Abstract
In vertebrates, stathmins form a family of proteins possessing two tubulin binding repeats (TBRs), which each binds one soluble tubulin heterodimer. The stathmins thus sequester two tubulins in a phosphorylation-dependent manner, providing a link between signal transduction and microtubule dynamics. In Drosophila, we show here that a single stathmin gene (stai) encodes a family of D-stathmin proteins. Two of the D-stathmins are maternally deposited and then restricted to germ cells, and the other two are detected in the nervous system during embryo development. Like in vertebrates, the nervous system-enriched stathmins contain an N-terminal domain involved in subcellular targeting. All the D-stathmins possess a domain containing three or four predicted TBRs, and we demonstrate here, using complementary biochemical and biophysical methods, that all four predicted TBR domains actually bind tubulin. D-stathmins can indeed bind up to four tubulins, the resulting complex being directly visualized by electron microscopy. Phylogenetic analysis shows that the presence of regulated multiple tubulin sites is a conserved characteristic of stathmins in invertebrates and allows us to predict key residues in stathmin for the binding of tubulin. Altogether, our results reveal that the single Drosophila stathmin gene codes for a stathmin family similar to the multigene vertebrate one, but with particular tubulin binding properties.
Keywords: Electron Microscopy, Microtubules, Protein-Protein Interactions, Surface Plasmon Resonance, Tubulin
Introduction
In vertebrates, stathmin is the generic element of a family of microtubule-regulating proteins comprising stathmin, SCG10, SCLIP, and RB3/RB3′/RB3″ coded by four different genes (stmn 1–4) (1–4). Stathmin is a ubiquitous cytosolic phosphoprotein highly expressed in early embryos, gonads, and in the nervous system (5–10). Stathmin-related proteins are enriched in the brain (11, 12) and targeted to vesicular and Golgi membranes via an N-terminal extension containing two close palmitoylated cysteines (13–19). Stathmin proteins are involved in the control of cell proliferation (20, 21), differentiation (10, 22, 23), and migration (24, 25). Cellular RNA interference knock-down experiments also revealed that SCG10 and SCLIP play essential roles in neuronal morphogenesis and, as predicted on the basis of the existence of different genes, that these roles are at least partially distinct (26, 27). In the stathmin knock-out mouse mild phenotypes were detected with a decreased innate fear response (28) and a minor axonopathy in aged animals (29).
Microtubules are key elements of the cytoskeleton involved in the cell cycle, cell shape, and intracellular organization and trafficking. They are composed of α/β-tubulin heterodimers, which are assembled or disassembled through phases of slow growth or rapid shrinkage separated by catastrophe and rescue events (30). This “dynamic instability” of microtubules is controlled by a variety of proteins that include, beside stabilizing and destabilizing proteins binding to microtubules, proteins of the stathmin family (31), which bind soluble tubulin.
Stathmin forms a complex and hence sequesters or releases two soluble tubulin molecules (32–37), thus favoring microtubule depolymerization or polymerization in a phosphorylation-dependent manner (32, 38–40). The nervous system-enriched stathmins also bind tubulin albeit with different kinetics and complex stabilities (41, 42). It has been also proposed that stathmin may be involved in microtubule depolymerization independently of its sequestering activity by promoting directly microtubule catastrophe (43, 44, for review see Ref. 45).
To decipher the in vivo roles of stathmin proteins and their important features conserved through evolution, we previously identified a unique stathmin-related gene (stai) in Drosophila predicted to code for at least two transcripts (46). The D-stathmin gene is expressed in germ cells and in the nervous system of the developing Drosophila embryos, and its role in tubulin sequestration is conserved. However the determinants of tubulin interaction in D-stathmin were not characterized, and its stoichiometry was not quantitatively analyzed. A striking physiological role of D-stathmins was revealed following RNA knockdown in Drosophila embryos, which resulted in germ cell migration defects as well as a major disorganization of nervous system development (46).
In the present study, we show that the single Drosophila stathmin gene codes for a family of four different D-stathmin proteins whose expression patterns are regulated during development. The stathmin gene is conserved in invertebrates and codes for isoforms with tubulin binding stoichiometries that are variable, higher than in vertebrates, and regulated by alternative splicing. Moreover, phylogenetic analysis allowed us to show the specificity of each tubulin binding region and predict key residues for the binding of tubulin.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
LD04103 clone (GenBankTM accession number) and clone 14 (46) were used for the construction of D-stathmin B2 and A1, respectively. To obtain the D-stathmin B1 construction, clone LD04103 and clone 19 (46) were digested by AflIII and BamHI and ligated together. The various D-stathmin derivatives used were: amino acids 55–196 of D-stathmin B2 (TBR 1-2-3/7),3 and 1–142 (TBR 1-2), 1–204 (TBR 1-2-3/6), or 146–257 (TBR 3/6-4) of D-stathmin A1. For eukaryotic expression, in vitro transcription/translation, prokaryotic expression, and surface plasmon resonance experiments D-stathmin cDNAs were amplified by PCR and inserted into the pcDNA3-Myc vector (47), the pSp64 vector (Promega, Madison, WI), pET-8c vector (Novagen, Madison, WI), or the pDW363-inducible expression vector (42), respectively. All cDNA constructs were checked by sequencing (Genome express, Meylan, France).
Recombinant Protein Expression
In Vitro Eukaryotic Protein Expression
1 μg of pSp64 plasmid containing D-stathmin A1, B1, and B2 coding sequences was used for in vitro transcription and translation with the TnTTM Coupled Reticulocyte Lysate System (Promega). 5 μl of 25 μl of total transcription/translation mix were analyzed by gel electrophoresis and Western blot analysis.
Recombinant Protein Production and Purification
h-stathmin was purified as previously described (32). The PET- 8C and the pDW363 cDNA clones were used to produce and purify the corresponding D-stathmin derivatives in the BL-21(DE3) Escherichia coli strain as described previously (42).
RNA Preparation, Northern Blot, and Reverse Transcription-PCR
Drosophila tissues were homogenized in TRIzol reagent (Invitrogen). S2R+ RNA was prepared using the RNeasy Minikit (Qiagen, Courtaboeuf, France) according to the manufacturer's protocol. Northern blots were performed as described (46). Multiprime-labeled fragments of PCR-amplified probes 1′-2′, 3-4-5, and 1-2 were added at 0.5 × 106 cpm/ml in the hybridization buffer, and hybridization was allowed to proceed overnight. The final wash was performed at 60 °C in 0.1× SSC, 0.1% SDS for 30 min. Reverse transcription-PCRs were performed on 1 μg of template RNA using 20 pmol of dT oligonucleotides, 1 h at 42 °C, with the ImProm-IITM reverse transcription system (Promega) followed by a PCR using appropriate oligonucleotides (1′, gagagctcgaggaaaccgtccgcgatataa; 7, ttgctaagctttgtgttggtgtattatgca; and 1, cattcgcttaattttcgccgacgacgcg).
RNA Interference
Templates for in vitro transcription were generated by PCR using the following pairs of oligonucleotides containing the sequence recognized by the T7 RNA polymerase: gagaattctaatacgactcactatagggagaaacacaatcaaaattgccgaaatcaaa and gagaattctaatacgactcactatagggagagagtttttgaactttttcaatttttttttgc, and gagaattctaatacgactcactatagggagaattgagcagaaacttaaggcggcc and gagaattctaatacgactcactatagggagacgtcttttcgatatcctgggcatg, to amplify exon 6 and exons 3-4-5, respectively. The corresponding double strand RNAs (dsRNAs) were then synthesized using the MEGAscript kit (Ambion, Inc., Austin, TX) according to the manufacturer's instructions. 37 nm of dsRNA was added to a 35-mm well culture plate containing 106 S2R+ cells in Schneider medium without serum, and the mixture was incubated at room temperature for 30 min after vigorous shaking. 2 ml of Schneider medium containing 10% serum was then added, and the cells were further grown for 3 or 7 days before analysis by Western blot.
