Abstract
Fibroblast growth factors are key proteins in many intercellular signaling networks. They normally remain attached to the extracellular matrix, which confers on them a considerable stability. The unrestrained accumulation of fibroblast growth factors in the extracellular milieu, either due to uncontrolled synthesis or enzymatic release, contributes to the pathology of many diseases. Consequently, the neutralization of improperly mobilized fibroblast growth factors is of clear therapeutic interest. In pursuing described rules to identify potential inhibitors of these proteins, gentisic acid, a plant pest-controlling compound, an aspirin and vegetarian diet common catabolite, and a component of many traditional liquors and herbal remedies, was singled out as a powerful inhibitor of fibroblast growth factors. Gentisic acid was used as a lead to identify additional compounds with better inhibitory characteristics generating a new chemical class of fibroblast growth factor inhibitors that includes the agent responsible for alkaptonuria. Through low and high resolution approaches, using representative members of the fibroblast growth factor family and their cell receptors, it was shown that this class of inhibitors may employ two different mechanisms to interfere with the assembly of the signaling complexes that trigger fibroblast growth factor-driven mitogenesis. In addition, we obtained evidence from in vivo disease models that this group of inhibitors may be of interest to treat cancer and angiogenesis-dependent diseases.
Keywords: Cell/Migration, Diseases/Cancer/Therapy, Extracellular Matrix/Heparan Sulfate, Growth Factors, Tumor/Therapy, Anti-angiogenesis, Heparin
Introduction
The fibroblast growth factors (FGFs)4 constitute one of the largest families of polypeptide growth factors. There are 22 FGFs in humans and mice that differ significantly in both size (17–20 kDa) and sequence, although each contains a core homology region encompassing 120–130 residues. Phylogenetic analyses suggest that the FGF genes can be arranged into seven subfamilies. All FGFs bind to heparin with high affinity (Kd between 1–2 nm), except for the members of the FGF-19 subfamily (i.e.: FGF-15, -19, -21, and -23) that have little or no affinity for these glycosaminoglycans (1). Apart from the family comprising FGF-11 to FGF-14, FGFs exert their diverse biological actions by binding to a series of membrane tyrosine kinase receptors (FGFRs) that are encoded by four genes (2–4). For this reason the FGF family is currently considered to be constituted by 18 members.
FGFs were first isolated in the 1980s from bovine brain extracts due to their mitogenic and angiogenic activities (5). The affinity of FGFs for heparin was recognized very soon after their discovery (6), although the physiological substrate for FGF in normal conditions is heparan sulfate, a proteoglycan whose glycoside moiety is a glycosaminoglycan like heparin. Although initially conceived as FGF traps and protectors, it was later shown that these proteoglycans also participate in FGF signaling, although they are not absolutely required (4, 7, 8).
In addition to the effects on cell replication and angiogenesis observed initially, FGFs regulate cell survival, apoptosis, proliferation, differentiation, matrix composition, chemotaxis, cell adhesion, and migration. Different cell types or even the same cell may display alternate and sometimes opposing responses to FGFs, depending on their state of differentiation, biochemical status, or the cellular, physical and chemical environment of the cell (4). These activities govern a wide variety of developmental and physiological processes, as practically all cell lineages derived from the embryonic mesoderm and neuroectoderm are under the control of these proteins. At the same time FGFs have been detected in most adult tissues derived from these embryonic cell types under physiological conditions. FGFs, at times expressed at very high levels, normally remains trapped in the extracellular matrix, from which they are released by heparanases or other specialized proteins when necessary (9–16). Obviously, the subversion of this powerful signaling system and any defects in its tight control, either through uncontrolled synthesis or the continuous mobilization of matrix bound FGFs, may cause very serious physiological disturbances (17). Indeed, it was recently shown that mutations in FGFs that decrease their affinity for heparan sulfate in the extracellular matrix and, consequently, that increase their diffusion through developing tissues led to the ectopic signaling responsible for the mouse Elbow knee synostosis syndrome (18).
Over the last 20 years, a wealth of information has accumulated regarding the involvement of different members of the FGF family in pathological conditions, much of which is related to cancer. Elevated levels of FGFs are detected in the serum of cancer patients (19). Sometimes FGFs seem to be directly involved in tumorigenesis by the autocrine, paracrine of juxtacrine induction of the growth of the cancer cells. In addition, FGF signaling may affect processes other than growth associated to tumor progression. For example, they can be involved in the creation of profuse blood irrigation networks to sustain the intense metabolism of the tumor cells, which subsequently favor their dissemination throughout the organism. Tumor metastasis is additionally favored by the activation of enzymes that degrade the basement membrane and the enhancement of cell motility by FGFs. Moreover, they are also involved in inhibiting apoptosis, enhancing the survival of tumor cells, promoting the resistance of tumors to chemotherapeutic drugs and radiation, controlling tumor dormancy (a step in tumor progression), and controlling the self-renewal of cancer stem cells. FGFs may also participate in paraneoplastic events in cancer patients (20–25).
Based on a relatively wide screening of naphthalene sulfonate derivatives and on three-dimensional structural studies, we recently presented a set of chemical leads to develop FGF inhibitors. The most promising inhibitor of this family of compounds was 5-aminonaphthalene-2-sulfonic acid (5A2NMS). These studies led us to propose some rules that can be applied to search for new FGF inhibitors (26). On the basis of these rules we have since searched for further potential candidates to inhibit FGFs, examining different libraries of natural products. We found that gentisic acid (GA; 2,5-dihydroxybenzoic acid), a widespread plant secondary metabolite involved in pest defense and a catabolite of aspirin, is a potential inhibitor of FGF (27–32). Here we show that GA does indeed define a new chemical group of FGF inhibitors, which includes homogentisic acid (HGA; 2-(2,5-dihydroxyphenyl)acetic acid), the toxic agent in alkaptonuria. Indeed, these compounds inhibit relevant activities attributed to the two prototypical members of the family in vitro and in vivo. The chemical constraints that define the efficiency of these aromatic derivatives in inhibiting FGF are also identified; the para conformation of the dihydroxyphenyl group and the functionalization of the aromatic ring by an acidic group. We also show that these compounds recognize both the growth factors and their receptors, displacing heparin from its binding site in these polypeptides, change the three-dimensional structure of the growth factor at their receptor recognizing site, and are capable of dissociating the receptor·growth factor signaling complex.
EXPERIMENTAL PROCEDURES
Protein Expression Purification and Analysis
The cDNAs encoding FGF-1 and FGF-2 were cloned into the pRAT-4 plasmid, expressed in Escherichia coli BL21(DE3), and purified by heparin-Sepharose chromatography (33, 34). The protocol to uniformly labeled 15N synthesis has already been described (35). C-LYT/aFGF was produced and purified by heparin-Sepharose chromatography as described previously (36). To express the extracellular domain (residues 25–361) of receptor 2 IIIc isoform (exdFGFR2IIIc), an expression cassette was constructed by PCR using pTK14 as the template (37) and using the nucleotide modifications proposed by ProteoExpert (Biomax Informatics AG) for optimal expression of the protein in E. coli. The cassette includes the T7 promoter of phage RNA polymerase, exdFGFR2IIIc with a His tag fused to its C terminus and optimized for Ni2+-chelating chromatography, and a 3′-untranslated region optimized for protein expression in which a T7 phage RNA polymerase terminator has been included (supplemental Fig. S1). The cassette was inserted into the 5′ end of the SmaI site of pUC19 vector by blunt end ligation to generate the expression plasmid pUCexdFGFR2IIIc. The E. coli BL21(DE3) strain C41 was used for expression (14). To synthesize exdFGFR2IIIc, 1 liter of LB medium was inoculated with an overnight culture of the transfected cells grown to an A660 of 0.05 and incubated with shaking at 37 °C. When the culture reached an A660 of around 0.7, protein expression was induced with isopropyl-β-d-thiogalactoside (1 mm). Five hours later the cells were harvested by centrifugation, resuspended in 50 mm Tris-HCl (pH 8), 0.5 m KCl, and 5 mm EDTA and lysed by sonication. The insoluble material was recovered by centrifugation (15,000 × g), and it was resuspended in wash buffer (50 mm Tris-HCl (pH 8.0), 2 mm dithiothreitol, 0.5 m KCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.5% Sulfobetaine-12) before recovering the insoluble material by centrifugation as above. The insoluble material recovered was dissolved in buffer (50 mm Tris-HCl (pH 8.0), 2 mm dithiothreitol, 5 m guanidinium HCl), loaded onto a 20-ml Ni2+-chelating column (HisTrapFF HP, GE Healthcare), and subjected to chromatography using a 0–0.5 m gradient of imidazole in 20 column volumes of the same buffer. The material eluted almost exclusively as a single peak that corresponded to pure exdFGFR2IIIc on SDS-PAGE analysis. A yield of 100 mg of protein per liter of culture was estimated at this stage. The purified exdFGFR2IIIc was diluted to an A280 of 0.24 in 50 mm Tris-HCl (pH 8.0), 2 mm dithiothreitol, and 5 m guanidinium HCl, and it was stored at −80 °C in 30-ml aliquots. For protein refolding, an aliquot (30 ml) was dialyzed against a linearly increasing volume (70–5000 ml, 17 h, 277 K) of 20 mm Hepes (pH 7.5), 150 mm NaCl, and 0.1 mm reduced and 1 mm oxidized glutathione. In this way the guanidine levels decreased slowly at the critical concentrations so that the protein could reshuffle and find its native conformation. The dialysis process was repeated a second time using the same buffer except that the glutathione was omitted. At the end of the whole process the guanidine concentration was ∼1 mm, and protein recovery after the refolding process was consistently >90%. ÄKTAprime and ÄKTAdesign chromatographers (GE Healthcare) were used for protein purification and analysis, respectively.
Proliferation Assays
The assays to test the inhibition of FGF-driven mitogenesis using murine Balb/c 3T3 fibroblasts have already been described (26). The FGF-1 and myo-inositol hexasulfate concentrations were 0.64 ng/ml and 100 μg/ml, respectively. When FGF-2 was tested, 1 ng of the growth factor and 10 μg of heparin per ml were used. Cells were counted by measuring the total amount of crystal violet fixed by cell nuclei by differential absorbance (38). The mitogenesis assays with FR1c-11 cells (kindly provided by D. M. Ornitz, Washington University) have been described previously, except that a single FGF-1 concentration was used (160 ng/ml) in the experiments reported here, and the cells were pretreated before the assay by overnight incubation in the culture conditions at 220,000 cell/ml, as detailed under “Results” (7). Just before the assay, the pretreated cells were diluted ∼10-fold with culture medium (without interleukin-3), they were packed by centrifugation to a volume of ∼50 μl, and they were again suspended in 10 ml of the same medium. After repeating the last washing step three times, the suspension was brought to a final concentration of 220,000 cells/ml. The cell number was determined at the end of the assay by monitoring the reduction of 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide (XTT; Roche Applied Science, cell proliferation kit II) by differential absorbance. All the mitogenesis assays were carried in quadruplicate. Deviation bars represent the S.E.
Migration Assay
Inhibition of FGF-induced Balb/c 3T3 fibroblast migration in culture was tested as described previously (39). Briefly, fibroblasts cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated calf serum, 2 mm l-glutamine, 103 units of penicillin/ml and 12 mg/ml gentamicin were seeded in the same medium at a density of 25 × 103 cells onto glass coverslips covering the bottom of 24-well culture plates. Once the cells reached confluence, a linear wound across the diameter of the coverslip was drawn with a rubber cell scrapper, and the medium was changed to Ham's F-12/Dulbecco's modified Eagle's medium (1:3) supplemented with a 1/100 (v/v) dilution of culture supplement ITS+ (Collaborative Research), 2.5 mm l-histidine, 2 mm l-glutamine, 50 mm ethanolamine, 0.1 mg/ml myo-inositol hexasulfate, 103 units/ml penicillin, 12 mg/ml gentamicin, and the doses of FGF and inhibitor detailed under “Results.” The cultures were fixed 48 h afterward in a 1% solution of glutaraldehyde, and they were stained with crystal violet as described (36). Each treatment was tested in three different cultures, and the total number of migrating cells in the entire wound was determined by manual counting under a light microscope. Only cells with a migratory phenotype were considered.
