Abstract
Ror2 is an orphan receptor tyrosine kinase with expression normally restricted to early stages of development. However, emerging evidence has placed aberrantly expressed Ror2, leading to an invasive phenotype, in several cancers including renal cell carcinoma (RCC). Although Ror2 is currently identified as up-regulated in an assortment of cancers, neither the regulatory role or mechanism of action have been delineated. We sought to place Ror2 in the most commonly mutated pathway of RCC, the loss of the tumor suppressor von Hippel-Lindau (VHL), which causes hypoxia-inducible factor (HIF)-1α and -2α stabilization and the transcriptional activation of a broad repertoire of response genes. We found that Ror2 was indeed associated with the pVHL loss in RCC as well as with VHL somatic mutations tightly coordinated with the induction of RCC. Additionally, knockdown and rescue analysis of HIF expression suggests that Ror2 is dependent on pathologic stabilization of either HIF-1α or HIF-2α. Subsequent evaluation of the ROR2 promoter suggests that HIF-2α and its dimerization partner, aryl hydrocarbon nuclear transferase localize to the ROR2 promoter via a cryptic transcriptional element. This data substantiates a unique regulation pattern for Ror2 in the VHL-HIF axis that has the potential to be applied to other cancer etiologies.
Keywords: Diseases/Cancer, Chromatin immunoprecipitation (ChIP), Hypoxia, Kidney, Receptor Tyrosine Kinase, HIF, RCC, Renal Cell Carcinoma, Ror2, VHL
Introduction
The tyrosine kinase Ror2 was initially identified as a homologue of the Trk neurotrophin receptors (1) and later as a member of the receptor tyrosine kinase superfamily (2). Ror2 is an orphan receptor with expression in the developing embryo identified in the embryonic limb buds, heart, primitive genitalia, developing somites, and mesenchymal cells in the developing lung, kidney, and cephalic regions (3–5). In the adult organism, Ror2 expression is present as a part of osteoblast differentiation, highly induced in the preosteoblast stage (6), and is suppressed as these cells terminally differentiate as osteocytes. This pattern of expression is inversely related to that of secreted frizzled related protein 1 (sFRP1), and can be transcriptionally suppressed by ectopic expression of sFRP1 in this cell type, but further insights into the major elements of Ror2 regulation are not known.
Aside from developmental programs regulating bone morphogenesis and primitive organ development, Ror2 has only recently been recognized to play a role in the adult organism. We have identified Ror2 expression as a characteristic of many renal cell carcinoma (RCC)2 cell lines and human tumors (7), where its expression is associated with increased cell migration and anchorage-independent growth. Ror2 also plays a prominent role in osteosarcoma (8) and has recently been identified in squamous cell carcinoma of the head and neck, where expression parallels the development of invasive features of these tumors (9). Furthermore, in a tumor genomic analysis of invasive gastric cancers, Ror2 was identified as a frequent target of mutagenesis (10). These findings place Ror2 as a frequently up-regulated feature of human cancers, and in each case, is associated with invasive tumor characteristics. Although Ror2 has been identified as a frequently overexpressed protein in a variety of tumor types, the regulatory or mechanistic events contributing to increased Ror2 expression in these tumors have yet to be deduced.
Ror2 was initially identified as a renal tumor antigen in cell lines derived from clear cell renal cell carcinomas that were known to have inactivating mutations of the von Hippel-Lindau (VHL) tumor suppressor. The majority of clear cell RCC tumors are characterized by mutation, methylation, or loss of the VHL gene (11–14). The most well documented function of the pVHL protein is to act as the substrate recognition component of an E3 ubiquitin ligase complex that includes Elongin C, Elongin B, Cullin 2, and ring box protein 1 (Rbx1 or Roc1) (15–18). The substrates of pVHL E3 ligase activity most tightly associated with RCC are the hypoxia-inducible factor (HIF)-α subunits (HIF-1α and HIF-2α), a family of transcription factors that coordinate much of the physiologic response to restricted oxygen availability (19–22). Under normal oxygen conditions, the prolyl hydroxylases (PHDs) hydroxylate the HIF subunits, which are subsequently recruited by pVHL to an E3 ubiquitin ligase complex for ubiquitylation, leading to proteasomal degradation (23–25). Under low oxygen conditions or as a result of VHL inactivation, one or both of these HIF factors are stabilized leading to the formation of a transcriptional complex with aryl hydrocarbon receptor nuclear translocator (ARNT, also known as HIF-1β), inducing the transcription of a large panel of hypoxia responsive genes including vascular endothelial growth factor, glucose transporter 1 (GLUT1), prolyl hydroxylase family member Egln3 (also known as PHD3), among many others (26, 27). This transcriptional response leads to the highly vascular phenotype of RCC and the transformed phenotype of the cells (28).
VHL mutation is an important event in the development of RCC, and activation of components of the HIF pathway can be detected early in pre-malignant cysts that precede the development of cancer in patients harboring a germline mutation in VHL (29). However, within the spectrum of the VHL mutation, tumors can demonstrate either stabilization of one or both of these HIF factors, and may promote the transcriptional activation of certain subsets of the repertoire of hypoxia response-induced genes (27). Additionally, the VHL mutational subtype may itself mediate HIF expression patterns based on studies in vitro (30), which parallel tumor growth in vivo (31). In particular, examination of human RCC tumors has demonstrated that the molecular profile of tumors is highly dependent on the expression of HIF-1 and HIF-2 in comparison to those tumors solely expressing HIF-2, with distinctions that correlate with divergent tumor activity (32).
Although VHL mutation and HIF dysregulation have been identified as major contributors to the RCC tumor phenotype, the specific molecular mechanisms associated with this pathway that contribute to RCC features of cell growth, invasion, or metastasis remain an active area of investigation. Thus, we sought to examine the potential that Ror2 regulation was occurring as a part of the VHL/HIF axis in RCC. We found that Ror2 expression was definitively associated with the loss of pVHL as well as with VHL mutations most tightly correlated with HIF-2α dysregulation. Knockdown analysis and rescue experiments suggest that Ror2 is dependent on the pathologic stabilization of either HIF-2α or HIF-1α expression, although it is not expressed as a component of the physiologic response to hypoxia. This finding prompted an examination of the ROR2 promoter, which suggests that HIF-2α and ARNT are localized to the ROR2 promoter, potentially utilizing a cryptic element for its interaction. Coupled with observations that Ror2 has been shown to be involved in the invasive tumor phenotype of RCC and other malignancies, this study places Ror2 in the VHL-associated HIF transcriptional pathway associated with VHL mutation-mediated stabilization of HIF factors providing support for a model where the spectrum of HIF target genes activated as a result of the VHL mutation may contribute substantially to the phenotype of an individual tumor.
