Abstract
Mutations in the mitochondrial encoded protein PTEN-induced putative kinase 1 (PINK1) cause autosomal recessive Parkinson disease (PD). In mammalian cells, mutant PINK1 has been reported to promote fission or inhibit fusion in mitochondria; however, the mechanism by which this process occurs remains elusive. Using an ecdysone-inducible expression system in mammalian dopaminergic neuronal cells, we report here that human mutant PINK1 (L347P and W437X) mediates an overall fission effect by increasing the ratio of mitochondrial fission over fusion proteins, leading to excessive dysfunctional fragmented mitochondria. Knocking down endogenous Pink1 produces similar effects. In contrast, overexpressing human wild type PINK1 produces a pro-fusion effect by increasing the ratio of mitochondrial fusion/fission proteins without resulting in functionally compromised mitochondria. Parkin knockdown blocks the imbalance in fission/fusion proteins. Furthermore, overexpressing parkin and ubiquitin increases degradation of the mitochondrial fission hFis1 protein, suggesting PINK1 and parkin maintain proper mitochondrial function and integrity via the fission/fusion machinery. Through genetic manipulations and treatment with the small molecule mitochondrial division inhibitor (mdivi-1), which inhibits DLP1/Drp1, both structural and functional mitochondrial defects induced by mutant PINK1 were attenuated, highlighting a potential novel therapeutic avenue for Parkinson disease.
Keywords: Mitochondria, Neurodegeneration, Neurological Diseases, Parkinson Disease, Ubiquitination, Drp1, hFis1, mdivi-1, mfn2, Mitochondrial Dynamics
Introduction
The discoveries of mutations in the mitochondrial protein PTEN-induced putative kinase 1 (PINK1)2 as a cause of autosomal recessive PD (1–3) have fueled the longstanding interest in the role of mitochondrial dysfunction in PD. Currently about 50 PINK1 mutations have been reported (4), making it the second most common causative gene for autosomal recessive PD after parkin, an E3 ubiquitin ligase that has been shown to function downstream of PINK1 and to affect mitochondrial morphology (5–8). Although PINK1 mutations are spread throughout the gene, they are most commonly found in the region encoding the functional serine/threonine kinase domain at the C terminus, leading to loss of PINK1 kinase activity. Recently this kinase domain has been shown to face the cytoplasm (9), providing evidence of spatial proximity to allow this mitochondrial protein to directly interact with cytosolic parkin.
Current consensus is that PINK1 is a protective protein. Supporting this role, PINK1 overexpression confers resistance to staurosporine, MPP+, and rotenone toxicity in cultured cells (10, 11) as well as to MPTP-induced dopaminergic neuronal loss in mice (12). Conversely, reducing PINK1 levels by RNAi in cultured cells leads to enhanced cell death in the presence of MPP+ and rotenone (12, 13). Mitochondrial defects leading to degeneration of flight muscles and loss of dopaminergic neurons have also been reported in Pink1-deficient Drosophila (5, 14). In recent years, PINK1 has gained significant attention for its role in mitochondrial dynamics (fission, fusion, and migration). In mammalian cell models (11, 15–17) and primary fibroblasts cultured from patients with PINK1 mutations (15, 18), PINK1 promotes fusion or inhibits fission of mitochondria. Because the imbalance in mitochondrial fission and fusion machinery has increasingly been linked to neurodegeneration (19, 20), it is critical to investigate further the mechanism by which PINK1 controls this vital mitochondrial process.
In the present study we seek to elucidate the mechanism by which PINK1 regulates mitochondrial fission/fusion in the mammalian dopaminergic system and to identify a potential protective therapy against mutant PINK1. By using an inducible system for greater temporal control of PINK1 expression levels and by knocking down endogenous Pink1 and parkin as well as by using various genetic and pharmacological manipulations of the fission/fusion machinery, we report here that PINK1 exerts an overall fusion effect on mitochondria by reducing fission (Drp1 and hFis1) as well as by concomitantly increasing fusion (Mfn2) protein levels. We also demonstrate that parkin acts downstream of PINK1 to mediate these changes, suggesting that one way by which PINK1 and parkin maintain proper mitochondrial function and integrity is through quality control of mitochondrial fission and fusion proteins. Last, we also present evidence that the structural and functional defects caused by excessive fission of the human relevant mutant PINK1 (as exemplified here by the L347P mutation) can be protected by the recently described mitochondrial division inhibitor (mdivi-1) (21), an inhibitor of Drp1-GTPase, thus, highlighting a novel treatment therapy for PD.
EXPERIMENTAL PROCEDURES
Generation of the Stable and Inducible PINK1 Cell Lines
The expression of various forms of PINK1 in the dopaminergic 1RB3AN27 (N27) neuronal cells, which were created from rat fetal mesencephalic cells (22, 23) (kindly provided by Dr. Anumantha Kanthasamy, Iowa State University), was accomplished by using The Complete Control® Inducible Mammalian Expression System (Stratagene, La Jolla, CA) according to company instructions and the following modifications.
Briefly, with this ecdysone-inducible system, two vectors are required to express the gene of interest: the pERV3 receptor vector and the pEGSH expression vector. First, using LipofectamineTM LTX and Plus, we stably transfected N27 cells with the repressor pERV3 vector to constitutively express the ecdysone receptor (EcR) and the retinoid X receptor heterodimer. Transfected cells were selected by the addition of G418 (500 μg/ml, Invitrogen). Two weeks later, surviving cells were collected, and single cells were seeded to 96-well plates and grown in the presence of G418. Twenty-three individual clones were subsequently isolated and expanded. The inducibility of these clones was assessed by transiently transfecting these cells with the pEGSH-Luciferase vector followed by ponasterone A (ponA, Alexis Biochemicals) treatment and luciferase assay. The most inducible clone was selected for the subsequent stable transfection of the modified pEGSH vector. This second vector contains the ecdysone-responsive element, which in the absence of ponA, tightly suppresses transcription. We modified the pEGSH vector from Stratagene by introducing a DNA fragment containing internal ribosome entry site-emerald GFP (kindly provided by Dr. Pradip Roy-Burman, University of Southern California) for two reasons; to avoid the concern that a fusion protein (PINK1-GFP) may alter the function of PINK1 and to enhance the GFP signal. The cDNAs of full-length human PINK1, PINK1L347P, and PINK1W437X, (kindly provided by Dr. Serge Przedborski, Columbia University) were subcloned into the multiple cloning site. Stably transformed cells were selected and maintained in RPMI containing 10% fetal bovine serum, G418 (500 μg/ml), and hygromycin (200 μg/ml). Inducible cells with high expression of GFP were selected by a FACSVantage cell sorter (BD Biosciences). Each cell type was sorted 3–4 times for the top 10% of cells with the highest GFP signal. The inducible expression of PINK1 was confirmed using immunocytochemistry, immunoblotting, and quantitative real-time PCR (with primers designed to amplify a segment that is common to both rat Pink1 (accession number NM_001106694) and human PINK1 (accession number NM_032409): forward (5′-CTGTCAGGAGATCCAGGCAATT-3′) and reverse (5′- GCATGGTGGCTTCATACACAGC-3′).
siRNA-mediated Pink1 Knockdown
Cultured cells were transfected with either siRNA against both rat and mouse Pink1 (Ambion, ID#180640 as previously validated (12)) or with a scrambled control siRNA that has no significant homology to any known gene sequences (Ambion, ID#4611) using LipofectamineTM 2000 according to company instructions. After a 5-h incubation, medium was replaced with RPMI plus 10% fetal bovine serum, and cells were grown for an additional 48 h before they were used. The efficiency of Pink1 knockdown was confirmed by isolating RNA (RNeasy Mini kit, Qiagen) from transfected cells and conducting real time RT-PCR analysis with the Bio-Rad iScriptTM One-Step RT-PCR kit with SYBR® Green. Oligonucleotide primers were designed using Primer Express® 1.5 (Applied Biosystems, Foster City, CA) to recognize a segment of rat Pink1 as indicated above; likewise, a portion of rat β-actin (accession number NM_031144) was amplified as an internal standard using forward (5′-ACCCTGTGCTGCTCACCGA-3′) and reverse (5′-CTGGATGGCTACGTACATGGCT-3′) primers. Reactions were conducted and analyzed on a Rotor-Gene 3000 real time light cycler from Corbett Robotics (San Francisco, CA), and expression levels are reported as a ratio to rat β-actin.
The extent of Pink1 knockdown was also visualized in cells grown on glass coverslips in which varying concentrations of Pink1 siRNA were transiently transfected together with mouse Pink1 tagged with GFP plasmid DNA (kindly provided by Dr. Xugang Xia, Thomas Jefferson University Medical College) using LipofectamineTM 2000. Of note, Pink1-siRNA used in the present study also recognizes 100% of this sequence in mouse Pink1. After 24 h, cells were fixed with 4% paraformaldehyde and observed for GFP signals using an inverted epifluorescence microscope.
siRNA Mediated parkin Knockdown
Pre-designed siRNA against rat parkin (ON-TARGET Plus SMARTpoolTM, catalog # L-090709-01) was purchased from Dharmacon Research, Inc. This is a mixture of 4 individual 19-mer siRNA duplexes that target 4 separate sequences of parkin to increase efficiency of gene silencing: GGAAGUGGUUGCUAAGCGA, GAGGAAAGUCACCUGCGAA, GAGGAAAAGUCACGAAACA, and CGCACGACCUCAUGGGAAA.