Angiogenesis Assays
Sterile gelatin sponges (1 cm3; Curaspon Dental, Clinimed Holding, Zwanenburg, The Netherlands) were implanted subcutaneously into the dorsal region of the neck in 10 intraperitoneally anesthetized Sprague-Dawley rats. The sponges were moistened with 200 μl of phosphate-buffered saline containing 25 μg/ml heparin and 10 μg/ml FGF-1. After implanting the sponges, the rats were randomly assigned to groups (n = 5) that received treatment by gavage with either the inhibitor (150 mg/kg twice daily) or the vehicle alone (tap water). After 7 days, the sponges were removed, photographed, fixed by immersion in 4% paraformaldehyde, embedded in paraffin, sliced in 6-μm sections, and stained with hematoxylin-eosin. The number of functional blood vessels (those containing erythrocytes) in six random fields (0.176 mm2) per sponge was determined by morphometric analysis (Motic Images Advanced 3.0, Xiamen, China) and averaged.
Progression of Subcutaneously Implanted Rat Gliomas
Rat C6 glioma cells were cultured as described previously (40). C6 cells cultured to confluence in 75-cm2 flasks were removed and implanted under the abdominal skin (5 × 105 cells in 10 μl of culture medium) of anesthetized rats according to the procedures described previously (41). Those rats that developed a tumor in the same area 5 days after implantation of the tumor cells were randomly assigned for daily intraperitoneal injections (300 μl) of either the inhibitor (100 mg per kg of animal; n = 6) or the vehicle alone (0.9% NaCl; n = 6). After 10 days of treatment, the subcutaneous gliomas were removed, and their volume was calculated according to the formula V = 4/3π(L/2)(A/2)2, where L is the larger diameter, and A is the smaller diameter, both expressed in mm.
Crystallization, Data Collection, Structure Determination, and Refinement
Crystals of the complex between FGF-1 and the inhibitors were grown using the sitting drop vapor diffusion method at 295 K. Equal volumes of protein (0.75 mm) and inhibitor (1.5 mm) solutions were mixed with drops containing 60% sodium/potassium tartrate buffered with 5 mm sodium phosphate (pH 7.8). The drops were equilibrated against 200 μl of 1.3 m Li2SO4, and typically the crystals grew to ∼0.7 × 0.5 × 0.2 mm within 2 weeks.
For the diffraction experiments, the crystals were flash-frozen in liquid N2 after transfer to a cryoprotectant buffer: 20% (v/v) glycerol and 60% sodium/potassium tartrate buffered with 5 mm sodium phosphate (pH 7.8). Complete data sets were collected on CCD detectors at 100 K at the European Synchrotron Radiation Facility (Grenoble), beamline BM14. The diffraction images were processed with MOSFLM (42) and scaled with the CCP4 suite (43). The crystals belong to the monoclinic system with space group P2. The structure was solved by molecular replacement with the AMoRe software (44) using the coordinates of the native FGF-1 (PDB code 1axm) as a search model (45). Using this method, the position of six protein molecules could be located by molecular replacement searches. With six protein molecules in the asymmetric unit, the Matthews coefficient was calculated to be 2.4 Å3 Da−1, and the solvent content was 48.0% (46). Electron density visualization and model building were carried out with COOT (47). After rigid-body and simulated-annealing torsion-angle refinement, the position of the inhibitors was clearly defined at the heparin binding site from the electron density maps. Furthermore, inspection of the initial sigma-A maps showed that a few regions were disordered, although some electron density was obvious. This problem was overcome by the use of non-crystallographic symmetry restraints that were then removed in the final refinement steps. The topology and parameter values for the 2,5-dihydroxyphenylsulfonate (2,5DHPS) and GA ligands were generated using the Dundee PRODRG2 server (48). Several rounds of simulated annealing and B factor refinement with CNS were combined with model rebuilding in COOT after inspection of both the 2Fo − Fc and Fo − Fc maps (49). The coordinates for the final model have been deposited at the Protein Data Bank under the codes 3JUT and 3K1X for FGF-1 bound to GS and 2,5DHPS, respectively.
Nuclear Magnetic Resonance Spectroscopy
Two-dimensional 1H,15N HSQC spectra were acquired at 298 K in a Bruker AVANCE 500 MHz spectrometer. The number of acquired complex points were 1024 in t2 (1H) and 128 in t1 (15N) with spectral widths of 6009.61 (12 ppm) and 2534.18 (50 ppm), respectively.
Saturation transfer difference (STD) experiments were carried out using the same equipment. The spectra were acquired at 298 K with 4096 scans, and selective protein saturation was achieved with a train of 40 Gauss-shaped pulses of 50 ms and a delay between pulses of 1 ms. The on-resonance irradiation of the protein was performed at 0 ppm, and the off-resonance irradiation was set at 50 ppm. Both on- and off-resonance spectra were subtracted through phase cycling. As a control, a STD experiment was carried out in which exdFGFR2IIIc was omitted from the solution.
RESULTS
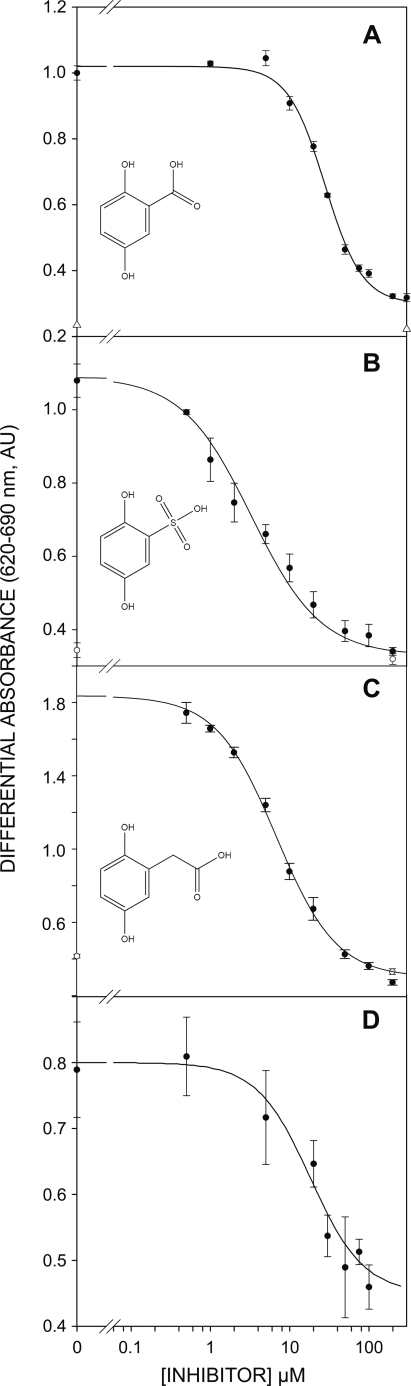
The potential inhibition of FGF mitogenic activity by GA was first assayed in vitro using FGF-1 and Balb/c 3T3 fibroblasts (26, 38), where it displayed a half-maximum inhibitory activity (I50) of ∼36 μm (Fig. 1A). This value was considerably lower than that reported for other inhibitors we have used as lead compounds (26, 38), the best of which was 5A2NMS with an I50 of ∼265 μm. In the absence of FGF, the number of quiescent cells did not vary at the concentrations of GA tested (Fig. 1A, open triangle). Hence, the decrease in cell number observed in the cultures challenged with FGF-1 did not seem to be due to the survival of the cells but, rather, to a decrease in the specific activity of the mitogen.
FIGURE 1.
Inhibition of the FGF-driven mitogenesis in cultures of Balb/c 3T3 fibroblasts by 2,5- dihydroxyphenyl acids. A, GA. B and D, 2,5DHPS. C, HGA. A–C, FGF-1. D, FGF-2. AU, absorbance units.
The full set of GA isomers is commercially available, and thus, we carried out a systematic exploration of the effects of the different relative positions of the two hydroxyls on the inhibition of FGF-1 mitogenic activity. Surprisingly, we found that the entire set of isomers tested was inactive at concentrations three times the I50 of GA. Neither inhibition nor toxicity was evident at the GA isomer concentrations tested (supplemental Fig. S2). Acetylsalicylic acid, a precursor of GA in animals (32), was also inactive at concentrations up to 1 mm. We also studied the effect of substituting the acidic group of GA. The effect of the substitution of the carboxylic group by an amino group could not be tested because the resulting compound was too unstable to use in these mitogenic assays. The elimination of the acidic group converts GA to hydroquinone, a compound with well documented toxicity that strongly reduces cell viability in these assays at low micromolar concentrations. Consequently, we could only test the substitution of the carboxylate group with other functional acidic groups. Substitution of carboxylate by phosphonate caused a consistent but not dramatic decrease in the I50 (typically 24 μm). Nevertheless, a considerable decrease in the I50 of approximately 1 order of magnitude (3 μm versus 36 μm) was observed when this group was replaced by sulfonate (2,5DHPS, Fig. 1B). HGA is an intermediate compound in tyrosine catabolism that reaches abnormally high levels in people affected by the recessive genetic disease known as alkaptonuria. In this compound an acetate group substitutes the carboxylate of GA. HGA inhibits FGF-1 with an I50 of 6.8 μm (Fig. 1C), similar to the activity of 2,5DHPS. With both compounds, the number of quiescent cells also remained relatively constant at the concentrations of inhibitor tested (Fig. 1, open symbols).
Atomic Structure of FGF-1 Bound to 2,5DHPS and GA
Inhibition of the mitogenic activity of FGF by 2,5-dihydroxyphenylic acids seems to be exquisitely tuned. To gain insight into the molecular basis of this interaction, FGF-1 was crystallized in the presence of two of these inhibitors, the least active GA and the most active 2,5DHPS. The structure of these complexes was then solved by x-ray diffraction analysis.
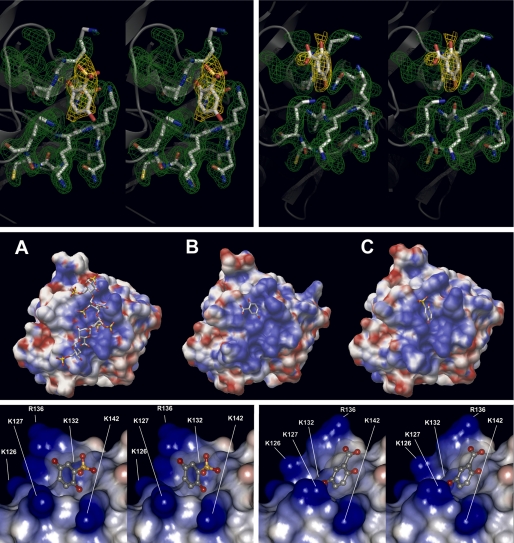
Monoclinic crystals of the complex between FGF-1 and these inhibitors were obtained in solutions containing sodium potassium tartrate as the main precipitant, which reached their maximum size in about 2 weeks. These crystals belong to the P2 space group, containing six molecules in the asymmetric unit, and they diffracted up to 2.0 Å under synchrotron radiation. The structures were solved by molecular replacement as described under “Experimental Procedures.” In the final models, the majority of the residues were located in the most favored regions of the Ramachandran plot. The data collection, structure determination, and refinement statistics are summarized in Table 1. As already observed for the 5A2NMS/FGF-1 structure (26), GA and 2,5DHPS were not detected in four of the six molecules of the asymmetric unit, probably due to steric clashes from the neighboring symmetrically related molecules. In the other two molecules a very clear electron density of about 2σ was observed, into which 2,5DHPS and GA could easily be modeled. After the final refinement both inhibitors and the neighboring protein residues were clearly defined and oriented in the 2Fo − Fc electron density maps (Fig. 2, top row).