EXPERIMENTAL PROCEDURES
Antibodies
Both monoclonal and polyclonal antibodies against Ror2 were obtained from R&D Systems (Minneapolis MN). Antibodies against HIF-2α and Ku80 (loading control) were obtained from Genetex (San Antonio, TX). The HIF-1α antibody was obtained from BD Transductions (Franklin Lakes, NJ) and the Glut1 and Egln3 antibodies were obtained from Novus Biologicals (Littleton, CO). For Figs. 3B and 5, a HIF-2α antibody obtained from R&D Systems was used for analysis.
FIGURE 3.
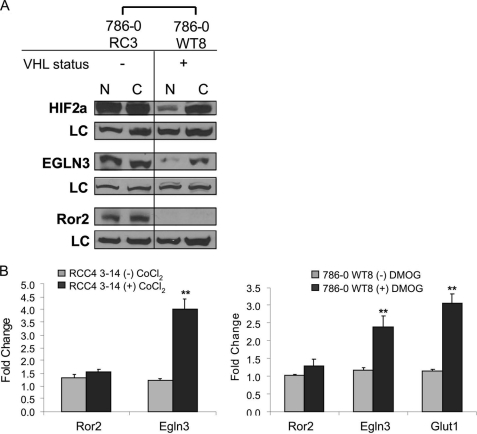
Minimal hypoxia induction of Ror2 expression. A, Ror2 expression is minimally induced after 48 h of exposure to cobalt chloride. Normoxic (N) and CoCl2 (C)-treated protein samples were immunoblotted with HIF-2α and Egln3 antibodies to show that HIF-2α and the HIF target Egln3 were induced in VHL(+) cell lines. Ror2 levels remained stable to this manipulation. Ku80 antibody was used as a loading control (LC). B, left, ROR2 mRNA levels are minimally induced under hypoxic-like conditions in RCC4 cells. qRT-PCR analysis of the hypoxia mimetic cobalt chloride (CoCl2) transcriptional induction after 24 h in the VHL expressing cell line RCC4 3-14 demonstrate induction of the HIF target gene EGLN3 (**, p = 0.0015) in response to treatment with hypoxia mimetic, confirming HIF transcriptional activity. Ror2 mRNA was not significantly induced under these conditions. Right, Ror2 mRNA levels are minimally induced under hypoxic-like conditions in 786-0 WT8 cells. qRT-PCR analysis of the hypoxia mimetic DMOG transcriptional induction after 24 h in the VHL expressing cell line 786-0 WT8 demonstrate induction of HIF target genes EGLN3 (**, p = 0.0091) and GLUT1 (**, p = 0.0015) in response to treatment with the hypoxia mimetic, confirming HIF transcriptional activity. Ror2 transcript levels show minimal enrichment upon treatment. Transcript values are shown as normalized to the β-actin RNA internal standard and relative to the unstimulated cells of each set of paired cells. Error bars represent ± S.E.
FIGURE 5.
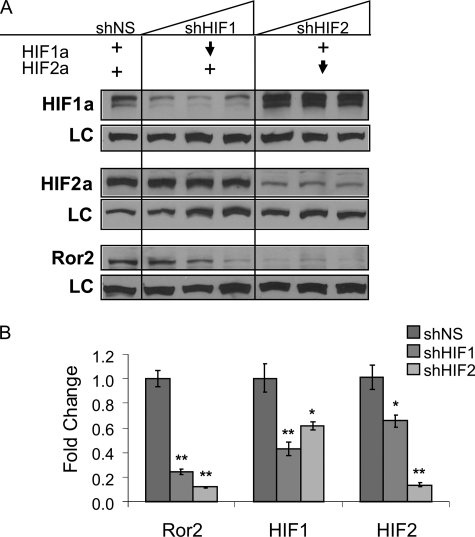
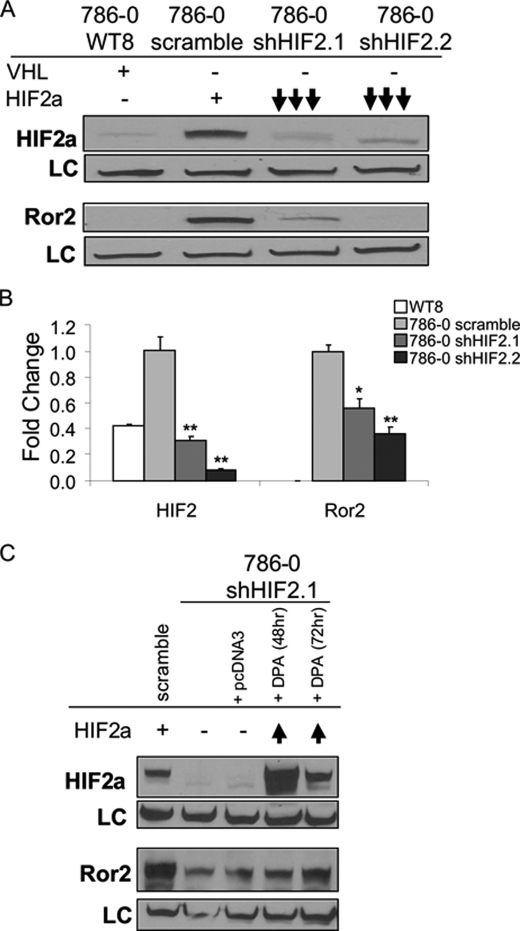
Ror2 is regulated by HIF-1α and HIF-2α expression. A, knockdown of either HIF-1α or HIF-2α subunits suppresses Ror2 expression. Cells constitutively expressing both HIF-1α and HIF-2α were subjected to lentiviral shRNA of each subunit and sorted by GFP expression. Three populations sorted for increasing GFP expression were assayed for each knockdown and compared with a nonspecific sequence (shNS). When HIF-2α levels were suppressed, HIF-1α levels remain constant, whereas Ror2 was down-regulated. Similarly, when HIF-1α levels were highly suppressed and HIF-2α was stable, Ror2 was also down-regulated. B, HIF-1α and HIF-2α knockdown each lead to a loss of ROR2 expression. qRT-PCR analysis demonstrated effective shRNA-mediated suppression of HIF-1α (**, p = 0.0074; *, p = 0.0165) or HIF-2α (**, p = 0.0002; *, p = 0.0277). Ror2 expression is regulated by both HIF factors at the transcriptional level (**, p = 0.0002 HIF-1α, p < 0.0001 HIF-2α). Error bars represent ± S.E.