The following modified “in-tube” transfection procedure (24) was used to increase transfection efficiency. LipofectamineTM 2000 (10 μl) was combined with 100 nm siRNA in RNase free microcentrifuge tubes for 20 min at room temperature. Subsequently, N27 cells (1.5 × 106) suspended in Opti-MEM were added to this preformed siRNA-lipid complex solution. After 1 h, this cell suspension was plated in 100-mm dishes. After 4 h, the medium was replaced with RPMI containing 10% fetal bovine serum with or without 10 μm ponA, and cells were harvested 48 h after transfection for immunoblotting. The efficiency of knocking down endogenous rat parkin was verified using immunoblotting (1:1000, Abcam).
Transient Transfection of Mitochondrial Fission and Fusion Constructs
Cells were transfected with plasmids (1 μg/1 × 105 cells) containing Drp1, DrpK38A, Mfn2-myc, or hFis-myc using LipofectamineTM LTX (Invitrogen) according to company instructions. Six hours later, medium was replaced with RPMI containing 10% fetal bovine serum and 10 μm ponA. After 24 h, cells were assessed for changes in mitochondrial morphology or ATP levels as described below. Transfected cells were visualized by immunostaining with monoclonal antibodies against Drp1 (1:250) or Myc (1:1000).
Mitochondrial Morphology Assessment
Cells were grown on poly-d-lysine-coated glass coverslips. Mitochondria were labeled with either DsRed-Mito (1 μg/60-mm dish, Clontech) or MitoTracker® Red (50 nm at 37 °C for 30 min, Invitrogen). Cells were fixed with 4% paraformaldehyde, and coverslips were mounted with Prolong Gold Antifade Reagent with DAPI (Invitrogen). Images were captured using either confocal microscopy (FV1000; Olympus, Tokyo, Japan) or an inverted epifluorescence microscope (Nikon, TE2000-U, with a mounted camera controlled by Spot® software). Measurements of mitochondrial size and shape were quantified blinded using ImageJ. More than 200 clearly identifiable mitochondria from randomly selected 10–15 cells per experiment were measured in 5 independent experiments.
Drp1 Translocation
Cells were grown on poly-d-lysine coated glass coverslips and then transfected with 1 μg each of DsRed-Mito and of Drp1-myc. After 24 h of 10 μm ponA addition, cells were fixed and immunostained with anti-Myc antibody followed by Alexa 633 secondary antibody. In the group with Pink1 knockdown, 50 nm siRNA-Pink1 was transfected at the same time. Images were captured and analyzed using confocal microscopy, and the Drp1 signal was pseudo-colored to green. About 50 cells were analyzed blinded per group in three independent experiments.
Mitochondrial Isolation
Mitochondria were isolated using a mitochondrial isolation kit (Pierce) according to the manufacturer's instructions. Briefly, cells were homogenized in a Dounce homogenizer and then centrifuged at 750 × g for 10 min at 4 °C. The supernatant was further centrifuged at 12,000 × g for 15 min at 4 °C. The pellet was then washed and kept as the mitochondrial fraction. The supernatant was further centrifuged at 100,000 × g for 1 h at 4 °C and designated as the cytosolic fraction.
Immunoprecipitation and Immunoblotting
For immunoprecipitation, HEK 293 cells were grown in 60-mm dishes and transiently transfected with 1 μg of each plasmid containing HA-ubiquitin (Addgene plasmid 17608, described in Lim et al. (25)), myc-parkin (Addgene plasmid 17612, described in Zhang et al. (26)), or hFis1 for 48 h in the presence or absence of 10 μm MG132 for ∼20 h. Cells were then collected and lysed as described (27) in immunoprecipitation buffer (1× phosphate-buffered saline, 0.5% Triton X-100, Complete Mini protease inhibitor (Roche Applied Science)) and rotated for 1 h at 4 °C. Supernatant was collected for immunoprecipitation after centrifugation at 17,500 × g for 15 min at 4 °C. Primary monoclonal anti-HA antibody (2 μg, Roche Applied Science) or 2 μg of mouse IgG (Santa Cruz) was bound to Dynabeads Protein G (Invitrogen) at room temperature for 30 min by rotation. The beads-antibody complexes were then washed in phosphate-buffered saline containing 0.01% Tween 20 before being incubated with the soluble cell lysates (500 μg) overnight at 4 °C with rotation. The beads-antibody-antigen complexes were then washed with citrate-phosphate buffer (pH 5.0). The immune complexes were then eluted by boiling in SDS-PAGE sample buffer for 5 min. Samples were then separated in gradient gel (5–15%) and immunoblotted with polyclonal anti-HA (1:1000, Zymed Laboratories Inc.) or hFis1 (1:3000, Alexis Biochemicals) antibodies.
For other immunoblotting studies, mitochondrial proteins (20 μg) or cytosolic proteins (40 μg) were separated in SDS-PAGE and then transferred to nitrocellulose membrane. The blots were then cut into two portions; the lower part was probed for hFis1 (1:3000), and the upper part was probed for larger proteins in the following order: PINK1 (1:1000, Novus Biologicals, catalog #BC100-494), Drp1 (1:2,000, BD Biosciences), Mfn2 (1:10,000, Sigma), and finally, Hsp60 (1:2000, Santa Cruz) or tubulin (1:5000, Sigma) as loading controls for mitochondrial or cytosolic proteins, respectively. After each development, the antibodies on the blots were gently removed using stripping buffer (Thermo Scientific, catalog #21059). Secondary antibodies conjugated with horseradish peroxidase were used, and immunoreactivity was visualized using chemiluminescence (SuperSignal Ultra, Pierce). Bands of interest were analyzed and quantified using Scion Image.
Immunocytochemistry
Cells grown on poly-d-lysine-coated glass coverslips were transiently transfected with plasmids containing Drp1, DrpK38A (Drp1 dominant negative), Drp1-myc, Mfn2-myc, or hFis1-myc followed by ponA (10 μm) for the expression of PINK1 for 24 h. Cells were fixed in 4% paraformaldehyde before being immunostained with monoclonal antibodies against Drp1 (1:250, BD Biosciences) or myc (1:1000, Sigma). Images were captured either by using confocal or epifluorescent microscopy as indicated above.
mdivi-1 Treatment
mdivi-1 (3-(2,4-dichloro-5-methoxyphenyl)-2-sulfanyl-4(3H)-quinazolinone) was purchased from Bionet through Key Organics Ltd. and dissolved in DMSO (50 mm) as a stock solution and diluted with culture medium to varying working concentrations.
Proteasomal Inhibition
MG132 (Sigma) was dissolved in DMSO as stock solution (5 mm) and diluted to appropriate concentrations in cell culture medium. Based on the dose-response studies of MG132 using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay for cell viability and immunoblotting for polyubiquitinated proteins (Biomol, catalog #PW8805-0500; Clone FK1), 100 nm was selected for the present study to inhibit proteasomal function. After 24 h of treatment with 100 nm MG132 and 10 μm PonA, cells were lysed in radioimmune precipitation assay buffer, and whole cell lysates were kept for immunoblotting.
Measurements of ATP Using HPLC
To minimize the quantity of constructs required for these experiments, N27 cells were grown in 24-well plates before being extracted for ATP as described (28). Briefly, culture medium was removed by aspiration followed by the immediate addition of liquid nitrogen. After evaporation on ice, 300 μl of ice-cold 0.4 m perchloric acid were added to the wells. Cells were scraped off and centrifuged at 14,000 × g for 15 min at 4 °C. The pellets were used for protein measurement. The supernatant was neutralized with 1 m K2CO3, kept at −80 °C to promote precipitation of the perchlorate, then centrifuged again. Supernatants were kept for HPLC assay. Samples (20 μl) were injected manually into a Rheodyne sample injector (with a fixed loop of 20 μl) attached to a 12-channel CoulArray 5600A (ESA Inc.) and eluted on a reverse-phase column (Lichrospher-100, Merck) of 250 × 3-mm inner diameter with a particle size of 5 μm at a flow rate of 0.4 ml/min using a mobile phase containing 215 mm KH2PO4, 2.3 mm tetrabutylammonium hydrogen sulfate, and 4% acetonitrile; pH 6 was adjusted with KOH, slightly modified from a previous study (29). Signals were detected using a UV detector (#526, ESA) at 260 nm and converted by a CoulArray Analog Input Adapter before being analyzed using the CoulArray® software. All peak areas were within the linear range of the standard curves.
Mitochondrial Membrane Potential (Δψm)
Cells were trypsinized for single cell suspension and washed with warm phosphate-buffered saline before being stained with 50 nm tetramethylrhodamine methyl ester (TMRM) for 20 min at 37 °C. Fluorescence signal was analyzed by BD FACSCantoTM II Flow Cytometry System (BD Biosciences) using the blue D 585/42-A channel. Non-cellular debris and dead cells were gated out, and ∼30,000 events were collected for each analysis. Cells were treated with 20 μm carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone for 20 min to collapse Δψm, and the obtained TMRM fluorescence was used to set the threshold. Data are expressed as the percentage of cells with signal above this threshold value as previously described (11).
Statistical Analyses
All values are expressed as mean ± S.E. Differences between means were analyzed using either one-way or two-way ANOVA followed by Newman-Keuls post hoc testing for pairwise comparison using SigmaStat Version 3.5®. The null hypothesis was rejected when the p value < 0.05.
