Abstract
The superfamily 2 vaccinia viral helicase nucleoside triphosphate phosphohydrolase-II (NPH-II) exhibits robust RNA helicase activity but typically displays little activity on DNA substrates. NPH-II is thus believed to make primary contacts with backbone residues of an RNA substrate. We report an unusual nucleobase bias, previously unreported in any superfamily 1 or 2 helicase, whereby purines are heavily preferred as components of both RNA and DNA tracking strands. The observed sequence bias allows NPH-II to efficiently unwind a DNA·RNA hybrid containing a purine-rich DNA track derived from the 3′-untranslated region of an early vaccinia gene. These results provide insight into potential biological functions of NPH-II and the role of sequence in targeting NPH-II to appropriate substrates. Furthermore, they demonstrate that in addition to backbone contacts, nucleotide bases play an important role in modulating the behavior of NPH-II. They also establish that processive helicase enzymes can display sequence selectivity.
Keywords: DNA Helicase, Molecular Motors, Protein-Nucleic Acid Interaction, RNA Helicase, Transcription Termination, DExH Helicase, NPH-II, Helicase Activity, Sequence Dependence
Introduction
Helicase superfamily 2 (SF2)4 is a ubiquitous class of enzymes involved in nearly every aspect of nucleic acid metabolism (1, 2). One feature that appears to distinguish SF2 and SF1 helicases is the mechanism of substrate binding and strand separation. SF1 helicases such as Rep, PcrA, and UvrD make extensive base contacts that have been proposed to aid in single strand translocation (3, 4). In contrast, crystal structures of several SF2 helicases (i.e. Hel308, HCV NS3, and RecG) reveal that the core enzyme makes extensive contacts with the sugar-phosphate backbone (5–7). Although specific base contacts are evident and appear to have some functional contribution, as in the Hel308 crystal structure, biochemical data suggest that backbone contacts are the dominant functional mode of interaction for SF2 helicases (8). Although SF1 helicases appear to interact more closely with substrate bases, neither SF1 nor SF2 helicases display sequence dependence. Indeed, the lack of sequence specificity would appear to be a general feature for most RNA and DNA helicases, because strict sequence specificity might hinder translocation of the enzyme along its substrate (9–11). Target specificity is thus apparently conferred with the aid of accessory proteins, cofactors, or other such means (12). RecBCD is one notable exception, where recognition of a specific sequence element (Chi) causes drastic changes in helicase behavior (13).
Biochemical studies on the viral helicase nucleoside triphosphate phosphohydrolase II (NPH-II) have been important for establishing the molecular mechanism and substrate recognition determinants for SF2 helicases. NPH-II is an essential protein from the vaccina pox virus and a prototypical member of the functionally distinct DEAD/DExH helicases (so named for the characteristic ATPase motif) that are involved in many known aspects of RNA metabolism (10). Although the specific function of NPH-II remains unknown, NPH-II is thought to play an important accessory role in transcriptional termination of early viral genes in vivo, perhaps by preventing the formation of secondary structures that might mask the early termination signal (14, 15). In vitro, NPH-II tracks along the backbone of single-stranded RNA in a processive, unidirectional 3′ → 5′ manner with a kinetic step size of six base pairs (16, 17). It efficiently unwinds RNA and RNA·DNA duplexes, and it can strip proteins from single-stranded RNA molecules (16, 18). Although NPH-II can bind DNA and RNA with equal affinity, NPH-II appears to have a clear preference for RNA as an unwinding substrate (19–21). NPH-II is tolerant of nicks and polyglycol linkers in the displaced strand but not in the tracking strand. Abasic residues can be traversed on either strand, albeit with greatly reduced efficiency on the tracking strand (17). These data support the conclusion that NPH-II is an RNA-specific translocase and helicase that primarily engages backbone contacts (such as phosphoryl oxygens and the ribose 2′-hydroxyl) as it moves along its single-stranded RNA track.
Here we demonstrate that although RNA is a preferable substrate in many respects, NPH-II can employ a DNA tracking strand when presented with an appropriate substrate. This surprising activity appears to be modulated by a sensitivity of the helicase to the composition of the substrate nucleobases in both RNA and DNA contexts. Specifically, NPH-II strongly prefers substrate tracking strands that are rich in purine nucleotides, whether RNA or DNA. These data suggest that NPH-II engages with the nucleobases in some important manner and that, contrary to an exclusive backbone tracking mechanism, the nucleobase component of recognition may help regulate and/or target helicase activity. These results establish the first known example of a SF2 helicase that is strongly influenced by substrate sequence. We further demonstrate how this sequence bias may be used in vivo to selectively target the activity of this helicase to specific locations within the viral genome.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A new protocol was optimized that allowed for adequate expression and purification from bacterial cell culture. Unless otherwise indicated, protein for this study was prepared in the following manner. In brief, His6-tagged protein was expressed in Rosetta2(DE3)pLysS cells (Novagen) at 16 °C for 12 h. The protein was first purified using nickel-nitrilotriacetic acid beads. The protein was then either subjected directly to gel filtration or was passed over a heparin column, followed by gel filtration, concentration, and a second heparin column. The aliquots were stored at −80 °C in 25 mm Tris, pH 7.4, 0.5 m NaCl, 2 mm dithiothreitol, 10% glycerol, and 0.2% CHAPS. Protein preps were normalized by measuring the ATPase activity of each prep. 1 unit is defined as the amount of protein in ng to hydrolyze 1 pmol of ATP in 1 min.
Synthesis of Nucleic Acid Substrates
RNA substrates used in the nucleotide analog interference mapping (NAIM) experiments were designed to have as little internal secondary structure as possible, as determined by M-Fold. The top strand consisted of sequence 5′-GGA GUG CAU GUC CUA GCG UCG UAU CGA UCU GGU CGU CUC C-3′. The bottom strand consisted of sequence 5′-GGA GAC GAC CAG AUC GAU ACG ACG CUA GGA CAU GCA CUC CAC UGA CUA ACA CGU ACU AAC AGG AUC AAC U-3′. These substrates were synthesized via in vitro transcription from plasmid or DNA oligonucleotide templates using T7 RNA polymerase or Y639F RNA polymerase. To create a pool of modified tracking strands, phosphorothioate ribonucleotides (NαS) or phosphorothioate deoxyribonucleotides (dNαS) (Trilink, San Diego, CA) were statistically incorporated (10%) into a transcription reaction for the loading strand RNA. Each transcription reaction was supplemented with only a single analog (ATPαS, CTPαS, dATPαS, and dCTPαS, etc.) for a total of eight different substrate pools. After gel purification, top strand transcripts were treated with Antarctic Phosphatase (New England Biolabs), 5′ end-labeled, and annealed to the tracking strand. The sequence and incorporation levels were checked by iodine cleavage and sequencing PAGE to ensure equal incorporation rates across all pools.