Embryo in Situ Hybridization and Immunohistochemical Staining
RNA in situ hybridization was performed as described before (48). Briefly, regions 1-2, 1′-2′, and 6 were PCR-amplified with 3′ oligonucleotides containing the sequence of the initiation of the T7 phage polymerase to directly synthesize digoxygenin-UTP-labeled RNA. Fixed embryos were hybridized with digoxygenin-UTP-labeled RNA overnight at 55 °C and then incubated with alkaline phosphatase-conjugated anti-digoxygenin antibodies. The signal was developed using the alkaline phosphatase reaction. For examination, embryos were mounted in Aqua-Polymount (Polysciences, Inc., Warrington, PA).
Polyacrylamide Gel Electrophoresis and Western Blotting
One-dimensional gel electrophoresis was performed on 12% BisTris polyacrylamide gels (NuPAGE, Invitrogen). The gels were transferred to nitrocellulose in a semi-dry electroblotting apparatus and probed with diluted antiserum (anti-peptide COOH-terminal antiserum 98 (1:10,000), anti-D-stathmin-DC antiserum 97 (1:10,000), or anti-Myc monoclonal antibody (1:2,000, Dako, A/S, Denmark)). Bound antibodies were detected with appropriate secondary antibodies and the chemiluminescent ECL kit (Amersham Biosciences), or by fluorescence (Odyssey, Li-COR Biosciences, Bad Homburg, Germany).
Cell Culture, DNA Transfection, and Immunofluorescence
Human HeLa cells were grown as monolayers in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal calf serum (Invitrogen) at 37 °C in 5% CO2. Transfections were performed using FuGENE (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's instructions. Cells were fixed with phosphate-buffered saline plus 2% paraformaldehyde and 30 mm saccharose for 10 min at 37 °C. Primary antibodies (monoclonal anti-α-tubulin N356, 1:300, Amersham Biosciences; polyclonal anti-Myc sc-789, 1:100, Tebu, Le Perray en Yvelines, France) were revealed with appropriate Alexa 488, 546-conjugated anti-rabbit (1:300) and anti-mouse (1:300) secondary antibodies (Jackson ImmunoResearch). The cells were mounted with Mowiol solution and examined with a Provis Olympus fluorescence photomicroscope equipped with a Princeton Instruments camera.
In Vitro Tubulin Polymerization Assay
Tubulin was purified from calf brain as described before (32). The effect of various D-stathmin variants or fragments on tubulin polymerization in polymerization buffer (50 mm 2-(N-morpholino)ethanesulfonic acid-KOH, pH 6.8, 30% glycerol, 0.5 mm EGTA, 6 mm MgCl2, and 0.5 mm GTP) was monitored turbidimetrically at 350 nm in an Ultrospec 3000 spectrophotometer (Amersham Biosciences) thermostatted at 37 °C as described before (41). Tubulin alone and h-stathmin were used as controls, with the results for D-stathmins being normalized on the basis of a 2:1 tubulin:h-stathmin reference ratio.
Gel-filtration Assay
The interaction of the D-stathmin derivatives with tubulin was studied by size-exclusion chromatography on a Superose 12 HR 10/30 column pre-equilibrated with buffer AB (80 mm Pipes/KOH/1 mm EGTA/5 mm MgCl2, pH 6.8) containing 1 m trimethylamine-N-oxide at 0.5 ml/min. Monitoring at 278 nm allowed us to observe the tubulin peaks mainly, because D-stathmin derivatives do not significantly absorb light at this wavelength. The interaction was favored by the addition of 1 m trimethylamine-N-oxide to the sample and elution buffers as previously described (42).
Surface Plasmon Resonance
BIAcore 2000 system, Sensorchip SA, and HBS buffer (0.01 m Hepes (pH 7.4)/0.15 m NaCl/3 mm EDTA/0.005% polyoxyethylenesorbitan) were from BIAcore AB (Uppsala, Sweden). The Sensorchip SA coated with streptavidin was preconditioned with three 10-μl injections of 50 mm NaOH, 1 m NaCl, and saturated with three 10-μl injections of 10 mg/ml bovine serum albumin. The first flow cell was used as a reference flow cell. The other three flow cells were coupled with the dialyzed S2 of the various D-stathmin derivatives that were specifically biotinylated on their NH2-terminal tag. To obtain surfaces with comparable molar densities, the amounts of the various proteins coupled were proportional to their molecular masses: ∼2000 resonance units of h-stathmin, 900 resonance units of TBR 1–2, and 370 resonance units of D-stathmin A1 were coupled at 10-μl/min in HBS buffer. Several runs of tubulin ranging from 0.5 to 10 μm were made at a constant flow rate of 30 μl/min, in buffer AB (80 mm Pipes/KOH/1 mm EGTA/5 mm MgCl2, pH 6.8) supplemented with 0.005% (v/v) P20 surfactant in the presence of 1 mm GDP. For the analysis, the reference flow cell sensorgram was subtracted from the corresponding sensorgrams.
Electron Microscopy and Tubulin-Stathmin Complex Size Measurements
Samples for glycerol spraying/low angle rotary metal shadowing were prepared as described (34, 37). Briefly, 20 μl of protein samples (0.1–0.3 mg/ml) were mixed with glycerol to a final concentration of 30%, sprayed onto freshly cleaved mica at room temperature, and rotary shadowed in a BA 511M freeze-etch apparatus (Balzers) with platinum/carbon at an elevation angle of 3–5°. Electron micrographs were taken in a Philips Morgagni transmission electron microscope operated at 80 kV equipped with a Megaview III charge-coupled device camera. The electron micrographs were used to calculate the length of the tubulin complexes formed with various stathmin constructs. If t is the length of the uncoated αβ-tubulin heterodimer, and e is the thickness of the platinum coating, the length of a coated tubulin heterodimer is T = t + 2e, and that of a platinum-coated T2S complex is T2 = 2t + 2e. Hence t = T2 − T, and e = (T − t)/2. With T = 17.5 nm, T2 = 28 nm, we found t = 10.5 nm, and e = 3.5 nm. One can then deduce from the measurements of the curved lengths (L) of the coated tubulin complexes the length (l) of the corresponding naked complexes: l = L − 2e = L − 7 nm.
In Silico Stathmin Gene and mRNAs Identification
To identify all stathmin sequences at the mRNA and genomic level, we ran the TBLASTN or BLASTN software on expressed sequence tag, non-redundant (nr), and genomic GenBankTM libraries using the D-stathmin A1, A2, B1, and B2 nucleotidic or amino acid sequences, as well as each individual exon as the query.
RESULTS
The Drosophila Stathmin Gene Codes for a Family of Proteins
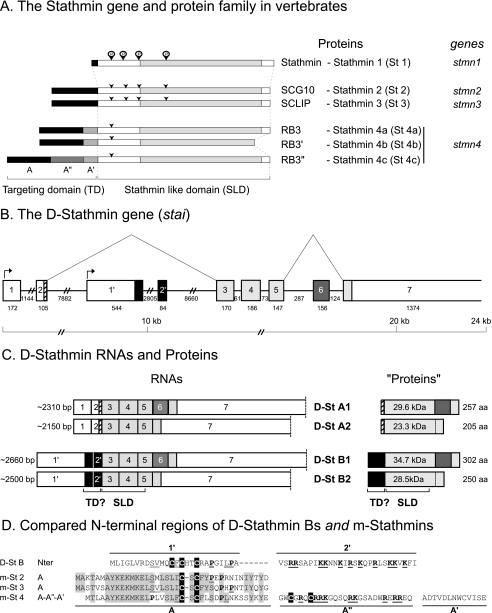
In most vertebrates, the six identified tubulin binding stathmin family proteins, stathmin (stathmin 1, St 1) and the mostly or exclusively neural proteins SCG10 (stathmin 2, St 2), SCLIP (stathmin 3, St 3), RB3 and its splice variants RB3′ and RB3″ (stathmin 4a, St 4a; stathmin 4b, St 4b; and stathmin 4c, St 4c), are encoded by four conserved genes (stmn1–4) (Fig. 1A).
FIGURE 1.