RESULTS
Generation of Stable Cells with Inducible Expression of Human Wild Type and Mutant PINK1
To characterize the role of PINK1 in mitochondrial dynamics, we created stable cell lines with inducible expression of human wild type PINK1, two recessive mutant forms (PINK1W437X and PINK1L347P), and a control group with only the empty vector. These two mutant forms were selected partly because, first, single heterozygous mutations of PINK1W437X (30) and PINK1L347P (31) have been reported in patients with PD. Second, overexpressing these PINK1 mutations in cells with endogenous PINK1/Pink1 background suggests they have dominant negative effects (32, 33). Third, in cell cultures, PINK1W437X confers a partial and PINK1L347P confers an almost complete loss of PINK1 kinase activity (32, 34), which is attributed to the loss of protective function of PINK1. These proteins were expressed in N27 (1RB3AN27) cells because these rat mesencephalic dopaminergic neuronal cells (22, 23) express the dopamine transporter and functional tyrosine hydroxylase, an enzyme involved in dopamine synthesis. Additionally, these cells display tubular mitochondrial morphology, which readily aids the identification of fragmented mitochondria.
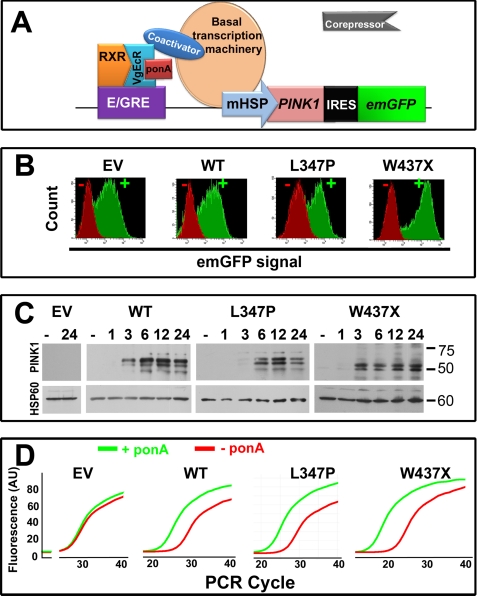
As depicted in Fig. 1A, in the presence of ponA, an ecdysone analogue inducer, the expression of PINK1 and emerald GFP were induced. Although GFP was not directly tagged to PINK1 (due to the concern that this fluorescent protein may alter the intrinsic function of PINK1), the expression of these two proteins in the same cells correlated well with each other. With GFP as a selectable marker, flow cytometry was used to sort cells with high transgene expression (Fig. 1B). Upon induction by ponA, the expression of PINK1 proteins was generally detectable as early as 3 h thereafter (Fig. 1C). Two prominent bands at ∼ 65 and ∼ 55 kDa were detected for wild type PINK1 and mutant PINK1L347P. These bands represent the full-length and the mature cleaved forms (32, 34, 35). The nonsense mutation W437X causes a truncation of the last 145 amino acids encoding the C terminus of the kinase domain, producing two smaller bands at ∼40–50 kDa. In the absence of ponA, PINK1 immunoreactivity was undetectable, indicating a tight regulation of this system because endogenous Pink1 is not detectable with currently available antibodies (9). To assess whether these stable clones had comparable levels of gene expression and to confirm further our immunoblotting data, quantitative real time RT-PCR was performed (Fig. 1D). Consistent with the increases in protein levels, comparable levels of PINK1 and mutant PINK1 transcript expression were induced 24 h after ponA addition (∼20-fold higher than endogenous Pink1). However, in control cells with empty vector, ponA did not change the level of endogenous Pink1, demonstrating the tight regulation of this inducible dopaminergic cell model.
FIGURE 1.
Inducible overexpression of PINK1 in rat dopaminergic N27 neuronal cells. A, first, stable cells containing a vector that constitutively expresses the ecdysone receptor (VgEcR) and the retinoid X receptor (RXR) were generated. These cells were subsequently stably transfected with a second vector that contains the ecdysone-responsive element (E/GRE) and a multiple cloning site for the insertion of either the human wild type or mutant PINK1 gene. The VgEcR-retinoid X receptor heterodimer binds to the E/GRE. In the presence of ponA, an ecdysone analogue inducer, the corepressors are released from VgEcR, and coactivators are recruited, resulting in gene activation via the minimal heat shock promoter (mHSP). Emerald green fluorescent protein (emGFP) was used as a selectable marker for cells with high inducible expression. IRES, internal ribosome entry site. B, histograms are shown of GFP intensity of various cell groups at the final sorting (either third or fourth sorting) using flow cytometry. EV, empty vector; WT, wild type. C, PINK1 expression was confirmed by immunoblotting. In the groups of cells with empty vector or without ponA(−), PINK1 immunoreactivity was undetectable. When induced, PINK1 was detectable by 3–6 h. D, using primers that recognize both endogenous rat Pink1 and human PINK1, quantitative real-time PCR confirmed the effects of ponA (24 h after addition) on the up-regulation of genes of interest, and comparable levels of inducibility were observed between these cell lines. AU, arbitrary units.
Alterations in PINK1 Status Induces Mitochondrial Fragmentation
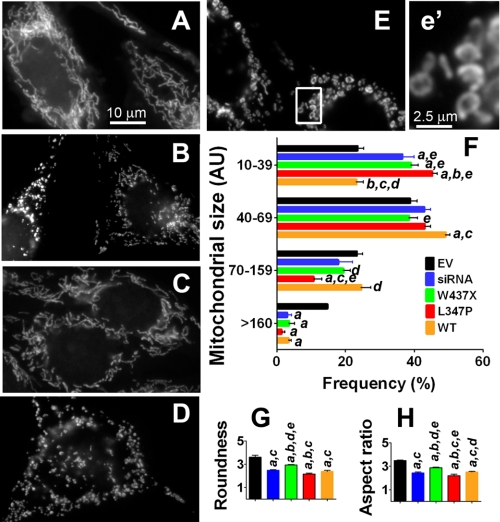
Our first step in determining the function of PINK1 in mitochondrial dynamics was to characterize the effects of various statuses of PINK1 on mitochondrial morphology. Using DsRed-Mito to label mitochondria, control N27 cells with empty vector displayed primary tubular and long mitochondria (Fig. 2A). However, when endogenous rat Pink1 was knocked down by ∼80% using siRNA (supplemental Fig. S1, A and B), severe mitochondrial fragmentation occurred (Fig. 2B), suggesting Pink1 is required for the maintenance of normal mitochondrial morphology. Consistent with this observation, overexpression of the mutant human PINK1W437X (Fig. 2C) and PINK1L347P (Fig. 2D) also produced fragmented mitochondria. To compare mitochondrial morphology more objectively between groups of cells, individual mitochondria were measured for size (Fig. 2F) and shape (Fig. 2, G and H). These quantitative data confirm a higher proportion of smaller and rounder mitochondria in the groups with Pink1 knockdown as well as with W437X and L347P mutations. Of note, although both mutations induced significant fragmentation, the effect was more pronounced with PINK1L347P than PINK1W437X. Next, we assessed mitochondrial morphology in cells overexpressing wild type PINK1. Interestingly, we also observed smaller mitochondria in these cells (Fig. 2, E and e′). High magnification images (Fig. 2e′) revealed that these mitochondria were primarily donut-shaped, which was strikingly distinct from those produced by Pink1 knockdown or mutant PINK1. The same results were obtained when mitochondrial morphology was visualized using MitoTracker Red (supplemental Fig. S2).
FIGURE 2.
Mitochondrial morphologies induced by various PINK1 functions. A–E, N27 cells were labeled with DsRed-Mito for the visualization of mitochondrial morphology. Control cells with empty vector (A) showed tubular and long mitochondria, in contrast to those 48 h after siRNA-mediated Pink1-knockdown (B) and to those overexpressing PINK1W437X (C), PINK1L347P (D), or wild type PINK1 (E). With the exception of the Pink1-knockdown group, cells were treated with 10 μm ponA for 24 h. Scale bars, 10 μm (A–E). F, quantitative measurement of mitochondrial perimeter (AU, arbitrary units, 1 AU = 0.066 μm) using ImageJ. Data represent the mean ± S.E., grouped into different size bins and analyzed by one-way ANOVA (Bin 10–39: F4,10 = 18.19, p < 0.001; Bin 40–69: F4,10 = 4.82, p < 0.05; Bin 70–159: F4,10 = 3.88, p < 0.05; Bin >160: F4,10 = 24.36, p < 0.001). Pairwise multiple comparisons were performed using the Newman-Keuls post hoc test. a, p < 0.05 compared with the empty vector (EV) group; b, p < 0.05 compared with the siRNA group; c, p < 0.05 compared with the PINKW437X group; d, p < 0.05 compared with the PINK1L347P group; e, p < 0.05 compared with the WT group. n = 3 independent experiments with >200 mitochondria per experiment from 10–15 cells measured. G and H, shown is quantification of mitochondrial shape. Roundness was calculated as perimeter2/4 π area. Aspect ratio is a measurement of major/minor axes. Both of these values approach 1 as the particle becomes circular. Data represent the mean ± S.E., analyzed by one-way ANOVA (G, F4,10 = 32.03, p < 0.001; H, F4,10 = 43.09, p < 0.001) followed by the Newman-Keuls post hoc test. n = 3 experiments with >200 mitochondria per experiment from 10–15 cells measured.
