Abstract
The Ser/Thr/Tyr kinase activity of human biliverdin reductase (hBVR) and the expression of Goodpasture antigen-binding protein (GPBP), a nonconventional Ser/Thr kinase for the type IV collagen of basement membrane, are regulated by tumor necrosis factor (TNF-α). The pro-inflammatory cytokine stimulates kinase activity of hBVR and activates NF-κB, a transcriptional regulator of GPBP mRNA. Increased GPBP activity is associated with several autoimmune conditions, including Goodpasture syndrome. Here we show that in HEK293A cells hBVR binds to GPBP and down-regulates its TNF-α-stimulated kinase activity; this was not due to a decrease in GPBP expression. Findings with small interfering RNA to hBVR and to the p65 regulatory subunit of NF-κB show the hBVR role in the initial stimulation of GPBP expression by TNF-α-activated NF-κB; hBVR was not a factor in mediating GPBP mRNA stability. The interacting domain was mapped to the 281CX10C motif in the C-terminal 24 residues of hBVR. A 7-residue peptide, KKRILHC281, corresponding to the core of the consensus D(δ)-Box motif in the interacting domain, was as effective as the intact 296-residue hBVR polypeptide in inhibiting GPBP kinase activity. GPBP neither regulated hBVR expression nor TNF-α dependent NF-κB expression. Collectively, our data reveal that hBVR is a regulator of the TNF-α-GPBP-collagen type IV signaling cascade and uncover a novel biological interaction that may be of relevance in autoimmune pathogenesis.
Keywords: Metabolism/Heme, Oxygen/Antioxidant, Phosphorylation/Kinases/Serine-Threonine, Signal Transduction/Protein Kinases/Calmodulin, Antioxidant, Antioxidant, Autoimmune Diseases, Bilirubin/Biliverdin, Goodpasture Syndrome, Heme Oxygenase
Introduction
Goodpasture syndrome (GPS)2 is a disorder mediated by autoantibody attack against the C-terminal noncollagenous-1 (NC1) domain of the α3 chain of the type IV collagen of basement membrane (α3(IV)NC1) (Goodpasture antigen (GPA)). The NC1 domain initiates the braiding of the collagenous domains into a triple helical structure (protomer) and then mediates the assembly of two individual protomers yielding a quaternary structure known as the hexamer. The autoantibody epitope is cryptic in the hexamer, and the mechanism for its immunological exposure remains unknown. The autoimmune reaction results in deposits of autoantibodies along alveolar and glomerular basement membranes, causing lung hemorrhage and rapidly progressive glomerulonephritis, the two cardinal clinical manifestations of GPS (1, 2).
Goodpasture antigen-binding protein (GPBP) is a nonconventional TNF-α-inducible Ser/Thr kinase that targets the α3(IV)NC1 domain and regulates basement membrane collagen organization (3–6). Cells express at least two GPBP isoforms of 77 and 91 kDa. The 77-kDa GPBP polypeptide (GPBP in this report) interacts with type IV collagen, whereas the 91-kDa isoform associates with the cellular membrane and regulates extracellular levels of the 77-kDa polypeptide (7). In the present study we concentrated on the interaction of hBVR with the 77-kDa form of GPBP by transfecting cells with an expression plasmid encoding this form and using the cytoplasmic fraction of the cell lysate; hBVR is a soluble protein. GPBP phosphorylates GPA at its N terminus (6). Alterations in protein phosphorylation affect processing and peptide presentation, which could lead to autoimmune response (8, 9).
Although the downstream effectors of GPBP have been well characterized, the upstream regulator(s) of GPBP activity and expression have remained unknown. The present study has identified the role of hBVR in such capacity. Among all proteins identified in the human cell, BVR arguably has the most expansive range of functions (for review, see Refs. 10 and 11). hBVR is a 296-residue polypeptide that was initially characterized in the context of its function as a reductase, with a unique dual pH/cofactor-dependent activity profile for the conversion of biliverdin to bilirubin in the heme (iron-protoporphyrin IX) metabolism pathway (12). hBVR has the distinction of being a dual specificity kinase (13), one of the rare forms of kinases that phosphorylate Ser/Thr/Tyr residues (14).