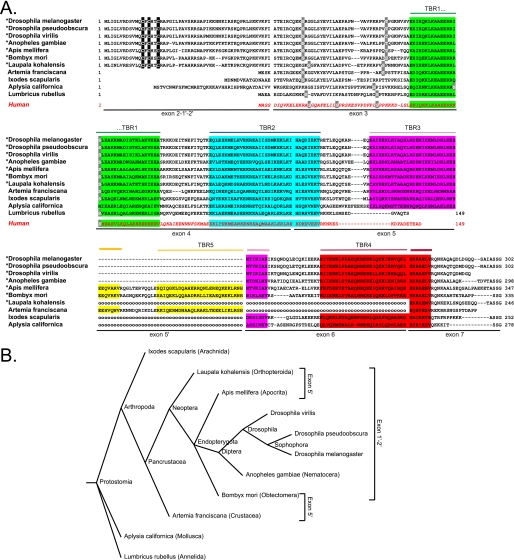
The stathmin family is conserved from vertebrates to Drosophila. A, stathmin genes and proteins in vertebrates. Stathmin, SCG10, SCLIP, and RB3 are designated stathmin 1, 2, 3 and 4, respectively, to be in agreement with the corresponding stmn1, -2, -3, and -4 gene nomenclature. The targeting domain (TD) specific of neural members of the family as well as the stathmin-like domain (SLD) involved in tubulin interaction are shown. Conserved consensus phosphorylation sites are indicated by arrows. B, the Drosophila stathmin gene (stai) organization. The organization and the size of the various uncoding (white boxes) and coding exons (shaded boxes) on chromosome 2L are represented. The exons expressed in all D-stathmin proteins are in light gray. Different mRNAs are transcribed from the stai gene depending on the use of alternative transcription initiations (arrows) and splicing (broken lines). C, the four mRNAs and predicted proteins expressed from the stai gene. As various polyadenylation sites were found on the DNA sequence as well as various ends on expressed sequence tag sequences, the size of exon 7 is undefined and thus delimited by a dashed line. The calculated molecular masses and amino acid lengths of the corresponding proteins are indicated, as well as the limits of their SLD and the putative N-terminal targeting domain (TD?) of D-stathmins B. D, comparison of the D-stathmin B N-terminal region with that of m-stathmins. The N-terminal domain present in D-stathmin B1 and B2 coded by exons 1′-2′ in Drosophila is aligned with the various N-terminal targeting domains of murine stathmins. In the 1′ domain, conserved residues are double underlined, cysteines are in black, and prolines are bold. In the 2′ domain, many basic residues (bold underlined) are present, similarly to the A″ domain expressed in m-stathmin 4c. Conserved residues among m-stathmins are shaded.
In Drosophila, a single stathmin gene (stai) (Fig. 1B) has been identified that we partially characterized previously (46). As deduced from expressed sequence tag sequences analysis, we now further identified exon 1′ between exons 2 and 2′, which corresponds to an alternate transcription initiation (Fig. 1C) (see below and under “Experimental Procedures”), and exon 6 that can be alternatively spliced out. For a systematic identification of all stai gene products, we performed reverse transcription-PCR using PCR primer couples targeting either exons 1 and 7 or 1′ and 7 (Fig. 2A). Altogether, four different D-stathmin mRNAs were identified which differ either by transcription initiation (exons 1-2 or 1′-2′) or by alternate splicing (of exons 1′-2′ and 6). D-stathmins A1 and A2 are corresponding to exons 1-2-3-4-5-6-7 and exons 1-2-3-4-5-7, and D-stathmin B1 and B2 to exons 1′-2′-3-4-5-6-7 and exons 1′-2′-3-4-5-7, respectively (Fig. 1C). The corresponding proteins share a stathmin-like domain (SLD) (41, 46) with C-terminal extensions of various lengths depending on the inclusion or not of exon 6 (Figs. 1C and 5D). Exons 1′-2′ encode an N-terminal extension in D-stathmins B (Figs. 1C and 5A). Exon 1′ codes for a sequence with no significant identity with the N-terminal targeting domain A of vertebrate neural stathmins 2–4 (Fig. 1D) (18, 41), but with three potential cysteine palmitoylation sites (C13, C15, and C18), which suggests that it may similarly be involved in subcellular targeting of D-stathmins B. Exon 2′ codes for a stretch rich in basic residues, in a way comparable to domain A″ of vertebrate stathmin 4c.
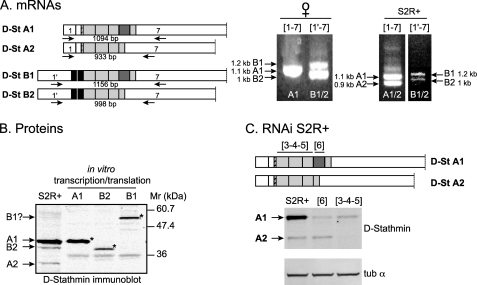
FIGURE 2.
Expression of endogenous D-stathmins. A, reverse transcription-PCR amplification of D-stathmin mRNAs in Drosophila. Left, primers specific of exons 1, 1′, and 7 were used, and the expected sizes of the corresponding PCR amplification products are shown. Right, the resulting amplified cDNAs from adult female flies or S2R+ cells are shown with their respective sizes following electrophoretic separation on a BET agarose gel. B, Western blot identification of D-stathmins expressed in S2R+ cells. S2R+ protein extracts and the in vitro transcription/translation products (*) of clones containing the sequences of D-stathmins A1, B1, or B2, were migrated on an SDS-PAGE gel and revealed by Western blotting with antiserum 98. C, RNA interference experiment on S2R+ cells. Drosophila S2R+ cells were treated with dsRNAs corresponding to exons 3-4-5 and 6, respectively, as indicated on the schema. Western blot analysis of D-stathmin proteins (antiserum 97, which recognizes form A better than B) showing the inhibition of D-stathmin A1 with both dsRNAs, and of the predicted A2 protein (see B and text) only with dsRNA 3-4-5 and not 6. The protein loads for each extract were checked with an anti-α-tubulin (tub α) monoclonal antibody.
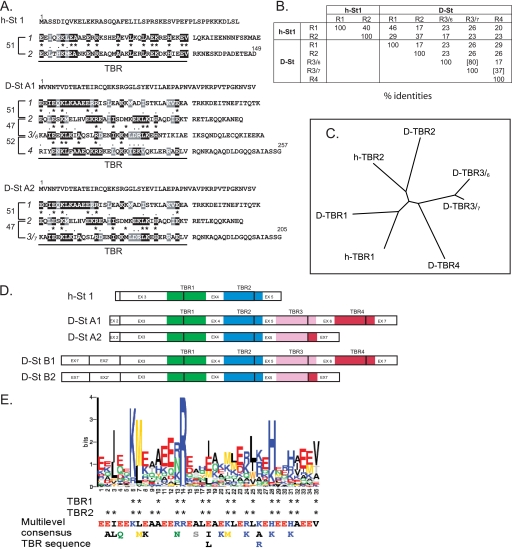
FIGURE 5.
Comparison of the various TBRs. A, TBR sequence analysis. Sequence identities and homologies with any (black, identity; gray, similarity) or adjacent (asterisk, identity; period, similarity) internal 35-amino acid TBRs in h-stathmin, D-stathmin A1, and D-stathmin A2 protein sequences. Distances between repeats are indicated with brackets, in numbers of amino acids between corresponding residues. TBR1, TBR2, TBR3, and TBR4 are the four TBRs identified in D-stathmin A1 from the N to C termini of the protein sequence. Alternate TBR3s are referred to as TBR3/6 or TBR3/7 with their C-terminal end coded either by exon 6 or 7, respectively, depending of the splicing or not of exon 6 (see also in D). B, percentage of amino acid identity of the various TBRs. C, ClustalW2 alignment-derived tree of the various D-stathmin TBRs showing their specific identity as compared with each other. D, schematic representation of the various TBRs with their exon coverage in h- and D-stathmins. E: Top, logos graphical representation resulting from the alignment of 86 stathmin TBRs (1–5) from vertebrates and invertebrates using Meme prediction software. At each position, the size of each residue is proportional to its frequency in that position, and the total height of all residues in the position is proportional to the conservation (information content) of the position. Residues of the TBR 1 and 2 regions pointing toward tubulin in the tubulin-stathmin 4a complex determined by crystallography are indicated by stars. Bottom, consensus TBR sequence derived from the alignment. In each column the residue with the highest probability is classified from top to bottom. All residues shown have a probability of being present higher than 0.2. Thus, the most likely sequence of the motif can be read from the top line.