TABLE 1.
Data collection and refinement statistics
ASU, asymmetric unit.
| FGF-1/2,5DHPS | FGF-1/GA | |
|---|---|---|
| Data collection statistics | ||
| Space group | P2 | P2 |
| Cell dimensions (Å, °) | ||
| a = 97.0 | a = 97.7 | |
| b = 47.3 β = 106.7 | b = 47.7 β = 106.4 | |
| c = 97.9 | c = 98.4 | |
| No. molecules in ASU | 6 | 6 |
| Wavelength | 0.979 | 0.979 |
| Resolution (Å)a | 30.01-2.0 (2.13-2.00) | 47.78-2.20 (2.33-2.20) |
| Measurements | 615,365 | 324,202 |
| Unique reflections | 63,239 | 42,342 |
| Rmerge (%)a | 5.9 (42.8) | 8.2 (32.8) |
| I/σ(I)a | 8.2 (2.5) | 11.2 (4.5) |
| Completenessa | 98.4 (88.6) | 95.4 (94.6) |
| Refinement statistics | ||
| Resolution range (Å) | 30.01-2.00 | 37.01-2.20 |
| Reflections (work/free) | 57,645/5,594 | 36,289/1,910 |
| Rwork/Rfree (%) | 22.7/27.31 | 21.88/27.40 |
| Root mean square deviation | ||
| Bond lengths | 0.021 | 0.026 |
| Bond angles | 1.88 | 2.01 |
| No. atoms | 6,366 | 6,272 |
| Protein | 6,226 | 6,226 |
| Ligand | 24 | 22 |
| Solvent | 116 | 24 |
| Average B factor (Å2) | 46.84 | 38.52 |
| Ramachandran analysis; favored regions/allowed regions/outliers (% of residues) | 93/6/1 | 94/5/1 |
a Values are for the last resolution shell.
FIGURE 2.
Binding of GA and 2,5DHPS to FGF-1. Top row, stereo diagrams of the 2,5DHPS (left) and GA (right) FGF-1 binding sites show the electron density maps for the inhibitors and their adjacent side chains. Electron densities, corresponding to the final 2Fo − Fc maps are shown at the 1σ contour level. Molecular models of the side chains and the inhibitors are represented by solid sticks (white, carbon; red, oxygen; blue, nitrogen), and the protein backbone is shown as a transparent gray schematic. The figure was produced using the PyMOL program (DeLano Scientific LLC). Middle row, shown are molecular surface representations of FGF-1 complexed with heparin (A) (45), GA (B), and 2,5DHPS (C). The ligands are shown as solid sticks colored as above. The electrostatic potential of the protein surface mapped in blue (positive) and red (negative) was generated using the same parameters in each of three cases with the APBS program (89). Bottom row, shown are stereoviews of the three-dimensional structure of the FGF-1 binding site for 2,5DHPS (left) and GA (right). Both inhibitors, shown as stick-and-ball models (gray, carbon; red, oxygen; yellow, sulfur) appear inside the semitransparent representation of their van der Waals volume. The electrostatic potential of the protein surface is mapped in blue (positive) and red (negative), and it was generated with the Discovery Studio Visualizer program (2.0.1.7347; Accelrys Software Inc.) using its internal parameters to calculate the electrostatic potential.
Inhibitor Binding Site
Both 2,5DHPS and GA bind in a 1:1 stoichiometric ratio to a positively charged cavity at the surface of FGF-1, part of the long cationic channel that constitutes the FGF-1 heparin binding site (Fig. 2, middle row) (45). The figure clearly suggests that strong steric hindrances prevent the simultaneous binding of more than one ligand to FGF-1. GA and 2,5DHPS dock at adjacent sites within a small groove that is partially occupied by the Nϵ of Lys-132 and that is formed by residues belonging to the β strand 10 and the loops between β strands 1 and 2 and strands 11 and 12 (Fig. 2, bottom row) (50). Two sulfate groups dwell in this pocket that somehow grasps Lys-132 in the reported crystallographic structure of the FGF-1·heparin complex, those belonging to O2-sulfoglucuronic acid at position 3 (from the reducing end; IDS-3) and the amino group of N,O-6-disulfoglucosamine at position 4 (SGN-4) (illustrated in supplement Fig. S3) (45, 51). This groove is highly conserved between FGF-1 and FGF-2 (at the primary structure level there is only one conservative substitution in eight residues: Lys-127 for Arg-129), and when the two proteins were crystallized in the absence of heparin, the position of the IDS-3 sulfate is occupied by strong anions like phosphate or selenate (illustrated in the supplement Fig. S4) (52–54). Lys-132 seems to be very important to create the specific physicochemical conditions at this groove that are critical for both heparin affinity and mitogenic activity of FGF-1. Indeed, these characteristics are strongly diminished when Lys-132 is either methylated or substituted by a non-cationic residue (8, 55). The aromatic ring of 2,5DHPS occupies an intermediate position between the binding sites for the two heparin sulfates that surround Lys-132, whereas that of GA is approximately situated at the position of the sulfate of SGN-4 (Fig. 2, bottom row).
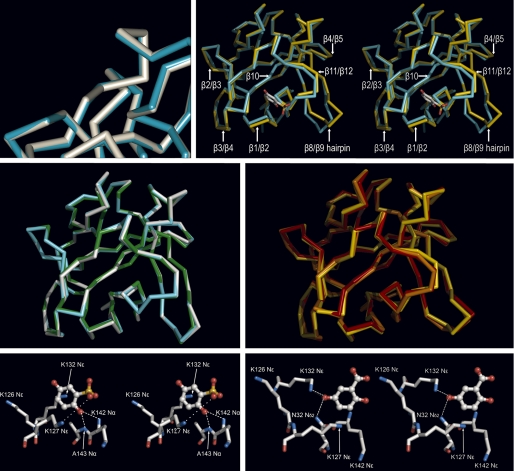
The binding of GA and 2,5DHPS causes specific rearrangements of the side chains of the residues at the binding site that do not affect to their amide backbone (Fig. 2, bottom row, and Fig. 3, top row, left). It should be noted that when the amide backbones of FGF-1 bound to 2,5DHPS and GA are superimposed, they match closely (Fig. 3, middle row, left). Furthermore, as shown in the same figure, a similar close match was observed when the backbone of FGF-1 bound to 5A2NMS was superimposed on those of the protein bound to the two former inhibitors. This result was unexpected, because although 5A2NMS binds to the same site than 2,5DHPS and GA, it is a compound chemically unrelated to them, and it docks in a totally different fashion (26).
FIGURE 3.
Binding of GA, 2,5DHPS, heparin, and 5A2NMS to FGF-1. Top row, left, shown is a superimposed three-dimensional structure of the FGF-1 backbone bound to 2,5DHPS (cyan) and GA (white) at the inhibitor binding site represented in Fig. 2; right, shown is a stereoview of the superimposed three-dimensional structure of the FGF-1 backbone bound to 2,5DHPS (cyan) and heparin (yellow) (45). For orientation purposes 2,5DHPS is represented at its binding site as a stick model, with the carbon, oxygen, and sulfur atoms colored white, red, and yellow, respectively. Representations were generated with the Discovery Studio Visualizer program in the first case and with the PyMOL program in the latter. In both cases the backbones of the superimposed molecules were reciprocally oriented to a minimal root mean square deviation with the program used to generate their representations. Displacement of the backbone at the level of the β3/β4 loop is not properly appreciated at the figure because the perspective of the drawing. Middle row, left, shown are superimposed three-dimensional structures of the amide backbone of FGF-1 bound to 2,5DHPS (cyan), GA (white), and 5A2NMS (green). Middle row, right, shown are superimposed three-dimensional structures of the amide backbone of FGF-1 bound to heparin (yellow) and incorporated into two different models of the FGF-1·FGFR2 complexes (orange, symmetric; red, asymmetric). The complex also includes heparin in the case of the asymmetric model (45, 51, 57). Superimposed molecules were reciprocally oriented to a minimal root mean square deviation and represented using PyMOL. Bottom row, shown is a network of non-covalent interactions between FGF-1 and 2,5DHPS (left) and GA (right). The ligands and the protein are shown as stick-and-ball models, respectively, with the atoms colored as above. Superimposed molecules were reciprocally oriented to a minimal root mean square deviation and represented using PyMOL.
Both 2,5DHPS and GA appear stabilized at their binding site by a network of non-covalent contacts. In the case of 2,5DHPS, the aromatic ring establishes a canonical π-cation interaction with the Nϵ of Lys-132 (56); the negatively charged sulfonate group establishes a salt bridge with the Nϵ of Lys-127; the hydroxyl group at position 2 establishes two simultaneous directional hydrogen bonds with the Nα of Lys-142 and the Nα of Ala-143, respectively (Fig. 3, bottom row, left). GA occupies a position with different coordinates despite its similarity with 2,5DHPS and although it docks at the same FGF-1 cleft as 2,5DHPS. There, its hydroxyl group at position 5 establishes simultaneous hydrogen bonds with the Nδ2 of Asn-32 and the Nϵ of Lys-132 (Fig. 3, bottom row, right).
Structural Differences between Heparin and 2,5DHPS-bound FGF-1
There are obvious differences between the structure of the protein complexed with either GA or 2,5DHPS and that activated by heparin (45). First, there is a general relocation of the lateral cationic chains that form the heparin binding site, an elongated channel that is probably partially induced by the even distribution of strong negative charges along the longitudinal axis of heparin. This channel practically disappeared in the case of the inhibitors, as those cationic side chains clustered around a more punctually localized charge (Fig. 2, middle row). When the structures of FGF-1 complexed to the inhibitor or heparin were superimposed (Fig. 3, top row, right), there were small displacements of the loops connecting β-strand 1 with 2 and strand 11 with 12, which pertain to the inhibitor binding site (β-strand 10, the third secondary structure element of the binding site, did not show any appreciable displacement). As shown in the same figure, alterations of regions unrelated to this site (e.g. loops β2/β3, β3/β4, β4/β5, β strand 8/9 hairpin) were also observed, more dramatically in certain loops. It should be noted that in the four molecules of the asymmetric unit where either 2,5DHPS or GA were not detected, the backbone of the β strand 8/9 hairpin adopted different positions intermediate to that of inhibitor or heparin-bound FGF-1. Moreover, a general loss of electron density in the side chains was observed that did not permit most of them to be traced. Consequently, it seems that the β strand 8/9 hairpin is disordered in the absence of ligands. A similar situation was observed when the structure of FGF-1 bound to 5A2NMS was resolved (26).