RNA (54 nt and shorter) and chimeric RNA-DNA oligonucleotides were purchased from Dharmacon, Inc. (Lafayette, CO). DNA oligonucleotides were purchased from Invitrogen. The substrates were prepared as described previously (16). The substrate duplexes contained a 5′-32P end label on the top strand. The substrates were stored at −20 °C in a buffer of 10 mm MOPS, pH 6.0, 1 mm EDTA, 30 mm NaCl. The sequence for the viral growth factor pseudo R-loop was taken from the C11R gene of the vaccinia virus genome (GenBankTM accession number AY243312, locus tag VACWR009). The pseudo R-loop substrate synthesized as above and purified on a native salt polyacrylamide gel (12% acrylamide, 0.5× TBE, 70 mm NaCl) and stored in a buffer of 10 mm MOPS, pH 6.0, 1 mm EDTA, and 154 mm NaCl. Trap RNA (24-bp duplex flanked by a 3′ single stranded tail of 18 nucleotides; top strand, 5′-GCC UCG CUG CCG UCG CCA GCA UAU-3′; bottom strand, 5′-AUA UGC UGG CGA CGG CAG CGA GGC AGA GGA GCA GAG GGA GCA-3′) was prepared as described previously(19).
Nucleotide Analog Interference Mapping
To identify optimal conditions for the NAIM experiment, the extent of unwinding was varied from 5 to 50%, and [NaCl] was varied from 10 to 120 mm. An optimal interference signal was detected with >50 mm NaCl and < 20% unwinding. A salt concentration of 70 mm was therefore chosen because it produced strong interferences but preserved relatively high levels of unwinding activity. The final reaction conditions were 40 mm Tris, pH 8.0, 4 mm Mg(OAc)2, 70 mm NaCl, 50 units/μl NPH-II at 25 °C, unless otherwise stated. NPH-II protein was preincubated with duplex substrate (0.5–2 nm) for 10 min at 25 °C and initiated by the simultaneous addition of 3.5 mm ATP and 400 nm Trap RNA (19). The reactions were quenched after 1 min, such that unwinding extent reached 15%. The duplex and unwound fractions were separated by native PAGE. Duplex, unwound, and untreated substrates were isolated and diluted to ∼30,000 cpm in 10 μl. The samples were then treated with iodine (final concentration, 1 mm) and incubated at room temperature for 5 min. The reactions were quenched by adding 30 μl of a 10 mm MOPS, pH 6.0, 1 mm EDTA solution, 10 μl of 3 m NaOAc, 1 μl of 10 mg/ml glycogen, and 1 pmol of cold top strand; precipitated; and resuspended in 4 μl of the MOPS/EDTA solution and 8 μl of denaturing loading dye (95% formamide, 25 mm EDTA, 0.05% bromphenol blue, 0.05% xylene cyanol). Each substrate pool (AαS, CαS, dAαS, dCαS, etc.) was treated and analyzed separately. Samples without iodine were also prepared in parallel. All of the products were loaded onto a 15% denaturing sequencing gel. The experiments were performed at least in triplicate.
Sequencing gels of NAIM experiments were visualized using a Storm PhosphorImager and analyzed using ImageQuant software. Peak areas for each band were determined using manual peak selection, and the intensity (I) of each band was calculated. Interferences at each position were calculated as described elsewhere (22). In brief, deoxy and sulfur interferences were calculated as: sulfur interference = Idplx(NαS)/Iunw(NαS) and deoxy interference = Idplx(dNαS)/Iunw(dNαS). For NAIM studies of NPH-II, interference values (κ) greater than 1.5 were scored as interferences. Only interferences seen in at least two independent experiments were considered significant. Positions that showed both phosphorothioate and deoxy interferences were scored as sulfur interferences unless the κdeoxy was significantly greater than κPT (>1.5-fold).
NPH-II Enzyme Unwinding Assays
All of the unwinding time courses were conducted using the same buffer and conditions as the NAIM assay, unless otherwise stated. The reactions were stopped with two volumes of quench buffer (25 mm EDTA, 0.4% SDS, 0.05% bromphenol blue, 0.05% xylene cyanol, 10% glycerol). Duplex and unwound products were normally resolved by 12% native PAGE. Unwinding of the pseudo R-loop substrate was resolved on 8% polyacrylamide gels containing 70 mm NaCl. Electrophoresis was performed at 4 °C for 2.5 h. The dried gels were visualized on a Storm PhosphorImager, and the reaction products were analyzed using ImageQuant software. All of the time courses were performed in triplicate.
RESULTS
A Chemogenetic Screen for Identifying Helicase-Substrate Contacts
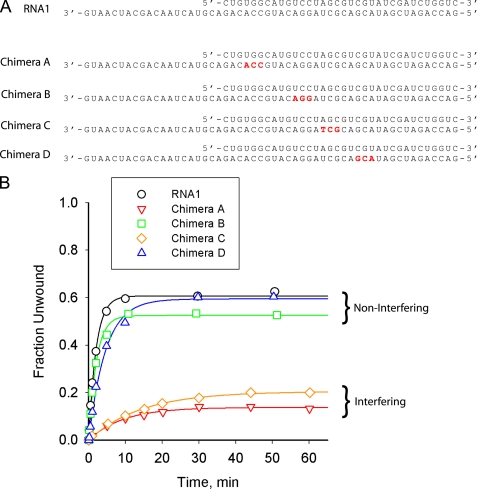
NPH-II is unable to unwind substrates that contain long stretches of DNA on the tracking strand except under highly permissive, low salt conditions (19, 23). To test whether NPH-II is only sensitive to 2′-deoxy substitutions at certain positions on the tracking strand, we examined the ability of NPH-II to unwind chimeric substrates that contain short, three nucleotide segments of DNA placed at different intervals on an otherwise all RNA tracking strand. Surprisingly, these results showed that these small patches could be tolerated at certain locations, but not at others (Fig. 1). We hypothesized that NPH-II may be making specific 2′-hydroxyl contacts at discrete loci within the duplex relative to the single strand/double strand (ss/ds) junction, thus explaining the RNA substrate specificity. We also theorized that if such chemical “stepping” behavior exists, it may be related to the six base pair kinetic stepping described by Jankowsky et al. (16).
FIGURE 1.
The effect of DNA patches in various positions throughout a tracking strand with random sequence. A, RNA1 is an all RNA substrate with random sequence. Chimeras A–D are chimeric substrates containing a 3-nt DNA patch (red letters) in an otherwise all RNA tracking strand. B, time courses show a decrease in unwinding rate constant and amplitude for chimeras A and C (A(A) = 0.15 ± 0.02; kobs(A) = 0.11 ± 0.02 min−1; A(C) = 0.20 ± 0.05; kobs(C) = 0.07 ± 0.01 min−1) when compared with substrates RNA1, B, and D (A(RNA1) = 0.61 ± 0.03; kobs(RNA1) = 0.50 ± 0.05 min−1; A(B) = 0.53 ± 0.07; kobs(B) = 0.48 ± 0.11 min−1; A(D) = 0.59 ± 0.03; kobs(D) = 0.22 ± 0.04 min−1). The protein was prepared as described previously (17).