PINK1 Overexpression Reduces the Ratio of Mitochondrial Fission/Fusion Proteins
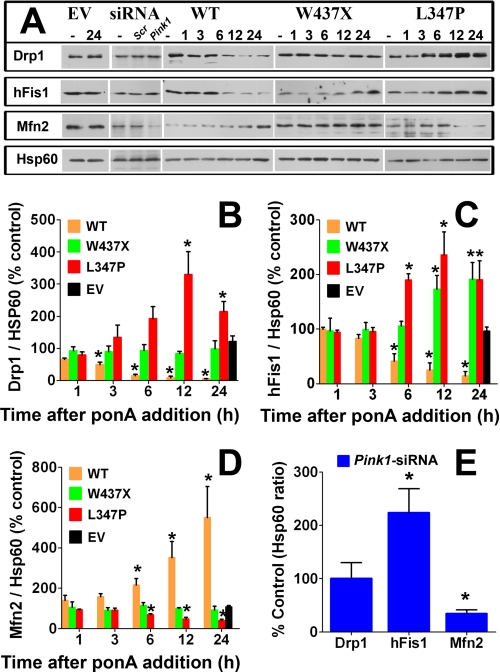
A balance of mitochondrial fission and fusion proteins is critical for the maintenance of proper mitochondrial morphology. Therefore, to investigate further the molecular mechanism behind mitochondrial fragmentation in the present study, mitochondria from different groups of N27 cells were isolated for the assessments of fission and fusion protein levels (Fig. 3). Twenty-four hours after the addition of ponA to the empty vector control cells, no changes were detectable in the mitochondrial fission proteins Drp1 and hFis nor in the fusion protein Mfn2, as illustrated in the immunoblots (Fig. 3A), whose band intensities were quantified (Fig. 3, B–E). However, in cells overexpressing wild type PINK1, the balance of fission/fusion machinery was heavily tipped toward fusion. As shown in Fig. 3, in a time-dependent fashion, PINK1 reduced Drp1 and hFis1 by more than 10-fold and increased Mfn2 to more than 5-fold of the base-line levels 24 h after ponA addition. No changes at the mRNA levels were detected when assessed by quantitative PCR (supplemental Fig. S3), indicating the alterations of these fission and fusion proteins did not originate from the transcript level. In the group of cells overexpressing mutant PINK1, both fission and fusion proteins levels were altered in a direction favoring mitochondrial fission. The most striking effects were observed in cells with PINK1L347P. In these cells, the levels of Drp1 and hFis1 increased in a time-dependent manner and peaked at 12 h after ponA addition, resulting in expression levels 3–4-fold higher than those in the empty vector control group and in the uninduced PINK1L347P cells. The levels of Mfn2, on the other hand, decreased over time by more than 2-fold 24 h post-induction of PINK1L347P. It is evident that PINK1L347P has a completely opposite effect on the levels of these proteins compared with wild type PINK1. Because Drp1 resides primarily in the cytosol, we also assessed the levels of this protein in the cytosolic fractions (supplemental Fig. S4). Neither wild type nor mutant PINK1 induced a change in Drp1 levels in this cellular compartment, suggesting total Drp1 levels were not altered.
FIGURE 3.
Alterations of the fission and fusion proteins induced by changes in PINK1 function. A, mitochondria were isolated from different groups of N27 cells without (−) or with 10 μm ponA at the indicated hours after induction and from empty vector (EV) cells transiently transfected with either Pink-siRNA, scrambled siRNA (scr.), or without siRNA (−). Alterations in the levels of the fission proteins Drp1 and hFis1 and the fusion protein Mfn2 from the ponA treatment (B–D) and from Pink1-siRNA (E) were quantified from individual band intensities and expressed as a percentage of the respective control groups without ponA or of the groups with scrambled siRNA. Data represent the mean ± S.E. from three-four independent experiments, analyzed by one-way ANOVA followed by the Newman-Keuls post hoc test. *, p < 0.05 compared with the respective control groups of each cell type. To avoid complex illustrations, extensive pairwise multiple comparisons between groups are not indicated here. WT, wild type.
To corroborate further the effects of mutant PINK1, endogenous rat Pink1 was knocked down using siRNA as described (supplemental Fig. S1, A and B). Isolated mitochondria from these cells displayed about a 2.5-fold increase in hFis1 and more than a 2-fold decrease in Mfn2 as compared with those obtained from the control cells transfected with scrambled siRNA (Fig. 3E). No apparent change in Drp1 levels was detectable. It is possible that the residual effect of incomplete knockdown of endogenous Pink1 prevented the changes in Drp1 levels. Consistent with this argument are our observations that PINK1W437X, which induces a less pronounced loss of PINK1 kinase activity and less severe dominant negative effects than PINK1L347P (32, 34), did not change Drp1 and Mfn2 levels. Thus, the differential effects observed between these groups of cells may be related to different degrees of loss of PINK1 function. These data also suggest that not all PINK1 mutations have the same extent of pathogenicity, an observation that has been reported in other studies as well.
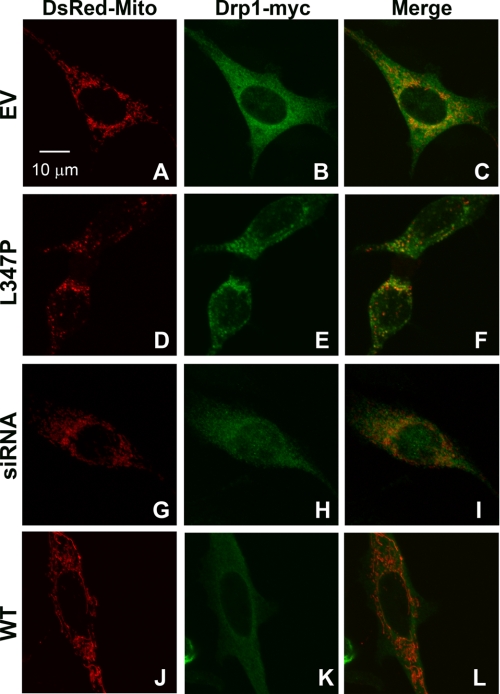
PINK1L347P Induces Drp1 Mitochondrial Translocation
Given that ∼3% of Drp1 is associated with mitochondria and the rest is cytosolic (36), the observations that PINK1L347P increased mitochondrial (, A and B), but not cytosolic (supplemental Fig. S4) Drp1, suggest that this mutant PINK1 does not increase total Drp1. To explore the possibility that this increase was a result of translocation from the cytosol to mitochondria, we co-transfected cells with Drp1-myc and DsRed-Mito. As shown in Fig. 4, after 24 h of induction, PINK1L347P increased both the number and size of Drp1 aggregates that co-localized with mitochondria. When counted blinded using confocal microscopy, this increase was significant as compared with the control empty vector cells (48.67% ± 1.76 versus 10.67% ± 1.76, t test, t15,23 = 4, p < 0.01, n = 3 independent experiments). No apparent changes in translocation were detected in cells with PINK1WT (10.67% ± 2.90), PINK1W437X (10.0% ± 2.31) or with siRNA-Pink1 (12.0% ± 1.15). These results support, first, the argument that PINK1L347P induced translocation, not increases in total levels of Drp1 and, second, explain the undetectable increase in mitochondrial Drp1 in cells with PINK1W437X and siRNA-Pink1 as assessed by immunoblotting (Fig. 3).
FIGURE 4.
PINK1L347P induces mitochondrial translocation of Drp1. N27 cells were co-transfected with DsRed-Mito and Drp1-myc for 6 h before the addition of ponA for an additional 24 h. Cells were then fixed and immunostained for Myc. In control cells with empty vector (EV, A–C), with PINK1WT overexpression (J–L), or with PINK1-knockdown after 48 h Drp1 appeared primarily diffused. However, in cells with PINK1L347P overexpression (D–F), the distribution of Drp1 was punctated and co-localized with mitochondria. WT, wild type.
Pro-fusion or Inhibiting Fission Also Forms Small Donut-shaped Mitochondria
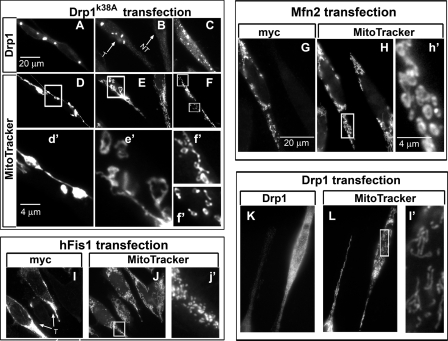
The increase in mitochondrial fission and reduction in fusion proteins as seen with mutant PINK1 and Pink1 knockdown led to a shift toward an overall increase in mitochondrial fission, which is consistent with the appearance of fragmented mitochondria in Fig. 1. On the other hand, overexpression of wild type PINK1 produced a dramatic reduction in both Drp1 and hFis1 proteins and a pronounced increase in Mfn2, which should lead to the fusion-dominant condition. It is possible that PINK1 induced the formation of the unique donut-shaped mitochondria through, at least in part, this fission/fusion imbalance. To investigate whether an overall fusion-favoring state would induce mitochondrial morphology similar to that produced by PINK1 overexpression, we transiently transfected cells with either Drp1 dominant negative (Drp1K38A) or Mfn2 to mimic the changes induced by PINK1. A collage of various images was constructed in Fig. 5 to represent different effects of fusion on mitochondrial morphology 24 h after transfection. In cells transfected with Drp1K38A (Fig. 5, A–C), some displayed remarkably long mitochondria with beads forming along the mitochondrial filaments (Fig. 5, D and d′). In other cells (Fig. 5, E and e′), it is apparent that donut-shaped structures started to break-off from the long mitochondrial networks, a process that perhaps eventually led to the collapse of the whole filamentous networks and resulted in individual circular donut-shaped mitochondrion (Fig. 5, F and f′). In cells with Mfn2 overexpression (Fig. 5, G–h′), in addition to those with elongated and clustered mitochondria (not shown), donut-shaped mitochondria were also detected. Our observations that Mfn2 and Drp1K38A induced the formation of multiple smaller mitochondria perhaps are similar to a previous study which reported that increasing the magnitude of mitochondrial fission inhibition would lead to the collapse and lysis of the mitochondrial network (36).