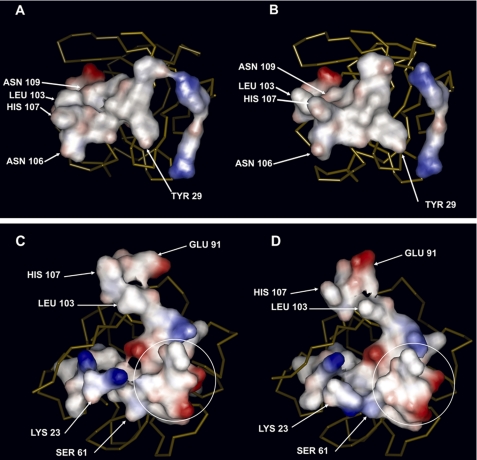
According to the three-dimensional structures currently available, FGFs interact directly with the D2 and D3 immunoglobulin domains of their FGFRs. In the case of FGF-1, two different architectures of the FGF·FGFR complex have been proposed, known as the symmetric and asymmetric models (51, 57), whereas only the symmetric one has been described for FGF-2 (58). Assembly of the asymmetric model necessarily requires heparin to integrate into the complex, which is not the case of the symmetric model (51, 57, 58). In the case of the symmetric model, studies carried out with FGF-2 show that the incorporation of heparin does not cause dramatic changes in the interaction between the growth factor and its receptor (59). However, although the three-dimensional structure of a symmetric FGF-1·FGFR·heparin complex is not yet available, if such a complex exists, it seems reasonable that the interactions between its polypeptide units would not be very different to those in the absence of heparin, given the strong similarities between the symmetric FGF-1·FGFR and FGF-2·FGFR models (60). The ensuing discussion will be carried out on the basis of the interaction of FGF-1 with FGFR2, as to date it constitutes the only complex for which data are available for both the symmetric and asymmetric models (51, 57). In both models, the interaction of FGF-1 with the D2 domain is the same. The tip of the β strand 8/9 hairpin is fundamental in this interaction of FGF-1 with the D2 domain, as 6 of the 13 amino acids that interface with the receptor belong to this region. As already mentioned, this region has a different relative position with respect to the rest of the molecule when FGF-1 is bound to heparin and to GA or 2,5DHPS. Accommodating this displacement necessarily affects the side chain of the neighboring residues, causing a clear distortion of the whole interface (Fig. 4, top row). In the case of the D3 domain, the FGF-1 interfaces that recognize the receptor are different in the symmetric and asymmetric models, although they share a common region at the vertex of what is the approximately a right angle defined in the FGF-1 surface by both interfaces when they are represented simultaneously (encircled in Fig. 4, bottom row). The general rearrangement imposed by the alteration of the heparin binding site when 2,5DHPS and GA bind to FGF-1, in addition to the displacement of the β strand 8/9 hairpin, also affects the interfaces of FGF-1 with the FGFR D3 domain in both the symmetric and asymmetric model (Fig. 4).
FIGURE 4.
Surface defined by the amino acids of FGF-1 that interface with FGFR2, in the case of the symmetric model of the complex and of the asymmetric model, when bound to heparin and 2,5DHPS, respectively. Top row, the surface is defined by the amino acids in FGF-1 that interact with the D2 immunoglobulin domain. These residues are the same in the asymmetric and symmetric three-dimensional structures of the FGF-1·FGFR2 complex (51, 57). A, FGF-1 bound to heparin is shown. B, FGF-1 bound to 2,5DHPS is shown. In both cases the protein is oriented identically, as the backbone trace (yellow) shows. The following amino acids of FGF-1 interact with the D2 immunoglobulin domain: Tyr-29 (β strand 1); Arg-49 (β strand 2); Arg-51 (loop β strands 2/3); Glu-101, Leu-103 (β strand 8); Asn-106, His-107 (loop β strands 8/9); Tyr-108, Asn-109 (β strand 9); Leu-147, Pro148; Leu-149, Pro-150 (β strand 12). Bottom row, shown is a three-dimensional representation of the surface defined by the whole set of amino acids of FGF-1 that interact with the D3 immunoglobulin domain in the asymmetric and in the symmetric three-dimensional structures of the FGF-1·FGFR2 complex. C, FGF-1 bound to heparin is shown. D, FGF-1 bound to 2,5DHPS is shown. In both cases the protein is identically oriented, as the backbone trace (yellow) shows. Circled residues are common to the interfaces of the symmetric and the asymmetric models. The interface with the D3 domain includes the residues circled plus those on top of them in the case of the asymmetric model and those to their left in the case of the symmetric one. The following amino acids are specific to the interface with the D3 domain in the asymmetric model: Leu-60, Arg-102, Leu-103, Glu-105, Asn-106, His-107, and Ile-112. Those specific to the symmetric model are: Tyr-22, Lys-23, Pro-25, Lys-26, Leu-28, and Tyr-69. The following residues are common to the interface in both models (circled): Ser-61, Ala-62, Glu-63, Ser-64, Val-65, Gly-67, Val-68, Ile-70, and Glu-101. Arrows point toward the residues most displaced when FGF-1 binds 2,5DHPS instead of heparin. The figure was generated, and the surfaces are colored according the electrostatic potential (red, negative; blue, positive) using Discovery Studio Visualizer and its internal parameters.
Inhibition of Other Members of the FGF Family
The crystallographic data clearly suggested that 2,5DHPS should also inhibit FGF-2-induced mitogenesis, as the 2,5DHPS binding site in FGF-1 is highly conserved in FGF-2 (see above), and they both show a high affinity for strong anions (52–54). Indeed, 2,5DHPS inhibits FGF 2-driven mitogenesis with an I50 of 19.1 μm (Fig. 1D). It should be pointed out that the rate of mitogenesis elicited by both FGF-1 and FGF-2 at inhibitory concentrations of 2,5DHPS corresponds to that elicited by the mitogen in the absence of heparin (that in the case of FGF-1 is practically unappreciable) (61). Thus, inhibition by 2,5DHPS cancels out the effect of sulfated glycosaminoglycans on FGF-driven mitogenesis (9, 62, 63). Given the homologies in primary structure, it would not be totally unexpected if FGF-3, FGF-4, FGF-8, FGF-9, FGF-16, and FGF-17 were also inhibited by 2,5DHPS and GA, as there is substantial conservation in the binding site between FGF-1 and each of these other family members, although not as tight as with FGF-2.
Stability of 2,5DHPS in the Presence of FGF-1
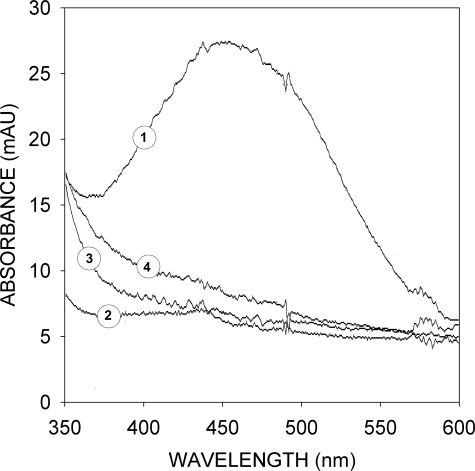
GA and 2,5DHPS bind to a site in FGF with a particularly high specific affinity for anions (Fig. 2). At the pH of the mitogenesis assays (pH 7.5), the solutions of both compounds swiftly turned yellow, an effect that could be reversed by acidification to a pH of ∼5.5. Subsequently, the solutions slowly acquire a slight brown color due to the appearance of a broad absorbance band reflecting the spontaneous appearance of a heterogeneous assortment of polyaromatic compounds due to quinone oxidation (Fig. 5). At pH values above 9, the process was considerably faster, and the solution became dark brown. Nevertheless the process slows down rapidly and spontaneously unless the solution is strongly buffered, as the formation of the brown pigment causes a quick drop in pH. Evidently, quinone oxidation would be accelerated considerably in a basic environment such as that at the FGF site to which GA and 2,5DHPS bind, unless their anionic form is stabilized by the opposing charges that surround them. This would seem to be the case for 2,5DHPS bound to FGF-1 as, in the presence of equimolar concentrations of this protein, degradation products do not accumulate in the 2,5DHPS solution after 48 h at the pH and ionic strength of the mitogenesis assays (Fig. 5). This would suggest that the ionized hydroquinone of GA and 2,5DHPS actually substitutes for the anions found at their binding site in the three-dimensional structures of both FGF-1 and FGF-2, either free or complexed to heparin (45, 51–54). Kinetic coupling between hydroquinone unprotonation and binding should make the later effect quite cooperative.
FIGURE 5.
Stabilization of 2,5DHPS in solution by FGF-1. Absorbance spectra are shown of 2,5DHPS (100 μm) in the presence (2 and 4) and absence (1 and 3) of an equimolar concentration of FGF-1, recorded just after the preparation of the solution (spectra 3 and 4) and 48 h later (spectra 1 and 2). Solutions were at the pH (7.5) and ionic strength (20 mm sodium phosphate, 75 mm NaCl) of the mitogenesis assay cultures. Approximately 10% of 2,5DHPS is lost by oxidation during the 48 h of incubation, according to the 1H NMR spectra of the initial and final solutions. mAU, milliabsorbance units.
Competition between 2,5DHPS and Heparin in Binding to FGF-1 and FGF-2
The analysis of the three-dimensional structure of FGF-1 bound to 2,5DHPS suggests that this inhibitor would compete with heparin for binding to FGF-1 and FGF-2. The pertinent studies confirmed these predictions.
Given the important structural similarities of the GA·FGF-1 and 2,5DHPS·FGF-1 complexes, studies were carried out with 2,5DHPS, the strongest of the two inhibitors. Competitive binding of 2,5DHPS and heparin to FGF was tested by heparin affinity chromatography. As expected, there was a decrease in the protein retained as the heparin concentration in the FGF-1 solution injected onto the column increased (Fig. 6A). Likewise, adding 2,5DHPS to the FGF solution produced an equivalent effect to heparin, both with FGF-1 and FGF-2, albeit at higher concentrations of the competitor in both cases (Fig. 6A). It should be pointed out that in all cases the ionic strength of the FGF solution in the presence of 2,5DHPS was considerably lower than that required to elute the protein from a heparin affinity chromatography column (12). Consequently, injection of FGF-1 onto the column (which has less affinity for heparin than FGF-2) in a solution containing 500 mm NaCl (a salt concentration that raised the conductivity of the solution to 30 millisiemens/cm and that was equivalent to that of the highest 2,5DHPS solutions used in the experiment) did not affect protein retention (data not shown). It should be also noted that the 2,5DHPS solution was prepared just before the mixture was to be injected onto the column. This competition between 2,5DHPS and heparin was then also tested in mitogenesis assays at subsaturating concentrations of heparin (Fig. 6B, first against eighth bars). The results show that 2,5DHPS decreased the rate of mitogenesis to a level similar to that of quiescent cells (bar labeled MM), which could be compensated by an increase in the heparin concentration in the assay.
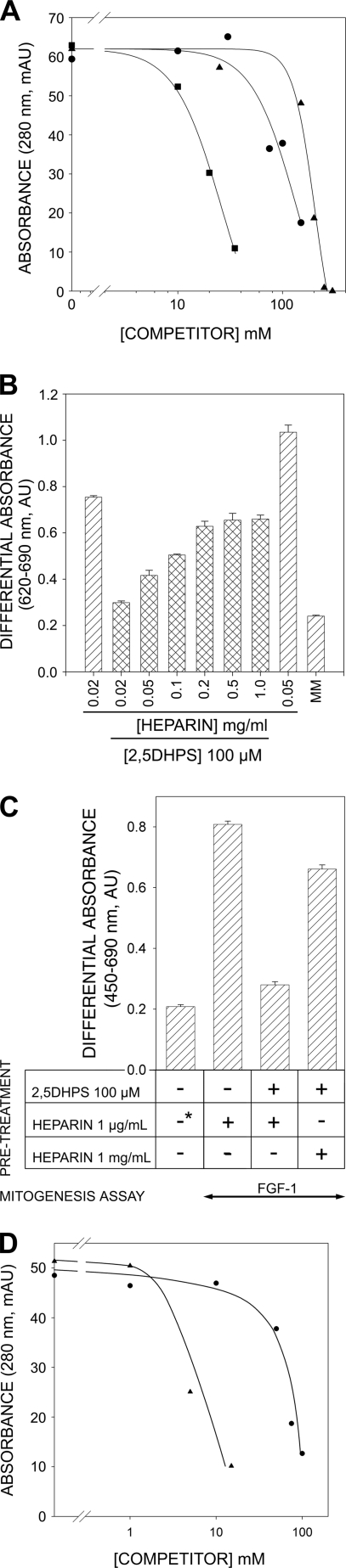
FIGURE 6.
