Abstract
Sterol regulatory element-binding protein (SREBP)-1 is a key transcription factor for the regulation of lipogenic enzyme genes in the liver. Polyunsaturated fatty acids (PUFA) selectively suppress hepatic SREBP-1, but molecular mechanisms remain largely unknown. To gain insight into this regulation, we established in vivo reporter assays to assess the activities of Srebf1c transcription and proteolytic processing. Using these in vivo reporter assays, we showed that the primary mechanism for PUFA suppression of SREBP-1 is at the proteolytic processing level and that this suppression in turn decreases the mRNA transcription through lowering SREBP-1 binding to the SREBP-binding element on the promoter (“autoloop regulatory circuit”), although liver X receptor, an activator for Srebf1c transcription, is not involved in this regulation by PUFA. The mechanisms for PUFA suppression of SREBP-1 confirm that the autoloop regulation for transcription is crucial for the nutritional regulation of triglyceride synthesis.
Keywords: Fatty Acid, Lipogenesis, Liver, Promoters, Transcription Factors, Polyunsaturated Fatty Acids, Sterol Regulatory Element-binding Protein
Introduction
Polyunsaturated fatty acids (PUFA)3 have been well established as negative regulators of hepatic lipogenesis (reviewed in Ref. 1). Allmann and Gibson (2) discovered that adding 2% linoleate to a high carbohydrate fat-free diet suppressed the rate of hepatic fatty acid biosynthesis and the activities of fatty-acid synthase and glucose-6-phosphate dehydrogenase by nearly 70% in mice. In contrast, supplementing the high carbohydrate diet with palmitate, oleate, or cholesterol had no effect on hepatic lipogenesis or the activity of lipogenic enzymes. Since then, a number of investigators have demonstrated that dietary PUFA of the n-6 and n-3 families suppress hepatic lipogenesis. This anti-lipogenic action of PUFA reflects decreases in mRNA levels of hepatic enzymes, including acetyl-CoA carboxylase, fatty-acid synthase, and stearoyl-CoA desaturase.
The fatty acid biosynthetic pathway, composed of some 25 enzymes, has been elucidated in detail (3). For the de novo synthesis of long chain saturated fatty acids, fatty-acid synthase, the main synthetic enzyme that catalyzes the condensation of malonyl-CoA to produce the 16-carbon saturated fatty acid palmitate, and acetyl-CoA carboxylase, which synthesizes malonyl-CoA from acetyl-CoA, are of particular importance. The regulation of these lipogenic enzymes has been revealed to be primarily controlled by a transcription factor sterol regulatory element-binding protein (SREBP)-1c (4, 5).
SREBPs are transcription factors that belong to the basic helix-loop-helix leucine zipper family and are considered to be profoundly involved in the transcriptional regulation of cholesterogenic and lipogenic enzymes (6, 7). Unlike other members of the basic helix-loop-helix leucine zipper family, SREBPs are synthesized as precursors bound to the endoplasmic reticulum and nuclear envelope. Upon activation, SREBPs are cleaved, and the N-terminal parts are released from the membrane into the nucleus as mature protein by a sequential two-step proteolytic processing. To date, three SREBP isoforms, SREBP-1a, -1c, and -2, have been identified and characterized. SREBP-1a and -1c are transcribed from the same gene, each by a distinct promoter, and the predominant SREBP-1 isoform in liver is 1c rather than 1a (8). It has been established by several lines of evidence, especially by those from transgenic and knock-out mouse models, that SREBP-1c controls hepatic lipogenesis, whereas SREBP-2 plays a crucial role in regulation of cholesterol synthesis (5, 9, 10).
In 1999, we and others reported that the PUFA-specific suppression of lipogenic enzymes is mediated by the reduction of nuclear SREBP-1c protein in the liver (11–14). Interestingly, PUFA selectively decreases SREBP-1, not affecting SREBP-2. The mechanism by which PUFA specifically suppresses SREBP-1c nuclear abundance, however, remains unclear, although several potential mechanisms have been implicated, including suppression of Srebf1c gene transcription and proteolytic processing as well as enhancement of proteasomal degradation and mRNA decay (11, 15–18). As for the suppression of Srebf1c gene transcription by PUFA, we have previously identified liver X receptor-binding element (LXRE) and SREBP-binding element (SRE) on the Srebf1c promoter region by a series of promoter analyses (19, 20), and we have also suggested that PUFA can antagonize LXR in an in vitro setting (16).
These situations prompted us to clarify the molecular mechanism underlying the suppressive effect of PUFA on nuclear SREBP-1 abundance, especially in the in vivo setting. Because the inhibitory effect of fatty acids on SREBP-1 was specific and clear for PUFA in in vivo experiments, whereas many previous reports using in vitro system have failed to show this specificity (21, 22), we adopted an approach of in vivo reporter assays utilizing the in vivo imaging system (IVISTM; Xenogen, Alameda, CA). First, to examine the transcriptional mechanism, in vivo promoter analyses were performed, and the responsible cis-element on the Srebf1c promoter was located at SRE, not at LXRE. Next, the mechanism by which PUFA decreases the nuclear form of SREBP-1 was explored by another reporter system detecting proteolytic activity for the precursor form of SREBP-1, demonstrating that PUFA suppresses the maturation of SREBP-1 through proteolytic processes. From these experiments, we concluded that the primary mechanism for PUFA suppression of SREBP-1 expression is at the proteolytic processing level and that this suppression in turn decreases the Srebf1c mRNA transcription through lowering SREBP-1 binding to SRE on the promoter (“autoloop regulation” (19)).
EXPERIMENTAL PROCEDURES
Materials
Eicosapentaenoic acid (EPA) ethyl ester (95% grade) was provided from Mochida Pharmaceutical (Tokyo, Japan) and GW532 (SCAP ligand) from GlaxoSmithKline (Les Ulis Cedex, France). The synthetic LXR agonist T0901317 was purchased from Cayman Chemical (Ann Arbor, MI). Standard laboratory diet (CRF-1, composed of 60% carbohydrate, 13% fat, and 27% protein on a caloric basis) and high carbohydrate fat-free diet (70% sucrose and 20% casein supplemented with methionine, vitamins, and minerals) were obtained from Oriental Yeast (Tokyo, Japan). Other materials were purchased from Sigma unless indicated otherwise.
Animals
Seven- to 9-week-old ICR male mice were purchased from CLEA (Tokyo, Japan). All animals were maintained in a temperature-controlled environment with a 12-h light/dark cycle and were given free access to standard laboratory diet and water. Four days before the start of indicated fatty acid administration, the basal diet was switched to a high carbohydrate fat-free diet. EPA or oleic acid ethyl ester (OLA) was administered orally once a day for 4 days. GW532 (0–15 mg/kg/day) or vehicle (0.9% carboxymethylcellulose, 9.95% polyethylene glycol 400, and 0.05% Tween 80) was administered orally once a day for 3 days. Mice were sacrificed in the light phase in a nonfasted state. All experiments were repeated at least twice. All animals studied were anesthetized and euthanized according to protocol approved by the Tokyo University Animal Care and Use Committee.