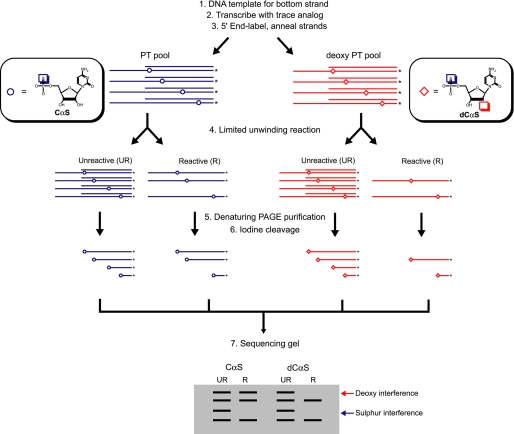
To more precisely define loci where NPH-II makes functional 2′-hydroxyl contacts, we developed a chemogenetic screen that is based on NAIM. NAIM involves the incorporation of phosphorothioate or deoxyphosphorothioate nucleotide analogs into RNA molecules during transcription by T7 RNA polymerase. When these pools of modified transcripts are subjected to a selection, such as unwinding by a helicase, the relative importance of individual functional groups can be assessed by quantifying how much a specific modification at a given position interferes with the success of the reaction (termed “interference effect” (24–26). Using this approach, we were able to simultaneously probe the 2′-hydroxyl of each RNA residue of the tracking strand and identify those functionally related to unwinding by NPH-II (Fig. 2; see also “Experimental Procedures”).
FIGURE 2.
Schematic diagram of nucleotide analog interference mapping of the NPH-II unwinding reaction. Steps 1 and 2, tracking strand oligonucleotides are synthesized via in vitro transcription. The reactions are doped with trace amounts (10%) of PT or 2′-deoxy PT nucleotide analogs. This results in a pool of randomly modified transcripts, each containing a single analog at a random position. The open circles represent positions of PT modifications; the open diamonds represent positions of deoxy PT modifications. Step 3, tracking strand transcripts are 5′ end-labeled and annealed to top strand. Step 4, oligonucleotides are subjected to limited unwinding (5%) by NPH-II under stringent (high salt) conditions, generating a population of unwound substrates (single-stranded; enriched with permissive substitutions) and substrates that failed to unwind (double-stranded; enriched with interfering substitutions). Step 5, following NPH-II unwinding, PAGE separates reacted (R) and unreacted populations (UR). Step 6, both pools are denatured and cleaved with iodine. Step 7, fragments are resolved on a sequencing gel revealing the locations of interfering PT analogs (sulfur interferences) and interfering deoxy PT analogs (deoxy interferences).
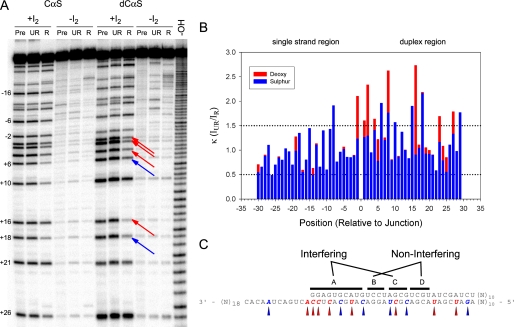
To generate substrates for NAIM analysis of NPH-II unwinding, a 70-nt tracking strand was randomly doped with phosphorothioate (PT) or deoxyphosphorothioate (deoxy PT) NTPs. These PT containing strands were annealed to a 40-nt RNA top strand, thereby creating a 40-bp duplex with a 30-nt 3′ single-strand overhang. The reaction conditions, set at a relatively high salt concentration of 70 mm NaCl, were chosen to challenge the helicase such that slight perturbations in the system would yield quantifiable interference effects (19, 20). The substrate pool was partially unwound by NPH-II, and a series of interference effects was observed (Fig. 3). Several phosphorothioate (sulfur) interferences were scattered throughout the duplex, indicating positions at which pro-Rp phosphoryl oxygens on the tracking strand make specific contributions to RNA unwinding by NPH-II. Deoxynucleotide interferences were also observed throughout the duplex, including a single interference in the 3′ ss region at position −1 relative to the ss/ds junction (Fig. 3B).
FIGURE 3.
Results of NAIM assay. A, a sample NAIM gel showing interference effects from the CαS and dCαS substrate pools. Additional substrate pools (AαS and dAαS, etc.) were analyzed on separate gels. Reacted (R) and unreacted (UR) substrate populations are shown. Precursor lanes (Pre) were not treated with NPH-II. Substrate pools are shown with (+I2) or without (−I2) iodine treatment. -OH indicates a size ladder generated by alkali hydrolysis. Sulfur interferences are indicated by blue arrows; deoxy interferences are indicated by red arrows. B, κ values measured for each position of the substrate tracking strand, depicted 3′ to 5′ from left to right. Position 0 indicates the ss/ds junction. No data were collected beyond position +30. The κ values were calculated as described under “Experimental Procedures.” Sulfur values (blue) are superimposed on top of deoxy values (red). κ values greater than 1.5 are scored as interferences. C, relevant interference effects projected on the substrate sequence. Specific interferences are shown as arrowheads below the sequence. The sequence is divided into four regions based on the relative frequency of observed interference effects: interfering regions A and C and noninterfering regions B and D.
Analysis of the interference patterns shows that interferences are generally spaced at various positions along the duplex. Between these regions are small patches of nucleotides that are relatively free of interference signals. Importantly, these noninterfering clusters coincide with the locations of the “permissive” DNA chimeric patches (Fig. 1A). These data confirm that NPH-II does not view each ribose sugar identically in this substrate. However, the interferences do not appear to occur with any specific spacing (e.g. 6 bp, corresponding to the observed kinetic step size) and are generally inconsistent with a pattern of regular, periodic contact with a specific subset of ribose sugars. Closer examination of the interference pattern reveals that with the exception of position −1, all deoxynucleotide interferences arose from pyrimidine nucleotides (Fig. 3C). Even the sequences of our preliminary chimeric substrates showed that the “permissive” DNA patches were purine rich, whereas the “nonpermissive” patches were rich in pyrimidines (Fig. 1A). This result implies that the interference pattern may be more related to the sequence of the substrate than the hypothesized chemical stepping mechanism.
Differential Effects of Purine and Pyrimidine Residues
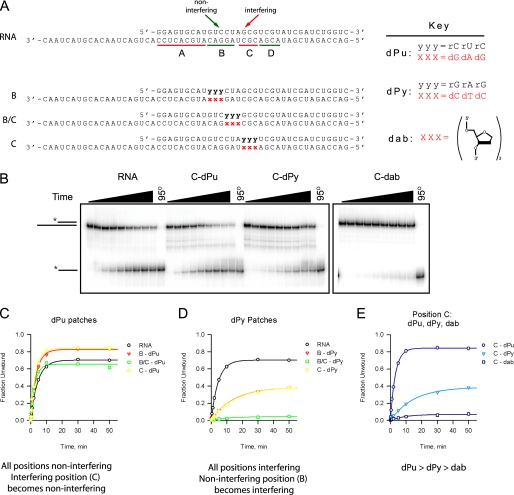
To differentiate between these possibilities, we created a truncated version of the NAIM substrate, and we defined three distinct regions that are based on the NAIM interference pattern: region B as a noninterfering region, C as an interfering region, and B/C as a transition region between the two (Fig. 4A). We then constructed a series of chimeric tracking strands based on the parental RNA sequence, each with a 3-nt DNA patch in either region B, B/C, or C. The sequence of the DNA patch was changed to either the purine sequence dG·dA·dG (B-dPu, B/C-dPu, C-dPu) or the pyrimidine complement dC·dT·dC (B-dPy, B/C-dPy, C-dPy), for a total of six chimeric substrates. Each of these substrates was subjected to unwinding by NPH-II under the same stringent conditions, and the resultant unwinding time courses were fit to a single exponential, yielding values for the apparent rate constant (kobs) and unwinding extent (amplitude, A) under single-cycle unwinding conditions (Fig. 4, B–D; see also “Experimental Procedures”). The unwinding parameters obtained for each of the chimeric substrates were then compared with those of the parental all-RNA substrate.
