 299/+1 region for TGF-β1-driven FXII expression. Further analysis of this promoter region demonstrated the presence of the SBE-containing consensus sequence at position −272 to −269. This is in line with the previously published reports demonstrating the interaction of the MH1 domain of receptor-associated Smad or the common mediator Smad4 with G/C-rich sequences of DNA, termed CAGA boxes (68, 69). Mutation or deletion of SBE-(−272/−269) further underscored its importance for the TGF-β1-mediated FXII expression in HLF. In addition, using different independent approaches, we demonstrated a direct interaction of Smad3 with SBE-(−272/−269). Interestingly, lower TGF-β1 inducibility was observed when longer portions of the FXII promoter were studied, a fact that is common to other inducible promoters as well (70). This initial observation resulted in the identification of a repressor element located at position −577/−541 bp indicating that initiation of FXII transcription in lung fibroblasts may only be achieved through the combinatorial interplay of various positive and negative regulatory elements located in different regions of FXII promoter. Thus, it is tempting to speculate that multiple DNA-protein interactions are required for cell type-specific regulation of FXII expression under normal and pathological conditions. In line with this assumption, previous reports demonstrated that cytokine, developmental, hormonal, and tissue- and cell type-specific regulation of gene expression may only be attained through the integration of a number of effects resulting from interactions of factors from multiple sites on the promoter (71–73).
Our findings raise the question of the role and function of FXII in fibroblasts and, more generally speaking, of the possible contribution of our observations to disease states. Our in vitro studies, showing a strong up-regulation of FXII expression in human lung fibroblasts in response to TGF-β1, suggest that our results may be particularly relevant in pulmonary diseases in which this cell population and this cytokine as well as an altered local coagulation system are thought to be critically involved, such as ARDS (24, 25, 49, 75–81). Accordingly, we observed increased FXII levels and a strong FXII co-localization with fibroblasts in the lungs of ARDS patients. These results support our in vitro observations, and they provide initial evidence that our findings may be clinically relevant. Several mechanisms exist by which fibroblast-derived FXII may contribute to pathological processes in the diseased lung. The procoagulant activities of FXII may contribute to excessive extravascular fibrin deposition in the injured lung, which in turn may serve as a reservoir of profibrotic growth factors and provide a provisional matrix on which fibroblasts can proliferate and secrete collagen (33–41). The possible role of lung fibroblasts as contributors to abnormal pulmonary fibrin turnover is underscored by the previous results showing that this cell population is an important source of other procoagulant and antifibrinolytic factors after stimulation with TGF-β1 and other cytokines that occur in the injured lung (18, 20, 42). However, recent studies reporting that targeted deletion of the fibrinogen gene provides no protection from experimental lung injury and subsequent pathological tissue remodeling have questioned the importance of fibrin in these processes (82). Furthermore, the pathophysiological significance of the FXII-triggered intrinsic pathway of coagulation for fibrin formation in vivo has been called into question by the observation that FXII deficiency in humans seems not to be associated with bleeding abnormalities (4, 83). In line with this observation, in this study we did not notice a significant contribution of FXII to the increased procoagulant activity in the ARDS BAL fluids. Together, these findings suggest that the hemostasis-independent cellular activities of FXII rather than its procoagulant effects may play an important role in pathological processes of the diseased lung.
In line with these considerations, preliminary in vivo studies from our group demonstrated by targeted deletion of the FXII gene and by blockage of FXII activity that FXII significantly contributes to pathological tissue remodeling in the bleomycin model of lung injury in mice and that FXII-induced proliferation of lung fibroblasts is critically involved in this process, thereby linking for the first time the activation of the contact phase of coagulation to the induction of fibroproliferation in the diseased lung.3 In ARDS, in particular, the mitogenic activities of FXII toward lung fibroblasts might be important for the progression of the early exudative stage to the proliferative and fibrotic phases. Fibroproliferation and fibrogenesis may occur very early in the course of this disease and are associated with a significantly increased risk of mortality (78, 80, 81). Our findings support the results from previous studies in which growth factor activities of FXII on other cell populations such as human hepatoma (HepG2) cells, fetal hepatocytes, endothelial cells, alveolar type II cells, and aortic smooth muscle cells have been described (74, 84). Mitogenic activities for fibroblasts have also been described for other coagulation factors such as factor X and thrombin, and similar to our observations, they have been found to be crucial for pathological tissue remodeling in the same bleomycin model of lung injury (37, 38). Whether our findings are also relevant for the pathogenesis of other lung disorders and for fibroproliferative processes in organs other than the lung needs to be investigated in future studies.