The in vitro translation products of D-stathmin clones A1, B1, and B2 migrated with higher apparent molecular masses than their calculated molecular masses, i.e. at 40, 51, and 38 kDa, respectively, at the same level as endogenous proteins from Drosophila S2R+ cells (expected low level detection of forms B1 and B2, as for their RNAs) (Fig. 2B). A 34-kDa stathmin-immunoreactive protein was also detected in S2R+ cells, which likely corresponds to D-stathmin A2. By RNA interference in S2R+ cells (Fig. 2C), the expression of D-stathmin A1 as well as of the 34-kDa A2 assigned protein was inhibited with the dsRNA directed against exons 3-4-5 but not with that directed against exon 6, which demonstrates that the 34-kDa protein is as predicted D-stathmin A2.
The Various D-stathmins Are Expressed Differently during Drosophila Embryogenesis
We characterized the developmental expression patterns of the various D-stathmin transcripts in vivo by in situ hybridization on 0- to 24-hour Drosophila embryos (Fig. 3A) with specific probes. D-stathmin A1/A2 mRNAs appeared highly accumulated at early stages of embryogenesis (stages 1–4) corresponding to maternal transcripts. Then the expression of D-stathmins A becomes restricted to the germ cells until the end of embryogenesis. On the other hand, the D-stathmins B-specific 1′-2′ probe labels the central and peripheral nervous systems but not the germ cells. First, D-stathmins B are highly expressed in neuroblasts at stage 12 and in the developing central nervous system after stage 13. Their expression remains high in the embryonic brain and the ventral cord after stage 12. The peripheral nervous system starts to express the D-stathmins B when the sensory organs begin to differentiate at stage 15. In situ hybridization with probe 6 (isoforms A1/B1) did not allow us to differentiate between D-stathmins B1 and B2 expression in the nervous system or between D-stathmins A1 and A2 in germ cells. We can thus observe a tissue segregation of the four transcripts during embryogenesis.
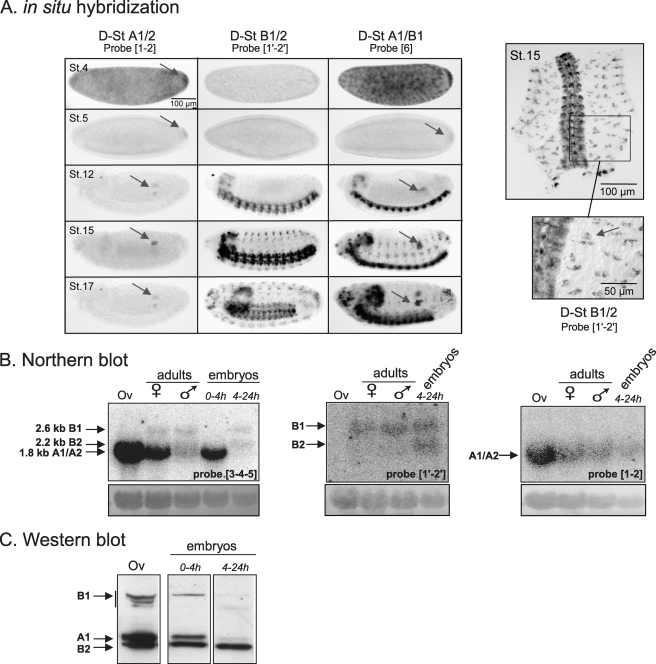
FIGURE 3.
Expression of the various D-stathmins during development. A, in situ hybridization with probes 1-2, 1′-2′, and 6 revealing various D-stathmin mRNA transcripts during embryonic development at different stages (St. 4, 5, 12, 15, and 17): D-stathmins A1/A2 are present in germ cells (arrows) and D-stathmin B1/B2 in the nervous system. Right panel: a higher magnification of the dissected nervous system at stage 15 labeled with probe 1′-2′ and showing the presence of D-stathmin B1/B2 in the sensory organs (arrow). B, Northern blot analysis of adult ovary (Ov) and adult and embryonic flies, with cDNA probes 3-4-5, 1′-2′, and 1-2, which detect all D-stathmin mRNAs, D-stathminS B1/B2, or D-stathmin A1/A2, respectively. The amount of RNA loaded on the gel was followed with methylene blue staining of rRNAs (bottom). C, D-stathmin Western blot analysis (antiserum 98) of adult ovary (Ov) and embryos.
By Northern blot analysis (Fig. 3B), we detected altogether 3 bands of ∼1.8, 2.2, and 2.6 kb, corresponding to forms A1/A2, B2, and B1, respectively. Only A1/A2 mRNAs were detected in ovaries. In whole adult flies, A1/A2 and B1 RNAs were detected, A1/A2 being more expressed in the female extract, which is ovary-enriched. In agreement with the in situ hybridization experiments, in 0- to 4-h embryonic maternal mRNA only A1/A2 forms were detected and in the 4- to 24-h embryo extract mostly the nervous system forms B1 and B2.
At the protein level, Western blot analysis (Fig. 3C) revealed that D-stathmin A1 is indeed the predominant form in adult ovaries, but D-stathmins B1 and B2 could also be visualized, suggesting that the protein expression/Western blot detection is more sensitive than in situ and Northern blot experiments. In 0- to 4-h embryos, forms A1, B1, and B2 and in 4- to 24-h embryos essentially only form B2 were detected. Form A2 was not clearly identified in these extracts.
All D-stathmin Forms Reduce the Microtubule Network in HeLa Cells
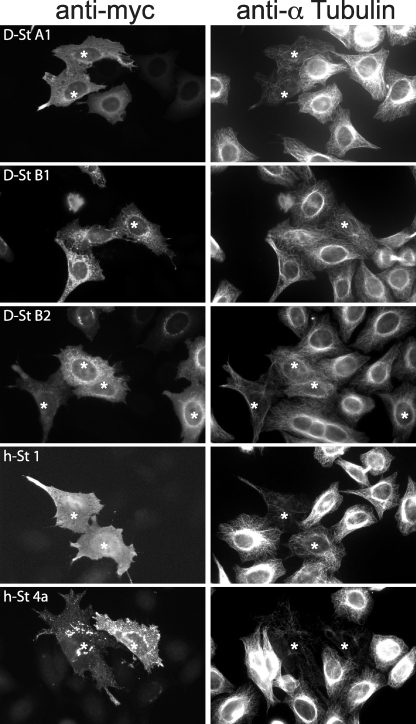
Stathmin in vertebrates is a potent microtubule polymerization inhibitor when overexpressed in cells. Despite the species distance, expression of either D-stathmin A1, B1, or B2 Myc-tagged proteins induced the depolymerization of the microtubule network in human HeLa cells (Fig. 4). Similarly to vertebrate stathmins, they did so in a subset of the transfected cells. This is likely due to different expression levels and post-translational regulation in individual cells with variable physiological states. D-stathmins seemed somewhat less potent than human stathmins 1 and 4a, possibly because of the lower affinity of D-stathmins for human tubulin. D-stathmin B1 and B2 affected mostly the dense perinuclear microtubule network, where they are localized. Immunostaining of Myc-tagged D-stathmin A1 revealed a cytosolic and diffuse distribution, similar to that of h-stathmin 1. Interestingly, that of D-stathmins B1 and B2 appeared punctuated and more dense in the perinuclear region, resembling that of h-stathmins 2–4 (h-stathmin 4a shown in Fig. 4). This may denote a specific membrane localization of D-stathmins B due to the presence of their putative subcellular membrane targeting N terminus domain encoded by exons 1′-2′ (see Fig. 1).
FIGURE 4.
Effect of D-stathmin expression on the microtubule network in HeLa cells. HeLa cells expressing the various Myc-tagged D-stathmins, h-stathmin 4a, or h-stathmin 1 were detected with an anti-Myc antibody (left), and the state of the microtubule network was assessed by co-labeling with an anti-α-tubulin antibody (right). Cells with a deficient microtubule network are shown with asterisks.
Tubulin Binding Repeats
The vertebrate stathmins sequences all contain a 35-amino acid internal repeat with a 51-residue distance between corresponding amino acids and 40% sequence identity (Fig. 5, A and B) (49) at the core of the two well characterized tubulin binding sites (33, 36, 41, 42). We therefore refer to each such repeat as a “tubulin binding repeat” (TBR).