Competition between heparin and 2,5DHPS. A, shown is the amount of FGF retained on a 1-ml HiTrapTM heparin HP column (GE Healthcare) when co-injected with either heparin (■) or 2,5DHPS (▴, ●). The amount of protein retained is represented by the height of the peak eluted with a 5-min linear gradient of 300 to 1.5 m NaCl in the chromatography buffer after post-injection column re-equilibration (20 mm sodium phosphate, 300 mm NaCl, pH 7.2). The flow rate was 1 ml/min. The protein (13 μm in 250 μl; ■, ▴, FGF-1; ●, FGF-2) was co-injected in the presence of different concentrations of the competitor, as plotted in the graph. mAU, milliabsorbance units; B, shown is reversion of the inhibition of the mitogenic activity of FGF-1 by 2,5DHPS at increasing heparin concentrations. The mitogenesis assay was carried out as described under “Experimental Procedures” using murine Balb/c 3T3 fibroblasts. C, FGF-1-driven mitogenesis of FR1c-11 cells preincubated as indicated in the figure and under “Experimental Procedures” (*, equivalent levels of mitogenesis were observed when 1 μg/ml of heparin was included in the pretreatment). D, shown is the amount of exdFGFR2IIIc retained in a HiTrapTM Heparin HP column (GE Healthcare) when co-injected with increasing concentrations of heparin (▴) or 2,5DHPS (●) using the chromatographic conditions described above.
2,5DHPS Competes with Heparin for Binding to FGFRs
The FGF receptors share a highly conserved heparin binding site that is essential for FGF-stimulated cell growth (64). Accordingly, we explored whether 2,5DHPS interacts with FGFRs and if this interaction interferes with FGF driven mitogenesis. A series of experiments were carried out with FR1c-11 cells, a heparin-less lymphoid cell line stably transfected with the full three-Ig domain murine FGF receptor 1 (splice variant IIIc (65–67)). Pretreatment with 2,5DHPS caused an almost complete inhibition of FGF-1-induced cell division, even after an extensive washing of the cells (“Experimental Procedures” and Fig. 6C). However, this effect was overridden by increasing the concentration of heparin during the preincubation with 2,5DHPS. It is obvious that a dual inhibitory target, the growth factor and its receptor, constitutes a kinetic feature of FGF-driven inhibition of mitogenesis inhibition by 2,5-dihydroxyphenylic acids that has to be taken into account. Moreover, given the highly conserved heparin binding sites in the four FGFRs, it would not be totally unexpected if all of them were capable of binding these acids, which would obviously considerably widen the spectrum of FGF-driven processes that these 2,5-dihydroxyphenyl derivatives can inhibit (64).
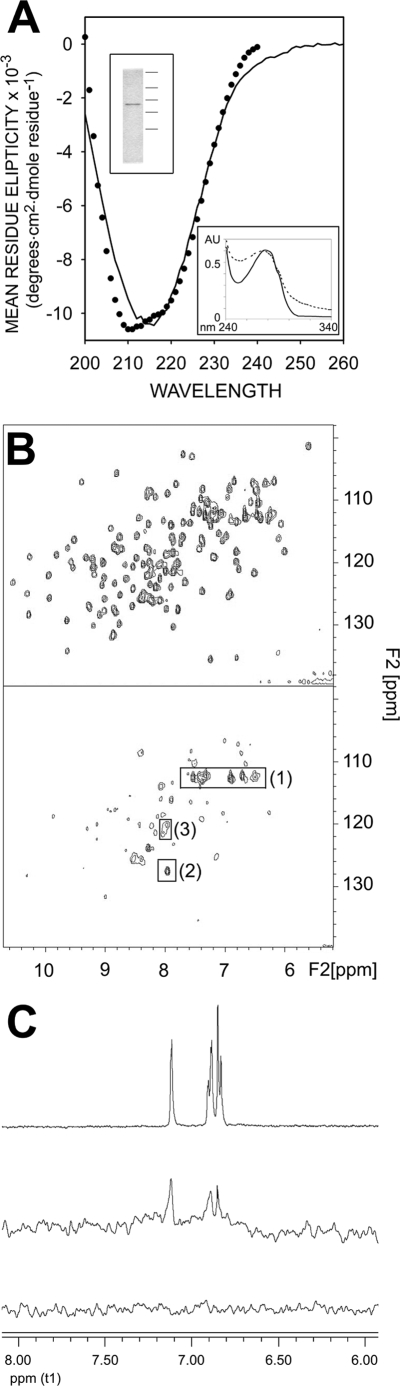
In vitro studies were carried out to further assess the competition between heparin and 2,5DHPS for binding to FGFRs. Thus, the complete extracellular domain of the IIIc isoform of receptor 2 (exdFGFR2IIIc) was synthesized and purified (upper inset of Fig. 7A). This isoform displays equivalent affinity for FGF-1 and FGF-2 (the FGFR variant present in FR1c-11 cells has higher affinity for FGF-1 than for FGF-2) (66). Evaluation of the secondary structure of exdFGFR2IIIc from its UV CD spectrum (Fig. 7A) using an unsupervised learning neural network yielded a β-sheet content of the polypeptide, in agreement with that computed from the crystallographic structures (∼41%; maximum error, 0.087) (51, 57, 60, 68). The low light scattering of the UV spectra at ∼0.5 mg/ml (Fig. 7A, lower inset) and the affinity for heparin (Fig. 6D) both also argue in favor of the correct folding of the synthesized exdFGFR2IIIc (64). Moreover, NMR spectroscopy showed that the exdFGFR2IIIc synthesized recognized FGF-1. The 1H,15N HSQC spectrum of the 15N-labeled FGF-1 backbone was determined in the presence and absence of 50 μm exdFGFR2IIIc (Fig. 7B). The two-dimensional distribution of cross-peaks, already fully characterized, represented a fingerprint of the protein (35). The intensity of the signals in these spectra is inversely related to the size of the protein, as the line width of the signal is inversely proportional to the transverse relaxation time (T2), which decreases as the efficiency of relaxation increases with the size of the molecule. This feature can be used to monitor the formation of complexes between polypeptides. As the lower panel shows, there was a significant line broadening of most of the FGF-1 signals upon the addition of exdFGFR2IIIc, which causes many of them to remain hidden below the spectral noise. The framed cross-peaks corresponded to groups belonging to highly flexible regions of the protein, whose transverse relaxation time remained largely independent of the overall size of the polypeptide (35). Because the position and intensity of these cross-peaks remained practically the same in the upper and lower panels of the figure, the overall decrease in intensity of the rest of the signals cannot be attributed to 15N-labeled FGF-1 coming out of solution upon the addition of exdFGFR2IIIc.
FIGURE 7.
Characterization of recombinant exdFGFR2IIIc. A, circular dichroism spectra in the far-UV region of exdFGFR2IIIc are shown. Closed circles represent the spectrum reconstructed on the basis of the percentage of the secondary structure components of exdFGFR2IIIc obtained by deconvolution of the experimental spectrum (DICHROWEB server (68, 90). Upper inset, Coomassie Brilliant Blue-stained SDS/polyacrylamide electrophoresis gel (15%) (50) of recombinant exdFGFR2IIIc after purification and refolding (the horizontal lines to the right indicate the position of molecular mass markers of 250, 75, 50, 37, and 25 kDa). Lower insert, UV spectra (∼0.5 mg/ml; 20 mm Hepes, 150 mm NaCl (pH 7.5)) of exdFGFR2IIIc before (dotted line) and after refolding are shown. AU, absorbance units. B, shown is a characteristic 1H,15N HSQC spectrum of the backbone of 15N-labeled FGF-1 (150 μm in 20 mm sodium phosphate (pH 7.2), 300 mm NaCl) in the presence (lower panel) and absence (upper panel) of 50 μm exdFGFR2IIIc. Framed 1H,15N cross-peaks correspond to Gln-54 and Gln-91 Nδ and His-138 Nα (1), Asp-154 Nα (2), and Lys-23 and Lys-24 Nα (3). C, shown is an STD spectrum of a solution of 2,5DHPS and exdFGFR2IIIc in 20 mm sodium phosphate, 150 mm NaCl (pH 7.2) at a molar ratio of 1 to 0.05 (middle spectrum). The top record is a 1H NMR spectrum of the aromatic region of a 1 mm solution of 2,5DHPS in the same buffer. The bottom trace is a STD spectrum equivalent to that of the middle trace, except that the protein was omitted. Circular dichroism spectra in the far UV region were obtained using a Jasco 710 spectropolarimeter at 20 °C in a 1-mm path length cuvette. The protein (0.13 mg/ml) was in 50 mm sodium phosphate (pH 7.0), and the spectra were averaged by accumulating four scans. A four-scan averaged spectrum of the buffer was routinely subtracted from the protein spectra.
STD NMR experiments (Fig. 7C) clearly demonstrated that exdFGFR2IIIc interacts with 2,5DHPS, as predicted from the experiments with FR1c-11 cells. Indeed, the characteristic signals of 2,5DHPS were clearly visible in the middle record, indicating that the excitation of the protein was transferred to the inhibitor, and consequently, that it binds to exdFGFR2IIIc (69).
The possibility that both heparin and 2,5DHPS compete for binding to FGFRs was explored using the same approach employed with FGF-1 and FGF-2 (Fig. 6A). Accordingly, 2,5DHPS clearly interfered with the binding of the external domain of the receptor to heparin (Fig. 6D), an effect that again could not be emulated by merely raising the conductivity of the buffer with NaCl to that of the highest 2,5DHPS solution used in the experiment (25 millisiemens/cm).
To determine whether 2,5DHPS could interfere with the binding of FGF to exdFGFR2IIIc, a hybrid C-LYT/aFGF polypeptide was used (36). The mitogenic activity and heparin affinity of this polypeptide was similar to that of FGF-1, whereas through a combination of cation-π and hydrophobic interactions, it also binds strongly to choline analogues, like the anion-exchange group of common chromatography solid phases (diethylaminoethyl- and trimethylammonium-) (36, 70). As hydrophobic interactions predominate in the binding of the C-LYT moiety to these choline analogues, once C-LYT/aFGF is loaded onto such columns, it can only be eluted in the presence of either choline or its analogues in the chromatography buffer but not by raising the ionic strength (36, 70, 71). Thus, C-LYT/aFGF enables common anion exchange columns to be converted into FGF affinity columns from which FGF complexes can be eluted with choline.