FIGURE 5.
Donut-shaped mitochondria induced by pro-fusion or fission inhibition. N27 cells were transfected with various indicated constructs and labeled with MitoTracker Red 24 h later for the visualization of mitochondrial morphology followed by immunocytochemistry for the identification of transfected cells. A–C, cells transfected with the dominant negative Drp1K38A (T, as indicated by the arrows) displayed elongated (D, d′) and donut-shaped mitochondria either loosely attached to the branches (E, e′) or completely detached from fragmented tubular mitochondria (F, f′). Non-transfected cells displayed the typical tubular mitochondria in this cell type (E). Cells transfected with Mfn2-myc (G–h′) displayed donut-shaped (H, h′), clustered, and elongated (not shown) mitochondria. Cells transfected with hFis1 (I–j′) or Drp1 (K–l′) displayed mitochondrial fragmentation but without the donut shape appearance. Scale bars, 20 μm for A–L, 4 μm for d′–f′, h′, j′, and l′, which are enlarged from the respective boxed images.
To demonstrate further that fragmented mitochondria produced by a fusion protein is morphologically distinct from that produced by a fission protein, we also transfected cells with the fission proteins hFis1 (Fig. 5, I–j′) and Drp1 (Fig. 5, K–l). These cells produced fragmented mitochondria without the donut shape. Of note, MitoTracker was used in Figs. 5 and 6 because it was difficult to locate cells with and without fission/fusion transfection adjacent to each other using DsRed-Mito because only some cells were transfected.
FIGURE 6.
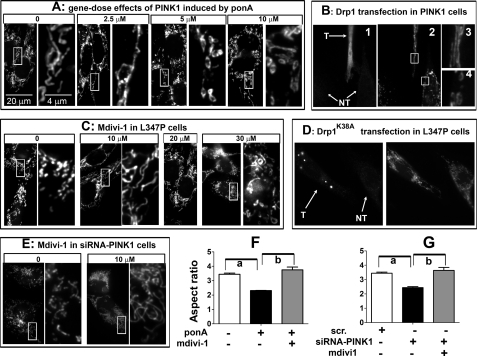
Abnormal mitochondrial phenotype induced by PINK1 and loss of PINK1 function can be prevented through the fission/fusion pathway. A, N27 cells with inducible expression of human PINK1 were induced with varying concentrations of ponA for 24 h before MitoTracker treatment. The right panels of each treatment group are enlarged from the adjacent left boxed images. B, N27 cells as used in A were transiently transfected with Drp1 for 6 h, then induced by 10 μm ponA for 24 h. Cells were then incubated with MitoTracker Red (B2) followed by immunocytochemistry for Drp1 to identify transfected (T) cells and non-transfected (NT) cells (B1). Cells with high Drp1 expression displayed tubular mitochondria (B3), whereas non-transfected cells retained donut-shaped morphology (B4). C, N27 cells with inducible PINK1L347P were treated concurrently with 10 μm ponA and varying concentrations of the Drp1 inhibitor mdivi-1. After 24 h, mitochondrial morphology was assessed using MitoTracker Red. D, N27 cells with inducible PINK1L347P were transiently transfected with Drp1K38A plasmid for 6 h, then induced with 10 μm ponA for 24 h. Cells were then incubated with MitoTracker Red followed by immunocytochemistry for Drp1 to confirm the transfection. Transfected cells (T) displayed tubular mitochondria, whereas non-transfected cells (NT) retained fragmented mitochondrial morphology. E, empty vector cells treated with PINK1-siRNA (50 nm) for 48 h displayed mitochondrial fragmentation that was prevented by 10 μm ponA. The protective effects of mdivi1 in C and D were quantified and expressed as aspect ratio in F and G. Scr., scrambled siRNA. Data represent the mean ± S.E., analyzed by one-way ANOVA (F, F2,8 = 38.8, p < 0.001; G, F2,8 = 23.55, p < 0.001) followed by Newman-Keuls post hoc test. n > 200 mitochondria per group from three independent experiments. a, p < 0.05 compared with the respective control group; b, p < 0.05 compared with the group with mdivi-1.
Abnormal Mitochondrial Morphology Induced by PINK1 and Loss of PINK1 Function Can Be Prevented by Modulations of the Fission/Fusion Pathway
To investigate further the effect of PINK1 on mitochondrial morphology, we first assessed the gene-dose effects of PINK1. Taking advantage of the inducible system in these cells, varying concentrations of ponA were added to induce different expression levels of PINK1, as assessed by quantitative real-time RT-PCR and immunoblotting (supplemental Fig. S5). With 2.5 μm ponA, which increased PINK1 mRNA about 10-fold and increased PINK1 protein to a low but detectable level after 24 h (supplemental Fig. S5), some elongation of mitochondrial tubules was detected (Fig. 6A). However, at concentrations of 5 and 10 μm ponA, PINK1 induced discrete mitochondria with a donut-shaped morphology (Fig. 6A). At 5 and 10 μm, ponA induced PINK1 mRNA ∼1.5 and 2.3-fold higher, respectively, as compared with the expression produced by 2.5 μm. However, these increases in PINK1 transcripts translated into ∼6- and 26-fold (full-length, top band) and ∼2- and 4-fold (mature form, lower band) higher levels in PINK1 protein (supplemental Fig. S5). Together, these results demonstrate that increasing levels of PINK1 induced changes in mitochondrial morphology that potentially could be secondary to increasing fusion.
If the donut shape mitochondria were caused by fusion induced by PINK1 overexpression, we reasoned that this abnormal mitochondrial morphology should be attenuated when the imbalance in mitochondrial fission/fusion machinery was corrected. To this end, we transiently transfected plasmids containing Drp1 into cells with overexpression of PINK1 induced by 10 μm ponA. In cells expressing high levels of Drp1 after transfection, as verified by Drp1 immunoreactivity (Fig. 6B1), mitochondrial morphology was reversed from donut-shaped to regular tubular structure (Figs. 6B, 2 and 3 and 4J). In contrast, in the adjacent lower cells, where Drp1 transfection did not take place, small mitochondria with donut- shaped morphology remained (Fig. 6B, 2 and 4). These results lend further support to the suggestion that PINK1 induced morphological changes in mitochondria through enhanced fusion.
In contrast to wild type PINK1, we hypothesized that PINK1L347P siRNA-Pink1 induced mitochondrial fragmentation by increased fission based on the observations that they increased the levels of fission proteins with a concomitant reduction in Mfn2. Using a small molecule, previously referred to as mdivi-1 (21), which blocks the GTPase activity of Drp1, we were able to block mitochondrial fragmentation induced by PINK1L347P (Fig. 6C) in a dose-dependent manner. The optimal concentration to confer this protective effect was 10 μm. Interestingly, as the concentration increased to 30 μm, small mitochondria with donut-shaped morphology appeared (Fig. 6C), perhaps due to excessive fission inhibition. At 10 μm, mdivi-1 also blocked mitochondrial fragmentation induced by siRNA-Pink1 (Fig. 6E). Transient transfection of cells with Drp1K38A (Fig. 6D) and Mfn2 (supplemental Fig. S6) also inhibited fragmentation induced by PINK1L347P. Together these results support a mitochondrial fusion role for PINK. Mutations in PINK1, as exemplified by PINK1L347P, and loss of Pink1 function mediated by siRNA-Pink1 lead to mitochondrial fragmentation due to excessive fission.
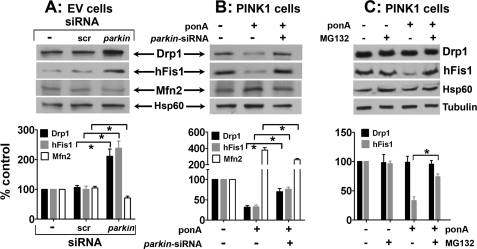
Alterations in Fission and Fusion Protein Levels Are Primarily Mediated by parkin
Parkin has been reported to function downstream of PINK1 (5–8). We, therefore, asked whether this E3 ubiquitin ligase mediated the effects of PINK1 on the alterations of mitochondrial fission/fusion proteins. When parkin was knocked down by ∼80% (supplemental Fig. S1C), we detected about a 2-fold increase in Drp1 and hFis1 levels and an ∼1.4-fold reduction in Mfn2 (Fig. 7A). Severe mitochondrial fragmentation was also detected (data not shown). Next, we used the same strategy to knock down parkin in N27 cells with PINK1 overexpression. Consistent with Fig. 3, 24 h after induction PINK1, the levels of Drp1 and hFis1 decreased, and the level of Mfn2 increased (Fig. 7B). However, when parkin was knocked down, these alterations were significantly attenuated (Fig. 7B). We also observed that knocking down parkin reduced the formation of donut-shaped mitochondria induced by PINK1 overexpression (supplemental Fig. S7).
FIGURE 7.