The pleiotropic functions result from hBVR possessing numerous consensus regulatory motifs with demonstrated function that are primarily contained in the regulatory carboxyl half of the protein; this region folds into a large six-stranded β-sheet, the putative protein-protein interactive domain (15). The 24-residue segment of hBVR that forms the C-terminal α-helix contains the Cys281-Xaa10-Cys292 configuration that is involved in divalent metal binding and protein-protein interactions and the D(δ)-Box motif. This motif is a common feature of kinases in the MAPK signaling cascade (16, 17). The hBVR D(δ)-Box core sequence is KKRILHC (aa 275–281). The secondary structure of hBVR (PDB accession 2H63), which resembles that of the rat enzyme (15, 18), is likely a significant factor in the molecular scaffolding activity and in the intracellular movement of the recruited kinases to target sites, as demonstrated for the cytoplasm-cell membrane transport of PKC-βII and PKC-ζ and for cytoplasm-nuclear transport of MEK-activated ERK1/2. The nuclear localization and export signals of hBVR play crucial roles in transcriptional activity of the MEK/ERK/Elk (19–21). hBVR also plays a role in energy-dependent transportation of extracellular heme into the nucleus (22).
GPBP and hBVR signaling pathways converge at the level of activation by TNF-α, the pro-inflammatory cytokine, which is an activator of hBVR kinase activity and an inducer of GPBP expression (4, 20). The cytokine is an upstream activator of the transcription factor NF-κB (23, 24). Activation of NF-κB by TNF-α is inhibited by biliverdin, whereas hBVR reverses the inhibition (25). Notably, a peptide corresponding to the hBVR D-Box motif blocks activation of PKC-ζ in response to TNF-α (20).
Activation of the two proteins, however, have opposing effects on cellular pathophysiology, with GPBP, as alluded to above, being a proinflammatory factor, whereas hBVR occupies a center stage in the battery of defense mechanisms against reactive oxygen species (ROS) available to the cell (26). In its capacity as a reductase, BVR converts biliverdin, the product of heme oxygenase-1 and -2, to bilirubin (27). Bilirubin is a quencher of ROS (28–30). ROS species have been implicated in the etiology of an assortment of inflammatory-linked diseases (31–34), including GPS (35). Induction of heme oxygenase-1 is considered to be a major component of the cellular defense mechanisms against ROS-mediated tissue damage (34, 36–39).
To understand whether the sharing of the signaling activator extends to a potential cross-talk between GPBP and hBVR, we undertook this study through which we reveal the occurrence of protein-protein interactions between the two kinases (down-regulation of GPBP kinase activity by hBVR) and that the TNF-α-NF-κB-dependent expression of GPBP is regulated by the reductase. The findings identify a previously unrecognized biological circuit controlled by TNF-α, which may be relevant in GPS pathogenesis.
EXPERIMENTAL PROCEDURES
Materials
N-Myristoylated peptides that correspond to the hBVR residues indicated in parenthesis were synthesized by EZBiolab (Westfield, IN). The specific sequences were KEVVGKDL (aa 134–141), KRNRYLSF (aa 224–231), KKRILHC (aa 275–281), and KYCCSRK (aa 290–296). TNF-α was obtained from Calbiochem, anti-FLAG M2-affinity beads and ATP were from Sigma, and [γ-32P]ATP and [32P]orthophosphate were from PerkinElmer Life Sciences. Rabbit anti-hBVR polyclonal antibody was isolated as done previously (12), and monoclonal antibodies against GPBP were prepared essentially as previously described (6). Polyvinylidene difluoride membrane was a product of Pall Science Corp. (Pensacola, FL). Goat anti-p65 polyclonal antibody and protein A/G beads were from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids and Mutants
The hBVR open reading frame was cloned in both pGEX-4T2 (for expression in Escherichia coli) and pcDNA3 (for mammalian cell expression). pEGFP-hBVR construct was described earlier (40). The truncated hBVR constructs, encoding aa 1–108, 1–272, 109–175, 158–296, 272–296, or 272–296 containing Cys281,292,293 changed to alanine, were prepared by PCR amplification of the wt cDNA using suitable primers and were cloned in pGEX-4T2. The plasmids were verified by sequencing. Plasmids encoding the 77-kDa FLAG-GPBP and the N-terminal peptide of GPA were generated as before (6). The plasmid pcDNA3-GPBP was generated by amplifying the open reading frame of GPBP using 5′-ctgaagatctatgtacccatacgatgttccagattacgctcttatgtcggataatcagagctggaactcgtcgggc and 5′-ctgactcgagctagaacaaaataggctttcctgcagttttttcttggacg primers, which also introduced a hemagglutinin epitope. The full-length GPBP sequence was cloned into the pcDNA3 vector, and the validity of the construct was confirmed by sequencing.