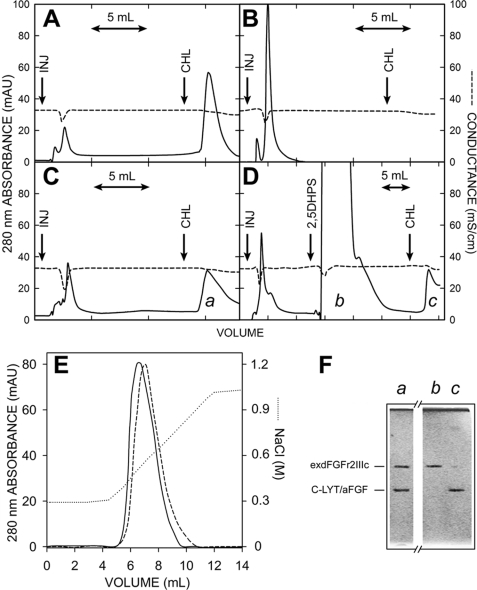
Accordingly, C-LYT/aFGF was eluted from the chromatography column when the choline concentration in the buffer was raised to 150 mm (Fig. 8A), whereas when exdFGFRIIIc was applied, the protein appeared in the flow-through, and nothing was eluted by choline under the same conditions (Fig. 8B). However, when this protein was injected in the presence of equimolar amounts of C-LYT/aFGF, it was retained, and both polypeptides were eluted together in a single peak (Fig. 8C, a) when the elution buffer contained 150 mm choline (see electrophoretogram of this fraction, Fig. 8F, lane a). When 1 ml of a 100 mm solution of 2,5DHPS (prepared just before it was applied to the column in Hepes 20 mm, adjusted to the same pH (pH 7.5) and conductivity (26 millisiemens/cm) as the chromatography buffer) was injected before the choline elution step, C-LYT/aFGF alone was detected in this last eluted fraction (peak c of Fig. 8D and Fig. 8F, lane c of the electrophoretogram). SDS-PAGE of the washout after 2,5DHPS injection confirmed that exdFGFR2IIIc was eluted in this fraction (Fig. 8D, peak b and Fig. 8F, lane b of the electrophoretogram). Similar results were obtained when the protein mixture injected onto the column contained an equimolar amount of heparin. The incorporation of heparin to the C-LYT/aFGF·exdFGFR2IIIc complex under these chromatographic conditions was evident by the higher ionic strength required to dissociate exdFGFR2IIIc in the presence of heparin (Fig. 8E). Such a moderate effect was predicted by the structural crystallography data given the minimal interaction of heparin with the receptor and also by the cell biology experiments (51, 57–59, 72). As C-LYT/aFGF cannot be eluted from the column by raising the ionic strength of the buffer, the chromatographic effect shown in Fig. 8E exclusively reflects the dissociation of the C-LYT/aFGF·exdFGFR2IIIc complex (36, 70).
FIGURE 8.
Chromatography of C-LYT/aFGF·exdFGFR2IIIc complexes. The protein (∼3.2 nmol in each case) was injected in 250 μl of the chromatography buffer at the arrows labeled INJ. The buffer contained 150 mm choline from the mark (arrow labeled CHL) to the end of the chromatogram. A, C-LYT/aFGF is shown. B, exdFGFRIIIc is shown. C, an equimolar mixture of C-LYT/aFGF and exdFGFRIIIc is shown. D, an equimolar mixture of C-LYT/aFGF and exdFGFRIIIc is shown as in C, except that 1 ml of a 100 mm 2,5DHPS was injected onto the column (arrow) before raising the choline concentration of the buffer to 150 mm (in C and D, the peak eluted with choline has a long tail only partially shown in the figure). E, shown is the chromatography of an equimolar mixture (∼3.5 nmol) of C-LYT/aFGF and exdFGFRIIIc (containing 5 mm 3-kDa heparin, in the case of the dashed line) using a NaCl gradient (dotted line; except for this NaCl gradient, the other chromatographic conditions are those used in the remainder of the figure). mAU, milliabsorbance units. F, shown is Coomassie Brilliant Blue-stained SDS/PAGE (15%) of the fractions a, b, and c of the chromatograms C and D. The horizontal lines to the left indicate the migration of C-LYT/aFGF (30.8 kDa) and exdFGFRIIIc (38.7 kDa). Chromatography was carried out on 1-ml HiTrapTM DEAE FF columns (GE Healthcare) at a flow rate of 2 ml/min and a pH of 7.5 (Hepes 20 mm, 300 mm NaCl), conditions chosen to avoid any appreciable ionic interaction of FGF-1 (pI 7.9) and exdFGFR2IIIc (pI 5.8), respectively, with the solid phase of the column.
2,5DHPS Inhibition of Cell Migration
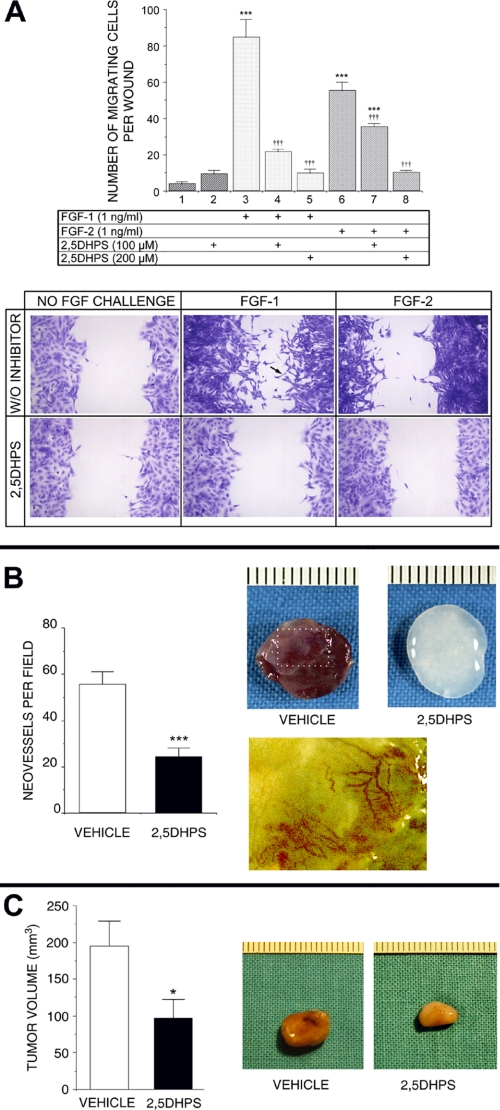
FGF signaling may affect important disease-associated processes other than growth. Given the fundamental importance of cell migration in the development of many important pathologies in which FGFs seem involved, whether 2,5DHPS could inhibit FGF-induced cell migration was evaluated (20–25). The capacity of FGF-2 to induce cell migration of bovine aortic endothelial cells from the edge of a denuded area in a confluent monolayer was demonstrated by Sato and Rifkin (73). This process, characterized by the disruption of the confluent monolayer structure of the culture, the appearance of cells with extended filopodia, and the invasion of the denuded plate area, could also be observed when Balb/c 3T3 fibroblasts cultures were exposed to either FGF-1 or FGF-2 (Fig. 9A). The figure also shows that, in effect, 2,5DHPS clearly inhibited FGF-1- and FGF-2-induced cell migration (I50 ∼100 μm). Complete inhibition was achieved by 2,5DHPS at concentrations close to those that produced a maximal effect in the mitogenesis assays (∼200 μm).
FIGURE 9.
Inhibition by 2,5DHPS of FGF-induced cell migration and of uncontrolled growth in vivo. A, top panel, the effect of different doses of 2,5DHPS on the migratory phenotype induced by either FGF-1 or FGF-2 in wounded confluent cultures of fibroblasts Balb/c 3T3 cells is shown. A, bottom panel, shown are representative photographs of the experiment (2,5DHPS, 200 μm). The black arrow points to a cell with a long filopodium, a characteristic of migrating cells colonizing the denuded area. Data are plotted as the mean ± S.E. of the number of migrating cells invading the wound made in the culture. ***, p < 0.001 versus column 1; †††, p < 0.001 versus columns 3 and 6 for the wells treated with FGF-1 and FGF-2, respectively, as assessed by one-factor analysis of variance followed by the Student-Newmann-Keuls test. B, left plot, shown is the effect of orally administered 2,5DHPS on blood vessel invasion of gelatin sponges soaked in FGF-1 and implanted subcutaneously in rats. B, right panel, shown are representative photographs of the experiment. The image in the second row is an amplification of the boxed area in the image above with the color hue manually modified to better appreciate of the blood vessels. Data are plotted as the mean ± S.E. ***, p < 0.001 versus vehicle by unpaired t test. C, left panel, shown are the effects of 2,5DHPS on the progress of subcutaneously implanted rat gliomas. C, right panel, shown are representative photographs of tumors with a size representing approximately the mean of the plot at the left. The data are expressed as the mean ± S.E. of the volume of excised gliomas at the end of the intraperitoneal treatment. *, p < 0.05 versus vehicle by unpaired t test.
2,5DHPS Inhibits Angiogenesis and Glioma Growth in Vivo
A more holistic preliminary approach to study the effects of 2,5DHPS in vivo was undertaken using two animal models of pathological angiogenesis and tumor growth.
Abnormal unrestrained angiogenesis can be evoked in vivo in rats by implanting collagen sponges soaked in inducers of angiogenesis subcutaneously (74), a model we have used repeatedly to evaluate FGF-induced angiogenesis (26, 75). The model was also used to evaluate the potential antiangiogenic activity of 2,5DHPS using gelatin sponges to permit a more precise histological evaluation. When removed 7 days after their implantation in untreated rats, sponges embedded with FGF-1 had an intensely bloody appearance (left sponge, Fig. 9B), contrasting with the sponges removed from rats to which 2,5DHPS was administered and that displayed no macroscopic evidence of angiogenesis (right sponge). Invasion of the sponge by tortuous blood vessels similar to those observed in tumor angiogenesis could be observed in amplified images after manually modifying the color hue to better appreciate the invading vessels. 2,5DHPS was administered orally in the cases shown in the figure, although the treatment was equally effective when the inhibitor was administered intraperitoneally and when FGF-2 was employed instead of FGF-1.
Prominent vascularization is an outstanding feature of gliomas, a tumor type that accounts for >60% of primary intracranial neoplasms, and this is considered a key characteristic in their malignancy (76). FGF-1 and FGF-2 are abundant in most glioblastomas, and although the former is associated to astrocytes, the latter is associated with the matrix surrounding proliferating blood vessels (77). In addition, they seem to constitute autocrine factors essential for the survival and proliferation of the malignant gliomal cells (78, 79). Combining anti-angiogenic and anti-mitotic agents have been proposed to treat gliomas because the efficacy of antiangiogenic treatments alone remains uncertain (80). The FGF inhibitors described here may fulfill both therapeutic goals. Consequently, gliomas seemed to be an appropriate pathological model to determine whether these compounds may be suitable for pharmacologically use in vivo. To this end, C6 rat glioma cells grown in vitro were implanted subcutaneously into 12 rats in accordance with a well known heterotopic model of glioma (41). Five days after implantation, when the tumors clearly protrude throughout the skin, the rats were randomly divided into two groups that received a daily intraperitoneal injection of either the vehicle alone or 2,5DHPS. The tumors were surgically removed 10 days after the onset of the treatment, and their volume was determined. In Fig. 9C, two tumors with the average size of those extirpated from the treated and untreated animals, respectively, clearly show that 2,5DHPS administration significantly impaired the development of the tumor. Moreover, the decrease in blood irrigation to the tumors extracted from the 2,5DHPS-treated animals was clearly evident from their color.
DISCUSSION
FGFs seem to be involved in a wide assortment of pathologies, and they promote the resistance of tumors to chemotherapy drugs and radiation. Considerable attention has been paid to their involvement in anomalous angiogenesis, as it has gradually become clear that uncontrolled angiogenesis causes or is necessary for the progress of many diseases, including cancer (81). Hence, the pharmacological inhibition of FGFs might be of considerable therapeutic interest.
The critical nature of the relative positions of the hydroxyl groups in the aromatic ring of the new family of FGF inhibitors described here could not have been foreseen in earlier studies. Furthermore, the orientation toward the solvent of the acidic group in GA and 2,5DHPS bound to FGF-1 was quite unexpected from both the three-dimensional structure of 5A2NMS bound to FGF-1, the inhibitor that led us to single out GA as a potential FGF inhibitor (26), and the affinity for strong anions of the groove at the FGF surface where GA and 2,5DHPS dock (45, 51–54). The high affinity for strong anions in this region of FGF clearly suggests that the hydroquinone moiety of both GA and 2,5DHPS fulfills this role in binding to FGF rather than acting as a hydrophobic group and is more suitable to that purpose than the smaller acidic groups of the inhibitors.