TBR1 and TBR2 sequences are conserved, with 46 and 37% sequence identity of Drosophila TBRs with their respective vertebrate counterparts (Fig. 5B). Within the C-terminal extension of D-stathmins A1 and B1, two additional TBRs can be identified, with a distance between all TBRs similar to that in vertebrates (Fig. 5, A and D). Interestingly, TBRs 3/6 and 4 are encoded by exons 5-6 and 6-7, respectively, in a way that splicing of exon 6 in D-stathmins A2 and B2 results in the loss of one TBR, a novel TBR 3/7 being encoded by the fusion of exons 5-7 (Fig. 5, A–D). The conservation tree of the two vertebrate and 4(/5) Drosophila TBRs (Fig. 5C) clearly shows the correspondence between h- and D-TBRs 1 and 2, respectively, and more distance with TBRs 3 and 4 (Fig. 5, B and C).
We then tried to identify the amino acid residues that are the most conserved throughout evolution in all TBR regions. The Meme prediction software (50), using 86 stathmin TBR sequences from vertebrates, Drosophila, and other Arthropods and invertebrates (supplemental Table 1 and Fig. 8) predicts a Multilevel 35-amino acid consensus sequence (Fig. 5E) for the TBR regions. Interestingly, the Logos graphical representation (51) of the pattern sequence prediction revealed 5 positions with highly conserved residues: 3 (I/L), 6 (K), 7 (M/L), 14 (R), and 28 (H/K). The corresponding residues have been previously shown to point toward tubulin in the 3.5-Å structure of T2S complex formed by tubulin with the SLD of h-stathmin 4a (36), which strengthens the prediction that each TBR belongs to a domain actually binding tubulin.
FIGURE 8.
Comparison of invertebrate stathmin sequences. A, alignment of selected invertebrate stathmin protein sequences (total sequence or part of it) together with that of h-stathmin (bottom sequence, red). TBRs (1–5) are color-shaded, exon limits are indicated, as well as potentially palmitoylated cysteines within the N-terminal targeting domain (black-shaded), and potential conserved phosphorylation sites (light gray). For the species marked with an asterisk, only the N-terminal domain coded by exon 1′-2′ and not the one coded by exon 2 is represented. The dashed lines represent a gap in the sequence and the “ooo” a gap corresponding to a missing exon that has not been detected but may exist. B, taxonomic tree obtained with the taxonomy browser at NCBI (www.ncbi.nlm.nih.gov/Taxonomy/CommonTree) showing selected invertebrate species with a stathmin-related sequence. The species with the exon 5′ coding for an additional TRB5 or with exon 1′-2′ are indicated.
Stoichiometry of Tubulin: D-stathmin Complexes in Vitro
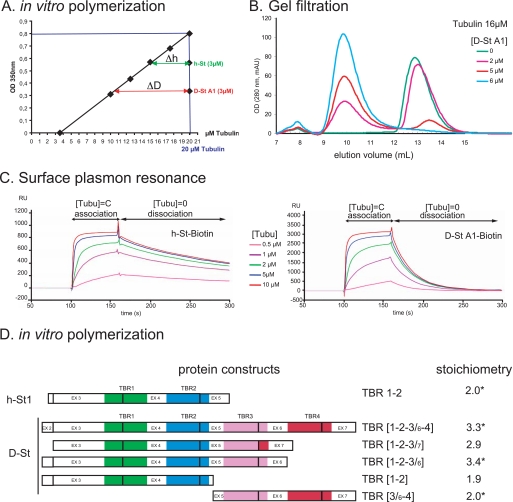
The identification of four TBRs in D-stathmins A1/B1 and three TBRs in A2/B2 strongly suggested the possibility that these Drosophila stathmins could bind up to four or three tubulin α/β heterodimers, respectively. To directly assess this hypothesis, we measured first the activity of D-stathmin A1 on tubulin (20 μm) polymerization in vitro (41, 42) by turbidimetry at 350 nm with or without stathmin (3 μm) (Fig. 6A). Bovine tubulin was used to probe both human and Drosophila stathmins binding, for practical reasons and because tubulin is highly conserved through evolution, over 95% sequence identity between bovine and Drosophila tubulins, with the exception of its variable C-terminal domain, which is not involved in stathmin interaction. Human stathmin, which was used as a reference for a known tubulin:stathmin stoichiometry of 2, yielded an experimental value of 1.7 ± 0.1 (n = 4). D-stathmin yielded an experimental stoichiometry of 2.85 ± 0.25 (n = 4), which clearly demonstrates that each D-stathmin A1 molecule is able to sequester at least three tubulin heterodimers, or more if one takes into account the apparent underestimation of the measured stoichiometry observed for h-stathmin.
FIGURE 6.
In vitro interaction of D-stathmin-A1 and various TBR combinations with tubulin. A, inhibition of tubulin polymerization by D-stathmin A1 compared with h-stathmin. The amount of microtubules formed is measured by turbidity at 350 nm. The addition of 3 μm h-stathmin or D-stathmin A1 to 20 μm bovine tubulin lowered the amount of polymerized microtubules as if they sequestered, respectively, Δh and ΔD concentrations of tubulin. The measured values correspond to the binding of 1.7 ± 0.1 and 2.85 ± 0.25 (n = 4) tubulins per stathmin, respectively. B, D-stathmin A1 forms a complex with tubulin revealed by gel filtration. Fast protein liquid chromatography gel-filtration profiles monitored at 280 nm of 16 μm tubulin incubated with increasing amounts of D-stathmin A1 in the presence of trimethylamine-N-oxide, showing the apparition of a peak at a smaller elution volume, corresponding to a tubulin-stathmin complex. C, surface resonance net sensorgrams revealing the direct binding of soluble tubulin to h-stathmin 1 or D-stathmin A1 coupled through an N-terminal biotin tag to streptavidin Sensorchips. Increasing concentrations of tubulin reach a binding saturation corresponding to the tubulin-stathmin stoichiometries of 1.7 and 2.7, respectively. D, tubulin interaction of D-stathmin A1 constructs containing different numbers and combinations of TBR regions. The tubulin binding potencies of the constructs schematized on the left were measured by tubulin sequestration in the in vitro tubulin polymerization assay, as well as by gel filtration when indicated (asterisk) and normalized to a tubulin:h-stathmin stoichiometry of 2.0.
To verify that the inhibition of tubulin polymerization was reflecting the actual binding of tubulin by D-stathmins, we examined the formation of a tubulin·D-stathmin complex by gel filtration. The addition of D-stathmin A1 to tubulin induced a slight shift of the eluted tubulin peak (monitored at 278 nm, because D-stathmins do not absorb at this wavelength), which suggested the formation of a tubulin·D-stathmin complex (not shown). However, the limited shift likely reflected a relatively weak interaction of bovine tubulin with D-stathmin A1 in the dilute chromatography conditions (41). We therefore repeated the same experiment in the presence of 1 m trimethylamine-N-oxide to stabilize the tubulin·D-stathmin complex (42). In those conditions, a clearly shifted tubulin peak reflecting the formation of a stable complex with D-stathmin was detected. The addition of increasing amounts of D-stathmin A1 to a fixed concentration of tubulin (16 μm) resulted in a progressive conversion of the free tubulin peak toward the tubulin·D-stathmin one (Fig. 6B). As the addition of 6 μm D-stathmin A1 but not 5 μm induced the complete complexation of free tubulin, the tubulin:D-stathmin ratio in the complex is between 2.6:1 and 3.2:1, which is in agreement with the values deduced from tubulin polymerization experiments.