FIGURE 4.
Differential effects of purine and pyrimidine nucleotides on the tracking strand. A, a truncated version of the NAIM substrate shows the noninterfering region B and the interfering region C. Chimeric substrates B, B/C, and C contain a DNA patch at the positions indicated with xxx. The sequence of the DNA patches are changed to either dG·dA·dG (dPu), dC·dT·dC (dPy), or dab residues. The sequence of the RNA top strand is changed to the respective complimentary sequence. B, sample PAGE showing unwinding time courses of RNA, C-dPu, C-dPy, and C-dab. C–E, plots of unwinding extent over time (empty symbols) are fit to a single exponential (solid lines). C, NPH-II unwinds all dPu chimeras comparable with the RNA control. Interfering block C is made noninterfering by the sequence substitution to purines (A(RNA) = 0.695 ± 0.004, kobs(RNA) = 0.225 ± 0.004; A(B-dPu) = 0.82 ± 0.01, kobs(B-dPu) = 0.29 ± 0.01; A(B/C-dPu) = 0.65 ± 0.01, kobs(B/C-dPu) = 0.37 ± 0.02; A(C-dPu) = 0.84 ± 0.01, kobs(C-dPu) = 0.31 ± 0.02). D, NPH-II unwinds all dPy chimeras with reduced efficiency. Noninterfering block B is made interfering by the sequence substitution to pyrimidines. Note that the time courses for B and C are superimposable (A(B-dPy) = 0.387 ± 0.008, kobs(B-dPy) = 0.069 ± 0.004; A(B/C-dPy) = 0.043 ± 0.002, kobs(B/C-dPy) = 0.09 ± 0.01; A(C-dPy) = 0.389 ± 0.008, kobs(C-dPy) = 0.066 ± 0.004). E, C-dab unwinds poorer than both C-dPu and C-dPy (A(C-dab) = 0.070 ± 0.003, kobs(C-dab) = 0.10 ± 0.01).
Intriguingly, each of the new dPy substrates was unwound with reduced efficiency when compared with the parental RNA (Fig. 4D). For unwinding of substrates B-dPy and C-dPy, a 3-fold reduction in unwinding amplitude and a nearly 4-fold reduction in rate constant were observed. Substrate B/C-dPy showed a 12-fold reduction in unwinding amplitude. In contrast, unwinding of the dPu substrates resulted in rate constants and amplitudes comparable with that of the parental RNA (Fig. 4C). These results are significant in that we were able to convert a permissive region, B, to nonpermissive merely by substituting three dPy in the place of dPu. Similarly, we converted a nonpermissive region, C, to permissive by substituting the dPu in place of the dPy. The pattern also held true for region B/C, which was intermediate between the two. This experiment demonstrates a clear preference for dPu over dPy regardless of position, which suggests that engagement with the bases may be relevant for the unwinding mechanism of NPH-II.
The demonstrable preference for dPu raises the question of whether purines and pyrimidines can both contribute to the mechanism of unwinding or whether purine residues play a uniquely important role in the process. To determine whether a dPy base is better than no base at all, we constructed a substrate that contains a patch of deoxyribose abasic (dab) residues in the C region (C-dab) to compare with the unwinding of substrates C-dPu and C-dPy. Unwinding of substrate C-dab resulted in a 10-fold reduction in unwinding amplitude, in contrast to the 3-fold reduction of substrate C-dPy (Fig. 4E). These data indicate that in the DNA context, all nucleobases make important contributions to the unwinding of substrates by NPH-II. They also indicate that although purine residues are preferable, pyrimidines retain some level of functionality.
Effect of Duplex Stability
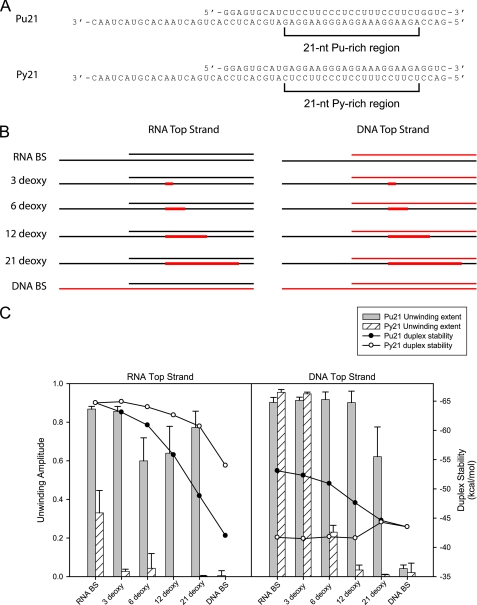
One potential explanation for the observed bias toward purine bases is that NPH-II is not sensitive to the base composition per se but is responding to differences in the thermodynamic stability of the substrates. For example, it has been demonstrated that rPu·dPy hybrid duplexes are more stable than the equivalent rPy·dPu duplexes (27). To address this question, we designed two new substrates, Pu21 and Py21, which contain 21-nt regions consisting entirely of either Pu or Py nucleotides, respectively (Fig. 5A). Based on these parental RNA substrates, we designed two sets of hybrid and chimeric substrates (Fig. 5B). Each set consists of six tracking strands: one all RNA, one all DNA, and four RNA chimeras with 3, 6, 12, or 21 DNA nucleotides within the 21-homonucleotide stretch. When annealed to either RNA or DNA, the thermodynamic stabilities of these substrates can be estimated using published tables (28–31). These values are summarized as a line plot in Fig. 5C. Generally, we observe that as we increase the DNA content of the substrates, the thermodynamic stability of the duplexes decrease. In particular we note a substantial energetic decrease when DNA replaces RNA as the top strand (10–20 kcal mol−1). Importantly, we observe that with an RNA top strand, the Py21 substrates are more stable, whereas the Pu21 substrates are more stable with the DNA top strands.
FIGURE 5.