Chromatin Immunoprecipitation Analysis
Cells were grown to 80% confluence before being cross-linked with 11% formaldehyde. The cross-linked DNA was then sonicated. Chromatin immunoprecipitation analysis was performed with HIF-1α (BD Transduction), HIF-2α (R&D Systems), and ARNT1 (Novus Biologicals). The resulting chromatin immunoprecipitated DNA was analyzed by PCR using primers targeting the known hypoxia response element region of Egln3/PHD3 (reverse primer, 5′-TAGGCACAGTAAACAGGCCC-3′; forward primer, 5′-AGCGTCCGTTCCCAGCTCAG-3′) as previously described (35) as positive controls to validate the system. Additionally, primers were designed to target the 1-kb region of the ROR2 promoter: primer A (reverse primer, 5′-ACGCGCTTGTCCCCACCGAC-3′, forward primer, 5′-CTGCACTGCGCACCGGGACA-3′); primer B (reverse primer, 5′-CGCTCTGGGCGCCTCTCCT-3′, forward primer, 5′-AGGAGCTGGGGCTGCAGCTGGG-3′), primer C (reverse primer, 5′-AGCCCGCGCCAAGGAACCTC-3′; forward primer, 5′-ACGATGCGTCCGCTCCTCCT-3′), and primer D (reverse primer, 5′-TCTGGCGTTCCGGCTTGTGC-3′; forward primer, 5′-CGTCGGGCGAGATGCGAATGG-3′).
Cell Culture and Cell Treatments
The 786-0 cell line and their paired derivatives, WT8 and RC3 (kindly provided by Dr. W. Kaelin, Boston MA), the 786-0 VHL point mutant knock-in cell lines and the RCC4 paired cell lines, 3-14 and 2-1 (kindly provided by Dr. M. C. Simon and Dr. B. Keith, Philadelphia, PA) were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, l-glutamine, and nonessential amino acids. For treated cells, the following conditions were utilized: cobalt chloride (CoCl2) (Thermo Sci Acros Organics, Geel, Belgium) was used at 100 μm for the specified times, dimethyloxalylglycine (DMOG) (Cayman Chemical Company, Ann Arbor, MI) at 1 mm for 24 h, or cells were incubated under conditions of 1% hypoxia for the specified times.
Immunoblot Analysis
Proteins were extracted from cultured cells using cell lysis buffer consisting of 20 mm Tris, 100 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, and a protease inhibitor mixture (Roche Diagnostics). Samples were quantitated using Bradford reagent to measure absorbance. 40–50 μg of protein samples were then separated on 8% SDS-polyacrylamide gels, transferred to nitrocellulose membrane (GE Healthcare), and the proper horseradish-conjugated secondary antibodies were used. Proteins were detected using the Amersham Biosciences ECL-Plus System and visualized on autoradiography film (ISC BioExpress, Kaysville, UT).
qRT-PCR
cDNA was made by reverse transcription from 0.5 μg of total RNA (5PRIME Perfect Pure RNA Cultured Cell Kit, Gaithersburg, MD) using oligo(dT) primers and the SuperScript II RT-PCR reagents (Invitrogen). Commercially available proprietary FAM-labeled primers (ROR2, EGLN3, GLUT1, HIF-1α, HIF-2α, and β-actin) were used for amplification and the samples analyzed on the 7900H Fast Real Time PCR System using the recommended cycle conditions (Applied Biosystems, Foster City, CA).
Transfections and shRNA
shRNA knockdown cells were generated by infecting cells with pRetroSuper (pRS) packaged shRNA for HIF-2α (kindly provided by Dr. W. Kaelin, Dana Farber Cancer Institute (33)). Cells were supplemented with puromycin in the medium to maintain the shRNA for stable suppression. Rescue of HIF-2α shRNA cell lines were generated transiently by using a HIF-2α (mutated double proline to alanine, DPA) expressing plasmid in a pcDNA3 backbone (kindly provided by Drs. B. Keith and M. C. Simon, University of Pennsylvania). RCC4 lentiviral knockdown cells were generated by infecting the cells with pLL 5.0 packaged shRNA (with a GFP-expressing tag) for HIF-1α and HIF-2α (kindly provided by Dr. W. Kim, University of North Carolina) and GFP-positive cells were selectively sorted via GFP expression levels by the University of North Carolina Flow Cytometry Core Facility.
Statistical Analysis
A one-way analysis of variance was used to analyze the standardized Ct values with the specified qRT-PCR target (ROR2, EGLN3, GLUT1, HIF-1α, and HIF-2α) as the fixed constant. Verification of the one-way analysis of variance model was performed using a Kruskal-Wallis test, a nonparametric statistical procedure, to compare the standardized Ct values. Error bars represent mean ± S.E. p values referenced as are followed: *, p < 0.05 and **, p < 0.01.
RESULTS
Ror2 Expression in Clear Cell RCC Is Dependent on VHL Status
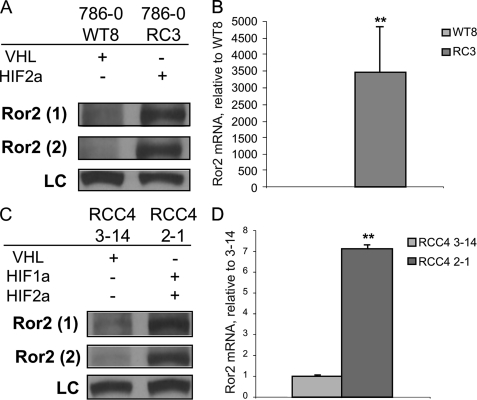
Our previous work has shown that Ror2 is expressed in RCC tumors and RCC cell lines. However, the tumorigenic mechanism that contributes to this aberrant regulation of Ror2 in RCC remained uncertain. Because pVHL expression is tightly associated with clear cell RCC, and the RCC cell lines in which Ror2 was initially identified lack functional pVHL expression, we sought to evaluate the expression of Ror2 in RCC cell lines in relation to the pVHL status. Whole cell extracts were examined by immunoblot for Ror2 expression in two independent sets of RCC cell lines. Ror2 was first examined in the VHL mutant cell line 786-0, stably transfected with empty expression vector (786-0 RC3), confirming Ror2 protein expression demonstrated by two independent antibodies targeting the Ror2 extracellular domain. However, when pVHL expression was restored by the addition of a HA-tagged VHL cDNA (786-0 WT8), Ror2 expression was undetectable using either antibody (Fig. 1A). To explore the nature of this regulation as a transcriptional event or as a consequence of protein stabilization, the VHL-dependent expression of ROR2 in RCC was assayed at the mRNA level using qRT-PCR. The VHL(−) 786-0 RC3 vector control cell line had significantly enhanced ROR2 expression of transcript compared with the VHL(+) wild type 786-0 WT8 derivative cell line, for which no transcript was detectable (Fig. 1B), paralleling the protein levels.