Yeast Two-hybrid Screen
The yeast two-hybrid screen used MATCHMAKER System 3 (Clontech). Full-length hBVR cDNA was amplified by PCR using the primers 5′-aatccatggcgaatgcagagcccgagaggaa and 5′-gatggatccctcctcctcttacttccttg. The PCR product was cloned into the bait vector, pGBKT7, and was used to transform the yeast strain AH109 using the lithium acetate method as described by the manufacturer. The resulting strain was mated to Y187 yeast cells transformed with a human kidney cDNA library. Yeast colonies from the mating were screened for hBVR-interacting proteins. Candidate clones were selected on plates lacking adenine, histidine, leucine, and tryptophan and were assayed for β-galactosidase activity. The target plasmids of positive clones were recovered from the yeast cells, and the sequences of the cDNA inserts were determined. Bait and purified positive plasmids were used to transform the yeast cells, and the phenotype of the transformants examined served as a confirmation of the interaction.
Cell Culture, Transfection, and Immunoprecipitation (IP)
HEK293A cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum and penicillin G/streptomycin for 24 h to achieve 70% confluence. Cells were transfected depending on the experiment with up to 1 μg of pcDNA3-hBVR and/or pcDNA3-GPBP plasmid or with pcDNA3-FLAG-GPBP per 10-cm culture dish using Transfectin (Bio-Rad). hBVR-based myristoylated peptides were introduced into cells as before (20). In experiments employing small interfering RNA (siRNA) for hBVR, viruses containing pSuper-sihBVR or sc-hBVR (control) were grown and packaged as described elsewhere (41). Cells were infected with four plaque-forming units/cell. Double-stranded siRNA for the p65 regulatory subunit of NF-κB was introduced by transfecting cells with a 10 μm stock solution (Santa Cruz Biotechnology) following the manufacturer's suggestions. In those experiments employing TNF-α, the cells were synchronized in medium containing 0.5% fetal bovine serum for 18 h, then treated with 20 ng/ml TNF-α for the times indicated. For IP experiments, cell lysate (500 μg of protein) was incubated with polyclonal anti-hBVR antibodies (generated against hBVR isolated from human kidneys), anti-FLAG affinity gel (Sigma), or normal rabbit serum overnight at 4 °C. Immunoprecipitated proteins were visualized by Western blotting using appropriate antibodies. IP after in vivo cross-linking was essentially performed as previously reported (7).
GST Pulldown Assay
The GST pulldown assay was performed as described previously (19). Lysate prepared from HEK293A cells transfected with pcDNA3-GPBP was subjected to GST pulldown assays using 10 μg of E. coli-expressed GST-hBVR fusion protein or GST-fused truncated hBVR proteins. Proteins bound to GST beads were resolved by SDS-PAGE followed by Western blotting.
Western Blot Analyses
Cell lysate was subjected to electrophoresis on 12 or 8% SDS-PAGE gels depending on the experiment, transferred to polyvinylidene difluoride or nitrocellulose membranes, and probed with the appropriate primary and secondary antibodies. The antigen was visualized by the ECL detection system (Amersham Biosciences). FLAG-tagged GPBP was detected using monoclonal mouse anti-FLAG antibody and visualized by ECL.
Metabolic Labeling
The experimental procedure was based on one described previously (20). Cells were starved and labeled with carrier-free, HCl-free [32P]H3PO4 (100 μCi/ml) for 4 h before treatment with TNF-α (20 ng/ml) for the duration appropriate to each experiment. Cell lysate was immunoprecipitated with immobilized-anti-FLAG or with polyclonal anti-hBVR antibodies. The phosphorylated and immunoprecipitated proteins were visualized by autoradiography.
Kinase Assay Reactions
GPBP kinase activity was measured in an assay system containing 10 mm MgCl2 at pH 7.0 and [γ32P]ATP, as previously described (6). hBVR kinase activity was determined in a system containing 20 mm MnCl2 and [γ32P]ATP at pH 8.0, as detailed before (13). Phosphorylated proteins were detected by SDS-PAGE and blotting to nitrocellulose membrane followed by autoradiography. After allowing radioactivity to decay, the identity of the proteins was confirmed by antibody detection.