The Pu/Py bias arises from the bases themselves and not from differences in duplex stability. A, substrates Pu21 and Py21 have a 21-nt sequence in the tracking strand (indicated by brackets) that is homopurine and homopyrimidine respectively. B, schematics illustrating the sets of chimeras and hybrids made based on these sequences. Black denotes RNA backbone; red indicates DNA. DNA nucleotides are incrementally substituted into the 21-nt homopurine/homopyrimidine region of the respective tracking strands, as indicated on the right. An RNA top strand set and a DNA top strand set is generated for each sequence. C, plot showing the duplex stabilities (line plot) and unwinding amplitude (bar chart) for each substrate. The error bars for the bar chart are standard deviations of three independent experiments. The measurable unwinding rate constants for Pu21 with RNA top strand are as follows: kobs(RNA) = 0.30 ± 0.02, kobs(3-deoxy) = 0.29 ± 0.12, kobs(6-deoxy) = 0.13 ± 0.02, kobs(12-deoxy) = 0.13 ± 0.01, kobs(21-deoxy) = 0.26 ± 0.07; with DNA top strand kobs(RNA) = 0.51 ± 0.09, kobs(3-deoxy) = 0.58 ± 0.14, kobs(6-deoxy) = 0.44 ± 0.19, kobs(12-deoxy) = 0.30 ± 0.01, kobs(21-deoxy) = 0.11 ± 0.03. The measurable unwinding rate constants for Py21 with RNA top strand are as follows: kobs(RNA) = 0.06 ± 0.02, kobs(3-deoxy) = 0.11 ± 0.05; with DNA top strand kobs(RNA) = 0.65 ± 0.07, kobs(3-deoxy) = 0.50 ± 0.06, kobs(6-deoxy) = 0.19 ± 0.18 min−1.
We tested NPH-II unwinding of these substrates using the same unwinding conditions as previously indicated. The extents of unwinding of each substrate are summarized as a bar chart in Fig. 5C. NPH-II unwinding was almost completely inhibited on all of the Py21 substrates paired with an RNA top strand. Intriguingly, this was true even on the RNA track. This result indicates that the Pu/Py bias is not limited to just the DNA context but is applicable for RNA substrates as well. When Py21 was paired with a DNA top strand (creating a weaker duplex), NPH-II performed somewhat better. It efficiently unwound through the RNA and 3-deoxy tracks, but as the DNA patch was extended, NPH-II unwinding was increasingly inhibited. This shows that although NPH-II is somewhat sensitive to the stability of its substrate, the helicase still responds negatively to the presence of dPy nucleotides. In contrast, NPH-II was able to efficiently unwind the Pu21 chimeric substrates in all cases, even when the duplexes were substantially more stable than their Py21 counterparts. It is also important to note that each of these hybrid chimeric substrates adopts a predominantly A-form conformation. These data confirm that the Pu/Py bias cannot be explained by differences in thermodynamic stabilities or helical geometries but arises from the identity of the nucleobases themselves.
Can NPH-II Act as a True DNA Helicase?
These data show that, once it is underway, NPH-II can displace a DNA top strand from a DNA track. Given the wealth of data demonstrating a relative lack of NPH-II activity on DNA (19, 21), the fact that NPH-II readily unwinds a substrate containing a 21-bp DNA duplex is surprising. Whether NPH-II can function completely as a DNA helicase is questionable, however, given that the Pu21 and Py21 substrates were not unwound by NPH-II when the entire tracking strand was composed of DNA (Fig. 5C). There are several possible explanations for the apparent inactivity of NPH-II on substrates that are completely composed of DNA. First, none of the substrates examined thus far contain exclusively purines on the tracking strand. It is possible that, given a homopurine sequence, NPH-II would be able to unwind a full DNA duplex. Second, it was previously shown that NPH-II unwinding can be inhibited by as little as four DNA nucleotides located on the single strand loading region immediately upstream of the single strand/double strand junction (19). This suggests that, although NPH-II may be able to translocate on a dPu tracking strand, it may require an RNA component to productively initiate unwinding. Finally, NPH-II may prefer a certain helical geometry, such as A-form instead of B-form, independent of the backbone chemistry or sequence (21).
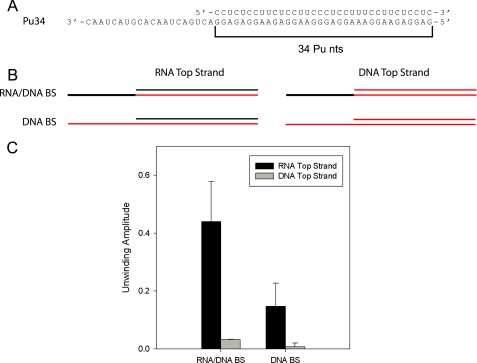
To address these questions, we designed substrate Pu34 (Fig. 6A). The tracking strand of this substrate contains a 34-nt purine-rich patch that spans the entire duplex region. The single-stranded overhang region was composed of either DNA or RNA, resulting in a chimeric tracking strand. These tracking strands were both annealed to either RNA or DNA top strands (Fig. 6B) and treated with NPH-II as before. NPH-II displayed the highest level of activity when displacing an RNA top strand from the chimeric tracking strand containing RNA in the ss overhang (44%, Fig. 6C). Whereas NPH-II showed significant activity when displacing RNA from an all-DNA tracking strand (15%), the helicase displayed almost no activity for displacement of a DNA top strand from either type of track.
FIGURE 6.
NPH-II unwinds DNA·RNA duplexes but not DNA·DNA duplexes. A, substrate Pu34 contains a 34 nt homopurine sequence that spans the entire duplex. B, schematics illustrating the chimeras and hybrids made based on this sequence. Black denotes RNA backbone; red indicates DNA. C, plot showing the unwinding amplitude for each substrate. The error bars are standard deviations of three independent experiments. The measurable rate constants with RNA top strand are as follows: kobs(RNA/DNA) = 0.19 ± 0.05, kobs(DNA) = 0.12 ± 0.05; with DNA top strand, kobs(RNA/DNA) = 0.08 ± 0.02 min−1.
These data provide some new insights into NPH-II substrate specificity. First, they suggest that the inability of NPH-II to unwind DNA·DNA duplexes may stem from inactivity toward B-form helices. NPH-II can clearly unwind hybrid duplexes (which are A-form) by tracking on either the RNA or DNA strand. However, the case for strong requirement for helical geometry is mitigated given that NPH-II can strip proteins from single-stranded RNA (18). Second, NPH-II is not as processive on a DNA track as on an RNA track. This is demonstrated by a greater dependence of the unwinding amplitude on the length of the DNA track (Figs. 4 and 5). This suggests that ribose, and perhaps the 2′-hydroxyl specifically, may form strong interactions with NPH-II. This may allow the helicase to maintain a stronger grip and thereby enhance processivity during strand displacement. Finally, given the higher activity displayed with an RNA ss overhang, the data suggest that NPH-II initiates unwinding more readily on RNA in vitro. Whatever the explanation, NPH-II may display a greater degree of DNA·RNA helicase activity under more physiologically relevant conditions, in the presence of partner proteins, or on substrates that more closely mimic the actual biological target.