FIGURE 1.
Ror2 is regulated by VHL status. A, Ror2 is expressed when VHL is mutated in the 786-0 paired cell line. Whole cell extracts from 786-0 cells expressing (786-0 WT8) or lacking expression (786-0 RC3) of pVHL were immunoblotted with monoclonal Ror2 antibody (Ror2(1)), polyclonal Ror2 antibody (Ror2(2)), and Ku80 antibody as a loading control (LC). B, Ror2 expression is regulated at the transcriptional level in the paired 786-0 cell lines. qRT-PCR analysis data are normalized to β-actin as an RNA internal standard and displayed as expression levels relative to the pVHL expressing member of the cell line pair. Significant differences were observed as ROR2 mRNA levels were suppressed in the VHL(+) cell line (light gray), relative to the paired vector-transfected VHL(−) control (dark gray), **, p < 0.0001. C, Ror2 is expressed when VHL is mutated in the RCC4 paired cell line. Whole cell extracts from RCC4 cells expressing (RCC4 3-14) or lacking expression (RCC4 2-1) of pVHL were immunoblotted with monoclonal Ror2 antibody (Ror2(1)), polyclonal Ror2 antibody (Ror2(2)), and Ku80 antibody as a LC. D, Ror2 expression is regulated at the transcriptional level in the paired RCC4 cell lines. qRT-PCR analysis data are normalized to β-actin as an RNA internal standard and displayed as expression levels relative to the pVHL expressing member of the cell line pair. Significant differences observed as ROR2 mRNA levels were suppressed in the VHL(+) cell line (light gray), relative to the paired vector-transfected VHL(−) control (dark gray), **, p = 0.0001. Error bars represent mean ± S.E.
To verify that the VHL-dependent regulation of Ror2 observed was not cell line specific, we utilized the RCC4 paired cell line for further analysis. Our results showed that VHL-null RCC4 cells stably transfected with vector (RCC4 2-1) expressed Ror2, whereas RCC4 cells stably expressing an HA-tagged wild type VHL cDNA (RCC4 3-14) displayed dramatically reduced levels of Ror2 protein as detected by immunoblot (Fig. 1C). This VHL dependence was also confirmed at the transcript level as ROR2 mRNA was suppressed to a significant degree in the VHL(+) cell line RCC4 3-14 compared with the VHL(−) counterpart RCC4 2-1 (Fig. 1D). These findings confirm that Ror2 expression is VHL-dependent at both the protein and mRNA levels suggesting a transcriptionally regulated mechanism of Ror2 expression is involved in multiple RCC cell lines.
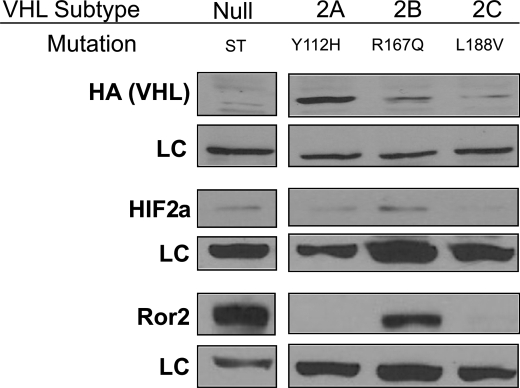
As regulation of Ror2 by wild type pVHL protein was evident at both the protein and mRNA levels, we also sought to evaluate the expression of Ror2 using the tight genotype/phenotype correlation seen with representative VHL mutations associated with the various VHL disease subtypes. We examined a panel of VHL somatic point mutants expressed in 786-0 cells, representative of the mutant VHL transgenes seen in the major subtypes of VHL disease associated with cancer predisposition. VHL Type 2B mutations are associated with the most diverse presentation of VHL disease and are thought to be more highly penetrant for the development of RCC. In these stable clonal cell lines expressing representative pVHL proteins, overexpression of Ror2 was detected by immunoblot only in the absence of pVHL or in the presence of a representative mutant pVHL of VHL disease, subtype type 2B (Fig. 2). This mutation, arginine 167 to glutamine (R167Q), has previously been associated with HIF-2α stabilization similar in extent to the VHL(−) cells (30). Furthermore, representative mutations of VHL disease subtypes 2A, which displays minimal deregulation of HIF factors and low risk for renal cell carcinoma and type 2C, which retains complete regulation of HIF factors and produces no risk for renal cell carcinoma, were completely able to reverse the aberrent Ror2 expression. This data ties the expression of Ror2 to VHL mutation and to correlative genotype/phenotype VHL regulation and its expression in RCC.
FIGURE 2.
Expression of Ror2 in VHL somatic mutation subtypes. A panel of VHL point mutant transgenes expressed in 786-0 cells, which recapitulate the mutant VHL genes representative of the major subtypes of VHL disease, was subjected to immunoblot analysis. There is graded HIF-2α expression associated with the VHL mutations, with the greatest HIF-2α levels present in cells expressing the VHL type 2B mutant and the vector-transfected cells that lack pVHL expression. Expression of Ror2 is detected outside of the VHL null cells only in the most penetrant subtype, VHL type 2B. Ku80 antibody used as a loading control (LC).
Minimal Induction of Ror2 under Hypoxic Conditions
The findings that Ror2 expression was associated with VHL loss or mutations associated with HIF deregulation suggested that Ror2 expression was correlated with HIF-2α stabilization. Thus, we asked if Ror2 may be regulated by the cellular hypoxic response. Multiple strategies were evaluated as stimulants of HIF stabilization, including exposure to 1% hypoxia and treatment with various hypoxia mimetics. We examined the Ror2 protein as a target of the hypoxic response by evaluating Ror2 protein levels by immunoblot in the 786-0 paired RCC cells after treatment a 16–96-h stimulation with 1% oxygen, or hypoxia mimetics CoCl2 and DMOG. HIF, Egln3, and Ror2 were highly expressed in cells lacking functional VHL expression levels, and were not further induced by CoCl2. In 786-0 cells rescued with wild type VHL (786-0 WT8), we observed an expected induction in HIF-2α levels and the concurrent induction of the well established hypoxia target Egln3 (a HIF-dependent prolyl hydroxylase). However, Ror2 protein levels were at most only minimally induced in the cell lines with functional VHL (an example, using CoCl2 is shown in Fig. 3A).