RNA Isolation and Northern Blotting
Total RNA from mouse liver was extracted using TRIzol reagent (Invitrogen), and a 7.5-μg RNA sample equally pooled among each group was run on a 1% agarose gel containing formaldehyde and transferred to a nylon membrane. The cDNA probes for mouse SREBP-1, SREBP-2, and 36B4 were cloned as described previously (11). The probes were labeled with [α-32P]dCTP using Megaprime DNA labeling system (Amersham Biosciences). The membranes were hybridized with the radiolabeled probe in Rapid-Hyb Buffer (Amersham Biosciences) and washed in 0.1× SSC, 0.1% SDS at 65 °C. Blots were exposed to BAS imaging plate for the BAS2000 BIO imaging analyzer (Fuji Photo Film). The quantification results obtained with the BAS2000 system were normalized to the signal generated from 36B4 mRNA.
Nuclear Protein Extraction from Liver
Nuclear extract protein from mouse or rat liver was prepared as described previously (23). Briefly, excised livers (0.5 g) were homogenized in a Polytron in 5 ml of buffer A, which consisted of 10 mm HEPES, pH 7.9, 25 mm KCl, 1 mm EDTA, 2 m sucrose, 10% glycerol, 0.15 mm spermine, and 2 mm spermidine, supplemented with protease inhibitors (6 μg/ml N-acetyl-leucyl-leucyl-norleucinal (ALLN, Calbiochem), 2.5 μg/ml pepstatin A, 2 μg/ml leupeptin, 0.1 mm phenylmethylsulfonyl fluoride and 2.5 μg/ml aprotinin). Pooled homogenate was then subjected to one stroke of a Teflon pestle in a Potter-Elvehjem homogenizer, followed by filtration through two layers of cheesecloth, and layered over 10 ml of buffer A. After centrifugation at 24,000 rpm on a Beckman SW28 rotor for 1 h at 4 °C, the resulting nuclear pellet was resuspended in a buffer containing 10 mm HEPES, pH 7.9, 100 mm KCl, 2 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, and 10% glycerol supplemented with protease inhibitors, after which 0.1 volume of 5 m NaCl was added. Each mixture was agitated gently for 30 min at 4 °C and then centrifuged at 89,000 rpm on a Himac S120AT2 rotor (Hitachi, Tokyo, Japan) for 30 min at 4 °C. The supernatant was used as nuclear extract.
Immunoblotting of SREBP Proteins
Aliquots of nuclear extract (10 μg) and total lysate (50 μg) proteins were subjected to SDS-PAGE. Immunoblot analysis was performed using the ECL Western blotting detection system (Amersham Biosciences) and exposed to XAR-5 film (Eastman Kodak Co.). The primary antibodies for SREBPs (rabbit polyclonal; number 931 for mouse SREBP-1 and number 528 for SREBP-2) were used as described previously (24). The primary antibody for LXRα/β (H-144; sc-13068) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Fatty Acid Composition of Liver
Fatty acid composition was measured as described previously (24). An aliquot (0.1 g) of liver samples snap-frozen by liquid nitrogen was homogenized in 1 ml of normal saline. The fatty acid composition was measured by gas chromatography at Bio-Medical Laboratories (Tokyo, Japan) (25) Briefly, total lipids in liver homogenates were extracted according to the Folch's procedure (26), followed by transesterification of fatty acids with boron trifluoride/methanol at 100 °C for 90 min. The methylated fatty acids were then extracted with hexane and analyzed using a GC-17A gas chromatograph (Shimadzu Corp., Kyoto, Japan) and BPX70 capillary column (0.25 mm inner diameter × 30 m, SGE International Ltd., Melbourne, Australia).
Transfection and Luciferase Assays
HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 25 mm glucose, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate supplemented with 10% fetal bovine serum. For luciferase assay, HEK293 cells were seeded in a 48-well plate and incubated until 80% confluent. The indicated amounts of expression plasmids, firefly luciferase reporter plasmid, and pSV40-Renilla luciferase plasmid were co-transfected into HEK293 cells using SuperFect transfection reagent (Qiagen) according to the manufacturer's protocol. Total amounts of transfected DNA were adjusted with empty vector. The luciferase activity in transfectants was measured on a luminometer. Renilla luciferase activities were used to normalize transfection efficiencies.
In Vivo Imaging of Luciferase Activity
In vivo imaging was performed as described previously (27). Mice were anesthetized with isoflurane/oxygen, and 3.0 mg of luciferin dissolved in 0.4 ml of phosphate-buffered saline (7.5 mg/ml) was injected into the intraperitoneal cavity. Mice were imaged from the ventral side using an In Vivo Imaging System (IVISTM, Xenogen) 15 min following the injection of luciferin. Relative photon emission over the liver region was quantified using LivingImageTM software (Xenogen).
Plasmid Construction
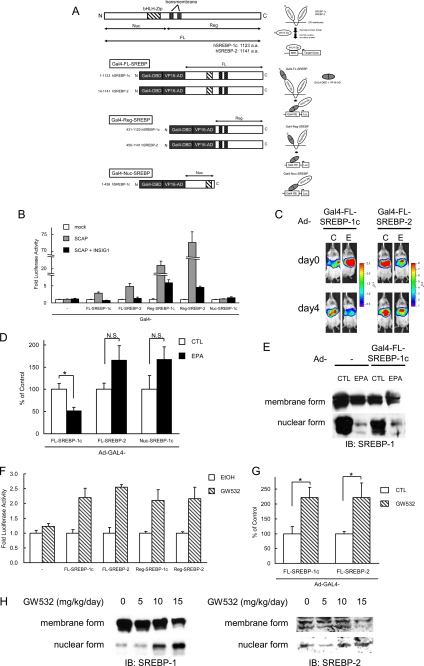
To construct expression plasmids for GAL4-DNA binding domain (GAL4-DBD) and VP16-transactivation domain (VP16-AD) fused to human SREBPs, VP16-AD from pACT vector (Promega) was inserted into pM vector (Clontech) with various lengths of DNA fragment of human SREBP-1c (amino acids 1–1123 (“FL” for full length), 1–436 (“Nuc” for N-terminal nuclear part), or 431–1123 (“Reg” for C-terminal regulatory domain) or human SREBP-2 (amino acids 14–1141 (FL) or 450–1141 (Reg)) retrieved by PCR from pTK-HSV-hSREBP-1c, pTK-HSV-hSREBP-2, and pcDNA3.1(+)-SREBP-1c (28, 29). Gal4-RE-Luc plasmid was described previously (30). For the construction of an expression plasmid for mouse Insig-1, cDNA fragment was amplified by PCR with primers 5′-GGATCCATGCCCAGGCTGCACGACCACG-3′ and 5′-CTCGAGTCAGTCACTGTGAGGCTTTTCCG-3′ and cloned into pcDNA3 vector with hemagglutin tag at the N terminus using BamHI and XhoI. The expression plasmid for SCAP is a kind gift from Dr. Nakakuki.