In Vivo Role for Pu/Py Bias and DNA·RNA Helicase Activity
It has been proposed that NPH-II aids in the transcriptional termination of early viral genes by disrupting or preventing undesirable secondary structures (14). Many early vaccinia genes contain a conserved sequence TTTTTNT in the sense DNA strand, which is transcribed into a cis-acting UUUUUNU termination signal (U5NU motif) as a part of the 3′-UTR of the nascent mRNA (32). It was previously proposed that NPH-II utilizes the nascent mRNA as the tracking strand, moving 3′ → 5′ away from the transcriptional fork. This activity has been proposed to reduce the formation of secondary structures, such as R-loops, that could potentially mask the U5NU motif (21).
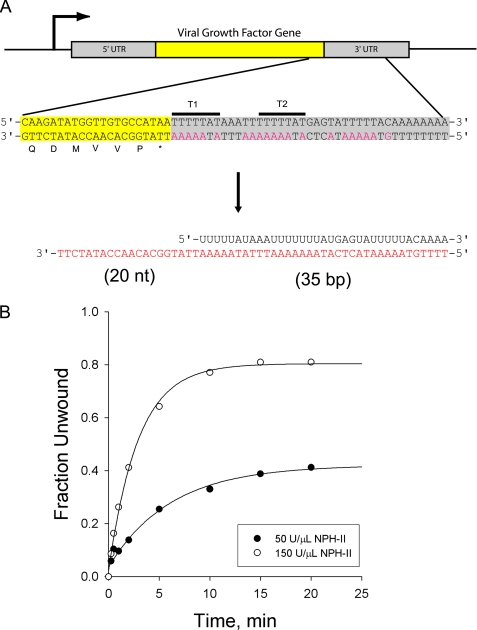
An alternative model for NPH-II activity on early genes is suggested by the purine bias and DNA tracking activity that we observe. At the U5NU termination signal, the complementary antisense DNA strand is a purine-rich sequence that would enable NPH-II to translocate in a 3′ → 5′ direction toward the transcription fork. Thus the DNA strand is a potential target for NPH-II (Fig. 7A). To test whether NPH-II can successfully unwind a DNA·RNA hybrid that mimics a hypothetical R-loop structure encountered by NPH-II during transcription of a vaccinia early gene, we constructed an unwinding substrate based on the sequence of the 3′-UTR of the vaccinia viral growth factor gene (see “Experimental Procedures”). This gene contains two tandem copies of the termination motif, terminating translation ∼50 bp downstream of the stop codon (33–35). The sequence of the DNA tracking strand for this new substrate was taken from the viral growth factor gene such that the overhang contains the last 20 nt of the coding region, whereas the duplex consists of the first 35 nt of the 3′-UTR (including both termination elements) annealed to a complementary RNA top strand (Fig. 7A). The stability of this construct is estimated to be approximately −20 kcal mol−1.
FIGURE 7.
Unwinding of a hypothetical DNA·RNA hybrid formed in vivo. A, schematic showing the coding region of the vaccinia viral growth factor gene. A blowup of the 3′-UTR sequence shows the two tandem termination motifs (T1 and T2) consisting of the conserved sequence TTTTTNT. The coding DNA strand in this region is rich in purine nucleotides (highlighted in magenta) presenting a possible DNA substrate for NPH-II. We construct a substrate (35 bp with 20-nt 3′ tail) representing a hypothetical R-loop based on this sequence. RNA nucleotides are in black, and DNA is in red. The duplex stability is estimated to be −20.1 kcal/mol. B, unwinding of the hypothetical R-loop using the same conditions as previously demonstrates a moderate level of unwinding. A = 0.40 ± 0.02, kobs = 0.19 ± 0.03 High unwinding is achieved by adding saturating levels of enzyme. A = 0.80 ± 0.01, kobs = 0.36 ± 0.01.
Interestingly, in contrast to the 15% unwinding of the DNA·RNA hybrid shown in Fig. 6C, NPH-II unwound the pseudo R-loop substrate to much greater extent (40%; Fig. 7B). In addition, unwinding efficiency was enhanced by increasing the concentration of NPH-II by 3-fold. Under these conditions NPH-II displayed robust unwinding of the pseudo R-loop substrate, equal to any previously unwound RNA, albeit at a slower rate (80%; Fig. 7B). Although unwinding of this substrate is probably aided by a lower duplex stability, this “natural” sequence more closely mimics the proposed in vivo target than any substrate previously tested. It might therefore be considered a more relevant test of NPH-II activity in vivo. We note that, lacking clear knowledge of exactly how and where NPH-II is utilized, this construct may introduce an artificial entry point for its helicase activity. In other words, we do not know how large the R-loop may be, where it might begin, how NPH-II is conveyed to the initiation site, or what other proteins might be involved to aide in the process. Nonetheless, these data show that, when considering the in vivo function of NPH-II, the helicase need not necessarily be considered to act solely or restrictively on RNA. Given the right context, NPH-II can effectively initiate and translocate on a DNA strand, thereby unwinding a biologically relevant duplex.
DISCUSSION
The data presented here reveal that the polymer specificity of NPH-II, and potentially other helicases, is more complicated than previously supposed. We have demonstrated that NPH-II displays a sequence bias, favoring purines over pyrimidines on both DNA and RNA tracking strands. Thus, contrary to earlier assessments, it appears that nucleobase identity plays an important role during duplex unwinding. Sequence recognition may play an important role in the mechanism and regulation of helicase function, and it may be a general feature that is common to many helicase families. This is supported by emerging work on the Rho helicase from superfamily 5. In that case, NAIM experiments indicate a significant influence of sequence on Rho function (36).
Although other helicases have shown a preference for unwinding certain kinds of sequences, the effect described here is mechanistically different from previous reports. For example, HCV NS3 and bacteriophage T7 helicases display a preference for unwinding duplexes that contain A·U base pairs (37, 38). This result reflects a sensitivity of the helicase to the thermodynamic stabilities of the substrate duplex (37, 38). In contrast, the sequence bias presented here derives from chemical differences between purine and pyrimidine nucleotides and not to the strength of base pairing or stacking interactions. We show that, whereas duplex stability can have some effect on the extent of unwinding by NPH-II, these differences are not responsible for the observed sequence bias. The effects reported here appear to arise specifically from the identity of the bases on the tracking strand alone.
It is a generally accepted axiom that helicases, and especially processive helicases, are not affected by sequence variations. The exception to this rule is the RecBCD holoenzyme, which is sensitive to the Chi sequence in DNA (13). However, Chi is recognized by the RecC subunit that lacks all the characteristic helicase motifs and is unable to unwind DNA on its own. Thus Chi does not represent an example of sequence specificity by a functional helicase. To our knowledge, NPH-II is the only helicase for which sequence bias has been shown to influence the mechanism and polymer specificity of unwinding. It is particularly striking that these effects are observed in the absence of accessory cofactors that might be expected to modulate the various functions of NPH-II.