As the primary event of hypoxia response signaling occurs at the transcriptional level, the effects of induction of the hypoxia transcriptional response were also examined using quantitative RT-PCR. The HIF target EGLN3, as well as another established HIF target, the glucose transporter (GLUT1), were used to demonstrate the traditional effect of these treatments on hypoxia response gene induction. In response to CoCl2 treatment, we observed an appropriate and significant increase in Egln3 mRNA after 24 h in RCC4 cells expressing a wild type VHL (RCC4 3-14), which are known to display intact hypoxia response signaling. However, no significant change was observed in the levels of ROR2 transcript in response to this treatment (Fig. 3B). A second, more potent, chemical mimetic of hypoxia, the prolyl hydroxylase inhibitor DMOG, was tested to determine whether maximal induction of this pathway may be sufficient to generate an induction in ROR2 levels after 24 h of treatment. Both control transcripts (GLUT1 and EGLN3) were robustly induced in response to this treatment within the 786-0 WT8 cell line (with functional VHL) demonstrating a functional hypoxia pattern of transcriptional response. Again, the hypoxia mimetic, DMOG, failed to show a statistically significant increase in total ROR2 transcript levels (Fig. 3B). Therefore, although VHL inactivation is associated with the dysregulated expression of Ror2, activation of the hypoxia response by either potent chemical inducers or profound exposure to low oxygen levels were not sufficient to engage either transcript or protein expression of Ror2.
Ror2 Is Regulated by HIF-2α Expression
Because HIFα-mediated transcriptional induction is critical to the role of VHL mutation in RCC (28) and Ror2 expression was increased at the transcriptional level in a VHL-dependent manner, we sought to further evaluate whether Ror2 expression shared any association with HIF-α dysregulation. As the VHL-deficient 786-0 cells express only HIF-2α, we used these cells to examine the linkage between HIF stabilization and HIF-dependent transcriptional regulation of Ror2 by suppressing HIF-2α in 786-0 cells with two independent shRNA constructs (shRNA constructs kindly provided by W. Kaelin, Dana Farber Cancer Institute (33)). Stable knockdown of the HIF-2α protein was achieved with both shRNA constructs compared with a scrambled control shRNA. Ror2 protein levels correlated linearly with HIF-2α, being markedly reduced when HIF-2α levels were suppressed by shRNA (Fig. 4A). We additionally evaluated the effects of HIF-2α knockdown on transcript levels. This experiment demonstrated a reduction in HIF-2α transcript by shRNA suppression at or below the physiologic levels, and a corresponding reduction in ROR2 mRNA with both shRNA vectors (Fig. 4B), further implicating this pathway in the mechanism of Ror2 expression in RCC.
FIGURE 4.
Ror2 is regulated by HIF-2α expression. A, Ror2 expression is suppressed in 786-0 HIF-2α shRNA knockdown cells. RCC cells expressing a VHL transgene (786-0 WT8) were used to control for VHL-induced suppression of Ror2. Cells lacking expression of VHL (786-0) were further transfected with a control scramble shRNA (scramble) or a HIF-2α short hairpin RNA retrovirus (shHIF-2) and immunoblotted with HIF-2α, Ror2, and Ku80 (loading control, LC) antibodies. B, ROR2 mRNA levels are suppressed in 786-0 HIF-2α shRNA knockdown cells. qRT-PCR analysis of the 786-0 HIF-2α shRNA knockdown cells verified effective HIF-2α knockdown in two independent knockdown cell lines (**, p = 0.0022, p = 0.0001). Ror2 expression was concordantly suppressed with HIF-2α knockdown (*, p = 0.0172; **, p = 0.0023). C, Ror2 expression can be rescued by HIF-2α overexpression. 786-0 cells stably expressing scramble shRNA and shHIF-2 were rescued with transient expression of empty vector (pcDNA) or proline hydroxylation-resistant double proline to alanine HIF-2α mutant (DPA) for 48 and 72 h. Induction of Ror2 expression is detected when HIF-2α expression is rescued with the DPA HIF-2α mutant in the shRNA cell line. Error bars represent ± S.E.
As we found Ror2 to be suppressed when HIF-2α was knocked down, we also asked if Ror2 expression can be rescued. Using a HIF-2α double proline to alanine (DPA) mutant cDNA plasmid, which is resistant to hydroxylation and VHL-mediated degradation (generously provided by B. Keith and M. C. Simon, University of Pennsylvania), we examined 786-0 cells stably expressing either of the HIF-2α shRNA constructs transiently transfected with this HIF-2α DPA mutant. We observed transient rescue of high levels of HIF-2α expression in the 786-0 HIF-2α knockdowns and saw a delayed partial rescue of Ror2 correlated with the highest levels of HIF-2α rescue (Fig. 4C) suggesting that Ror2 is expressed downstream of HIF-2α and its expression is directly tied to HIF-2α expression in the context of VHL inactivation.
Ror2 Is Regulated by Both HIF-1α and HIF-2α Expression
Observing that Ror2 could be regulated by HIF-2α, we also explored the HIF-2α-dependent regulation in the context of concurrent HIF-1α stabilization and examined the more ubiquitously expressed HIF-1α subunit as a potential regulator of Ror2 expression. Although many HIF-1α and HIF-2α targets overlap, some are dependent on only one or the other subunit, HIF-1α or HIF-2α, deriving the assumption that each of these subunits may have unique or context-specific functions (27). Utilizing the RCC4 cell line, which expresses both HIF-1α and HIF-2α subunits, we were able to examine this proposition by suppressing HIF-1α and HIF-2α independently using a lentiviral shRNA system, which included an iRES GFP for stable selection (lentiviral constructs kindly provided by W. Kim, University of North Carolina). Transfecting cells with lentiviral shRNA viral particles with suppression titrated by increasing GFP expression, we observed that effective suppression of HIF-1α and HIF-2α could be achieved without nonspecific suppression of the complementary HIF proteins. However, as has been observed previously, some positive compensation in levels of the alternate HIF factor was observed (32). In particular, when HIF-2α was suppressed, an increase in the basal levels of HIF-1α were detected, although the net cellular levels of HIF appeared to be reduced. In this system, which preserved the exclusive deregulation of one HIF factor, we observed that Ror2 protein levels were highly suppressed when HIF-2α levels were reduced (Fig. 5A) and HIF-1α were sustained, or even enhanced, further implicating the HIF-2α pathway in the role of Ror2 regulation. However, when HIF-1α protein levels were suppressed, particularly at the upper end of HIF-1α suppression, there was also a measurable reduction in Ror2 protein expression (Fig. 5A), suggesting that the regulation of Ror2 can also be influenced by HIF-1α expression. Utilizing the most highly suppressed cell line for each knockdown (based on selection for the highest level of GFP expression), this phenomenon was recapitulated at the transcript level, demonstrating suppression of the ROR2 transcript with shRNA knockdown of either HIF-1α or HIF-2α (Fig. 5B). Additionally, simultaneous knockdown of both factors produced an additive effect on ROR2 transcript (supplemental Fig. S1). This observation that ROR2 transcription is clearly linked to HIF stabilization, but that the maintenance of one stabilized HIF factor fails to convey a functional redundancy, suggests that the overall amount of HIFα present in the cells influences the levels of Ror2 that are expressed. These results imply that Ror2 expression is deregulated in cancer as a result of VHL inactivation and is dependent on sustained or pathologically high level expression of either HIF subunit, HIF-1α or HIF-2α.