In conclusion, we demonstrated that TGF-β1-induced FXII production in HLF is mediated by JNK and Smad3 signaling pathways. Moreover, we identified an SBE at position −272 to −269 within the FXII promoter that is crucial for TGF-β1-induced FXII promoter activity, and we show the importance of JNK in Smad3 binding to SBE-(−272/−269). Our findings provide new insights into the molecular mechanism responsible for the regulation of FXII expression in HLF and implicate its possible role in pathological conditions characterized by elevated TGF-β1 levels and fibroproliferation.
Supplementary Material
Acknowledgments
We thank Gisela Mueller and Horst Thiele for excellent technical assistance.
This work was supported by German Research Foundation Grant KFO 118 and the Excellence Cluster “Cardiopulmonary System.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
E. Jablonska, P. Markart, K. T. Preissner, and M. Wygrecka, unpublished observations.
- FXII
- coagulation factor XII
- TGF-β1
- transforming growth factor-β1
- JNK
- c-Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- PI3K
- phosphoinositide 3-kinase
- IL
- interleukin
- HLF
- human lung fibroblasts
- TβRI
- TGF-β receptor type I
- SBE
- Smad-binding element
- ChIP
- chromatin immunoprecipitation
- TF
- tissue factor
- CTI
- corn trypsin inhibitor
- ARDS
- acute respiratory distress syndrome
- BAL
- bronchoalveolar lavage
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- siRNA
- small interfering RNA.
REFERENCES
- 1.Colman R. W., Schmaier A. H. (1986) Crit. Rev. Oncol. Hematol. 5, 57–85 [DOI] [PubMed] [Google Scholar]
- 2.Colman R. W., Schmaier A. H. (1997) Blood 90, 3819–3843 [PubMed] [Google Scholar]
- 3.Renné T., Pozgajová M., Grüner S., Schuh K., Pauer H. U., Burfeind P., Gailani D., Nieswandt B. (2005) J. Exp. Med. 202, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colman R. W. (1999) Thromb. Haemost. 82, 1568–1577 [PubMed] [Google Scholar]
- 5.Chien P., Pixley R. A., Stumpo L. G., Colman R. W., Schreiber A. D. (1988) J. Clin. Invest. 82, 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toossi Z., Sedor J. R., Mettler M. A., Everson B., Young T., Ratnoff O. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11969–11972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farsetti A., Misiti S., Citarella F., Felici A., Andreoli M., Fantoni A., Sacchi A., Pontecorvi A. (1995) Endocrinology 136, 5076–5083 [DOI] [PubMed] [Google Scholar]
- 8.Citarella F., Misiti S., Felici A., Farsetti A., Pontecorvi A., Fantoni A. (1996) Steroids 61, 270–276 [DOI] [PubMed] [Google Scholar]
- 9.Farsetti A., Moretti F., Narducci M., Misiti S., Nanni S., Andreoli M., Sacchi A., Pontecorvi A. (1998) Endocrinology 139, 4581–4589 [DOI] [PubMed] [Google Scholar]
- 10.Gordon E. M., Johnson T. R., Ramos L. P., Schmeidler-Sapiro K. T. (1991) J. Lab. Clin. Med. 117, 353–358 [PubMed] [Google Scholar]
- 11.Gordon E. M., Douglas J. G., Ratnoff O. D., Arafah B. M. (1985) Blood 66, 602–605 [PubMed] [Google Scholar]
- 12.Briseid K., Hoem N. O., Johannesen S., Fossum S. (1991) Thromb. Res. 61, 123–133 [DOI] [PubMed] [Google Scholar]
- 13.Gordon E. M., Ratnoff O. D., Saito H., Donaldson V. H., Pensky J., Jones P. K. (1980) J. Lab. Clin. Med. 96, 762–769 [PubMed] [Google Scholar]
- 14.Gordon E. M., Williams S. R., Frenchek B., Mazur C. A., Speroff L. (1988) J. Lab. Clin. Med. 111, 52–56 [PubMed] [Google Scholar]