The observed reduction in unwinding amplitude in response to pyrimidine nucleotides, DNA duplex length, and increased duplex stability indicates that all of these factors are likely to influence the behavior of the enzyme. Our results suggest that NPH-II makes important interactions with nonbridging phosphoryl oxygens, ribose 2′-hydroxyls, and the nucleobases themselves. When one mode of interaction becomes unavailable (e.g. ribose 2′-hydroxyls), the helicase may compensate by relying more heavily on another (e.g. nucleobases). However, a specific physical or chemical basis for the observed purine bias remains unclear. It is possible that the protein makes contacts with the major groove and establishes unique interactions with purine atoms, such as N7. Alternatively, specific NPH-II side chains may stack against the bases, and purines are more likely to provide an optimal stacking interface than pyrimidines. This latter hypothesis is consistent with the observation that although purines are better than pyrimidines, pyrimidines can be better than no base at all. It will be of interest to identify the specific types of interactions that stabilize the NPH-II-nucleic acid interface.
Perhaps most importantly, these data have implications for the function of NPH-II in pox viruses. NPH-II has long been suspected to be crucial for proper transcriptional termination of early viral genes in vivo (14). Classic features of the transcriptional phenotype of NPH-II-deficient virions are 1) accurate but reduced initiation at early promoters; 2) abnormally long mRNA products; and 3) inefficient release of mRNAs from the viral core (14). Early transcriptional termination is facilitated by the presence of a termination signal U5NU found on the nascent mRNA (32, 39) (Fig. 8A). This U5NU motif is recognized by the vaccinia termination factor, which then affects proper termination 20–50 bp downstream of the termination signal in conjunction with NPH-I, a DExH protein with no detectable helicase activity (34, 39–41). Importantly, defects in this termination system (e.g. base pairing caused by secondary structures or substituted uracil residues in the mRNA) result in phenotypes similar to those displayed by NPH-II-deficient virions (42). One proposed role for NPH-II is that it acts to prevent the formation of R-loops (extraneous RNA·DNA hybrids formed during transcription) that could mask the U5NU motif (14, 21, 43). In theory, this could be accomplished by either translocating on the DNA (3′ → 5′ toward the RNA polymerase) or on the nascent mRNA (3′ → 5′ away from the RNA polymerase). Because earlier experiments indicated that NPH-II could not use DNA as a tracking strand, it was concluded that NPH-II moved along the mRNA. However, substrates used to determine substrate specificity were often designed both to offer an energetically flat unwinding landscape and to minimize unwanted off pathway secondary structures (16, 17, 19). These pseudo-random sequences have little or no direct relevance to the proposed in vivo substrate of the enzyme. In the present work we show that when presented with a biologically relevant sequence, such as a DNA·RNA hybrid within the 3′-UTR of the viral growth factor gene, the behavior of the helicase was radically altered from what was expected. Not only do we show that it is possible for NPH-II to act on the purine-rich DNA strand, but our data imply that the pyrimidine-rich RNA strand may not be as hospitable a substrate as previously supposed.
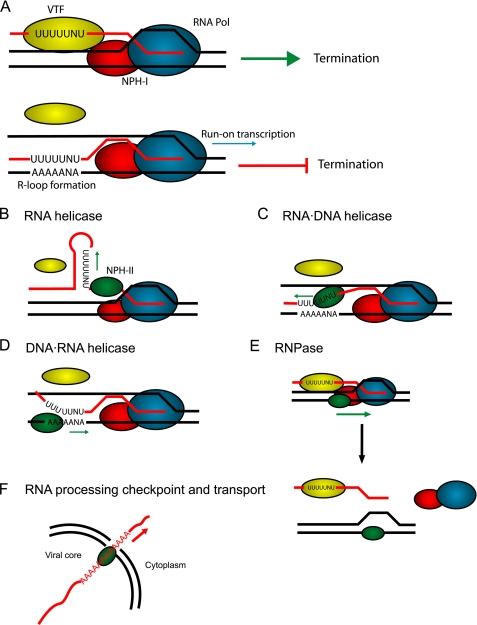
FIGURE 8.
Potential roles for NPH-II in vaccinia virions relating to transcriptional termination and mRNA export. A, transcriptional termination is accomplished through recognition of the U5NU termination motif on the nascent mRNA (red strand) by the viral vaccinia termination factor. The vaccinia termination factor then interacts with the DExH helicase NPH-I, which acts as an energy coupling factor to effect release of the nascent mRNA by the RNA polymerase ∼50 nt downstream of the termination signal. The termination signal can be masked by secondary structures (by formation of R-loops for example) preventing vaccinia termination factor signaling and generating run-on transcripts. Possible biological roles for NPH-II (based on biochemical data) include B, RNA helicase activity to disrupt masking RNA structures. C, RNA·DNA helicase activity to prevent R-loop formation. D, DNA·RNA helicase activity to prevent R-loop formation. E, RNPase activity to aide NPH-I in dislodging the RNA polymerase. F, single strand translocase activity to check for proper mRNA polyadenylation and mRNA export.
Knowing which strand NPH-II tracks along should influence our thinking about how NPH-II works and how it is utilized by the virus. If it acts on the RNA as previously proposed, NPH-II is less likely to be hampered by sequence effects or initiation problems. It would also be in a position to disrupt hairpins or remove proteins that might also mask the termination signal (Fig. 8, B and C). However, this requires that NPH-II be either constantly reloaded as the nascent mRNA is elongated, or we must invoke some sort of bi-directional shuttling that has yet to be observed for this particular helicase. On the other hand, if NPH-II acts on DNA, it need only be loaded once. It is then poised to follow directly behind the translocating polymerase and prevent possible R-loop formation (Fig. 8D). If its activity is only required to ensure that the termination signal remains clear, it need not necessarily be especially processive, thus obviating the need for long purine-rich DNA sequences. Additionally, in this orientation, NPH-II would now be in a position to use its RNPase activity to potentially aid in dislodging the RNA polymerase complex (18). However, this model is complicated by the issues of entry (where does it start) and initiation (how does it start) on the DNA substrate. Experiments testing the ability of NPH-II to translocate independent of unwinding on DNA and RNA substrates of various sequence compositions may help clarify these issues.
It is unlikely that NPH-II exerts its effect on DNA·DNA duplexes in any significant way. Rather, the data seem to support the notion that its activity is probably restricted to RNA hairpins or hybrid duplexes such as those proposed for R-loops. Our data do not discount the possibility that NPH-II could be acting simultaneously in both capacities. In addition, it is important to note that the U5NU motif is almost exclusively confined to the 3′-UTR and is rarely found in the coding regions of early genes. Thus the virus may have evolved a unique mechanism to spatially regulate the DNA·RNA helicase activity of NPH-II to the 3′-UTR. We also speculate that the purine bias of NPH-II might additionally target it to the polyadenylated ends of mature mRNAs, enabling NPH-II to act as a checkpoint for full processing as well as a transporter molecule (Fig. 8F). Indeed, given the need for compact genomes, viral proteins are often required to carry out multiple, even disparate enzymatic roles (44).
Overall, the data presented in this study demonstrate that the behavior and substrate specificity of NPH-II can be modified by the nucleobase component of its nucleic acid substrates. The sequence bias for NPH-II challenges the model of sequence neutrality invoked for processive helicases and the predominant backbone-tracking model of SF2 helicases. We show that NPH-II, once considered a strict RNA helicase, need not be limited to RNA helicase roles in vivo. In addition, our results underscore the importance of accounting for in vivo substrate configurations, where possible, when designing biochemical experiments on helicase enzymes.