RT-PCR Experiments
RT-PCR was performed using total RNA isolated from cells transfected or infected with retrovirus expressing siRNA as described above. Synchronized cells were then treated with TNF-α (20 ng/ml). In experiments testing RNA stability, cells were treated with TNF-α for 6 h then with actinomycin D (2.5 μg/ml) for the indicated times before RNA extraction. Total RNA was purified from cells lysed with TRIzol reagent (Invitrogen) following the manufacturer's recommendation. 2.5 μg of RNA was used as a template for cDNA synthesis using random hexamer primers. The cDNA was used as a template for quantitative RT-PCR using gene-specific primers and Taq polymerase in an ABI-fast 7500 PCR system. Products were detected by SYBR Green; specificity was determined from melting curves, and in some samples the reaction products were resolved by gel electrophoresis. Specific signals were normalized on 18 S rRNA and quantified by the ΔΔCT method. The S.E. in ΔΔCT was also determined in individual experiments; this error was included in the calculated relative mRNA level as error = mRNA(ΔΔCT) ± mRNA(ΔΔCT±S.E.).
Luciferase Reporter Assay
HEK293A cells seeded in 6-well plates were cotransfected with a reporter plasmid containing five copies of the NF-κB responsive element fused to a downstream luciferase gene (pNF-κB, Stratagene, La Jolla, CA), either empty pEGFP or pEGFP-hBVR, and pCMV-β-gal plasmid reporter. After 12 h of transfection, cells were treated with TNF-α for an additional 12 h and then harvested, washed, and lysed. Five μl of lysate from each sample was used for the luciferase assay (Promega). β-Galactosidase activity was used to assess transfection efficiency, and luciferase activity was normalized against β-galactosidase activity.
Statistical Analysis
Data as presented in bar graphs are the means with S.D. of three experiments, each with triplicate samples. Statistical analyses used GraphPad prism software (GraphPad Software, San Diego). Data were analyzed by one-way analysis of variance from which Student's t was calculated for all sample pairs. Differences were considered to be significant if p ≤ 0.05; it should be noted that differences between experiments were not significant. All experiments were performed three times unless otherwise indicated.
RESULTS
hBVR Binds to Goodpasture Antigen-binding Protein
Because a number of correlative phenomena suggested a possible link between hBVR and GPBP and the finding that in every instance that hBVR modulates activity of a cell signaling component protein-protein interaction is involved, we examined in a yeast two-hybrid screen system whether hBVR and GPBP interact.
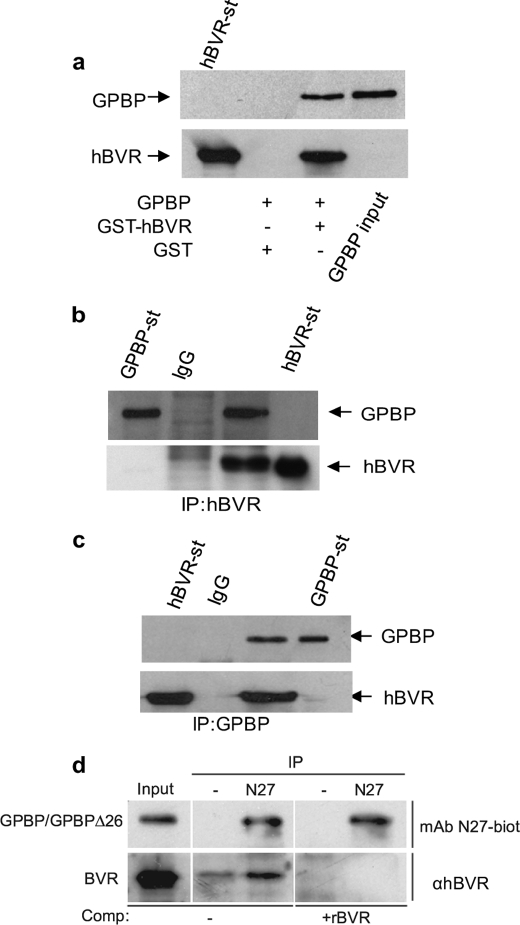
To explore this phenomenon, full-length hBVR was used as bait in a yeast two-hybrid screen system using a human kidney cDNA library. Approximately 17 million yeast colonies were screened for hBVR-interacting protein(s). Among 31 positive clones, 11 exhibited sequence identity to the C-terminal domain (aa 466–624) and 1 encoded amino acid residue 150–292, including part of the heptad repeat of the coil-coil structure containing the bipartite nuclear localization of GPBP (6). To confirm the results of the yeast two-hybrid screen, full-length GPBP was cloned into pcDNA3, a mammalian expression vector, and tested in an in vitro binding assay using GST-hBVR (Fig. 1a). In a GST pulldown assay, GST-hBVR, but not GST alone, precipitated GPBP from cell lysate. This interaction was confirmed in a follow-up IP experiment