HIF Complex Localizes to the ROR2 Promoter
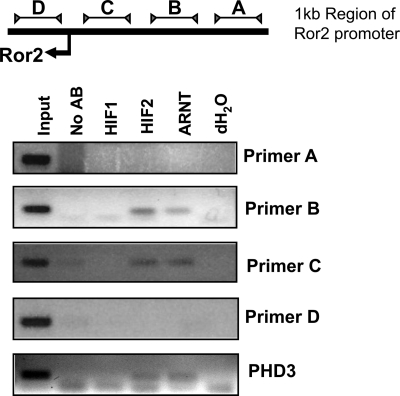
Observing that Ror2 could be regulated by both HIF-1α and HIF-2α, we examined the ROR2 genomic sequence and untranslated regions for evidence of a hypoxia response element that could account for HIF-mediated transcriptional activation. None of the previously reported canonical hypoxia response element binding sites (5′-CACGTA-3′, or the degenerate sequence 5′-BACGTSSK-3′) were detected in the immediate proximity of the ROR2 gene. Although isolated matching sites were found throughout the sequence 10 kb upstream and 2 kb downstream, we decided to assess if HIF proteins could specifically interact with the immediate 5′ region of the Ror2 promoter. The HIF-2α only expressing cell line 786-0 was utilized for our studies. Chromatin immunoprecipitation, using HIF-1α, HIF-2α, and ARNT/HIF-1β antibodies, was performed to analyze HIF interactions with the ROR2 promoter, utilizing four primer sets covering a 1-kb region overlapping the transcriptional start site. HIF-1α immunoprecipitation in this experiment was used as a control, as this factor is not expressed in 786-0 cells. As a positive control, known primers targeting the hypoxia response element of Egln3 (also known as PHD3) were utilized to confirm HIF complex/chromatin interactions (35). We consistently observed positive interactions with HIF-2α and ARNT in both the EGLN3 control, as well as with primer set B and less consistently with primer set C in the ROR2 promoter (Fig. 6). A second set of primers encompassing the same general region as primer set B were designed, and also consistently demonstrated a positive interaction (data not shown). Over multiple evaluations, the A and D primers did not demonstrate an interaction with HIF-2α or ARNT in these flanking segments of the ROR2 promoter (Fig. 6). These data suggest that we may have identified a cryptic site suitable of HIF complex binding in the immediate promoter region of ROR2.
FIGURE 6.
HIF-2α and ARNT interact with the ROR2 promoter. Cells lacking expression of VHL and overexpressing only HIF-2α (786-0) were subjected to chromatin immunoprecipitation with HIF-1α (used as a negative control), HIF-2α, and ARNT antibodies. Primers were designed targeting the 1-kb region of the Ror2 promoter that overlaps the Ror2 start site. Using primers targeted against the promoter region containing a known hypoxic response element of PHD3, also known as EGLN3, we show that HIF-2α and its dimerization partner, ARNT, interact with the PHD3 promoter as expected. HIF-1α interaction is not detected, as this cell line does not express HIF-1α. When the ROR2 promoter was examined, HIF-2α and ARNT were localized to ROR2 promoter regions B and C similarly to the control promoter. Interaction was not observed for flanking regions A and D.
DISCUSSION
Ror2 is a developmentally regulated kinase normally expressed in mesenchymal cells of organs such as the brain, lung, heart, and kidney but is normally repressed in most adult tissues, including the kidney (7). However, this kinase is derepressed in a subset of human RCC tumors (7) as well as other cancers (8–10), where it is emerging as a major factor promoting tumor cell migration and tumor invasion phenotypes. We sought to identify the mechanism of regulation for Ror2, contributing specifically to its stable expression in RCC. The most commonly referenced mutation in RCC pathology is of the VHL gene, with over 70% of RCC tumors displaying inactivation of VHL leading to the subsequent stabilization of one or more HIF factors, notably HIF-1α and HIF-2α. The stabilization of these potent transcriptional activators leads to the expression of multiple hypoxia response genes responsible for such phenotypes of renal tumors as prolific angiogenesis, cell invasion, and metastasis (34).
We first observed that Ror2 was expressed in a VHL-dependent manner in RCC. This was further corroborated by our analysis of the VHL somatic point mutations that recapitulate the VHL mutant transgenes that represent the major subtypes of VHL disease. This analysis demonstrated that Ror2 is expressed most highly in the subtype that maintains expression of the mutant VHL, but promotes the most significantly deregulated level of HIF-2α and is thought to lead to a highly penetrant form of RCC, type 2B. Furthermore, our work has previously shown that the VHL type 2B mutation, which was unable to suppress Ror2 expression of VHL mutant cells, is dysregulated for HIF-2α both in vitro (30) and in vivo (31). This ties Ror2 to an important aspect of RCC tumorigenesis, as a potential contributor to invasive forms of this cancer. Furthermore, this work linked Ror2 expression indirectly to dysregulated stabilization of HIF, as Ror2 expression was observed in cell culture VHL somatic mutation models that produce either stabilization of both HIF factors (VHL null), or the exclusive stabilization of HIF-2α (VHL 2B).