Acknowledgments
We thank members of the Pyle lab, particularly O. Federova, N. Zingler, V. Serebrov, A. Kohlway, C. Matranga, S. Ding, and D. Rawling, for technical advice and helpful discussions about this work. We also thank M. Sehorn for helpful advice on protein expression and purification and S. Shuman for helpful discussions on experimental design.
This work was supported, in whole or in part, by National Institutes of Health Grant GM60620 (to A. M. P.).
This work is dedicated to the memory of Bruce Carmichael.
- SF2
- superfamily 2
- SF1
- superfamily 1
- NPH-II
- nucleoside triphosphate phosphohydrolase II
- ss
- single strand
- ds
- double strand
- NAIM
- nucleotide analog interference mapping
- nt
- nucleotide
- PT
- phosphorothioate
- dPu
- deoxy purine
- dPy
- deoxy pyrimidine
- dab
- deoxyribose abasic
- UTR
- untranslated region
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MOPS
- 4-morpholinepropanesulfonic acid
- deoxy PT
- deoxyphosphorothioate.
REFERENCES
- 1.Koonin E. V., Senkevich T. G. (1992) J. Gen. Virol. 73, 989–993 [DOI] [PubMed] [Google Scholar]
- 2.Bork P., Koonin E. V. (1993) Nucleic Acids Res. 21, 751–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soultanas P., Wigley D. B. (2000) Curr. Opin. Struct. Biol. 10, 124–128 [DOI] [PubMed] [Google Scholar]
- 4.Lee J. Y., Yang W. (2006) Cell 127, 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Büttner K., Nehring S., Hopfner K. P. (2007) Nat. Struct. Mol. Biol. 14, 647–652 [DOI] [PubMed] [Google Scholar]
- 6.Kim J. L., Morgenstern K. A., Griffith J. P., Dwyer M. D., Thomson J. A., Murcko M. A., Lin C., Caron P. R. (1998) Structure 6, 89–100 [DOI] [PubMed] [Google Scholar]
- 7.Singleton M. R., Scaife S., Wigley D. B. (2001) Cell 107, 79–89 [DOI] [PubMed] [Google Scholar]
- 8.Pyle A. M. (2008) Annu. Rev. Biophys. 37, 317–336 [DOI] [PubMed] [Google Scholar]
- 9.Rocak S., Linder P. (2004) Nat. Rev. Mol. Cell Biol. 5, 232–241 [DOI] [PubMed] [Google Scholar]
- 10.Tanner N. K., Linder P. (2001) Mol. Cell 8, 251–262 [DOI] [PubMed] [Google Scholar]
- 11.Tuteja N., Tuteja R. (2004) Eur. J. Biochem. 271, 1835–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman E., Edwalds-Gilbert G., Lin R. J. (2003) Gene 312, 1–16 [DOI] [PubMed] [Google Scholar]
- 13.Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B. (2004) Nature 432, 187–193 [DOI] [PubMed] [Google Scholar]
- 14.Gross C. H., Shuman S. (1996) J. Virol. 70, 8549–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss B. (2001) in Fields Virology (Knipe D. M., Howley P. M. eds) 4th Ed., pp. 2850–2882, Lippincott Williams & Willkins, New York [Google Scholar]
- 16.Jankowsky E., Gross C. H., Shuman S., Pyle A. M. (2000) Nature 403, 447–451 [DOI] [PubMed] [Google Scholar]
- 17.Kawaoka J., Jankowsky E., Pyle A. M. (2004) Nat. Struct. Mol. Biol. 11, 526–530 [DOI] [PubMed] [Google Scholar]
- 18.Jankowsky E., Gross C. H., Shuman S., Pyle A. M. (2001) Science 291, 121–125 [DOI] [PubMed] [Google Scholar]
- 19.Kawaoka J., Pyle A. M. (2005) Nucleic Acids Res. 33, 644–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shuman S. (1993) J. Biol. Chem. 268, 11798–11802 [PubMed] [Google Scholar]
- 21.Gross C. H., Shuman S. (1996) J. Virol. 70, 2615–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedorova O., Boudvillain M., Kawaoka J., Pyle A. M. (2005) in Handbook of RNA Biochemistry (Hartmann R. K., Bindereif A., Schön A., Westhof E. eds) pp. 259–293, Wiley, Weinheim, Germany [Google Scholar]
- 23.Bayliss C. D., Smith G. L. (1996) J. Virol. 70, 794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudvillain M., de Lencastre A., Pyle A. M. (2000) Nature 406, 315–318 [DOI] [PubMed] [Google Scholar]
- 25.Ryder S. P., Strobel S. A. (1999) Methods 18, 38–50 [DOI] [PubMed] [Google Scholar]
- 26.Boudvillain M., Pyle A. M. (1998) EMBO J. 17, 7091–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung S. H., Yu Q., Gray D. M., Ratliff R. L. (1994) Nucleic Acids Res. 22, 4326–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.SantaLucia J., Jr., Hicks D. (2004) Annu. Rev. Biophys. Biomol. Struct. 33, 415–440 [DOI] [PubMed] [Google Scholar]
- 30.Nakano S., Kanzaki T., Sugimoto N. (2004) J. Am. Chem. Soc. 126, 1088–1095 [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M. (1995) Biochemistry 34, 11211–11216 [DOI] [PubMed] [Google Scholar]
- 32.Shuman S., Moss B. (1988) J. Biol. Chem. 263, 6220–6225 [PubMed] [Google Scholar]
- 33.Venkatesan S., Gershowitz A., Moss B. (1982) J. Virol. 44, 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuen L., Moss B. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6417–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohrmann G., Yuen L., Moss B. (1986) Cell 46, 1029–1035 [DOI] [PubMed] [Google Scholar]
- 36.Schwartz A., Rabhi M., Jacquinot F., Margeat E., Rahmouni A. R., Boudvillain M. (2009) Nat. Struct. Mol. Biol. 16, 1309–1316 [DOI] [PubMed] [Google Scholar]
- 37.Cheng W., Dumont S., Tinoco I., Jr., Bustamante C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13954–13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donmez I., Rajagopal V., Jeong Y. J., Patel S. S. (2007) J. Biol. Chem. 282, 21116–21123 [DOI] [PubMed] [Google Scholar]
- 39.Shuman S., Broyles S. S., Moss B. (1987) J. Biol. Chem. 262, 12372–12380 [PubMed] [Google Scholar]
- 40.Deng L., Shuman S. (1998) Genes Dev. 12, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piacente S., Christen L., Dickerman B., Mohamed M. R., Niles E. G. (2008) Virology 376, 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuman S., Moss B. (1989) J. Biol. Chem. 264, 21356–21360 [PubMed] [Google Scholar]
- 43.Gross C. H., Shuman S. (1998) J. Virol. 72, 4729–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beran R. K., Serebrov V., Pyle A. M. (2007) J. Biol. Chem. 282, 34913–34920 [DOI] [PubMed] [Google Scholar]