shown in Fig. 1, b and c, which used HEK293A cells co-transfected with pcDNA-hBVR and pcDNA-GPBP expression plasmids. In this experiment cell lysate was obtained and subjected to IP using either anti-hBVR (Fig. 1b) or anti-GPBP antibody (Fig. 1c). The precipitated complexes were resolved on SDS-PAGE followed by Western blotting with either anti-GPBP or anti-hBVR antibodies. As shown under both conditions, GPBP and hBVR were present in the precipitate. To address whether the interactions may have arisen as a consequence of overexpressing one or both proteins, the experiment was repeated in normal cells that had been cross-linked with formaldehyde before lysis and IP with monoclonal antibody to GPBP (N27) (Fig. 1d). Under in vivo cross-linking conditions hBVR was efficiently precipitated by N27, although there was a minor interaction of hBVR with the protein A-Sepharose beads.
FIGURE 1.
GPBP binds to hBVR in HEK293A cells. a, a GST pulldown assay is shown. HEK293A cells were transfected with the pcDNA3-expression plasmid encoding GPBP. One day after DNA addition, cell lysates were prepared and subjected to a GST pulldown assay using either GST-hBVR or GST alone. The membrane was sequentially probed with anti-GPBP and anti-hBVR antibodies. b, co-IP of GPBP and hBVR detected by hBVR antibody is shown. Lysate prepared from cells co-transfected with pcDNA3-hBVR and pcDNA3-GPBP was immunoprecipitated with an anti-hBVR antibody or with control rabbit IgG. The precipitate was subjected to Western blot analysis by sequentially probing the membrane with anti-GPBP and anti-hBVR antibodies. c, co-IP of GPBP and hBVR as detected by GPBP antibody is shown. The same cell lysates were immunoprecipitated using anti-GPBP antibody or control rabbit IgG and analyzed as in b. d, cross-linking of GPBP and hBVR is shown. HEK293A cells were cross-linked with formaldehyde, and cell extracts were immunoprecipitated with protein A-Sepharose after incubation with or without the indicated GPBP-specific antibodies. The input lane shows the presence of both GPBP (and/or the closely migrating GPBPΔ26) and BVR. The immunoprecipitates were resolved by gel electrophoresis, and the Western blot was probed with biotinylated N27 antibody or anti-BVR. In the competition experiment, anti-BVR binding was blocked by the addition of recombinant hBVR. In a–d, detection was by secondary antibody conjugate followed by ECL. Purified preparations of GST-hBVR (BVR-st) and GPBP were included as controls (GPBP-st).
GPBP Phosphorylation Is Attenuated in the Presence of hBVR
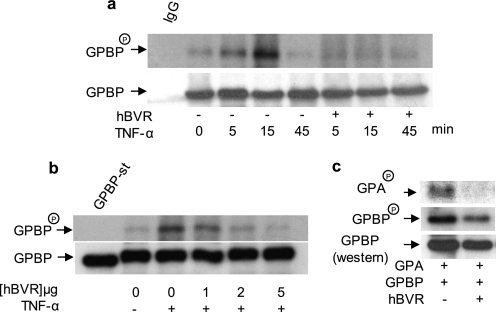
hBVR functions in TNF-α-NF-κB signaling, wherein it is activated by the cytokine and activates the transcription factor (20, 25). GPBP is also activated by TNF-α (4). Accordingly, in the following experiments, whether GPBP phosphorylation would be affected by hBVR was examined. First, the effect of TNF-α on phosphorylation of GPBP in the presence or absence of hBVR was tested (Fig. 2a). Cells overexpressing both proteins or GPBP alone were synchronized, metabolically labeled with [32P]H3PO4, and treated with TNF-α. Cell lysates were obtained and immunoprecipitated with FLAG-specific antibodies and used for analysis of phosphorylated GPBP. As displayed in the composite, TNF-α treatment produced a time-dependent increase in GPBP phosphorylation peaking at 15 min and returning to basal level by 45 min. TNF-α-stimulated GPBP phosphorylation, however, was markedly reduced when hBVR was co-expressed with GPBP.