Although the HIF-α subunits share certain similarities, they also have divergent responsibilities (27). As Ror2 has been shown to be VHL-dependent and our data suggested a role for a correlation with HIF stabilization, it provided a natural extension to analyze the independent effects of HIF-1α and HIF-2α subunits on Ror2 expression. Normally, transcriptional targets that are unique to one of the HIF factors are not affected by genetic repression of the uninvolved HIF factor. Additionally, transcriptional targets that are mutual to HIF-1α and HIF-2α, such as GLUT1 or vascular endothelial growth factor, typically require the depletion of both factors, due to transcription factor compensation, to impact expression. We wanted to tease out the difference in expression and analyze if exclusively one or both HIF-α subunits had an effect on Ror2 expression. Our investigations point toward a clear correlation between HIF-2α stabilization and Ror2 expression in 786-0 cells, in which genetic knockdown and rescue experiments confirm this linear association. In cells expressing both HIF-1α and HIF-2α, the depletion of either factor was sufficient to reduce Ror2 expression at the transcript and protein levels. This suggests that with graded suppression of either HIF-1α or HIF-2α, Ror2 is coordinately expressed in a similar manner, and that functional redundancy between the factors may be dose-dependent. Such a pattern of regulation is not typical of most transcriptional components of the hypoxia response pathway, although similar patterns of atypical regulation have been previously reported for HIF targets (27). In the case of Ror2, the depletion of either factor from the maximally stabilized state causes the loss of Ror2 expression, indicative of a dosage effect of HIF factors on transcriptional activation of ROR2.
Prior hypoxia target investigations suggest that the Ror family ortholog cam-1 is a HIF-dependent hypoxia target in Caenorhabditis elegans (35). However, although the HIF pathway has been studied extensively, Ror2 has yet to be identified as a target of the hypoxia response pathway in mammalian cell screens. Activation of the hypoxia response pathway via stabilization of one or both of the HIF-α subunits is a commonly observed phenotype in RCC tumors, and physiologic up-regulation of HIF factors as a result of strict decreases in oxygen availability is an important cellular process in a variety of disease states. We thus extended our analysis to address this issue. Despite the observation of clear linear connections between HIF stabilization and Ror2 expression, we observed minimal induction of Ror2 expression in these studies, even after prolonged exposure to hypoxia, or chemical poisoning of the prolyl hydroxylation activity with potent inhibitors. These data suggest that within the context of this tumor cell system, physiologically relevant exposure to hypoxia may not be sufficient to induce changes in Ror2 expression levels compared with normoxic controls. This surprising result may shed further light on the mechanism of Ror2 regulation as a cancer-associated kinase. Sustained oncogenic levels of HIF factors are not routinely attained in response to hypoxic insult. However, in renal cell carcinoma, as a result of VHL inactivation, or in other tumors, notably squamous cell carcinoma of the head and neck, HIF is induced to high constitutive levels, which may account for the identification of a novel HIF target associated with cancer.
Whether it is the distinctly high levels of HIF factors associated with VHL loss or the maintenance of high levels for sustained periods of time that contributes to Ror2 expression is not certain. However, constitutive stabilization of HIF factors occurs broadly across many types of tumors, such that Ror2 as a tumor-associated kinase may be encountered even more frequently. One potential reason hypoxia may fail to up-regulate Ror2 expression is that ROR2 transcriptional regulation may be steps removed from HIF induction. Certainly several factors downstream of HIF activation could be considered as intermediate transcriptional regulators of the expression of ROR2, although which of these factors may contribute to regulation of Ror2 expression either developmentally or in the tumor physiology scenario remains to be determined. However, in our analysis of the ROR2 promoter, we found that HIF-2α and ARNT can be localized to a small region of the immediate ROR2 promoter region, which lacks any known hypoxia response element binding sequence. This interaction of the HIF complex at a cryptic ROR2 promoter site is suggestive of a direct mechanism of HIF regulation. However, this finding does not preclude the possibility that one or more additional transcriptional co-factors may be required to coordinate activation in a manner that is dependent on VHL loss or HIF stabilization, or that more distant conserved hypoxia response element sites may be utilized for HIF-dependent transcriptional regulation.
An additional possibility exists to link the dependence on VHL inactivation and HIF stabilization in clear cell renal cell carcinoma, and the failure of Ror2 to exhibit hypoxic induction. VHL mutation also mediates alternate effects on cell signaling external to the well studied effects on HIF regulation. For example, pVHL is known to affect the stabilization of a non-HIF-associated protein, JADE-1 (36), and in a sophisticated genetic screen in C. elegans, at least four HIF-independent but VHL-dependent genes were identified (37). Thus, our results may reflect the activation of a gene that is HIF-dependent only in the setting of a VHL mutation because of other context specific cues. Furthermore, patterns of tumor cell invasion have been suggested to be dependent on aspects of VHL loss that are not clearly dependent on HIF stabilization. The extent to which Ror2 may play a role in promoting these invasive features of tumors will need to be further investigated.
RCC is a difficult to treat cancer that has few effective options for treatment. We previously reported that Ror2 expression has a distinct role in RCC tumor cell biology (7) and has the potential to be important for components of tumor invasion. We have since demonstrated a mechanism of Ror2 regulation that, with respect to clear cell RCC, can be linked to VHL loss and is a component of the HIF-dependent transcriptional program in clear cell RCC cell lines. This sustained stabilization of HIF is a feature of many tumors, some of which are independent of VHL mutation and are associated with advanced disease. Here we offer a unique mechanism of regulation that offers insight into how Ror2 is deregulated in RCC that has the potential for application in many other cancers.
Supplementary Material
Acknowledgments
We thank members of the Rathmell laboratory for many helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health NRSA Program Grants F31CA132543 (to T. M. W.) and R01 CA121781 (to W. K. R.) from the NCI and a grant from the Doris Duke Charitable Fund (to W. K. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- RCC
- renal cell carcinoma
- DMOG
- dimethyloxalylglycine
- Egln3
- Egl nine homolog 3
- Glut1
- glutamate transporter 1
- HIF
- hypoxia-inducible factor
- qRT
- quantitative reverse transcriptase
- Ror2
- receptor tyrosine kinase-like orphan receptor 2
- shRNA
- short hairpin RNA interference
- VHL
- von Hippel-Lindau tumor suppressor gene
- PHD
- prolyl hydroxylase
- ARNT
- aryl hydrocarbon nuclear transferase
- GFP
- green fluorescent protein
- DPA
- mutated double proline to alanine.