Finally, we examined the association and dissociation kinetics of tubulin to and from D-stathmin A1 in comparison with h-stathmin by surface plasmon resonance (Fig. 6C). To allow homogeneous fixation of proteins on the chip, we produced and purified N-terminal-biotinylated tagged h-stathmins or D-stathmins that were fixed to streptavidin chips allowing the same accessibility for the interaction experiments (42). A constant tubulin concentration flow on chips resulted in an increased mass, visualizing directly the binding of tubulin to the immobilized stathmins. The association kinetics of tubulin to D-stathmin A1 could not be distinguished from that to h-stathmin, whereas the dissociation in tubulin free buffer was faster with D-stathmin A1 than with h-stathmin (Fig. 6C). This latter observation is consistent with the lower affinity observed in the gel-filtration experiments. With increasing concentrations of tubulin injected on the stathmin-bound chip surface, the mass increase reached a saturation which allowed us to estimate the maximal stoichiometry of tubulin binding. The tubulin:stathmin ratios determined by this method were 1.7 for h-stathmin and 2.7 for D-stathmin A1, the latter which corresponds to a ratio of 3.2 for D-stathmin A1 when normalized to the known 2:1 ratio for h-stathmin, in good agreement with values obtained with the tubulin polymerization or gel filtration assays. Altogether, the three different methods are in agreement with a direct interaction of D-stathmin A1 with at least three tubulin heterodimers, the experimental results being, however, lower than the predicted maximal value of four tubulins for one D-stathmin A1.
The Predicted Drosophila TBR Regions Are All Able to Interact with Tubulin
We therefore assayed whether each TBR corresponds to an actual tubulin binding site. We produced various D-stathmin derivatives containing either 2 (1-2 and 3/6-4) or 3 (1-2-3/6 and 1-2-3/7) TBR regions. We compared their tubulin interaction properties with those of full-length D-stathmin A1 (TBR regions 1-2-3/6-4) and h-stathmin (h-TBR regions 1-2) by in vitro tubulin polymerization assay and checked their direct interaction by gel filtration (Fig. 6D). Both D-stathmin derivatives containing two TBRs displayed as expected potencies to inhibit tubulin polymerization with similar stoichiometries, close to the 2:1 ratio of h-stathmin, indicating that all four regions containing TBRs 1, 2, 3/6, and 4 of D-stathmin A1 are indeed able to bind one tubulin molecule when associated two by two. The constructions containing three TBRs displayed a stoichiometry of ∼3:1, indicating that, even when associated by three, the TBR-containing regions are able to interact with one tubulin heterodimer each, including the 3/6 and 3/7 variants of TBR 1-2-3. The latter observation demonstrates that deletion of exon 6 results in the formation of an efficient tubulin binding region containing the novel “composite” TBR 3/7.
Visualization of the Tubulin-Stathmin Complexes by Electron Microscopy
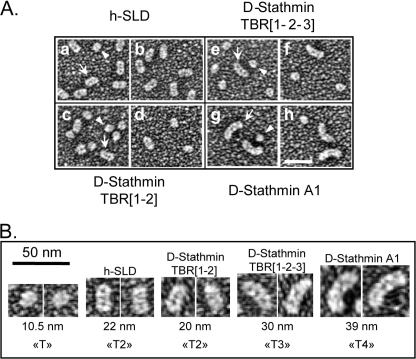
To further ascertain the number of tubulin molecules actually bound by D-stathmins and their TBRs, we visualized the complexes between tubulin and different D-stathmin constructs by electron microscopy after glycerol spraying and subsequent rotary metal shadowing (Fig. 7). The morphologies of these complexes were compared with that of the T2S complex between tubulin and the SLD of h-stathmin 4a used as a reference. With h-SLD, the T2S complex (arrow) appeared very similar in shape and size to previously published T2S complexes (34, 37), and a few, presumably uncomplexed single tubulin molecules (arrowhead) were also visible (Fig. 7, A (panels a and b) and B). Complexes between D-stathmin TBR1–2 and tubulin displayed shapes and dimensions similar to those of the T2S complex (Fig. 7, A (panels c and d) and B), but with fewer complexes (arrow) and a higher proportion of free tubulin (arrowhead), in good agreement with the lower stability of the complex between mammalian tubulin and the Drosophila stathmin construct. Interestingly, complexes of tubulin with full-length D-stathmin TBR 1-2-3 appeared longer (Fig. 7, A (panels e and f) and B), and even longer with D-stathmin A1 (Fig. 7, A (panels g and h) and B). These complexes display a curvature similar to that of the mammalian T2S complex (33, 34, 36, 37), their increased lengths revealing this curvature very strikingly. Whereas a single tubulin heterodimer had an average length of 10.5 nm, the T2S complex formed with the SLD of h-stathmin 4a was estimated at 22 nm, in good agreement with the inclusion of two sequestered tubulins. Interestingly, tubulin complexes with the D-stathmin constructs spanning two, three, or four TBRs had estimated lengths of 20, 30, and 39 nm, in good agreement also with them containing two, three, and four aligned tubulins, respectively. These results clearly show that D-stathmin A1 is able to form, as predicted, a complex with up to four αβ-tubulin heterodimers. In this case shorter complexes with three or two tubulins could be also distinguished (not shown), likely explaining the slightly lower than 4:1 stoichiometry measured by biochemical methods.
FIGURE 7.
Electron microscopy visualization of tubulin complexes with stathmin constructs. A, rotary metal shadowed electron micrographs of tubulin in the presence of the SLD of h-stathmin 4a (a and b), the TBR1-2 (c and d), and TBR1-2-3 (e and f) of D-stathmin constructs and D-stathmin A1 (g and h). In each field, uncomplexed individual tubulin molecules can be seen (arrowheads) as well as elongated complexes of increasing sizes (arrows). B, enlarged views of the various tubulin complexes shown in A, with their calculated “uncoated” lengths (see “Experimental Procedures”) and deduced tubulin stoichiometries, clearly showing the formation of T4S complexes with D-stathmin A1.
Phylogenetic Conservation of the Stathmin Gene and TBR Regions in Arthropods and Invertebrates
The comparison of invertebrate stathmin-related gene and expressed sequence tag sequences available in public databases (Fig. 8 and supplemental Table 1) revealed a high conservation of the gene sequences and intron/exon organization with those of vertebrates and Drosophila.
In Pancrustacea, we identified an additional exon, 5′, between exons 5 and 6, which codes for a domain lacking in Drosophila and more generally in Diptera. Interestingly, we identified the N-terminal 1′-2′ extension domain in most of the Neoptera (Fig. 8, A and B, and supplemental Table 1), all presenting a high level of sequence identity especially in the domain coded by exon 1′. The three cysteines possibly palmitoylated are conserved in all sequences analyzed, which suggests that they may play an important role in particular for subcellular targeting (Fig. 8A).
The tubulin interaction domain is also well conserved, being longer than in vertebrates, and is thus characteristic of Arthropods and other invertebrates. The number and the type of TBR is variable: in addition to the four TBR regions previously described in Drosophila, we identified a new one coded by exon 5′. So, for example, stathmin in Bombyx mori is predicted to have up to five TBR regions and thus to bind up to five dimers of tubulin, in a way likely regulated by differential splicing (Fig. 8A). This phylogenetic analysis allowed us to predict that the number and type of tubulin binding sites in stathmin are important for its function.
DISCUSSION
Microtubule dynamics is regulated by a diversity of proteins, including those of the stathmin family. In vertebrates, a family of four genes codes for stathmin proteins, which all bind tubulin and whose functional invalidation yields at least partially distinct phenotypes (26–28). We previously identified a single stathmin gene in Drosophila (46). In the present work, we show that it codes for a protein family similar to the one in vertebrates, that the corresponding proteins bind tubulin with higher and regulated stoichiometry, and that this diverse family is conserved in Arthropods and other invertebrates.
In contrast to the situation in vertebrates where four genes code for the stathmin protein family, the single Drosophila stathmin gene produces, either by differential splicing (exons 1′-2′ or 6) or by distinct transcription initiations (at exon 1 or exon 1′), all the distinct isoforms of the stathmin family that could be detected at the nucleic and proteic expression levels. D-stathmins A1 and/or A2 are predominantly expressed in the Drosophila embryo germ cells and in ovaries, which are also known to be enriched in stathmin 1 in vertebrates (5, 6), whereas the N-terminally extended D-stathmins B1 and/or B2 are restricted to the nervous system as is mostly the case for the corresponding “long” stathmins 2–4 in vertebrates. D-stathmins A1/A2 and D-stathmins B1/B2 could thus be considered as the paralogues of vertebrate stathmin 1 and stathmins 2–4, respectively. The N-terminal extensions of the neural specific forms have been shown, thanks in part to the palmitoylation of two close cysteines, to address the proteins to subcellular membrane compartments in vertebrates (14, 17, 19), suggesting that this localization is important for their function in the nervous system. Three close cysteines are also present in the N-terminal Drosophila domain and are evolutionary conserved and may thus be able to address the proteins to subcellular compartments in Drosophila. In agreement with this hypothesis, D-stathmins B expressed in HeLa cells are localized in a punctuate localization close to the nucleus. In the Drosophila embryo, we were actually able to show that D-stathmins B are recovered mostly in the mitochondrial fraction, a subcellular compartment also targeted by the vertebrate stathmins N-terminal A domains in some instances (19). It will be of interest to analyze further the targeting properties and regulations of N-terminal domains 1′ and 2′ in Drosophila.