FIGURE 2.
hBVR suppresses the kinase activity of GPBP. a, TNF-α-stimulated phosphorylation of GPBP is suppressed by hBVR. HEK293A cells transfected with GPBP alone or co-transfected with hBVR-pcDNA expression plasmids were metabolically labeled with [32P]H3PO4. Cell lysates were obtained, and GPBP was immunoprecipitated. The precipitate was resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and autoradiographed. The blot was subsequently probed with anti-GPBP antibody followed by ECL detection as the control for loading. Immunoprecipitates with mouse IgG and anti-mouse IgG-ECL was used as the control for specificity of binding. b, hBVR decreases GPBP autophosphorylation in a concentration-dependent manner. Cells were transfected with FLAG-GPBP expression plasmid together with increasing concentrations of pcDNA3-hBVR. One day later cells were starved overnight and thereafter treated with TNF-α (20 ng/ml, 15 min). GPBP was isolated with anti-FLAG beads and assayed in the kinase reaction as described under “Experimental Procedures” (6). c, hBVR suppresses GPBP-dependent phosphorylation of its substrate, the GPA-derived peptide. Cells were transfected with expression plasmid for FLAG-GPBP alone or co-transfected with pcDNA3-hBVR followed 24 h later by starvation overnight, treatment with TNF-α as in b, and subsequent lysis. GPBP was isolated from the lysates using anti-FLAG beads. The N-terminal peptide of GPA was isolated as a GST fusion polypeptide from E. coli and used as the substrate in the GPBP kinase reaction assay. The reaction mixture was resolved by SDS-PAGE and transferred to nitrocellulose membrane, and phosphorylated GPBP and GPA (GPBPP and GPAP) was visualized by autoradiography. The membrane was subsequently probed with anti-GPBP antibody followed by ECL detection.
Next, to examine whether expression and subsequent binding of hBVR to GPBP also modulates the kinase activity of GPBP, GPBP was isolated from TNF-α-induced cells expressing increasing amounts of hBVR and assayed for autophosphorylation under optimal conditions for GPBP phosphate transfer (pH 7.0 in the presence of Mg 2+) (6). As shown in Fig. 2b, hBVR suppressed GPBP autophosphorylation in a concentration-dependent manner. The inhibitory effect of hBVR on GPBP kinase activity extended to its ability to phosphorylate its substrate, GPA-derived peptide (Fig. 2c). As expected, GPBP isolated from cells not expressing recombinant hBVR efficiently phosphorylated GPA-derived peptide, whereas peptide phosphorylation was markedly suppressed when assaying GPBP isolated from cells co-expressing hBVR.
hBVR Regulation of GPBP Expression
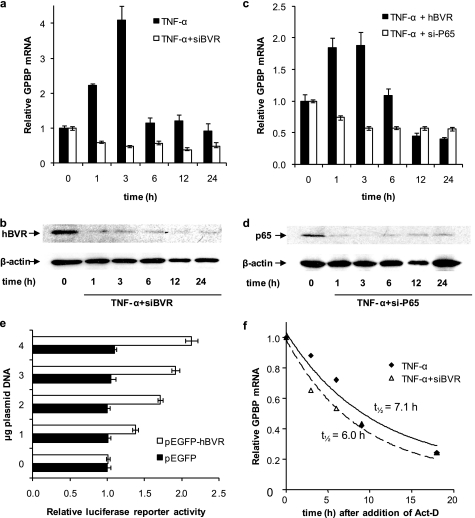
Next, we questioned whether hBVR alters expression of GPBP. The following experiments were carried out to examine the effect of hBVR on TNF-α stimulation of GPBP expression; conditions in which hBVR was overexpressed as well as decreased using pcDNA-hBVR expression plasmid and si-hBVR, respectively, were used. In the first experiment the levels of GPBP mRNA were measured at 1, 3, 6, 12, and 24 h after TNF-α treatment using si-hBVR to reduce expression of hBVR. As we showed in previous studies (20, 21, 41) the si-hBVR, under conditions used in this experiment, effectively caused a reduction in the cellular level of hBVR (Fig. 3b). As shown in Fig. 3a, in normal cells a notable increase in GPBP mRNA was observed at 1- and 3-h time points that returned to the zero time levels at 6 h. This transient induction was blocked by the presence of si-hBVR, suggesting that hBVR is necessary for induction of GPBP transcription. Because the transcriptional activation of GPBP by TNF-α is mediated by the transcription factor NF-κB and hBVR is an upstream regulator of NF-κB (25), the consequence of overexpression of hBVR on TNF-α-mediated transcriptional activation of GPBP was examined. Increasing hBVR by overexpression did not notably alter the outcome of the TNF-α treatment (Fig. 3c). The extent of induction differed somewhat between experiments, as reflected in the difference in scales between Figs. 3, a and c. In addition, si-p65 was used to block transcription of the p65 regulatory subunit of NF-κB as shown in Fig. 3d. p65 is the regulatory subunit of the Janus-faced transcription factor, NF-κB, which regulates expression of cell survival as well as cell death genes (42, 43). The presence of si-p65, as with si-hBVR, blocked the TNF-α-mediated increase in GPBP mRNA (Fig. 3c).