REFERENCES
- 1.Masiakowski P., Carroll R. D. (1992) J. Biol. Chem. 267, 26181–26190 [PubMed] [Google Scholar]
- 2.Forrester W. C. (2002) Cell Mol. Life Sci. 59, 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwabe G. C., Trepczik B., Süring K., Brieske N., Tucker A. S., Sharpe P. T., Minami Y., Mundlos S. (2004) Dev. Dyn. 229, 400–410 [DOI] [PubMed] [Google Scholar]
- 4.Al-Shawi R., Ashton S. V., Underwood C., Simons J. P. (2001) Dev. Genes Evol. 211, 161–171 [DOI] [PubMed] [Google Scholar]
- 5.Mikels A., Minami Y., Nusse R. (2009) J. Biol. Chem. 284, 30167–30176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billiard J., Way D. S., Seestaller-Wehr L. M., Moran R. A., Mangine A., Bodine P. V. (2005) Mol. Endocrinol. 19, 90–101 [DOI] [PubMed] [Google Scholar]
- 7.Wright T. M., Brannon A. R., Gordan J. D., Mikels A. J., Mitchell C., Chen S., Espinosa I., van de Rijn M., Pruthi R., Wallen E., Edwards L., Nusse R., Rathmell W. K. (2009) Oncogene 28, 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morioka K., Tanikawa C., Ochi K., Daigo Y., Katagiri T., Kawano H., Kawaguchi H., Myoui A., Yoshikawa H., Naka N., Araki N., Kudawara I., Ieguchi M., Nakamura K., Nakamura Y., Matsuda K. (2009) Cancer Sci. 100, 1227–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi M., Shibuya Y., Takeuchi J., Murata M., Suzuki H., Yokoo S., Umeda M., Minami Y., Komori T. (2009) Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 107, 398–406 [DOI] [PubMed] [Google Scholar]
- 10.Kubo T., Kuroda Y., Shimizu H., Kokubu A., Okada N., Hosoda F., Arai Y., Nakamura Y., Taniguchi H., Yanagihara K., Imoto I., Inazawa J., Hirohashi S., Shibata T. (2009) Carcinogenesis 30, 1857–1864 [DOI] [PubMed] [Google Scholar]
- 11.Gnarra J. R., Tory K., Weng Y., Schmidt L., Wei M. H., Li H., Latif F., Liu S., Chen F., Duh F. M., et al. (1994) Nat. Genet. 7, 85–90 [DOI] [PubMed] [Google Scholar]
- 12.Gnarra J. R., Ward J. M., Porter F. D., Wagner J. R., Devor D. E., Grinberg A., Emmert-Buck M. R., Westphal H., Klausner R. D., Linehan W. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9102–9107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo K., Yao M., Yoshida M., Kishida T., Shuin T., Miura T., Moriyama M., Kobayashi K., Sakai N., Kaneko S., Kawakami S., Baba M., Nakaigawa N., Nagashima Y., Nakatani Y., Hosaka M. (2002) Genes Chromosomes Cancer 34, 58–68 [DOI] [PubMed] [Google Scholar]
- 14.Shuin T., Kondo K., Torigoe S., Kishida T., Kubota Y., Hosaka M., Nagashima Y., Kitamura H., Latif F., Zbar B., et al. (1994) Cancer Res. 54, 2852–2855 [PubMed] [Google Scholar]
- 15.Iwai K., Yamanaka K., Kamura T., Minato N., Conaway R. C., Conaway J. W., Klausner R. D., Pause A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12436–12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamura T., Koepp D. M., Conrad M. N., Skowyra D., Moreland R. J., Iliopoulos O., Lane W. S., Kaelin W. G., Jr., Elledge S. J., Conaway R. C., Harper J. W., Conaway J. W. (1999) Science 284, 657–661 [DOI] [PubMed] [Google Scholar]
- 17.Kibel A., Iliopoulos O., DeCaprio J. A., Kaelin W. G., Jr. (1995) Science 269, 1444–1446 [DOI] [PubMed] [Google Scholar]
- 18.Pause A., Lee S., Worrell R. A., Chen D. Y., Burgess W. H., Linehan W. M., Klausner R. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iliopoulos O., Levy A. P., Jiang C., Kaelin W. G., Jr., Goldberg M. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10595–10599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 21.Ohh M., Park C. W., Ivan M., Hoffman M. A., Kim T. Y., Huang L. E., Pavletich N., Chau V., Kaelin W. G. (2000) Nat. Cell Biol. 2, 423–427 [DOI] [PubMed] [Google Scholar]
- 22.Benita Y., Kikuchi H., Smith A. D., Zhang M. Q., Chung D. C., Xavier R. J. (2009) Nucleic Acids Res. 37, 4587–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivan M., Haberberger T., Gervasi D. C., Michelson K. S., Günzler V., Kondo K., Yang H., Sorokina I., Conaway R. C., Conaway J. W., Kaelin W. G., Jr. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13459–13464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 25.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim Av., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y., Zhang W., Kondo K., Klco J. M., St. Martin T. B., Dufault M. R., Madden S. L., Kaelin W. G., Jr., Nacht M. (2003) Mol. Cancer Res. 1, 453–462 [PubMed] [Google Scholar]
- 27.Hu C. J., Wang L. Y., Chodosh L. A., Keith B., Simon M. C. (2003) Mol. Cell Biol. 23, 9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo K., Klco J., Nakamura E., Lechpammer M., Kaelin W. G., Jr. (2002) Cancer Cell 1, 237–246 [DOI] [PubMed] [Google Scholar]
- 29.Mandriota S. J., Turner K. J., Davies D. R., Murray P. G., Morgan N. V., Sowter H. M., Wykoff C. C., Maher E. R., Harris A. L., Ratcliffe P. J., Maxwell P. H. (2002) Cancer Cell 1, 459–468 [DOI] [PubMed] [Google Scholar]
- 30.Hacker K. E., Lee C. M., Rathmell W. K. (2008) PLoS One 3, e3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C. M., Hickey M. M., Sanford C. A., McGuire C. G., Cowey C. L., Simon M. C., Rathmell W. K. (2009) Oncogene 28, 1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordan J. D., Lal P., Dondeti V. R., Letrero R., Parekh K. N., Oquendo C. E., Greenberg R. A., Flaherty K. T., Rathmell W. K., Keith B., Simon M. C., Nathanson K. L. (2008) Cancer Cell 14, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo K., Kim W. Y., Lechpammer M., Kaelin W. G., Jr. (2003) PLoS Biol. 1, E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordan J. D., Simon M. C. (2007) Curr. Opin. Genet. Dev. 17, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen C., Nettleton D., Jiang M., Kim S. K., Powell-Coffman J. A. (2005) J. Biol. Chem. 280, 20580–20588 [DOI] [PubMed] [Google Scholar]
- 36.Zhou M. I., Wang H., Foy R. L., Ross J. J., Cohen H. T. (2004) Cancer Res. 64, 1278–1286 [DOI] [PubMed] [Google Scholar]
- 37.Bishop T., Lau K. W., Epstein A. C., Kim S. K., Jiang M., O'Rourke D., Pugh C. W., Gleadle J. M., Taylor M. S., Hodgkin J., Ratcliffe P. J. (2004) PLoS Biol. 2, e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.