- 15.Citarella F., Felici A., Brouwer M., Wagstaff J., Fantoni A., Hack C. E. (1997) Blood 90, 1501–1507 [PubMed] [Google Scholar]
- 16.Bonniaud P., Margetts P. J., Ask K., Flanders K., Gauldie J., Kolb M. (2005) J. Immunol. 175, 5390–5395 [DOI] [PubMed] [Google Scholar]
- 17.Gauldie J., Bonniaud P., Sime P., Ask K., Kolb M. (2007) Biochem. Soc. Trans. 35, 661–664 [DOI] [PubMed] [Google Scholar]
- 18.Felts S. J., Stang M. T., Getz M. J. (1997) Oncogene 14, 1679–1685 [DOI] [PubMed] [Google Scholar]
- 19.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idell S., Zwieb C., Boggaram J., Holiday D., Johnson A. R., Raghu G. (1992) Am. J. Physiol. 263, L487–L494 [DOI] [PubMed] [Google Scholar]
- 21.Idell S., Kumar A., Zwieb C., Holiday D., Koenig K. B., Johnson A. R. (1994) Am. J. Physiol. 267, L693–L703 [DOI] [PubMed] [Google Scholar]
- 22.Miyazono K., ten Dijke P., Heldin C. H. (2000) Adv. Immunol. 75, 115–157 [DOI] [PubMed] [Google Scholar]
- 23.Nakao A., Imamura T., Souchelnytskyi S., Kawabata M., Ishisaki A., Oeda E., Tamaki K., Hanai J., Heldin C. H., Miyazono K., ten Dijke P. (1997) EMBO J. 16, 5353–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idell S., James K. K., Levin E. G., Schwartz B. S., Manchanda N., Maunder R. J., Martin T. R., McLarty J., Fair D. S. (1989) J. Clin. Invest. 84, 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idell S., Koenig K. B., Fair D. S., Martin T. R., McLarty J., Maunder R. J. (1991) Am. J. Physiol. 261, L240–L248 [DOI] [PubMed] [Google Scholar]
- 26.Kotani I., Sato A., Hayakawa H., Urano T., Takada Y., Takada A. (1995) Thromb. Res. 77, 493–504 [DOI] [PubMed] [Google Scholar]
- 27.Günther A., Mosavi P., Ruppert C., Heinemann S., Temmesfeld B., Velcovsky H. G., Morr H., Grimminger F., Walmrath D., Seeger W. (2000) Thromb. Haemost. 83, 853–860 [PubMed] [Google Scholar]
- 28.Imokawa S., Sato A., Hayakawa H., Kotani M., Urano T., Takada A. (1997) Am. J. Respir. Crit. Care Med. 156, 631–636 [DOI] [PubMed] [Google Scholar]
- 29.Guo W., Shan B., Klingsberg R. C., Qin X., Lasky J. A. (2009) Am. J. Physiol. Lung Cell. Mol. Physiol. 297, L864–L870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fine A., Goldstein R. H. (1987) J. Biol. Chem. 262, 3897–3902 [PubMed] [Google Scholar]
- 31.Pelaia G., Gallelli L., D'Agostino B., Vatrella A., Cuda G., Fratto D., Renda T., Galderisi U., Piegari E., Crimi N., Rossi F., Caputi M., Costanzo F. S., Vancheri C., Maselli R., Marsico S. A. (2007) J. Cell. Physiol. 210, 489–497 [DOI] [PubMed] [Google Scholar]
- 32.Goulet S., Bihl M. P., Gambazzi F., Tamm M., Roth M. (2007) J. Cell. Physiol. 210, 167–176 [DOI] [PubMed] [Google Scholar]
- 33.Burkhardt A. (1989) Am. Rev. Respir. Dis. 140, 513–524 [DOI] [PubMed] [Google Scholar]
- 34.Seeger W., Elssner A., Günther A., Krämer H. J., Kalinowski H. O. (1993) Am. J. Respir. Cell Mol. Biol. 9, 213–220 [DOI] [PubMed] [Google Scholar]
- 35.Grinnel F. (1980) Experientia 36, 505–507 [DOI] [PubMed] [Google Scholar]
- 36.Senior R. M., Skogen W. F., Griffin G. L., Wilner G. D. (1986) J. Clin. Invest. 77, 1014–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanc-Brude O. P., Archer F., Leoni P., Derian C., Bolsover S., Laurent G. J., Chambers R. C. (2005) Exp. Cell Res. 304, 16–27 [DOI] [PubMed] [Google Scholar]
- 38.Chambers R. C., Dabbagh K., McAnulty R. J., Gray A. J., Blanc-Brude O. P., Laurent G. J. (1998) Biochem. J. 333, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogatkevich G. S., Tourkina E., Silver R. M., Ludwicka-Bradley A. (2001) J. Biol. Chem. 276, 45184–45192 [DOI] [PubMed] [Google Scholar]