Parkin mediates the downstream effects of PINK1 on fission and fusion protein levels. A, N27 cells with empty vector (EV) control (−) were transfected with either siRNA targeting rat parkin or non-targeting scrambled control. After 48 h, mitochondrial were isolated and immunoblotted for the proteins of interest. Data represent the mean ± S.E. from three-four independent experiments and are expressed as either the ratio of Drp1, hFis1, or Mfn2 to Hsp60 and normalized as the percentage control to the group without siRNA treatment (−). scr, scrambled siRNA, Differences between means were analyzed by a two-tailed Student's t test (Drp1, t4.19 = 6; *, p = 0.006; hFis1, t5.37 = 6; *, p < 0.01; Mfn2, t4.12 = 4; *, p < 0.01). B, N27 cells were induced with 10 μm ponA for PINK1 expression and transfected with parkin siRNA for 48 h. Data represent the mean ± S.E. from four-five independent experiments and are expressed as indicated above in A. Differences between means were analyzed by a two-tailed Student's t test (Drp1, t4.40 = 8; *, p < 0.01; hFis1, t6.12 = 8; *, p < 0.001; Mfn2, t3.34 = 6; *, p < 0.01). C, N27 cells induced with 10 μm ponA for PINK1 expression were treated with or without 100 nm MG132 for 24 h before extraction of whole cell proteins. Data represent the mean ± S.E. from four independent experiments and are expressed as either the ratio of Drp1 to tubulin or hFis1 to Hsp60 and normalized as percentage control to the control group without ponA and without MG132 treatments. Differences between means were analyzed by a two-tailed Student's t test (t5.09 = 6; *, p < 0.01).
To investigate whether Drp1 and hFis1 reduction induced by PINK1 was mediated by parkin degradation through the ubiquitin-proteasomal pathway, we treated cells with MG132, an inhibitor. Using whole cell lysates, the results indicate that MG132 blocked the reduction of hFis1 induced by PINK1 (Fig. 7C), demonstrating the intriguing possibility that parkin-mediated proteasomal degradation acts on the outer surface of mitochondria to control the levels of hFis1. However, MG132 treatment did not change the total level of Drp1, suggesting that cytosolic Drp1 is not a direct target for this pathway. This result also lends further support to our argument that it is the translocation of Drp1, not its total level, that is affected.
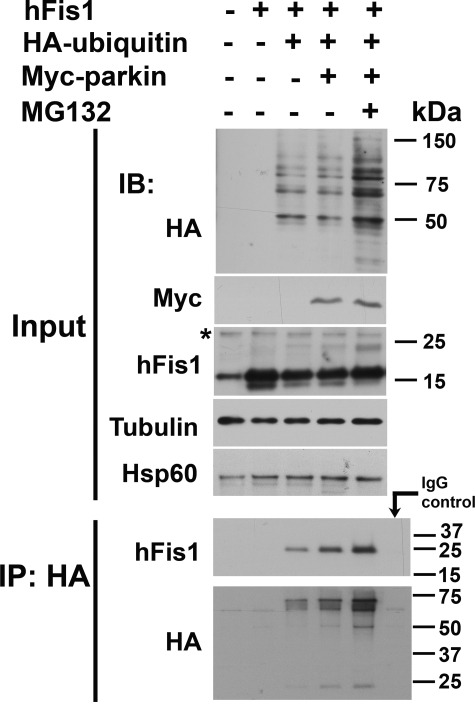
hFis1 Is Ubiquitinated by Parkin
Although our results (Fig. 7) support the role of parkin in the degradation of hFis1 through the proteasomal pathway, we seek to provide a more direct evidence for this mechanism. To this end, we used HEK 293 cells (due to their high transfection efficiency) and transfected them with myc-parkin, HA-ubiquitin, and hFis1 constructs in the presence or absence of MG132 followed by immunoprecipitation (Fig. 8). First, we analyzed total cell lysates (Input panel) using immunoblotting and confirmed the overexpression of these proteins. When ubiquitin and parkin were overexpressed, the total level of hFis1 was reduced, and MG132 blocked this reduction. Similar results were previously observed in Fig. 7C where PINK1 was overexpressed and, thus, consistent with our proposal that parkin functions downstream of PINK1 in the proteasomal degradation of hFis1. Additionally, MG132 increased the accumulation of a band at ∼24 kDa, suggesting parkin adds a single ubiquitin moiety (∼8 kDa) to hFis1. No additional bands specific to MG132 treatment were consistently detectable at higher molecular mass (not shown). When total cell lysates were immunoprecipitated with anti-HA and immunoblotted with hFis1 antibodies, parkin increased the level of pulled down hFis1 at a higher molecular size of ∼24 kDa, suggesting this E3 ubiquitin ligase monoubiquitinates hFis1. Interestingly, although this form of ubiquitination does not direct a protein to proteasomal degradation, the level of hFis1 was dramatically increased in the presence of MG132. In the control immunoprecipitation using mouse IgG pulled down, no hFis1 or HA-ubiquitin was detected from triple-transfected cell lysates treated with MG132. In combination, results from Figs. 7 and 8 support the role of parkin and proteasome in the degradation of hFis1. However, the immunoprecipitation data suggest that parkin may not mediate this degradation through its direct ubiquitination of hFis1.
FIGURE 8.
hFis1 is ubiquitinated by parkin and degraded by proteasome. Total cell lysates from HEK 293 cells transfected with indicated plasmids for ∼48 h in the presence or absence of 10 μm of MG132 for ∼24 h were immunoblotted (IB) with anti-HA, anti-Myc, and hFis1 antibodies (Input panel). Tubulin and Hsp60 were used as the loading control. Immunoprecipitation (IP) was performed with a mouse monoclonal anti-HA antibody. When immunoblotted with a polyclonal anti-hFis1 antibody, a single band of hFis1, shifted from ∼16 to ∼24 kDa was detected in cells transfected with ubiquitin and dramatically increased when parkin and MG132 were also present. * denotes nonspecific band. Experiments were repeated twice.
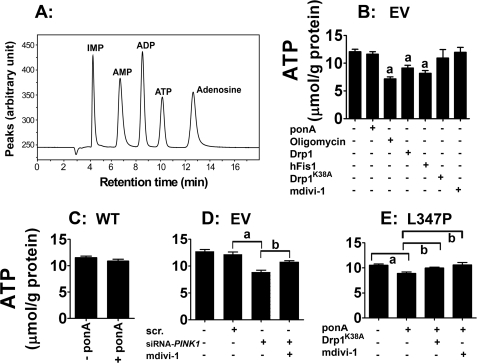
mdivi-1 Is Protective against Bioenergetic Defects Induced by PINK1L347P
As demonstrated, both wild type and mutant PINK1 overexpression induced morphological changes in mitochondria by inducing opposite effects on the mitochondrial fission and fusion machinery. It was not clear, however, whether both forms of abnormal mitochondria would lead to the same functional bioenergetic defect and, more importantly, from a clinically relevant perspective, whether the defect induced by mutant PINK1 can be prevented by manipulating the fission and fusion machinery. To these ends, HPLC was used to measure ATP (Fig. 9A). To validate our model system, we first treated control N27 cells containing the empty vector with oligomycin. As expected, this inhibitor of the electron transport chain complex V significantly reduced ATP levels by ∼40% (Fig. 9B). Transient overexpression of Drp1 and hFis1 also reduced ATP levels (∼20 and ∼30%, respectively). However, in cells either transfected with Drp1K38A or treated with 10 μm mdivi-1, no reduction in ATP was detectable, suggesting that decreasing mitochondrial fission does not reduce ATP production. Consistent with these observations, in cells overexpressing wild type PINK1, no ATP reduction was detected after 24 h of ponA addition (Fig. 9C) despite the formation of small donut-shaped mitochondria under this condition (Fig. 2E).
FIGURE 9.
mdivi-1 prevents bioenergetic defects induced by loss of PINK1 function. A, an HPLC chromatogram illustrates the separation of ATP and its related adenine nucleotides. B, N27 control cells containing the empty vector (EV) were used to assess the effects of mitochondrial fission/fusion on ATP levels. As a positive control, oligomycin (10 μm, a complex V inhibitor) significantly reduced ATP level. Transient transfection of Drp1 and hFis1 also reduced ATP production. In contrast, neither transfection with Drp1K38A nor treatment with mdivi-1 affected ATP levels. C, overexpression of human wild type (WT) PINK1 did not affect ATP levels. D, reduction in ATP was detected in cells transfected with siRNA-Pink1 but not in those with scrambled siRNA or without siRNA (−). E, after 24 h of ponA addition, PINK1L347P caused ATP reduction, which was abolished when cells were transfected with Drp1K38A 6 h before ponA addition or concurrently treated with 10 μm mdivi-1 and ponA for 24 h. Data represent the mean ± S.E. from six independent experiments, analyzed by one-way ANOVA (panel B, F6,29 = 7.99, p < 0.001; panel D, F2,12 = 5.32, p = 0.02; panel E, F3,20 = 5.81, p < 0.01) followed by the Newman-Keuls post hoc test (panel B, a, p < 0.01 compared with the control group without ponA; panel D, a, p < 0.05; panel E, a, p < 0.01, b, p < 0.05).
In cells transfected with siRNA against endogenous rat Pink1, we observed an ∼20% reduction in ATP production (Fig. 9D), suggesting that basal endogenous Pink1 is required for proper ATP production. Last, in cells overexpressing PINKL347P, after 24 h of induction by ponA, an ∼20% reduction in ATP was also detected (Fig. 9E). This metabolic deficit was completely abolished when mitochondrial fission was blocked by either transfection with Drp1K38A or by treatment with mdivi-1. This latter set of data indicates that genetic or pharmacological treatments of cells harboring pathogenic human mutant PINK1 would lead to a positive functional bioenergetic outcome.