Preparation of Recombinant Adenoviruses
To construct various lengths of mouse Srebf1c promoter luciferase reporter plasmids, DNA fragments retrieved from pGL2 vectors constructed previously (19, 20, 31) were inserted into pGL3 basic vector plasmids (Promega). The fragments including promoter region linked to luciferase reporter gene were inserted into the Gateway entry vector pENTR4 (Invitrogen) and generated by homologous recombination between the entry vector and the pAd promoterless vector (Invitrogen). The fragments of human SREBPs GAL4-DBD and VP16-AD fusion protein from various GAL4-SREBP vectors were inserted into pENTR4 and generated by homologous recombination with the pAd/CMV/V5-DEST vector (Invitrogen). Adenoviruses encoding SREBP-1-specific and LacZ-specific shRNA for RNA interference (SREBP1i and LacZi, respectively) were described previously (32). Adenovirus construct encoding SREBP-2-specific shRNA (SREBP2i) targeting 5′-GGAGCAGTCTCAACGTCAACG-3′ sequence on SREBP-2 was subcloned into U6 entry vector (Invitrogen) and generated by homologous recombination with the pAd promoterless vector. Adenovirus construct encoding both LXRα and LXRβ shRNA (LXRi) targeting 5′-ACAGCTCCCTGGCTTCCTA-3′ sequence on LXRα (33) and 5′-CTACAACCACGAGACAGAA-3′ sequence on LXRβ, respectively, was subcloned into U6 entry vector (Invitrogen) and generated by homologous recombination with the pAd promoterless vector. Recombinant adenoviruses were propagated in HEK293 cells and purified by CsCl gradient centrifugation as described previously (34).
RESULTS
PUFA Selectively Decreases SREBP-1, Not Affecting SREBP-2
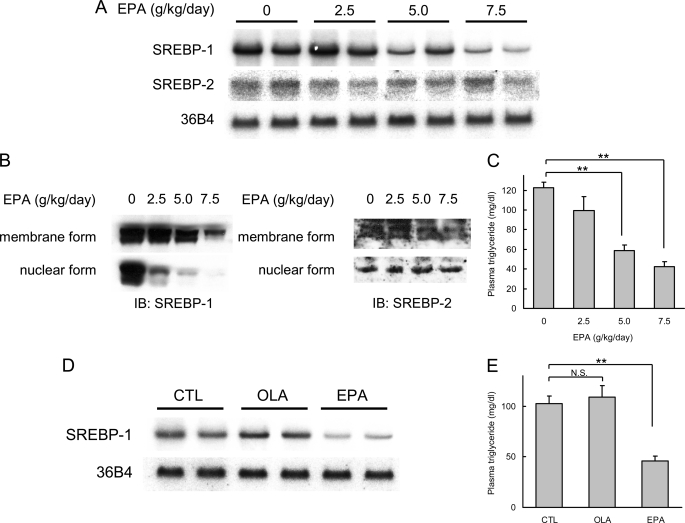
In our first series of experiments, we verified the effects of PUFA on SREBP-1 and -2 expression in the liver. As expected, administration of EPA, one of the major n-3 PUFA in mammals, down-regulated SREBP-1 mRNA and protein expression (Fig. 1, A, B, D, and E), leading to a decrease in plasma triglycerides concentration (Fig. 1, C and F) as compared with control or oleate. The delivery of orally administered EPA to the liver was confirmed by gas chromatography analysis (Table 1). EPA administration did not affect the body weights of mice (data not shown). The dose-response relationship data suggested that the nuclear form is more sensitive to PUFA than the membrane form of protein or mRNA. In contrast, SREBP-2 mRNA and protein levels were not altered by PUFA.
FIGURE 1.
PUFA selectively decreases SREBP-1, not affecting SREBP-2. A, Northern blot analysis of SREBP-1 and SREBP-2 from livers. Total RNA (7.5 μg) from livers pooled equally from two mice for each group was subjected to Northern blotting to determine SREBP-1, SREBP-2, and 36B4 (used as a loading control) mRNA levels. ICR male mice were fed a high carbohydrate fat-free diet and treated orally with EPA at indicated doses once a day for 4 consecutive days. Control mice were treated orally with 7.5 g/kg water. Mice were sacrificed in a nonfasted state. B, immunoblot (IB) analysis of mature and precursor SREBP-1 and SREBP-2 proteins from livers. Aliquots of nuclear extracts (10 μg) and total proteins (50 μg) from livers pooled equally from four male mice for each group were subjected to immunoblot analysis. The primary antibodies used were polyclonal anti-mouse SREBP-1 and polyclonal anti-mouse SREBP-2. C, plasma triglycerides in EPA-treated mice. D, Northern blot analysis of SREBP-1 from livers. ICR male mice were fed a high carbohydrate fat-free diet and treated orally with 7.5 g/kg OLA or EPA once a day for 4 days. Total RNA (7.5 μg) from livers pooled equally from two mice for each group was subjected to Northern blotting to determine SREBP-1 and 36B4 (used as a loading control (CTL)) mRNA levels. E, plasma triglycerides in OLA- and EPA-treated mice. These data are representative of at least two independent experiments (n = 4 mice/group). Results are means ± S.E. **, p < 0.01; N.S., not significant.
TABLE 1.
Fatty acid composition in the liver
Fatty acid composition in the liver was analyzed by gas chromatography (n = 4 mice/group). Results are means ± S.E.