- 40.Chambers R. C., Leoni P., Blanc-Brude O. P., Wembridge D. E., Laurent G. J. (2000) J. Biol. Chem. 275, 35584–35591 [DOI] [PubMed] [Google Scholar]
- 41.Scotton C. J., Krupiczojc M. A., Königshoff M., Mercer P. F., Lee Y. C., Kaminski N., Morser J., Post J. M., Maher T. M., Nicholson A. G., Moffatt J. D., Laurent G. J., Derian C. K., Eickelberg O., Chambers R. C. (2009) J. Clin. Invest. 119, 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olman M. A., Mackman N., Gladson C. L., Moser K. M., Loskutoff D. J. (1995) J. Clin. Invest. 96, 1621–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wygrecka M., Morty R. E., Markart P., Kanse S. M., Andreasen P. A., Wind T., Guenther A., Preissner K. T. (2007) J. Biol. Chem. 282, 21671–21682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernard G. R., Artigas A., Brigham K. L., Carlet J., Falke K., Hudson L., Lamy M., LeGall J. R., Morris A., Spragg R. (1994) J. Crit. Care 9, 72–81 [DOI] [PubMed] [Google Scholar]
- 45.Wygrecka M., Markart P., Ruppert C., Kuchenbuch T., Fink L., Bohle R. M., Grimminger F., Seeger W., Günther A. (2004) Thromb. Haemost. 92, 529–540 [DOI] [PubMed] [Google Scholar]
- 46.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 47.Wygrecka M., Markart P., Fink L., Guenther A., Preissner K. T. (2007) Thorax 62, 880–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wrighton K. H., Feng X. H. (2008) Cell. Signal. 20, 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Günther A., Mosavi P., Heinemann S., Ruppert C., Muth H., Markart P., Grimminger F., Walmrath D., Temmesfeld-Wollbrück B., Seeger W. (2000) Am. J. Respir. Crit. Care Med. 161, 454–462 [DOI] [PubMed] [Google Scholar]
- 50.Ratnoff O. D. (1991) J. Lab. Clin. Med. 117, 343. [PubMed] [Google Scholar]
- 51.Citarella F., Misiti S., Felici A., Aiuti A., La Porta C., Fantoni A. (1993) Biochim. Biophys. Acta 1172, 197–199 [DOI] [PubMed] [Google Scholar]
- 52.Cichon S., Martin L., Hennies H. C., Müller F., Van Driessche K., Karpushova A., Stevens W., Colombo R., Renné T., Drouet C., Bork K., Nöthen M. M. (2006) Am. J. Hum. Genet. 79, 1098–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neth P., Arnhold M., Nitschko H., Fink E. (2001) Thromb. Haemost. 85, 1043–1047 [PubMed] [Google Scholar]
- 54.Utsugi M., Dobashi K., Ishizuka T., Masubuchi K., Shimizu Y., Nakazawa T., Mori M. (2003) Am. J. Respir. Cell Mol. Biol. 28, 754–761 [DOI] [PubMed] [Google Scholar]
- 55.Yu L., Hébert M. C., Zhang Y. E. (2002) EMBO J. 21, 3749–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kutz S. M., Hordines J., McKeown-Longo P. J., Higgins P. J. (2001) J. Cell Sci. 114, 3905–3914 [DOI] [PubMed] [Google Scholar]
- 57.Das F., Ghosh-Choudhury N., Venkatesan B., Li X., Mahimainathan L., Choudhury G. G. (2008) J. Cell. Physiol. 214, 513–527 [DOI] [PubMed] [Google Scholar]
- 58.Engel M. E., McDonnell M. A., Law B. K., Moses H. L. (1999) J. Biol. Chem. 274, 37413–37420 [DOI] [PubMed] [Google Scholar]
- 59.Liu Q., Mao H., Nie J., Chen W., Yang Q., Dong X., Yu X. (2008) Perit. Dial. Int. 28, S88–S95 [PubMed] [Google Scholar]