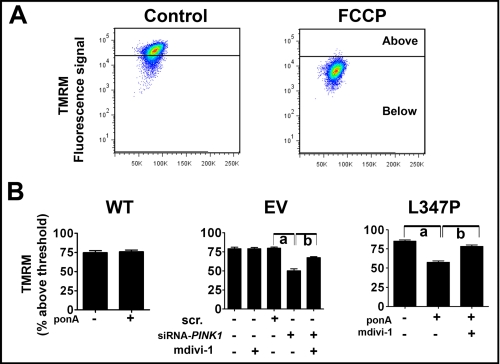
mdivi-1 Protects against the Collapse of Mitochondrial Membrane Potential (Δψm) Induced by PINK1L347P
Cells with PINK1 deficiency display reduced Δψm (11, 15, 37). To further characterize functional defects and recovery of mitochondria in the present study, we used fluorescence-activated cell sorting to measure Δψm in live cells using TMRM (Fig. 10). As expected, the protonophore carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone significantly collapsed Δψm. Consistent with the role of PINK1 in maintaining Δψm, either knocking down or overexpressing PINKL347P significantly reduced Δψm, a defect that was attenuated by mdivi-1. Collectively our data suggest that mdivi-1 confers protection against mitochondrial functional deficits (such as ATP and Δψm reduction) and mitochondrial structural integrity (mitochondrial fragmentation) induced by mutant PINK1.
FIGURE 10.
mdivi-1 prevents collapse of Δψm induced by loss of PINK1 function. A, Δψm of N27 cells was measured using fluorescence-activated cell sorting based on the intensity of the mitochondrial accumulation of 50 nm TMRM. As an illustration, cells with empty vector were treated with 20 μm carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) for 20 min to collapse Δψm, and the obtained TMRM fluorescence was used to set the threshold. B, cells with siRNA-PINK1 (50 nm), but not those with scrambled siRNA (scr) or without transfection, displayed reduced Δψm. After 24 h of 10 μm ponA addition, overexpression of PINK1L347P, but not PINK1WT, also reduced Δψm, a perturbation that was prevented by the treatment of 10-μm mdivi-1. Data are expressed as percentage of cells with signal above the threshold established by FCCP as shown in A. Data represent the mean ± S.E. from six independent experiments, analyzed by one-way ANOVA (Panel EV, F2,17 = 30.64, p < 0.001; panel L347P, F2,17 = 68.35, p = 0.001 followed by Newman-Keuls post hoc test). Panel EV, a, p < 0.01; panel L347P, a, p < 0.01 and b, p < 0.001).
DISCUSSION
In the present study, we provide morphological, functional, and mechanistic evidence to demonstrate that PINK1 has pro-fusion and inhibition of fission functions on mitochondria in dopaminergic mammalian neuronal cells. Furthermore, we also show that mutant PINK1, with primary emphasis on the L347P mutation, induced mitochondrial fragmentation and dysfunction, which could be prevented by a recently described novel molecule mdivi-1.
Morphologically, cells with either endogenous Pink1 knockdown or with mutant PINK1 overexpression displayed severe mitochondrial fragmentation, which can be blocked by mdivi-1 and by transient transfection of dominant negative Drp1K38A or of Mfn2. On the other hand, cells with inducible overexpression of wild type PINK1 displayed gene-dose effects on mitochondrial morphology ranging from elongated to multiple smaller donut-shaped mitochondria. This abnormal morphology was abolished when these cells were transfected with Drp1, suggesting a fusion process was involved. Although additional studies are necessary to fully understand how PINK1 overexpression forms these donut-shaped mitochondria, our data indicate enhanced fusion or inhibition of fission is a possible mechanism. Similar mitochondrial morphology has also been shown in HeLa cells overexpressing parkin and Drp1K38A (38), Mfn2 (39), and siRNA-Drp1 (40). Additionally, when van der Bliek and co-workers (36) expressed different dominant negative mutations of Drp1 in COS7 cells, they demonstrated that mitochondrial phenotypes range from highly interconnected to collapsed and fragmented networks under gradient conditions ranging from weak to strong fission inhibition.
Functionally, in cells with PINK1 overexpression, no collapse of mitochondrial membrane potential or bioenergetic defect was detectable. However, the fragmented mitochondria resulting from loss of PINK1 function exhibited collapsed membrane potential and reduced ATP levels. Although the reduction in ATP levels was modest (15–20%), we believe these results do not undermine the role of PINK1 on mitochondrial function for the following reasons. First, immortalized cells, such as N27 in this study, rely heavily on glycolysis, not oxidative phosphorylation. Hence, perturbation in the mitochondrial electron transport chain may not significantly reduce ATP levels. Consistent with this property of such cell lines, we had to apply 10 μm oligomycin and 10 μm rotenone (data not shown) to reduce ∼40% of ATP level in the present study. In isolated mouse brain mitochondria, 100 nm rotenone reduces ∼70% ATP levels (41). Second, because this is a genetic model that does not directly target the electron transport chain, it is not surprising to see the drop in ATP not as dramatic as the acute toxic models. This scenario of modest bioenergetic defects could be relevant to the chronic slow onset state as seen in patients with PD. More importantly, as a proof of concept, mdivi-1 and Drp1K38A rescue this defect, highlighting a potential novel therapeutic avenue for PD.
Mechanistically, we have identified one pathway by which PINK1 induces its fusion effect on mitochondria. In a time-dependent manner, we demonstrated that overexpression of PINK1 dramatically decreased mitochondrial Drp1 and hFis1 as well as concomitantly increased Mfn2 levels. This synchronous change tips the balance of fission/fusion machinery toward fusion. Opposite effects were detected in cells with Pink1 knockdown or with mutant PINK1. Parallel time-course studies of mitochondrial morphology and Δψm (data not shown) indicate that the changes in fission/fusion proteins precede morphological and functional defects. We discovered that PINK1 mediated these changes through parkin, as knocking down endogenous parkin significantly attenuated the imbalance caused by overexpressed PINK1. We further demonstrated that parkin enhanced the degradation of hFis1 through the proteasomal pathway, as evidenced by either knocking down parkin or adding MG132 increased the levels of hFis1, and conversely, overexpressing parkin plus ubiquitin increased hFis1 degradation, which is blocked by MG132.
As an outer mitochondrial membrane protein, hFis1 has been reported to be a target for the proteasome through ubiquitination, as overexpressing the E3 ubiquitin ligase March-V/MITOL reduced the levels of endogenous hFis1 and MG132 blocked this reduction (42). However, our results suggest that the enhanced degradation of hFis1 via parkin is not likely a direct result of ubiquitination of hFis1. Only monoubiquitinated hFis1 was detected in our immunoprecipitation study; however, polyubiquitination is required for proteasomal degradation. It is possible that parkin indirectly mediates the degradation of hFis1 via polyubiquitination and subsequent proteasomal degradation of an unidentified protein, which stabilizes hFis1. In addition, although well known for its role in polyubiquitination and degradation, parkin has been shown to be a multifunctional E3 ligase (43, 44) capable of performing degradation-independent monoubiquitination (27), a post-translational modification that confers biological functions to the protein. Thus, parkin may have a dual role on hFis1 by indirectly modulating its expression level and by directly affecting its function through a single tagged ubiquitin moiety. The functional significance of monoubiquitinated hFis1 requires additional studies.
Although we detected an increase in the mitochondrial fraction of Drp1 in cells with parkin knockdown, our results indicate parkin does not play a role in Drp1 degradation because MG132 did not increase the total levels of Drp1. Other studies show that ubiquitination of Drp1 by overexpression of MITOL/March-V does not reduce the levels of Drp1 (42, 45). Overexpressing wild type or mutant PINK1 also did not change the total levels of Drp1; however, confocal analysis showed PINK1L347P dramatically increased the translocation of Drp1 from the cytosol to mitochondria. This translocation was not detectable in cells with PINK1W437X or siRNA-Pink1, consistent with the lack of an increase in mitochondrial Drp1 as assessed by immunoblotting (Fig. 3). This lack of effect could be due to the residual effects of incomplete knockdown of endogenous Pink1 and less severe dominant negative effects of PINK1W437X than PINK1L347P (32, 34). The mechanism by which PINK1L347P induced Drp1 translocation remains to be explored.
In contrast to Drp1 and hFis1, parkin exerts an opposite effect on Mfn2. That is, knocking down parkin reduced the level of Mfn2. It is not clear whether this observation was due to a direct lack of parkin function on Mfn2 or a secondary consequence of interactions between Mfn2 and hFis, Drp1, or other proteins that affect the stability of Mfn2. This observation is consistent with a study where overexpression of March V/MITOL in COS7 cells increases mitochondrial fusion in an Mfn2-dependent mechanism (46).
Dysfunction in mitochondria and the ubiquitin-proteasomal pathway represent two major pathogenic mechanisms in PD. Together, mutations in parkin and PINK1 are the two most common causes of recessive familial PD. The coordinated interactions between these two proteins, with parkin being downstream of PINK1, to maintain proper mitochondrial morphology and function have been a topic of great interest in recent years. Parkin has recently been demonstrated to be recruited by PINK1 to dysfunctional mitochondria where it induces mitophagy (38, 47–49). In the present study, we demonstrate that alterations in the level of mitochondrial fission and fusion proteins induced by PINK1 are mediated at least in part through parkin. This is a novel and significant finding because although the degradation of cytosolic proteins through the ubiquitin-proteasomal pathway has been well described, the involvement of this machinery in quality control of mitochondrial proteins has been largely unexplored, especially in the context of PD. Thus, these results suggest that one way by which PINK1 and parkin maintain proper mitochondrial integrity is through quality control of mitochondrial fission and fusion proteins.
In summary, our study provides a mechanism to support the role of PINK1 as a promoter of fusion and an inhibitor of fission in mammalian dopaminergic neuronal cells. The overall effect of these changes is consistent with the mitochondrial fusion effect of PINK1 reported in studies in which other mammalian cell models (11, 15–17) and primary fibroblasts cultured from patients with PINK1 mutations (15, 18) were used. More importantly, we demonstrate that the excessive mitochondrial fragmentation and dysfunction caused by human relevant mutant PINK1 can be rescued by a small molecule that inhibits the mitochondrial fission pathway. Because mutant PINK1 and loss of PINK1 function have a pro-fission effect, molecules such as mdivi-1 may be a significant novel therapeutic avenue for PD and other neurodegenerative disorders such as Alzheimer disease (50, 51) and Huntington disease (52), where excessive mitochondrial fragmentation has been reported.