The hydroquinone ring of 2,5DHPS slowly oxidizes at the physiological pH of the mitogenesis assays, although it is stabilized in the presence of FGF. It would seem reasonable to assume that the hydroquinone group should remain partially unprotonated under such pH conditions, which when added to the negative charge of its acidic group, should direct 2,5DHPS toward its FGF binding site. Once positioned in this highly basic region, the hydroquinone ring must become fully unprotonated and acquire a markedly anionic character, which should contribute decisively to displacing the acidic group toward the solvent. Encapsulation of the benzene ring as a quinone-like structure inside this highly cationic shell seems to be sufficient to account for the oxidation shielding observed under these conditions. This model could reasonably be extended to the other 2,5-dihydroxyphenylic inhibitors described in this study. The ortho and meta conformations are probably much less appropriate than the para isomer for the dihydroxyphenyl ring to acquire such strong anionic character, which may account for the striking inactivity of the rest of the GA isomers as FGF inhibitors. The differences in the docking of GA and 2,5DHPS at their FGF binding sites were unanticipated. It seems plausible that the cloud of π electrons of the aromatic ring loses the characteristic donut-shape conformation required for canonical π-stacking that is established for 2,5DHPS, due to the close proximity of the resonant carboxylic group in GA (56).
Although initially conceived as a FGF trap and protector, sulfated glycosaminoglycans were later shown to participate in FGFR binding and activation (82). Accordingly, a quite straightforward model can be proposed to explain the inhibitory effects of the dihydroxyphenyl derivatives described here based on the hindrance of heparin interaction with FGFs and FGFRs. However, the crystallographic data and results published elsewhere (26) might lead us to envisage a more sophisticated scenario.
Free FGF-1 is a quite flexible polypeptide (83), a feature further substantiated by the crystallographic data reported here and elsewhere (26). However, upon binding to heparin and other functional analogues of heparin, this dynamic free-energy conformational landscape, with shallow transition barriers and minor populated energy wells, clearly alters, as the apoFGF-1 conformation most appropriate for binding to these ligands accumulates in the solution (45, 83). This also seems to be the case when apoFGF-1 faces 5A2NMS and the family of 2,5-dihydroxyphenylic inhibitors described here (26).
The backbone trace of FGF-1 bound to heparin in solution (45) closely superimposes onto those of this polypeptide incorporated into either the asymmetric (containing heparin) or symmetric FGF-1·FGFR2 complexes (Fig. 3 middle row, right) (45, 51, 57). This tight match suggests that the binding of heparin·FGF-1 to FGFRs predominantly occurs by a lock-and-key rather than by an induced fitting mechanism (in the absence of the activating ligand, induced fitting should predominate, with the subsequent entropic cost and the well documented requirement of higher growth factor concentrations for activity). Differences in the conformations that preferentially accumulate in the presence of heparin and the inhibitors (Fig. 3) would necessarily have dramatic consequences on receptor binding in a predominant lock-and-key kinetic landscape of receptor recognition. Accordingly, inhibition of FGF by 5A2NMS and the family of 2,5-dihydroxyphenylic inhibitors described here is probably caused by the accumulation of the protein in a conformation incapable of fitting on the FGFR interface in addition to the displacement of heparin from the membrane signaling complex. It is obvious that further research is required to ascertain whether this dual inhibitory system also acts on other members of the FGF family.
Both the activators (heparin/heparin sulfate) and the inhibitors referred to in this report have a common chemical feature, that of being strong anions. Nevertheless, both groups clearly differ in terms of plasticity; the activators are flexible, whereas the inhibitors are predominantly stiff. This difference may be decisive in the mechanisms sustaining their binding to FGFs, as in the first case, the ligand easily adapts to the binding site, but in the second, it is the binding site that has to adapt to the ligand. This would be feasible for a flexible protein that would easily sample within its free energy conformational landscape for the most appropriated conformation for efficient ligand-binding interaction. The discrete nature of this well landscape probably explains why sorts of chemically distinct inhibitors like 5A2NMS and 2,5-dihydroxyphenylic acids induce the same protein conformation, whereas they also show different inhibitory efficiencies at the same time. Methylation and point-directed mutagenesis of Lys-132 have already highlighted the relevance for FGF-1 activity of maintaining the integrity of the architectural conformation of the site where 5A2NMS and the family of 2,5-dihydroxyphenylic inhibitors described in this study bind (8, 55). The adaptation necessary to bind these stiff inhibitors may constitute another way of altering such architecture. Finally, the existence of a well defined ligand-inducible inhibited state of FGF-1 suggests that physiological ligands may also exist that are responsible for such allosteric inhibition. The results reported here obviously point toward HGA, although any other compounds of similar chemical nature that may exist in the organism should also be considered. By bearing in mind that moderate levels of HGA may constitute a defense against pathologies caused by the uncontrolled synthesis and cell matrix release of FGFs, perhaps we could better understand the prevalence of the recessive genetic disease alkaptonuria. On the other hand, the inhibition of FGFs should probably be taken into account when explaining the symptomatology of this pathology.
FGF-1 and FGF-2 were the first two pure polypeptides shown to promote angiogenesis (84, 85), although there are currently more than a dozen factors that promote angiogenesis and that target the different cell types involved in vessel formation, most of them identified in tumor proliferation studies (86, 87). The formation of blood vessels requires a synergy between FGFs and vascular endothelial cell growth factors (VEGFs), probably the only ones that directly promote the proliferation of endothelial cells (86). Thus, it is not surprising that increased expression of FGFs and VEGFs is observed in many vascularized tumors when compared with their normal tissue counterparts. Consequently, inhibiting the mitogenic activity of FGF could be a potential target for the development of antiangiogenic therapies. Perhaps such an approach might serve as an alternative to VEGF inhibition worthy of exploration once cell biology studies have presented data clearly substantiating much emerging clinical evidence that VEGF inhibition is not successful as a long term anti-tumor therapy (88).
GA and 2,5DHPS seem to be very convenient compounds as they can be taken orally and they are very safe. Administering appropriate doses of a physiological metabolite, such as HGA, might also be considered. In addition to being a catabolite of aspirin, GA is present in extracts of gentian roots that have been used for centuries as anti-inflammatory agents and in bitter spirits. Moreover, GA accumulates in the organism of subjects on vegetarian diets. 2,5DHPS is the active principle of Doxium®, a drug that is scarcely used and that was employed for many years without any known side effects as a vasculotropic agent, an effect apparently opposed to that reported here. Indeed, our data might also help to explain some of the therapeutic effects of aspirin, salicylate, and vegetarian diets.
The 2,5-dihydroxyphenylic acids described here define a new class of FGF inhibitors, and they constitute new leads to search for drugs to treat diseases caused by uncontrolled synthesis/release of FGFs. As their action could be based on an already existing physiological mechanism of FGF inhibition, they might be expected to cause fewer problems than those produced by antiangiogenic drugs that target VEGF activity (88).
Supplementary Material
Acknowledgments
We thank Margarita Carrascosa, Concepción Fernández-Cabrera, Begoña Cuevas, Argentina Fernández-Ayerdi, and the staff of BM14 at the European Synchrotron Radiation Facility (France) for technical assistance.
This work was supported by BFU2008-02595 from the Ministerio de Educación y Ciencia (Spain).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 3JUT and 3K1X) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FGF
- fibroblast growth factor
- FGFR
- plasmalemmal tyrosine kinase FGF receptor
- 2,5DHPS
- 2,5-dihydroxyphenylsulfonate
- 5A2NMS
- 5-aminonaphthalene-2-sulfonic acid
- exdFGFR2IIIc
- complete extracellular domain of the IIIc isoform of FGFR2
- I50
- half-maximum inhibitory activity
- IDS-3
- O2-sulfoglucuronic acid
- GA
- gentisic acid (2,5-dihydroxybenzoic acid)
- HGA
- homogentisic acid (2-(2,5-dihydroxyphenyl) acetic acid)
- SGN-4
- N,O6-disulfoglucosamine
- STD
- saturation transfer difference NMR spectroscopy
- HSQC
- heteronuclear single quantum correlation
- VEGF
- endothelial cell growth factor.