| EPA (g/kg/day) | ||||
|---|---|---|---|---|
| 0 | 2.5 | 5.0 | 7.5 | |
| mg/g liver weight | ||||
| C12:0 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| C14:0 | 0.22 ± 0.05 | 0.09 ± 0.03 | 0.05 ± 0.01a | 0.07 ± 0.02a |
| C14:1n-5 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| C16:0 | 10.11 ± 1.07 | 8.06 ± 0.60 | 6.48 ± 0.40a | 7.97 ± 0.58 |
| C16:1n-7 | 2.06 ± 0.26 | 1.10 ± 0.18a | 0.70 ± 0.04b | 0.75 ± 0.13b |
| C18:0 | 3.12 ± 0.09 | 3.72 ± 0.17 | 3.13 ± 0.18 | 4.15 ± 0.18b |
| C18:1n-9 | 14.77 ± 2.96 | 7.76 ± 1.82a | 3.70 ± 0.54a | 4.88 ± 1.43a |
| C18:2n-6 | 1.76 ± 0.32 | 1.74 ± 0.23 | 1.92 ± 0.27 | 1.65 ± 0.30 |
| C18:3n-6 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.03 ± 0.00 | 0.03 ± 0.00 |
| C18:3n-3 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.03 ± 0.00 | 0.00 ± 0.00 |
| C20:0 | 0.04 ± 0.01 | 0.07 ±0.02 | 0.08 ± 0.02 | 0.08 ± 0.02 |
| C20:1n-9 | 0.64 ± 0.17 | 0.22 ± 0.09 | 0.10 ± 0.02a | 0.12 ± 0.04a |
| C20:2n-6 | 0.07 ± 0.01 | 0.05 ± 0.00 | 0.05 ± 0.01 | 0.04 ± 0.00a |
| C20:3n-9 | 0.93 ± 0.06 | 0.18 ± 0.10b | 0.07 ± 0.01b | 0.08 ± 0.02b |
| C20:3n-6 | 0.44 ± 0.09 | 0.19 ± 0.02a | 0.17 ± 0.03a | 0.15 ± 0.01a |
| C20:4n-6 | 2.37 ± 0.33 | 1.79 ± 0.05 | 1.64 ± 0.15 | 1.54 ± 0.08a |
| C20:5n-3 | 0.12 ± 0.02 | 3.15 ± 0.31b | 3.06 ± 0.17b | 4.42 ± 0.35b |
| C22:0 | 0.09 ± 0.02 | 0.14 ± 0.03 | 0.16 ± 0.03 | 0.17 ± 0.04 |
| C22:1n-9 | 0.04 ± 0.01 | 0.00 ± 0.00 | 0.03 ± 0.01 | 0.03 ± 0.01 |
| C22:4n-6 | 0.11 ± 0.02 | 0.07 ± 0.01 | 0.06 ± 0.01a | 0.06 ± 0.01a |
| C22:5n-3 | 0.05 ± 0.01 | 1.21 ± 0.33a | 1.50 ± 0.23b | 2.01 ± 0.44b |
| C22:6n-3 | 2.88 ± 0.23 | 3.60 ± 0.32 | 3.33 ± 0.34 | 3.65 ± 0.09a |
| C24:0 | 0.07 ± 0.01 | 0.10 ± 0.02 | 0.12 ± 0.01a | 0.13 ± 0.02a |
| C24:1n-9 | 0.18 ± 0.01 | 0.18 ± 0.01 | 0.12 ± 0.02a | 0.15 ± 0.01 |
a p < 0.05.
b p < 0.01.
PUFA Suppresses Srebf1c Promoter Activity through SRE Site
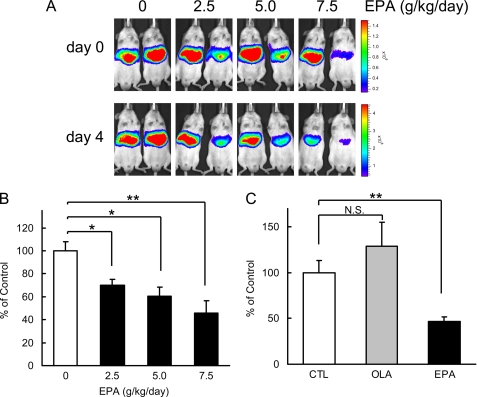
Next, we attempted to estimate the Srebf1c promoter activity in the liver with an in vivo luciferase reporter analysis. The luciferase reporter gene driven by the 2200-bp Srebf1c promoter was adenovirally transduced into mouse liver, and the transcriptional activity was assessed by measuring luciferase activity with the IVIS imaging system. The physiological activity of this promoter had previously been confirmed by transgenic mice (31). As shown in Fig. 2, EPA decreased the Srebf1c promoter activity in a dose-dependent manner.
FIGURE 2.
PUFA suppresses Srebf1c promoter activity. 2.2-kbp Srebf1c-Luc adenovirus (Ad-2.2k- Srebf1c-1c-Luc) (6.0 × 106 pfu/body) was intravenously injected into ICR male mice. After 4 days, mice (n = 4 for each group) were treated orally with EPA, OLA, or water (CTL) at indicated doses once a day for 4 days. A, on day 0 (before) and day 4 from the first treatment of EPA, luciferin was injected intraperitoneally in nonfasted mice, and the luminescence from liver was captured with IVIS. The color overlay on the image represents the photons/s emitted from the animal with a range of 1.5 × 105–1.5 × 106 photons/s (day 0), 5.0 × 105–5.0 × 106 photons/s (day 4), as indicated by the color scale next to the images. B and C, quantification of luciferase activity with LivingImage software. Fold changes of luciferase activity on day 4 versus day 0 are shown. These data are representative of at least two independent experiments (n = 4 mice/group). Results are means ± S.E. *, p < 0.05; **, p < 0.01, respectively. N.S., not significant.
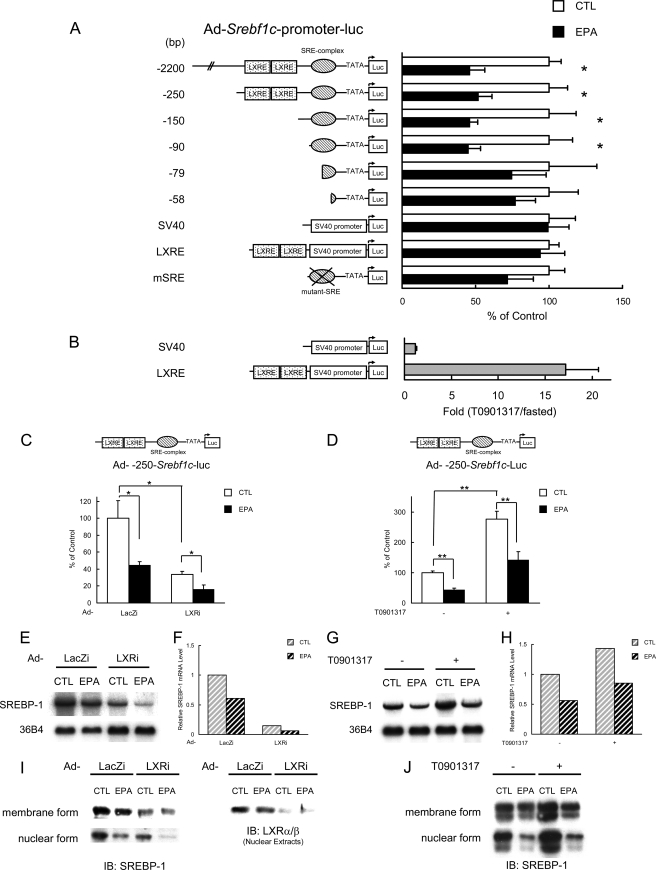
To determine the responsible region for PUFA suppression, a promoter deletion study was performed; six adenovirus constructs containing different lengths of the Srebf1c promoter (ranging from 2200 to 58 bp) were transduced into mouse livers, and the suppressive effect of EPA on promoter activity was assessed with IVIS (Fig. 3A). From this experiment, the responsible element was located at the SRE site within −90 to −60 bp upstream of the transcription start site. This result was confirmed by another experiment using a mutant SRE construct.
FIGURE 3.