In vertebrates, stathmins are known to be major phosphorylation-dependent microtubule dynamics-regulating proteins. The SLD domain binds directly and sequesters two tubulin heterodimers in a phosphorylation-dependent fashion, thus controlling the amount of tubulin available to polymerize into microtubules (32, 35). It has been also proposed that stathmins promote catastrophes by binding to microtubule ends (for reviews see Refs. 45, 52). Our present analysis reveals the conservation of tubulin-binding domains in Drosophila stathmins, as well as in other invertebrates. However, stathmins in Drosophila possess four (D-stathmin A1/B1) or three (D-stathmin A2/B2) TBR regions, depending on the presence or not of exon 6, whereas an additional repeat is present in the sequence of some arthropod species as a result of the insertion of an additional exon 5′. It is interesting to note, in terms of TBR regulation and evolution, that TBRs are overlapping two adjacent exons, and that insertion of exon 6 or exon 5′ introduces an additional TBR by replacing the C-terminal portion of the upstream TBR, which then completes the inserted N-terminal portion of the additional TBR. It is also interesting to note that this allows the conservation of the distance between TBRs, which is a constitutive feature of them being part of actual tubulin binding sites. Indeed, because effective tubulin binding to stathmins requires the binding of at least two adjacent tubulin heterodimers, the distance between two adjacent TBRs should allow and favor the interaction interface between adjacent tubulins.
Extensive biochemical and microscopic analysis revealed that each tubulin binding repeat indeed corresponds to a tubulin binding site. Although forming less stable complexes than their mammalian counterparts, D-stathmin binds quite well to mammalian tubulin, meaning a high degree of functional conservation through evolution. The fact that the maximal predicted binding of four tubulins for one D-stathmin could not be fully reached by biochemical experimentation despite its visualization by electron microscopy might be due to a lower stability of the complex with four tubulins, a hypothesis supported by the microscopic visualization of complexes with stoichiometries of four, but also three or two. The functional conservation of both stathmins and tubulins is also illustrated by the capacity of D-stathmins to alter the microtubule network in human HeLa cells. The fact that this was observed only in a subset of D-stathmin-expressing cells, as in similar experiments with vertebrate stathmins, is likely due to the necessity to reach a threshold expression level for efficient tubulin binding and to the diversity of physiological states of the cells within the culture.
It was previously demonstrated that in vertebrate stathmins TBR1 and TBR2 contribute differentially to the stability of the T2S complex, TBR2 being mostly responsible for the difference in stability of the tubulin complexes formed with stathmins 1 and 4, respectively (42). The evolution tree of all the identified vertebrate and invertebrate TBRs shows that TBRs of a given type (1–5) cluster in separate branches (see Fig. 5C for D-stathmin TBRs) and hence have a specific identity. This clustering likely reflects also specific tubulin-interacting properties, as with TBRs 1 and 2 in vertebrates. The combination of more than two TBRs could thus determine not only the level of tubulin binding, but also the nature (affinity and complex stability) of the interaction. Sequence comparison of a large number of TBRs throughout evolution allowed us to derive a consensus sequence with some highly conserved residues, which happen to be at positions oriented toward tubulin in the described mammalian crystal structure (36), and hence likely essential for the binding of tubulin. This observation further confirms the role of TBRs in tubulin binding in a way similar to the one described in vertebrates. The amino acids that have a strong conservation might then also be considered as targets to disrupt the interaction between stathmin and tubulin, whereas the consensus sequence could be used as a basis to design peptides as potential tubulin traps in cancer therapy.
Interestingly, among the identified invertebrate stathmin sequences, only Lumbricus rubellus has two TBRs, whereas other invertebrates such as molluscs (Aplysia) and arachnids also have at least four TBRs. Whereas TBRs 1 and 2 presumably result from very early duplication in evolution, the additional TBRs (3–5) seem to result from further exon duplications before the appearance of molluscs and Arthropods. Interestingly, the fifth TBR further identified in some Arthropods seems to have been acquired during evolution in Pancrustacea, and lost later in Diptera species. One can speculate that the additional exons and the regulation of their expression were acquired in invertebrates to yield a “single gene” protein family, whereas in vertebrates gene duplication and further evolution generated the well characterized “multigene” stathmin family.
The biological significance of the high tubulin binding stoichiometry of many invertebrate stathmins as compared with vertebrates is intriguing. One can note that it strongly argues for the tubulin-sequestering model of microtubule dynamics regulation, because binding of more tubulins by each stathmin molecule makes this process even more efficient in terms of sequestration than in vertebrates. The high and post-transcriptionally regulated stoichiometry may also compensate for a simpler genome in invertebrates, whereas a likely more sophisticated regulation through multiple gene regulation has evolved in vertebrates. It will be of interest to determine which biological contexts, during development and in the adult, and which molecular and signaling mechanisms in invertebrates control the stathmin isoforms expression at the mRNA levels (initiation of transcription or splicing) or at the protein level by phosphorylation on conserved or additional phosphorylation sites (46), and by palmitoylation of their N-terminal extension.
In conclusion, the stathmins being a family of proteins conserved from invertebrates to mammals argues for major biological roles and importance in diverse biological processes, from development to differentiated tissues and cells. Further characterization of these roles and of the associated regulations should open ways to understand some dysregulations, such as in cancer and in the nervous system, and to interfere with the processes involved with potential therapeutic perspectives.
Supplementary Material
Acknowledgments
We thank A. Maucuer for suggestions and discussions, J. Cartaud for initial electron microscopy experiments, Thibault Griseri for initial constructs, and I. Jourdain for the production of biotinylated tagged human-stathmin. We are indebted to B. Maco for assistance with the rotary metal-shadowing experiments. Fluorescence and confocal microscopy were performed in the Institut du Fer à Moulin Imaging facility. We are grateful to the Interdisciplinary Center for Microscopy of the University of Basel for access to the transmission electron microscope.
This work was supported by INSERM, the Université Pierre et Marie Curie (UPMC), the Association pour la Recherche Contre le Cancer, the Association Française Contre les Myopathies, the Association pour la Recherche sur la Sclérose en Plaque, and the Agence Nationale pour la Recherche.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- TBR
- tubulin binding repeat
- dsRNA
- double-stranded RNA
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SLD
- stathmin-like domain.