Supplementary Material
Acknowledgments
We thank Dr. Tianzheng Yu for the initial assistance in the analysis of mitochondrial morphology, Drs. Yanxin Zhao, Raju Ilangovan, and Jacqueline Tibbett for technical assistance, and Dr. Linda Callahan of the University of Rochester School of Medicine Confocal and Conventional Microscopy Core for assistance in using the FV1000 Olympus confocal microscope.
This work was supported, in whole or in part, by National Institutes of Health Grant ES014899 and ES17470 (to K. T.), DK078618 and DK061991 (to Y. Y.), and P30 ES01247 and T32 ES07026.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- PINK1
- PTEN-induced putative kinase 1
- PD
- Parkinson disease
- mdivi-1
- mitochondrial division inhibitor
- EcR
- ecdysone receptor
- ponA
- ponasterone A
- TMRM
- tetramethylrhodamine methyl ester
- RT
- reverse transcription
- GFP
- green fluorescent protein
- siRNA
- small interfering RNA
- HA
- hemagglutinin
- HPLC
- high performance liquid chromatography
- ANOVA
- analysis of variance.
REFERENCES
- 1.Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W. (2004) Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 2.Hatano Y., Li Y., Sato K., Asakawa S., Yamamura Y., Tomiyama H., Yoshino H., Asahina M., Kobayashi S., Hassin-Baer S., Lu C. S., Ng A. R., Rosales R. L., Shimizu N., Toda T., Mizuno Y., Hattori N. (2004) Ann. Neurol. 56, 424–427 [DOI] [PubMed] [Google Scholar]
- 3.Rohé C. F., Montagna P., Breedveld G., Cortelli P., Oostra B. A., Bonifati V. (2004) Ann. Neurol. 56, 427–431 [DOI] [PubMed] [Google Scholar]
- 4.Mills R. D., Sim C. H., Mok S. S., Mulhern T. D., Culvenor J. G., Cheng H. C. (2008) J. Neurochem. 105, 18–33 [DOI] [PubMed] [Google Scholar]
- 5.Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 6.Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 7.Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J., Lee G., Chung J. (2009) Biochem. Biophys. Res. Commun. 378, 518–523 [DOI] [PubMed] [Google Scholar]
- 9.Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E. A., Przedborski S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12022–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petit A., Kawarai T., Paitel E., Sanjo N., Maj M., Scheid M., Chen F., Gu Y., Hasegawa H., Salehi-Rad S., Wang L., Rogaeva E., Fraser P., Robinson B., St. George-Hyslop P., Tandon A. (2005) J. Biol. Chem. 280, 34025–34032 [DOI] [PubMed] [Google Scholar]
- 11.Sandebring A., Thomas K. J., Beilina A., van der Brug M., Cleland M. M., Ahmad R., Miller D. W., Zambrano I., Cowburn R. F., Behbahani H., Cedazo-Mínguez A., Cookson M. R. (2009) PLoS One 4, e5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque M. E., Thomas K. J., D'Souza C., Callaghan S., Kitada T., Slack R. S., Fraser P., Cookson M. R., Tandon A., Park D. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng H., Jankovic J., Guo Y., Xie W., Le W. (2005) Biochem. Biophys. Res. Commun. 337, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 14.Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., Lu B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Exner N., Treske B., Paquet D., Holmström K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H. H., Gasser T., Krüger R., Winklhofer K. F., Vogel F., Reichert A. S., Auburger G., Kahle P. J., Schmid B., Haass C. (2007) J. Neurosci. 27, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagda R. K., Cherra S. J., 3rd, Kulich S. M., Tandon A., Park D., Chu C. T. (2009) J. Biol. Chem. 284, 13843–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutz A. K., Exner N., Fett M. E., Schlehe J. S., Kloos K., Lämmermann K., Brunner B., Kurz-Drexler A., Vogel F., Reichert A. S., Bouman L., Vogt-Weisenhorn D., Wurst W., Tatzelt J., Haass C., Winklhofer K. F. (2009) J. Biol. Chem. 284, 22938–22951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grünewald A., Gegg M. E., Taanman J. W., King R. H., Kock N., Klein C., Schapira A. H. (2009) Exp. Neurol. 219, 266–273 [DOI] [PubMed] [Google Scholar]
- 19.Detmer S. A., Chan D. C. (2007) Nat. Rev. Mol. Cell Biol. 8, 870–879 [DOI] [PubMed] [Google Scholar]
- 20.Knott A. B., Perkins G., Schwarzenbacher R., Bossy-Wetzel E. (2008) Nat. Rev. Neurosci. 9, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassidy-Stone A., Chipuk J. E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M. J., Shaw J. T., Hinshaw J. E., Green D. R., Nunnari J. (2008) Dev. Cell 14, 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad K. N., Clarkson E. D., La Rosa F. G., Edwards-Prasad J., Freed C. R. (1998) Mol. Genet. Metab. 65, 1–9 [DOI] [PubMed] [Google Scholar]
- 23.Prasad K. N., Carvalho E., Kentroti S., Edwards-Prasad J., Freed C., Vernadakis A. (1994) In Vitro Cell Dev. Biol. Anim. 30A, 596–603 [DOI] [PubMed] [Google Scholar]
- 24.Halterman M. W., Giuliano R., Dejesus C., Schor N. F. (2009) J. Neurosci. Methods 177, 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Gao J., Chung K. K., Huang H., Dawson V. L., Dawson T. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore D. J., West A. B., Dikeman D. A., Dawson V. L., Dawson T. M. (2008) J. Neurochem. 105, 1806–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manfredi G., Yang L., Gajewski C. D., Mattiazzi M. (2002) Methods. 26, 317–326 [DOI] [PubMed] [Google Scholar]
- 29.Volonté M. G., Yuln G., Quiroga P., Consolini A. E. (2004) J. Pharm. Biomed. Anal. 35, 647–653 [DOI] [PubMed] [Google Scholar]
- 30.Criscuolo C., Volpe G., De Rosa A., Varrone A., Marongiu R., Mancini P., Salvatore E., Dallapiccola B., Filla A., Valente E. M., De Michele G. (2006) Movement Disorders 21, 1265–1267 [DOI] [PubMed] [Google Scholar]
- 31.Rogaeva E., Johnson J., Lang A. E., Gulick C., Gwinn-Hardy K., Kawarai T., Sato C., Morgan A., Werner J., Nussbaum R., Petit A., Okun M. S., McInerney A., Mandel R., Groen J. L., Fernandez H. H., Postuma R., Foote K. D., Salehi-Rad S., Liang Y., Reimsnider S., Tandon A., Hardy J., St. George-Hyslop P., Singleton A. B. (2004) Arch. Neurol. 61, 1898–1904 [DOI] [PubMed] [Google Scholar]
- 32.Pridgeon J. W., Olzmann J. A., Chin L. S., Li L. (2007) PLoS Biol. 5, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W., Vives-Bauza C., Acín-Peréz R., Yamamoto A., Tan Y., Li Y., Magrané J., Stavarache M. A., Shaffer S., Chang S., Kaplitt M. G., Huang X. Y., Beal M. F., Manfredi G., Li C. (2009) PLoS One 4, e4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D. W., Petsko G. A., Cookson M. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5703–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E. M., Casari G. (2005) Hum. Mol. Genet. 14, 3477–3492 [DOI] [PubMed] [Google Scholar]
- 36.Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. (2001) Mol. Biol. Cell 12, 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood-Kaczmar A., Gandhi S., Yao Z., Abramov A. Y., Abramov A. S., Miljan E. A., Keen G., Stanyer L., Hargreaves I., Klupsch K., Deas E., Downward J., Mansfield L., Jat P., Taylor J., Heales S., Duchen M. R., Latchman D., Tabrizi S. J., Wood N. W. (2008) PLoS. One 3, e2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rojo M., Legros F., Chateau D., Lombès A. (2002) J. Cell Sci. 115, 1663–1674 [DOI] [PubMed] [Google Scholar]
- 40.Möpert K., Hajek P., Frank S., Chen C., Kaufmann J., Santel A. (2009) Exp. Cell Res. 315, 2165–2180 [DOI] [PubMed] [Google Scholar]
- 41.Tieu K., Perier C., Caspersen C., Teismann P., Wu D. C., Yan S. D., Naini A., Vila M., Jackson-Lewis V., Ramasamy R., Przedborski S. (2003) J. Clin. Invest. 112, 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H., Inatome R., Yanagi S. (2006) EMBO J. 25, 3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore D. J. (2006) Biochem. Soc. Trans. 34, 749–753 [DOI] [PubMed] [Google Scholar]
- 44.Tai H. C., Schuman E. M. (2008) Nat. Rev. Neurosci. 9, 826–838 [DOI] [PubMed] [Google Scholar]
- 45.Karbowski M., Neutzner A., Youle R. J. (2007) J. Cell Biol. 178, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. (2006) EMBO Rep. 7, 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Su B., Siedlak S. L., Moreira P. I., Fujioka H., Wang Y., Casadesus G., Zhu X. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19318–19323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., Su B., Lee H. G., Li X., Perry G., Smith M. A., Zhu X. (2009) J. Neurosci. 29, 9090–9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H., Lim P. J., Karbowski M., Monteiro M. J. (2009) Hum. Mol. Genet. 18, 737–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.