PUFA suppresses Srebf1c promoter activity through SRE site. A, adenoviruses encoding various lengths of Srebf1c promoter, as well as two LXRE sites on Srebf1c promoter (−239 to −165) with SV40 promoter and SRE mutant version of 90-bp Srebf1c promoter, attached with luciferase (Luc) (6.0 × 106 pfu/body) were injected intravenously into ICR male mice. After 4 days, mice (n = 4–7 for each group) were treated orally with 7.5 g/kg EPA or water (CTL) once a day for 4 days. On day 0 and 4 after the first treatment of EPA, luciferin was injected intraperitoneally, and the luminescence from liver was captured with IVIS. B, Ad-LXRE-Luc injected mice were administered orally with 50 mg/kg T0901317 or vehicle (0.9% carboxymethylcellulose, 9.95% polyethylene glycol 400, and 0.05% Tween 80) after fasting overnight. At 0 and 16 h following T0901317 treatment in fasted states, luciferin was injected intraperitoneally, and the luminescence from liver was captured with IVIS. Fold change of luciferase activity at 16 versus 0 h was presented. C, E, F, and I, knockdown of hepatic LXRα/β by adenoviral expression of shRNA. 250-bp Srebf1c-Luc adenovirus (Ad-250-bp- Srebf1c-Luc; 6.0 × 106 pfu/body) plus adenovirus expressing LXRα/β-specific or LacZ-specific shRNA (Ad-LXRi or Ad-LacZi, respectively; 2.5 × 108 pfu/body) were intravenously injected into ICR male mice. After 4 days, the mice (n = 4 for each group) were treated orally with 7.5 g/kg EPA or water (CTL) once a day for 4 days. On day 0 and 4 after the first treatment of EPA, luciferin was injected intraperitoneally, and the luminescence from liver was captured with IVIS. D, G, H, and J, inhibitory effect of PUFA on Srebf1c gene transcription is not affected by LXR agonist. 250-bp Srebf1c-Luc adenovirus (Ad-250bp- Srebf1c-Luc) (6.0 × 106 pfu/body) was intravenously injected into ICR male mice. After 6 days, the mice (n = 4 for each group) were treated orally with 7.5 g/kg EPA or water (CTL) once a day for 4 days. At day 3 of EPA treatment, the mice were administered orally with 10 mg/kg T0901317 or vehicle. On days 0 and 4 after the first treatment of EPA, luciferin was injected intraperitoneally, and the luminescence from liver was captured with IVIS. C and D, quantification of luciferase activity with LivingImage software. Fold changes of luciferase activity on day 4 versus day 0 are shown. E and G, Northern blot analysis of SREBP-1 from livers. Total RNA (7.5 μg) from livers pooled equally from mice of each group was subjected to Northern blotting to determine SREBP-1 and 36B4 (used as a loading control) mRNA levels. F and H, quantification of the data shown in E and G. The fold change is the relative ratio of each signal versus the control mice. I and J, immunoblot (IB) analysis of mature and precursor SREBP-1 proteins and LXRα/β in livers. Aliquots of nuclear extracts (10 μg) and total proteins (50 μg) from livers pooled equally from four male mice of each group were subjected to immunoblot analysis. The primary antibodies used were polyclonal anti-mouse SREBP-1 and polyclonal anti-mouse LXRα/β. These data are representative of at least two independent experiments (n = 4–7 mice/group). Results are means ± S.E. *, p < 0.05, and **, p < 0.01 versus controls.
LXR Is Not Involved in the PUFA Suppression of Srebf1c Gene Expression in Vivo
Furthermore, it was clarified from the series of deletion studies that EPA did not suppress the promoter activity of a construct containing only LXRE (Fig. 3A), demonstrating that EPA does not antagonize LXR binding to LXRE on the Srebf1c promoter at least in the in vivo setting. Moreover, as shown in Fig. 3, C, E, F, and I, it was demonstrated that the simultaneous knockdown of both LXRα and -β did not affect the suppressive effect of PUFA on Srebf1c promoter activity. Furthermore, the stimulation of LXR by an LXR agonist T0901317 was also shown to be independent of the inhibitory effect of PUFA, as shown in Fig. 3, D, G, H, and J. Based on these findings, we concluded that LXR is not the direct target of PUFA regulation in the in vivo setting, although LXR is a determinant of the expression level of SREBP-1 mRNA.
Inhibitory Effect of PUFA on Srebf1c Gene Transcription Is Mediated through SREBP-1 Itself
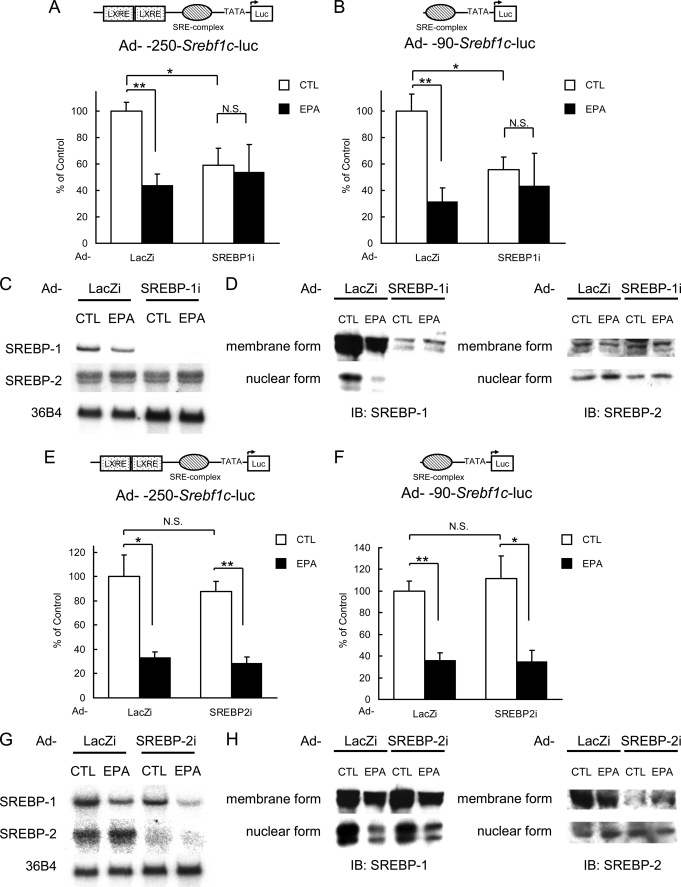
To determine whether the trans-acting factor for SRE is SREBP-1 or -2 or both, SREBP-1/2 was knocked down by RNA interference. As shown in Fig. 4, A and B, the effects of PUFA administration and knocking down SREBP-1 were essentially equal, and when SREBP-1 was knocked down, the promoter activity of Srebf1c gene was reduced by half, and PUFA showed no additive effects. In contrast, knocking down SREBP-2 did not affect the promoter activity of Srebf1c as assessed by luciferase reporter expression (Fig. 4, E and F), demonstrating that SREBP-2 is not involved in the transcriptional regulation of Srebf1c gene expression. From these findings, it was concluded that the trans-acting factor for SRE on the Srebf1c promoter is SREBP-1 and not SREBP-2. This indicates that SREBP-1 constitutes an autoloop regulatory circuit.
FIGURE 4.