REFERENCES
- 1.Maucuer A., Moreau J., Méchali M., Sobel A. (1993) J. Biol. Chem. 268, 16420–16429 [PubMed] [Google Scholar]
- 2.Stein R., Orit S., Anderson D. J. (1988) Dev. Biol. 127, 316–325 [DOI] [PubMed] [Google Scholar]
- 3.Ozon S., Byk T., Sobel A. (1998) J. Neurochem. 70, 2386–2396 [DOI] [PubMed] [Google Scholar]
- 4.Ozon S., Maucuer A., Sobel A. (1997) Eur. J. Biochem. 248, 794–806 [DOI] [PubMed] [Google Scholar]
- 5.Amat J. A., Fields K. L., Schubart U. K. (1991) Brain Res. Dev. Brain Res. 60, 205–218 [DOI] [PubMed] [Google Scholar]
- 6.Koppel J., Boutterin M. C., Doye V., Peyro-Saint-Paul H., Sobel A. (1990) J. Biol. Chem. 265, 3703–3707 [PubMed] [Google Scholar]
- 7.Doye V., Kellermann O., Buc-Caron M. H., Sobel A. (1992) Differentiation 50, 89–96 [DOI] [PubMed] [Google Scholar]
- 8.Schubart U. K., Xu J., Fan W., Cheng G., Goldstein H., Alpini G., Shafritz D. A., Amat J. A., Farooq M., Norton W. T., et al. (1992) Differentiation 51, 21–32 [DOI] [PubMed] [Google Scholar]
- 9.Guillaume E., Evrard B., Com E., Moertz E., Jégou B., Pineau C. (2001) Mol. Reprod. Dev. 60, 439–445 [DOI] [PubMed] [Google Scholar]
- 10.Koppel J., Rehák P., Baran V., Veselá J., Hlinka D., Manceau V., Sobel A. (1999) Mol. Reprod. Dev. 53, 306–317 [DOI] [PubMed] [Google Scholar]
- 11.Wuenschell C. W., Mori N., Anderson D. J. (1990) Neuron 4, 595–602 [DOI] [PubMed] [Google Scholar]
- 12.Ozon S., El Mestikawy S., Sobel A. (1999) J. Neurosci. Res. 56, 553–564 [DOI] [PubMed] [Google Scholar]
- 13.Stein R., Mori N., Matthews K., Lo L. C., Anderson D. J. (1988) Neuron 1, 463–476 [DOI] [PubMed] [Google Scholar]
- 14.Di Paolo G., Lutjens R., Pellier V., Stimpson S. A., Beuchat M. H., Catsicas S., Grenningloh G. (1997) J. Biol. Chem. 272, 5175–5182 [DOI] [PubMed] [Google Scholar]
- 15.Lutjens R., Igarashi M., Pellier V., Blasey H., Di Paolo G., Ruchti E., Pfulg C., Staple J. K., Catsicas S., Grenningloh G. (2000) Eur. J. Neurosci. 12, 2224–2234 [DOI] [PubMed] [Google Scholar]
- 16.Maekawa S., Morii H., Kumanogoh H., Sano M., Naruse Y., Sokawa Y., Mori N. (2001) J. Biochem. 129, 691–697 [DOI] [PubMed] [Google Scholar]
- 17.Charbaut E., Chauvin S., Enslen H., Zamaroczy S., Sobel A. (2005) J. Cell Sci. 118, 2313–2323 [DOI] [PubMed] [Google Scholar]
- 18.Gavet O., El Messari S., Ozon S., Sobel A. (2002) J. Neurosci. Res. 68, 535–550 [DOI] [PubMed] [Google Scholar]
- 19.Chauvin S., Poulain F. E., Ozon S., Sobel A. (2008) Biol. Cell 100, 577–589 [DOI] [PubMed] [Google Scholar]
- 20.Melhem R. F., Strahler J. R., Hailat N., Zhu X. X., Hanash S. M. (1991) Biochem. Biophys. Res. Commun. 179, 1649–1655 [DOI] [PubMed] [Google Scholar]
- 21.Rubin C. I., Atweh G. F. (2004) J. Cell. Biochem. 93, 242–250 [DOI] [PubMed] [Google Scholar]
- 22.Balogh A., Mège R. M., Sobel A. (1996) Exp. Cell Res. 224, 8–15 [DOI] [PubMed] [Google Scholar]
- 23.Ohkawa N., Fujitani K., Tokunaga E., Furuya S., Inokuchi K. (2007) J. Cell Sci. 120, 1447–1456 [DOI] [PubMed] [Google Scholar]
- 24.Baldassarre G., Belletti B., Nicoloso M. S., Schiappacassi M., Vecchione A., Spessotto P., Morrione A., Canzonieri V., Colombatti A. (2005) Cancer Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 25.Giampietro C., Luzzati F., Gambarotta G., Giacobini P., Boda E., Fasolo A., Perroteau I. (2005) Endocrinology 146, 1825–1834 [DOI] [PubMed] [Google Scholar]
- 26.Poulain F. E., Chauvin S., Wehrlé R., Desclaux M., Mallet J., Vodjdani G., Dusart I., Sobel A. (2008) J. Neurosci. 28, 7387–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulain F. E., Sobel A. (2007) Mol. Cell Neurosci. 34, 137–146 [DOI] [PubMed] [Google Scholar]
- 28.Shumyatsky G. P., Malleret G., Shin R. M., Takizawa S., Tully K., Tsvetkov E., Zakharenko S. S., Joseph J., Vronskaya S., Yin D., Schubart U. K., Kandel E. R., Bolshakov V. Y. (2005) Cell 123, 697–709 [DOI] [PubMed] [Google Scholar]
- 29.Liedtke W., Leman E. E., Fyffe R. E., Raine C. S., Schubart U. K. (2002) Am J. Pathol. 160, 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchison T., Kirschner M. (1984) Nature 312, 237–242 [DOI] [PubMed] [Google Scholar]
- 31.Curmi P. A., Gavet O., Charbaut E., Ozon S., Lachkar-Colmerauer S., Manceau V., Siavoshian S., Maucuer A., Sobel A. (1999) Cell Struct. Funct. 24, 345–357 [DOI] [PubMed] [Google Scholar]
- 32.Curmi P. A., Andersen S. S., Lachkar S., Gavet O., Karsenti E., Knossow M., Sobel A. (1997) J. Biol. Chem. 272, 25029–25036 [DOI] [PubMed] [Google Scholar]
- 33.Gigant B., Curmi P. A., Martin-Barbey C., Charbaut E., Lachkar S., Lebeau L., Siavoshian S., Sobel A., Knossow M. (2000) Cell 102, 809–816 [DOI] [PubMed] [Google Scholar]
- 34.Honnappa S., Cutting B., Jahnke W., Seelig J., Steinmetz M. O. (2003) J. Biol. Chem. 278, 38926–38934 [DOI] [PubMed] [Google Scholar]
- 35.Jourdain L., Curmi P., Sobel A., Pantaloni D., Carlier M. F. (1997) Biochemistry 36, 10817–10821 [DOI] [PubMed] [Google Scholar]
- 36.Ravelli R. B., Gigant B., Curmi P. A., Jourdain I., Lachkar S., Sobel A., Knossow M. (2004) Nature 428, 198–202 [DOI] [PubMed] [Google Scholar]
- 37.Steinmetz M. O., Kammerer R. A., Jahnke W., Goldie K. N., Lustig A., van Oostrum J. (2000) EMBO J. 19, 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amayed P., Pantaloni D., Carlier M. F. (2002) J. Biol. Chem. 277, 22718–22724 [DOI] [PubMed] [Google Scholar]
- 39.Honnappa S., Jahnke W., Seelig J., Steinmetz M. O. (2006) J. Biol. Chem. 281, 16078–16083 [DOI] [PubMed] [Google Scholar]
- 40.Larsson N., Segerman B., Howell B., Fridell K., Cassimeris L., Gullberg M. (1999) J. Cell Biol. 146, 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charbaut E., Curmi P. A., Ozon S., Lachkar S., Redeker V., Sobel A. (2001) J. Biol. Chem. 276, 16146–16154 [DOI] [PubMed] [Google Scholar]
- 42.Jourdain I., Lachkar S., Charbaut E., Gigant B., Knossow M., Sobel A., Curmi P. A. (2004) Biochem. J. 378, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manna T., Thrower D., Miller H. P., Curmi P., Wilson L. (2006) J. Biol. Chem. 281, 2071–2078 [DOI] [PubMed] [Google Scholar]
- 44.Howell B., Larsson N., Gullberg M., Cassimeris L. (1999) Mol. Biol. Cell 10, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cassimeris L. (2002) Curr. Opin. Cell Biol. 14, 18–24 [DOI] [PubMed] [Google Scholar]
- 46.Ozon S., Guichet A., Gavet O., Roth S., Sobel A. (2002) Mol. Biol. Cell 13, 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawler S., Gavet O., Rich T., Sobel A. (1998) FEBS Lett. 421, 55–60 [DOI] [PubMed] [Google Scholar]
- 48.Tautz D., Pfeifle C. (1989) Chromosoma 98, 81–85 [DOI] [PubMed] [Google Scholar]
- 49.Maucuer A., Doye V., Sobel A. (1990) FEBS Lett. 264, 275–278 [DOI] [PubMed] [Google Scholar]
- 50.Bailey T. L., Elkan C. (1994) Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 51.Schneider T. D., Stephens R. M. (1990) Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinmetz M. O. (2007) J. Struct Biol. 158, 137–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.