- 60.Yoshida K., Matsuzaki K., Mori S., Tahashi Y., Yamagata H., Furukawa F., Seki T., Nishizawa M., Fujisawa J., Okazaki K. (2005) Am. J. Pathol. 166, 1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mori S., Matsuzaki K., Yoshida K., Furukawa F., Tahashi Y., Yamagata H., Sekimoto G., Seki T., Matsui H., Nishizawa M., Fujisawa J., Okazaki K. (2004) Oncogene 23, 7416–7429 [DOI] [PubMed] [Google Scholar]
- 62.Liberati N. T., Datto M. B., Frederick J. P., Shen X., Wong C., Rougier-Chapman E. M., Wang X. F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hua X., Liu X., Ansari D. O., Lodish H. F. (1998) Genes Dev. 12, 3084–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qing J., Liu C., Choy L., Wu R. Y., Pagano J. S., Derynck R. (2004) Mol. Cell. Biol. 24, 1411–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen X., Hu P. P., Liberati N. T., Datto M. B., Frederick J. P., Wang X. F. (1998) Mol. Biol. Cell 9, 3309–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Piwien-Pilipuk G., MacDougald O., Schwartz J. (2002) J. Biol. Chem. 277, 44557–44565 [DOI] [PubMed] [Google Scholar]
- 67.Buck M., Poli V., van der Geer P., Chojkier M., Hunter T. (1999) Mol. Cell 4, 1087–1092 [DOI] [PubMed] [Google Scholar]
- 68.Inagaki Y., Truter S., Ramirez F. (1994) J. Biol. Chem. 269, 14828–14834 [PubMed] [Google Scholar]
- 69.Kim S. J., Jeang K. T., Glick A. B., Sporn M. B., Roberts A. B. (1989) J. Biol. Chem. 264, 7041–7045 [PubMed] [Google Scholar]
- 70.Lindahl G. E., Chambers R. C., Papakrivopoulou J., Dawson S. J., Jacobsen M. C., Bishop J. E., Laurent G. J. (2002) J. Biol. Chem. 277, 6153–6161 [DOI] [PubMed] [Google Scholar]
- 71.Savagner P., Miyashita T., Yamada Y. (1990) J. Biol. Chem. 265, 6669–6674 [PubMed] [Google Scholar]
- 72.Cao S. X., Gutman P. D., Dave H. P., Schechter A. N. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5306–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garzon R. J., Zehner Z. E. (1994) Mol. Cell. Biol. 14, 934–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmeidler-Sapiro K. T., Ratnoff O. D., Gordon E. M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 4382–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dhainaut J. F., Charpentier J., Chiche J. D. (2003) Crit. Care Med. 31, S258–S264 [DOI] [PubMed] [Google Scholar]
- 76.Fahy R. J., Lichtenberger F., McKeegan C. B., Nuovo G. J., Marsh C. B., Wewers M. D. (2003) Am. J. Respir. Cell Mol. Biol. 28, 499–503 [DOI] [PubMed] [Google Scholar]
- 77.Meduri G. U., Tolley E. A., Chinn A., Stentz F., Postlethwaite A. (1998) Am. J. Respir. Crit. Care Med. 158, 1432–1441 [DOI] [PubMed] [Google Scholar]
- 78.Chesnutt A. N., Matthay M. A., Tibayan F. A., Clark J. G. (1997) Am. J. Respir. Crit. Care Med. 156, 840–845 [DOI] [PubMed] [Google Scholar]
- 79.Fukuda Y., Ishizaki M., Masuda Y., Kimura G., Kawanami O., Masugi Y. (1987) Am. J. Pathol. 126, 171–182 [PMC free article] [PubMed] [Google Scholar]
- 80.Martin C., Papazian L., Payan M. J., Saux P., Gouin F. (1995) Chest 107, 196–200 [DOI] [PubMed] [Google Scholar]
- 81.Meduri G. U., Eltorky M., Winer-Muram H. T. (1995) Semin. Respir. Infect. 10, 154–175 [PubMed] [Google Scholar]
- 82.Hattori N., Degen J. L., Sisson T. H., Liu H., Moore B. B., Pandrangi R. G., Simon R. H., Drew A. F. (2000) J. Clin. Invest. 106, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmaier A. H. (2008) J. Clin. Invest. 118, 3006–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gordon E. M., Venkatesan N., Salazar R., Tang H., Schmeidler-Sapiro K., Buckley S., Warburton D., Hall F. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.