Inhibitory effect of PUFA on Srebf1c gene transcription is mediated through SREBP-1 itself. A–H, knockdown of hepatic SREBP-1 (A–D) or SREBP-2 (E–H) by adenoviral expression of shRNA. 250- or 90-bp Srebf1c-Luc adenovirus (Ad-250bp- Srebf1c-Luc or Ad-90bp- Srebf1c-Luc; 6.0 × 106 pfu/body) plus adenovirus expressing SREBP-1-specific, SREBP-2-specific, or LacZ-specific shRNA (Ad-SREBP1i, Ad-SREBP2i, or Ad-LacZi; 2.5 × 108 pfu/body) were intravenously injected into ICR male mice. After 4 days, the mice (n = 3–6 for each group) were treated orally with 7.5 g/kg EPA or water (CTL) once a day for 4 days. On days 0 and 4 after the first treatment of EPA, luciferin was injected intraperitoneally, and the luminescence from liver was captured with IVIS. A, B, E, and F, quantification of luciferase activity with LivingImage software. Fold changes of luciferase activity on day 4 versus day 0 are shown. C and G, Northern blot analysis of SREBP-1 and -2 in the liver. Total RNA (7.5 μg) from livers pooled equally from mice for each group was subjected to Northern blotting to determine SREBP-1, -2, and 36B4 (used as a loading control) mRNA levels. D and H, immunoblot (IB) analysis of mature and precursor SREBP-1 and -2 proteins in the liver. Aliquots of nuclear extracts (10 μg) and total proteins (50 μg) from livers pooled equally from four male mice of each group were subjected to immunoblot analysis. The primary antibodies used were polyclonal anti-mouse SREBP-1 and polyclonal anti-mouse SREBP-2. These data are representative of at least two independent experiments (n = 3–6 mice/group). Results are means ± S.E. *, p < 0.05, and **, p < 0.01, respectively.
PUFA Suppresses the Proteolytic Activation of SREBP-1, Not Affecting SREBP-2
SREBP-1 is synthesized as a precursor bound to the endoplasmic reticulum and nuclear envelope and is released from the membrane into the nucleus as a mature protein by a cleavage process (7). To explore the molecular mechanism by which PUFA decreases the nuclear form of SREBP-1, another reporter system to detect proteolytic activity for the precursor form of SREBPs was constructed (Fig. 5A). In this system, Gal4-DNA binding domain and VP16 activation domain were attached to the N terminus of SREBPs, and the nuclear entry of this N-terminal fragment was measured by the Gal4-UAS system to assess the proteolytic release of the SREBP N termini. Using this cleavage-detecting reporter system, the proteolytic activities for precursor forms of SREBP-1 or -2 were tested in vitro with SCAP and insulin-induced gene (INSIG)-1 overexpressions as an activator and an inhibitor for cleavage of SREBP, respectively. As expected, the SCAP overexpression drastically enhanced the cleavage of N-terminal fragments of both SREBP-1 and -2, and the co-expression of INSIG1 reversed the effect of SCAP overexpression (Fig. 5B), indicating that this reporter system reflects the physiological regulation of SREBP activation processes by proteolysis. Next, we transduced the reporter genes into mouse livers using adenoviruses and examined the effect of PUFA in the in vivo setting. As shown in Fig. 5, C and D, EPA suppressed only the SREBP-1 cleavage-detecting reporter but did not affect the SREBP-2 reporter. Additionally, EPA did not decrease the reporter activity from the construct that contains only the N terminus (designated as Nuc-SREBP-1c) and enters the nucleus without cleavage, demonstrating that EPA did not accelerate the degradation of reporter fragment. In contrast, GW532, a SCAP activator, enhanced the cleavage of both SREBP-1 and -2 in the in vitro (Fig. 5F) and in vivo (Fig. 5H) situations.
FIGURE 5.
PUFA suppresses the proteolytic activation of SREBP-1, not affecting SREBP-2. A, various regions of human SREBP-1c (amino acids (a.a.) 1–1123 (FL), 431–1123 (Reg), 1–436 (Nuc)) and human SREBP-2 (amino acids 14–1141 (FL), 450–1141 (Reg)) were fused to Gal4-DNA binding domain and VP16-transactivation domain. FL, full length; Nuc, nuclear; Reg, regulatory; bHLH-Zip, basic helix-loop-helix leucine zipper. GAL4-VP16-SREBP fusion protein is attached to the endoplasmic reticulum (ER) similarly as endogenous SREBP protein. Upon activation, its N-terminal region is released from the membrane into the nucleus by a cleavage process. The GAL4-VP16 promotes the luciferase (Luc) reporter gene expression by binding to GAL4-RE containing eight copies of upstream activation sequence, Gal4-binding site. B, regulation of cleavage of GAL4-VP16 fusion SREBP protein by SCAP and INSIG-1. HEK293 cells in a 48-well plate were co-transfected with GAL4-RE-Luc plasmid (100 ng/well), expression plasmids of GAL4-VP16 fusion SREBP protein (100 ng/well), SCAP (250 ng/well), INSIG-1 (100 ng/well), and pSV40-Renilla plasmid (50 ng/well). Cells were harvested 24 h after transfection. The firefly luciferase activity was measured and normalized by Renilla luciferase activity. All experiments were performed in triplicate. C–E, regulation of cleavage of GAL4-VP16 fusion SREBP protein by EPA in vivo. GAL4-RE-Luc adenovirus (Ad-GAL4-RE-Luc, 2.0 × 108 pfu/body) plus adenovirus expressing GAL4-VP16 fusion SREBP protein (Ad-Gal4-FL-SREBP-1c, Ad-Gal4-FL-SREBP-2 or Ad-Gal4-Nuc-SREBP-1c, 1.0 × 108 pfu/body) were intravenously injected into ICR male mice. After 2 days, the mice (n = 7–9 for each group) were treated orally with 7.5 g/kg EPA or water (CTL) once a day for 4 days. C, 0 and 4 days after the fast treatment of EPA, luciferin was injected intraperitoneally, and the luminescence from liver was captured with IVIS. The color overlay on the image represents the photons/s emitted from the animal with a range of 4.0 × 105–2.5 × 106 photons/s (Ad-Gal4-FL-SREBP-1c) and 1.0 × 106–8.0 × 106 photons/s (Ad-Gal4-FL-SREBP-2), as indicated by the color scale next to the images. D, quantification of luciferase activity with LivingImage software. Fold changes of luciferase activity on day 4 versus day 0 are shown. E, immunoblot analysis of mature and precursor SREBP-1 proteins from livers. Aliquots of nuclear extracts (10 μg) and total proteins (50 μg) from livers pooled equally from male mice for each group were subjected to immunoblot analysis. The primary antibodies used were polyclonal anti-mouse SREBP-1. N.S., not significant. F–H, SCAP ligand GW532 accelerates SREBP-1 cleavage. F, regulation of cleavage of GAL4-VP16 fusion SREBP protein by GW532. HEK293 cells in a 48-well plate were co-transfected with GAL4-RE-Luc plasmid (100 ng/well), expression plasmids for GAL4-VP16 fusion SREBP protein (100 ng/well), and pSV40-Renilla plasmid (50 ng/well). 3 h after transfection, GW532 (1 μm) or EtOH was added to media, and cells were harvested 24 h after transfection. The firefly luciferase activity was measured and normalized by Renilla luciferase activity. All experiments were performed in triplicate. G and H, GAL4-RE-Luc adenovirus (Ad-GAL4-RE-Luc, 2.0 × 108 pfu/body) plus adenovirus expressing GAL4-VP16 fusion SREBPs protein (Ad-Gal4-FL-SREBP-1c or Ad-Gal4-FL-SREBP-2, 1.0 × 108 pfu/body) were intravenously injected into ICR male mice. After 2 days, the mice (n = 8 for each group) were administered orally with GW532 or vehicle at indicated doses. 6 h after the last treatment, luciferin was injected intraperitoneally, and the luminescence from liver was captured with IVIS. G, quantification of luciferase activity with LivingImage software. Fold changes on day 3 versus day 0 are shown. H, immunoblot analysis of mature and precursor SREBP-1 and SREBP-2 proteins from livers. Aliquots of nuclear extracts (10 μg) and total proteins (50 μg) from livers pooled equally from four male mice for each group were subjected to immunoblot (IB) analysis. The primary antibodies used were polyclonal anti-mouse SREBP-1 and anti-mouse SREBP-2. These data are representative of at least two independent experiments (n = 3–8 mice/group). Results are means ± S.E. *, p < 0.05.