REFERENCES
- 1.Asada M., Shinomiya M., Suzuki M., Honda E., Sugimoto R., Ikekita M., Imamura T. (2009) Biochim. Biophys. Acta 1790, 40–48 [DOI] [PubMed] [Google Scholar]
- 2.Olsen S. K., Garbi M., Zampieri N., Eliseenkova A. V., Ornitz D. M., Goldfarb M., Mohammadi M. (2003) J. Biol. Chem. 278, 34226–34236 [DOI] [PubMed] [Google Scholar]
- 3.Itoh N., Ornitz D. M. (2004) Trends Genet. 20, 563–569 [DOI] [PubMed] [Google Scholar]
- 4.Szebenyi G., Fallon J. F. (1999) Int. Rev. Cytol. 185, 45–106 [DOI] [PubMed] [Google Scholar]
- 5.Thomas K. A., Giménez-Gallego G. (1986) Trends Biochem. Sci. 11, 81–84 [Google Scholar]
- 6.Klagsbrun M., Shing Y. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 805–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lozano R. M., Redondo-Horcajo M., Jimenez M. A., Zilberberg L., Cuevas P., Bikfalvi A., Rico M., Giménez-Gallego G. (2001) J. Biol. Chem. 276, 35723–35734 [DOI] [PubMed] [Google Scholar]
- 8.Burgess W. H., Shaheen A. M., Ravera M., Jaye M., Donohue P. J., Winkles J. A. (1990) J. Cell Biol. 111, 2129–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baird A., Böhlen P. (1990) in Handbook of Experimental Pharmacology (Sporn M. B., Roberts A. B. eds) Vol. 95, pp. 369–418, Springer-Verlag, Berlin [Google Scholar]
- 10.Cuevas P., García-Calvo M., Carceller F., Reimers D., Zazo M., Cuevas B., Muñoz-Willery I., Martínez-Coso V., Lamas S., Giménez-Gallego G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11996–12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuevas P., Carceller F., Reimers D., Fu X., Giménez-Gallego G. (1994) Neurol. Res. 16, 310–312 [DOI] [PubMed] [Google Scholar]
- 12.Cuevas P., Reimers D., Giménez-Gallego G. (1996) Neurosci. Lett. 221, 25–28 [DOI] [PubMed] [Google Scholar]
- 13.Cuevas P., Reimers D., Carceller F., Xiaobing F., Giménez-Gallego G. (1996) in Cell Signal Transduction, Second Messengers, and Protein Phosphorylation in Health and Disease (Martín Municio A., Miras-Portugal M. T. eds) pp. 161–167, Plenum Press, New York [Google Scholar]
- 14.Abuharbeid S., Czubayko F., Aigner A. (2006) Int. J. Biochem. Cell Biol. 38, 1463–1468 [DOI] [PubMed] [Google Scholar]
- 15.Vlodavsky I., Fuks Z., Ishai-Michaeli R., Bashkin P., Levi E., Korner G., Bar-Shavit R., Klagsbrun M. (1991) J. Cell. Biochem. 45, 167–176 [DOI] [PubMed] [Google Scholar]
- 16.Finetti F., Solito R., Morbidelli L., Giachetti A., Ziche M., Donnini S. (2008) J. Biol. Chem. 283, 2139–2146 [DOI] [PubMed] [Google Scholar]
- 17.Powers C. J., McLeskey S. W., Wellstein A. (2000) Endocr. Relat. Cancer 7, 165–197 [DOI] [PubMed] [Google Scholar]
- 18.Harada M., Murakami H., Okawa A., Okimoto N., Hiraoka S., Nakahara T., Akasaka R., Shiraishi Y., Futatsugi N., Mizutani-Koseki Y., Kuroiwa A., Shirouzu M., Yokoyama S., Taiji M., Iseki S., Ornitz D. M., Koseki H. (2009) Nat. Genet. 41, 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dirix L. Y., Vermeulen P. B., Pawinski A., Prové A., Benoy I., De Pooter C., Martin M., Van Oosterom A. T. (1997) Br. J. Cancer 76, 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffers M., LaRochelle W. J., Lichenstein H. S. (2002) Expert Opin. Ther. Targets 6, 469–482 [DOI] [PubMed] [Google Scholar]
- 21.Greber B., Lehrach H., Adjaye J. (2007) BMC Dev. Biol. 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dvorak P., Dvorakova D., Hampl A. (2006) FEBS Lett. 580, 2869–2874 [DOI] [PubMed] [Google Scholar]
- 23.Song S., Wientjes M. G., Gan Y., Au J. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8658–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Indraccolo S., Stievano L., Minuzzo S., Tosello V., Esposito G., Piovan E., Zamarchi R., Chieco-Bianchi L., Amadori A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4216–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J. T., Herlyn M. (2007) J. Cell. Physiol. 213, 603–609 [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Tornero C., Lozano R. M., Redondo-Horcajo M., Gómez A. M., López J. C., Quesada E., Uriel C., Valverde S., Cuevas P., Romero A., Giménez-Gallego G. (2003) J. Biol. Chem. 278, 21774–21781 [DOI] [PubMed] [Google Scholar]
- 27.Griffiths L. A. (1958) Nature 182, 733–734 [Google Scholar]
- 28.Santiago R., Malvar R. A., Baamonde M. D., Revilla P., Souto X. C. (2005) J. Econ. Entomol. 98, 1349–1356 [DOI] [PubMed] [Google Scholar]
- 29.Fisher D. C., Kogan M., Paxton J. (1990) J. Entomol. Sci. 25, 230–238 [Google Scholar]
- 30.Campbel W. F., Haws B. A., Asay K. H., Hansen J. D. (1964) Journal of Range Management 37, 365–369 [Google Scholar]
- 31.Todd G. W., Getahun A., Cress D. C. (1971) Ann. Entomol. Soc. Am. 64, 718–722 [Google Scholar]
- 32.Needs C. J., Brooks P. M. (1985) Clin. Pharmacokinet. 10, 164–177 [DOI] [PubMed] [Google Scholar]
- 33.Cuevas P., Carceller F., Lozano R. M., Crespo A., Zazo M., Giménez-Gallego G. (1997) Growth Factors 15, 29–40 [DOI] [PubMed] [Google Scholar]
- 34.Peränen J., Rikkonen M., Hyvönen M., Kääriäinen L. (1996) Anal. Biochem. 236, 371–373 [DOI] [PubMed] [Google Scholar]
- 35.Canales A., Lozano R., López-Méndez B., Angulo J., Ojeda R., Nieto P. M., Martín-Lomas M., Giménez-Gallego G., Jiménez-Barbero J. (2006) FEBS J. 273, 4716–4727 [DOI] [PubMed] [Google Scholar]
- 36.Ortega S., García J. L., Zazo M., Varela J., Muñoz-Willery I., Cuevas P., Giménez-Gallego G. (1992) Biotechnology 10, 795–798 [DOI] [PubMed] [Google Scholar]
- 37.Houssaint E., Blanquet P. R., Champion-Arnaud P., Gesnel M. C., Torriglia A., Courtois Y., Breathnach R. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8180–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lozano R. M., Jiménez M., Santoro J., Rico M., Giménez-Gallego G. (1998) J. Mol. Biol. 281, 899–915 [DOI] [PubMed] [Google Scholar]
- 39.Cuevas P., Díaz-González D., Dujovny M. (2004) Neurol. Res. 26, 273–275 [DOI] [PubMed] [Google Scholar]
- 40.Cuevas P., Díaz-González D., Giménez-Gallego G., Dujovny M. (2005) Neurol. Res. 27, 797–800 [DOI] [PubMed] [Google Scholar]
- 41.Rodts G. E., Jr., Black K. L. (1994) Neurol. Res. 16, 184–186 [DOI] [PubMed] [Google Scholar]
- 42.Leslie A. (1991) in Crystallographic Computing (Moras D., Podjarny A. D., Thierry J. C. eds) Vol. 5, pp. 27–38, Oxford University Press, Oxford [Google Scholar]
- 43.(1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 44.Navaza J. (1994) Acta Crystallogr. Sect. A 50, 157–163 [Google Scholar]
- 45.DiGabriele A. D., Lax I., Chen D. I., Svahn C. M., Jaye M., Schlessinger J., Hendrickson W. A. (1998) Nature 393, 812–817 [DOI] [PubMed] [Google Scholar]
- 46.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 47.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 48.van Aalten D. M., Bywater R., Findlay J. B., Hendlich M., Hooft R. W., Vriend G. (1996) J. Comput. Aided Mol. Des. 10, 255–262 [DOI] [PubMed] [Google Scholar]
- 49.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 50.Pineda-Lucena A., Jiménez M. A., Nieto J. L., Santoro J., Rico M., Giménez-Gallego G. (1994) J. Mol. Biol. 242, 81–98 [DOI] [PubMed] [Google Scholar]
- 51.Pellegrini L., Burke D. F., von Delft F., Mulloy B., Blundell T. L. (2000) Nature 407, 1029–1034 [DOI] [PubMed] [Google Scholar]
- 52.Blaber M., DiSalvo J., Thomas K. A. (1996) Biochemistry 35, 2086–2094 [DOI] [PubMed] [Google Scholar]
- 53.Romero A., Pineda-Lucena A., Giménez-Gallego G. (1996) Eur. J. Biochem. 241, 453–461 [DOI] [PubMed] [Google Scholar]
- 54.Eriksson A. E., Cousens L. S., Matthews B. W. (1993) Protein Sci. 2, 1274–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harper J. W., Lobb R. R. (1988) Biochemistry 27, 671–678 [DOI] [PubMed] [Google Scholar]
- 56.Scrutton N. S., Raine A. R. (1996) Biochem. J. 319, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stauber D. J., DiGabriele A. D., Hendrickson W. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plotnikov A. N., Schlessinger J., Hubbard S. R., Mohammadi M. (1999) Cell 98, 641–650 [DOI] [PubMed] [Google Scholar]
- 59.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Linhardt R. J., Mohammadi M. (2000) Mol. Cell 6, 743–750 [DOI] [PubMed] [Google Scholar]
- 60.Plotnikov A. N., Hubbard S. R., Schlessinger J., Mohammadi M. (2000) Cell 101, 413–424 [DOI] [PubMed] [Google Scholar]
- 61.Gimenez-Gallego G., Conn G., Hatcher V. B., Thomas K. A. (1986) Biochem. Biophys. Res. Commun. 135, 541–548 [DOI] [PubMed] [Google Scholar]
- 62.Rapraeger A. C., Krufka A., Olwin B. B. (1991) Science 252, 1705–1708 [DOI] [PubMed] [Google Scholar]
- 63.Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991) Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- 64.Kan M., Wang F., Xu J., Crabb J. W., Hou J., McKeehan W. L. (1993) Science 259, 1918–1921 [DOI] [PubMed] [Google Scholar]
- 65.Ornitz D. M., Leder P. (1992) J. Biol. Chem. 267, 16305–16311 [PubMed] [Google Scholar]
- 66.Ornitz D. M., Xu J., Colvin J. S., McEwen D. G., MacArthur C. A., Coulier F., Gao G., Goldfarb M. (1996) J. Biol. Chem. 271, 15292–15297 [DOI] [PubMed] [Google Scholar]
- 67.Wang H., Toida T., Kim Y. S., Capila I., Hileman R. E., Bernfield M., Linhardt R. J. (1997) Biochem. Biophys. Res. Commun. 235, 369–373 [DOI] [PubMed] [Google Scholar]
- 68.Andrade M. A., Chacón P., Merelo J. J., Morán F. (1993) Protein Eng. 6, 383–390 [DOI] [PubMed] [Google Scholar]
- 69.Mayer N., Meyer B. (1999) Angew. Chemie Int. Ed. 38, 1784–1788 [DOI] [PubMed] [Google Scholar]
- 70.Fernández-Tornero C., López R., García E., Giménez-Gallego G., Romero A. (2001) Nat. Struct. Biol. 8, 1020–1024 [DOI] [PubMed] [Google Scholar]
- 71.Fernández-Tornero C., García E., de Pascual-Teresa B., López R., Giménez-Gallego G., Romero A. (2005) J. Biol. Chem. 280, 19948–19957 [DOI] [PubMed] [Google Scholar]
- 72.McKeehan W. L., Wu X., Jang J. H., Kan M. (1997) In Vitro Cell Dev. Biol. Anim. 33, 727–730 [DOI] [PubMed] [Google Scholar]
- 73.Sato Y., Rifkin D. B. (1988) J. Cell Biol. 107, 1199–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pesenti E., Sola F., Mongelli N., Grandi M., Spreafico F. (1992) Br. J. Cancer 66, 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cuevas P., Lozano R. M., Giménez-Gallego G. (1999) Neurol. Res. 21, 191–194 [DOI] [PubMed] [Google Scholar]
- 76.Plate K. H., Breier G., Weich H. A., Risau W. (1992) Nature 359, 845–848 [DOI] [PubMed] [Google Scholar]
- 77.Stefanik D. F., Rizkalla L. R., Soi A., Goldblatt S. A., Rizkalla W. M. (1991) Cancer Res. 51, 5760–5765 [PubMed] [Google Scholar]
- 78.Okumura N., Takimoto K., Okada M., Nakagawa H. (1989) J. Biochem. 106, 904–909 [DOI] [PubMed] [Google Scholar]
- 79.Aoki T., Kato S., Fox J. C., Okamoto K., Sakata K., Morimatsu M., Shigemori M. (2002) Int. J. Oncol. 21, 629–636 [PubMed] [Google Scholar]
- 80.Gerstner E. R., Duda D. G., di Tomaso E., Sorensen G., Jain R. K., Batchelor T. T. (2007) Expert Opin. Investig. Drugs 16, 1895–1908 [DOI] [PubMed] [Google Scholar]
- 81.Carmeliet P. (2005) Nature 438, 932–936 [DOI] [PubMed] [Google Scholar]
- 82.Ornitz D. M., Herr A. B., Nilsson M., Westman J., Svahn C. M., Waksman G. (1995) Science 268, 432–436 [DOI] [PubMed] [Google Scholar]
- 83.Pineda-Lucena A., Jiménez M. A., Lozano R. M., Nieto J. L., Santoro J., Rico M., Giménez-Gallego G. (1996) J. Mol. Biol. 264, 162–178 [DOI] [PubMed] [Google Scholar]
- 84.Thomas K. A., Rios-Candelore M., Giménez-Gallego G., DiSalvo J., Bennett C., Rodkey J., Fitzpatrick S. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 6409–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Esch F., Baird A., Ling N., Ueno N., Hill F., Denoroy L., Klepper R., Gospodarowicz D., Böhlen P., Guillemin R. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 6507–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 87.Nissen L. J., Cao R., Hedlund E. M., Wang Z., Zhao X., Wetterskog D., Funa K., Bråkenhielm E., Cao Y. (2007) J. Clin. Invest. 117, 2766–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loges S., Mazzone M., Hohensinner P., Carmeliet P. (2009) Cancer Cell 15, 167–170 [DOI] [PubMed] [Google Scholar]
- 89.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.