DISCUSSION
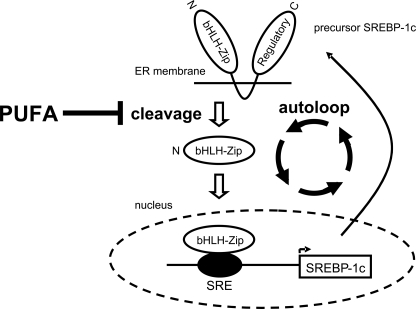
This study has clearly demonstrated that the primary mechanism of the inhibitory effect of PUFA is the suppression of the proteolytic activation of SREBP-1 and that the transcriptional regulation is secondary to this post-translational suppression of mature SREBP-1 that itself binds to the SRE site on the Srebf1c promoter (autoloop regulation (19)), whereas LXR is not involved in the transcriptional regulation by PUFA (Fig. 6).
FIGURE 6.
Schematic representation of molecular mechanisms for inhibitory effects of PUFA on SREBP-1. The primary mechanism for PUFA suppression of SREBP-1 is at the proteolytic processing level, and this suppression in turn decreases the mRNA transcription through lowering SREBP-1 binding to SRE on the promoter (autoloop regulatory circuit), although LXR is not involved in this regulation.
This is the first report that has clearly demonstrated the inhibitory effect of PUFA on the SREBP-1 proteolytic processing in vivo. This result is consistent with our previous report showing that PUFA decreases nuclear SREBP-1, although it does not affect the nuclear abundance of the truncated form of SREBP-1 expressed from a transgene, suggesting that PUFA does not accelerate the degradation of nuclear SREBP-1 protein (11).
We have shown that two mechanisms are involved in the PUFA regulation on SREBP-1; one is at the proteolytic processing level, and the other is at the transcription level. What is the physiological role of this two-step regulation? As we have shown in Fig. 1, the proteolytic mechanism is high sensitive, although the transcriptional mechanism is low sensitive, consistent with the previous report by Ezaki and co-workers (35). The combination of these two steps of regulation with different sensitivity connected in series probably helps to achieve the broader responsive range of the amount of PUFA.
One of our conclusions is that LXR is not involved in the transcriptional regulation by PUFA, but controversy exists over this point; several previous reports suggested the involvement of LXR in this regulation (15, 16), whereas others did not (36). These previous studies have been all performed in in vitro settings, and therefore we evaluated the contribution of the LXR pathway in the in vivo setting for the first time, and we have concluded that the involvement of LXR is not detectable, although LXR is an important determinant of the SREBP-1 expression level.
Because an in vitro model is always only a small part of the whole in vivo system, how large or small the contribution of a regulatory pathway elucidated in vitro is in the whole in vivo system cannot be estimated until it is assessed in the in vivo setting. To address these issues, our approach of the extension of in vitro reporter assays to in vivo settings will be very useful in various situations.
We have demonstrated that PUFA selectively suppresses the proteolytic processing of SREBP-1, but the molecular mechanism underlying this SREBP-1-specific regulation is currently unknown. SCAP escorts both SREBP-1 and -2 to the Golgi, and SREBP-1-specific adaptor protein has not been reported yet. Recently, an endoplasmic reticulum membrane protein TRC8 (translocation in renal cancer from chromosome 8) has been documented to hamper endoplasmic reticulum-to-Golgi transport of SREBP-2/SCAP and reduce SREBP-2 cleavage specifically (37). Perhaps there might be some adaptor molecule that specifically interacts with SREBP-1 and mediates the suppressive effect of PUFA, although we have no evidence. If the molecular mechanism underlying this SREBP-1-specific effect of PUFA is clarified in the future, it will be a potential molecular target for new lipid-lowering drugs. In conclusion, the primary mechanism for PUFA suppression of SREBP-1 expression is at the proteolytic processing level and that this suppression in turn decreases the transcription of Srebf1c through lowering SREBP-1 binding to SRE on the promoter.
This work was supported by grants-in-aid from the Ministry of Science, Education, Culture and Technology of Japan (to N. Y. and H. S.) and research grants from The Uehara Memorial Foundation, ONO Medical Research Foundation, Takeda Science Foundation, Suzuken Memorial Foundation, Japan Heart Foundation, Kanae Foundation for the Promotion of Medical Science, Senri Life Science Foundation, Japan Foundation for Applied Enzymology, and Okinaka Memorial Institute for Medical Research (to N. Y.).
- PUFA
- polyunsaturated fatty acid
- EPA
- eicosapentaenoic acid
- LXR
- liver X receptor
- LXRE
- liver X receptor-binding element
- SREBP
- sterol regulatory element-binding protein
- SRE
- SREBP-binding element
- pfu
- plaque-forming unit
- OLA
- oleic acid ethyl ester
- shRNA
- short hairpin RNA
- SCAP
- SREBP cleavage-activating protein.
REFERENCES
- 1.Clarke S. D., Jump D. B. (1994) Annu. Rev. Nutr. 14, 83–98 [DOI] [PubMed] [Google Scholar]
- 2.Allmann D. W., Gibson D. M. (1965) J. Lipid Res. 6, 51–62 [PubMed] [Google Scholar]
- 3.Goodridge A. G. (1991) in Biochemistry of Lipids, Lipoproteins, and Membranes (Vance D. E., Vance J. eds) pp. 111–139, Elsevier Science Publishers, Amsterdam [Google Scholar]
- 4.Shimomura I., Shimano H., Korn B. S., Bashmakov Y., Horton J. D. (1998) J. Biol. Chem. 273, 35299–35306 [DOI] [PubMed] [Google Scholar]
- 5.Shimano H., Yahagi N., Amemiya-Kudo M., Hasty A. H., Osuga J., Tamura Y., Shionoiri F., Iizuka Y., Ohashi K., Harada K., Gotoda T., Ishibashi S., Yamada N. (1999) J. Biol. Chem. 274, 35832–35839 [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama C., Wang X., Briggs M. R., Admon A., Wu J., Hua X., Goldstein J. L., Brown M. S. (1993) Cell 75, 187–197 [PubMed] [Google Scholar]
- 7.Brown M. S., Goldstein J. L. (1997) Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 8.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. (1997) J. Clin. Invest. 99, 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton J. D., Shimomura I., Brown M. S., Hammer R. E., Goldstein J. L., Shimano H. (1998) J. Clin. Invest. 101, 2331–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimano H., Horton J. D., Shimomura I., Hammer R. E., Brown M. S., Goldstein J. L. (1997) J. Clin. Invest. 99, 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yahagi N., Shimano H., Hasty A. H., Amemiya-Kudo M., Okazaki H., Tamura Y., Iizuka Y., Shionoiri F., Ohashi K., Osuga J., Harada K., Gotoda T., Nagai R., Ishibashi S., Yamada N. (1999) J. Biol. Chem. 274, 35840–35844 [DOI] [PubMed] [Google Scholar]
- 12.Xu J., Nakamura M. T., Cho H. P., Clarke S. D. (1999) J. Biol. Chem. 274, 23577–23583 [DOI] [PubMed] [Google Scholar]
- 13.Kim H. J., Takahashi M., Ezaki O. (1999) J. Biol. Chem. 274, 25892–25898 [DOI] [PubMed] [Google Scholar]
- 14.Mater M. K., Thelen A. P., Pan D. A., Jump D. B. (1999) J. Biol. Chem. 274, 32725–32732 [DOI] [PubMed] [Google Scholar]
- 15.Ou J., Tu H., Shan B., Luk A., DeBose-Boyd R. A., Bashmakov Y., Goldstein J. L., Brown M. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshikawa T., Shimano H., Yahagi N., Ide T., Amemiya-Kudo M., Matsuzaka T., Nakakuki M., Tomita S., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Takahashi A., Sone H., Osuga Ji J., Gotoda T., Ishibashi S., Yamada N. (2002) J. Biol. Chem. 277, 1705–1711 [DOI] [PubMed] [Google Scholar]
- 17.Xu J., Teran-Garcia M., Park J. H., Nakamura M. T., Clarke S. D. (2001) J. Biol. Chem. 276, 9800–9807 [DOI] [PubMed] [Google Scholar]
- 18.Botolin D., Wang Y., Christian B., Jump D. B. (2006) J. Lipid Res. 47, 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amemiya-Kudo M., Shimano H., Yoshikawa T., Yahagi N., Hasty A. H., Okazaki H., Tamura Y., Shionoiri F., Iizuka Y., Ohashi K., Osuga J., Harada K., Gotoda T., Sato R., Kimura S., Ishibashi S., Yamada N. (2000) J. Biol. Chem. 275, 31078–31085 [DOI] [PubMed] [Google Scholar]
- 20.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Osuga J., Harada K., Gotoda T., Kimura S., Ishibashi S., Yamada N. (2001) Mol. Cell. Biol. 21, 2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worgall T. S., Sturley S. L., Seo T., Osborne T. F., Deckelbaum R. J. (1998) J. Biol. Chem. 273, 25537–25540 [DOI] [PubMed] [Google Scholar]
- 22.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. (2001) J. Biol. Chem. 276, 4365–4372 [DOI] [PubMed] [Google Scholar]
- 23.Sheng Z., Otani H., Brown M. S., Goldstein J. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekiya M., Yahagi N., Matsuzaka T., Najima Y., Nakakuki M., Nagai R., Ishibashi S., Osuga J., Yamada N., Shimano H. (2003) Hepatology 38, 1529–1539 [DOI] [PubMed] [Google Scholar]
- 25.Sattler W., Puhl H., Hayn M., Kostner G. M., Esterbauer H. (1991) Anal. Biochem. 198, 184–190 [DOI] [PubMed] [Google Scholar]
- 26.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 27.Contag P. R., Olomu I. N., Stevenson D. K., Contag C. H. (1998) Nat. Med. 4, 245–247 [DOI] [PubMed] [Google Scholar]
- 28.Hua X., Sakai J., Brown M. S., Goldstein J. L. (1996) J. Biol. Chem. 271, 10379–10384 [DOI] [PubMed] [Google Scholar]
- 29.Inoue N., Shimano H., Nakakuki M., Matsuzaka T., Nakagawa Y., Yamamoto T., Sato R., Takahashi A., Sone H., Yahagi N., Suzuki H., Toyoshima H., Yamada N. (2005) Mol. Cell. Biol. 25, 8938–8947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto T., Shimano H., Nakagawa Y., Ide T., Yahagi N., Matsuzaka T., Nakakuki M., Takahashi A., Suzuki H., Sone H., Toyoshima H., Sato R., Yamada N. (2004) J. Biol. Chem. 279, 12027–12035 [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi Y., Yahagi N., Nakagawa Y., Matsuzaka T., Shimizu R., Sekiya M., Iizuka Y., Ohashi K., Gotoda T., Yamamoto M., Nagai R., Kadowaki T., Yamada N., Osuga J., Shimano H. (2007) Biochem. Biophys. Res. Commun. 363, 329–335 [DOI] [PubMed] [Google Scholar]
- 32.Kumadaki S., Matsuzaka T., Kato T., Yahagi N., Yamamoto T., Okada S., Kobayashi K., Takahashi A., Yatoh S., Suzuki H., Yamada N., Shimano H. (2008) Biochem. Biophys. Res. Commun. 368, 261–266 [DOI] [PubMed] [Google Scholar]
- 33.Seo J. B., Moon H. M., Kim W. S., Lee Y. S., Jeong H. W., Yoo E. J., Ham J., Kang H., Park M. G., Steffensen K. R., Stulnig T. M., Gustafsson J. A., Park S. D., Kim J. B. (2004) Mol. Cell. Biol. 24, 3430–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ide T., Shimano H., Yahagi N., Matsuzaka T., Nakakuki M., Yamamoto T., Nakagawa Y., Takahashi A., Suzuki H., Sone H., Toyoshima H., Fukamizu A., Yamada N. (2004) Nat. Cell Biol. 6, 351–357 [DOI] [PubMed] [Google Scholar]
- 35.Nakatani T., Kim H. J., Kaburagi Y., Yasuda K., Ezaki O. (2003) J. Lipid Res. 44, 369–379 [DOI] [PubMed] [Google Scholar]
- 36.Deng X., Cagen L. M., Wilcox H. G., Park E. A., Raghow R., Elam M. B. (2002) Biochem. Biophys. Res. Commun. 290, 256–262 [DOI] [PubMed] [Google Scholar]
- 37.Irisawa M., Inoue J., Ozawa N., Mori K., Sato R. (2009) J. Biol. Chem. 284, 28995–29004 [DOI] [PMC free article] [PubMed] [Google Scholar]