FIGURE 3.
Both NF-κB and hBVR are required for TNF-α induction of GPBP. a, treatment of cells with si-hBVR prevents TNF-α induction of GPBP expression. Cells were infected with si-hBVR retrovirus 16 h before the addition of 20 ng/ml TNF-α; RNA was prepared after the indicated intervals and used as a template for random hexamer-primed cDNA synthesis. The GPBP mRNA content was determined by quantitative RT-PCR by the ΔΔCt method using 18 S rRNA as control. A representative experiment is shown; the errors were determined from the differences between the relative mRNA calculated by ΔΔCT and ΔΔCT ± S.E. (“Experimental Procedures”). b, Western blot of cells treated with siRNA for hBVR. Total cell lysates shown in a were subjected to immunoblotting. The nitrocellulose membrane was sequentially probed with anti-hBVR and anti β-actin antibodies. c, treatment of cells with si-p65 prevents TNF-α induction of GPBP expression. Cells were transfected with hBVR or with siRNA against the p65 NF-κB subunit. Eighteen hours later the cells were transferred to low serum medium and treated with 20 ng/ml TNF-α. Sample preparation and RT-PCR analysis were as in a. d, a Western blot of cells treated with siRNA for p65 is shown. Total cell lysates shown in c were subjected to immunoblotting. The nitrocellulose membrane was sequentially probed with anti-p65 and anti β-actin antibodies. e, activation of NF-κB is enhanced by elevated hBVR in the cell. HEK293A cells seeded in 6-well plates were co-transfected with the pNF-κB luciferase reporter plasmid, β-galactosidase reporter, and increasing amounts of either pEGFP-hBVR or the empty pEGFP vector as indicated. 12 h after DNA addition the cells were treated with TNF-α, and after a further 12 h the cells were harvested and lysed, and the luciferase and β-galactosidase activities were measured. Luciferase activity was normalized to that of β-galactosidase to correct for differences in transfection efficiency. f, hBVR does not alter GPBP mRNA stability. Cells were pretreated with si-hBVR retrovirus for 12 h followed by 20 ng/ml TNF-α for 6 h. The cells were then treated with 2.5 μg/ml actinomycin D (Act-D), and samples were withdrawn at the indicated times. Quantification of GPBP mRNA was as in a; the data were fitted by nonlinear regression to the first order exponential decay equation, enabling calculation of the half-life of the mRNA.
To demonstrate the effect of hBVR on the activity of NF-κB, cells were cotransfected with the pNF-κB reporter construct together with either pEGFP-FLAG-hBVR or empty pEGFP plasmids. The cells were then treated with TNF-α for 12 h beginning 12 h after the addition of DNA. Luciferase activity is dependent upon activation of NF-κB; as shown in Fig. 3e, the luciferase activity was increased by the presence of elevated levels of hBVR in a dose-dependent manner. No induction was seen with the empty vector, indicating that hBVR enhances the activation of NF-κB.
Whether a decrease in GPBP mRNA was the consequence of an accelerated rate of decay due to the presence of si-hBVR was investigated by blocking synthesis of new message with actinomycin D in TNF-α-treated cells. The finding shown in Fig. 3f indicates little change in the stability of GPBP mRNA in the presence of si-hBVR. As noted in supplemental Fig S1b, there was no apparent adverse effect of GPBP on hBVR protein levels or phosphorylation.
GPBP Binds to the C-terminal Segment of hBVR
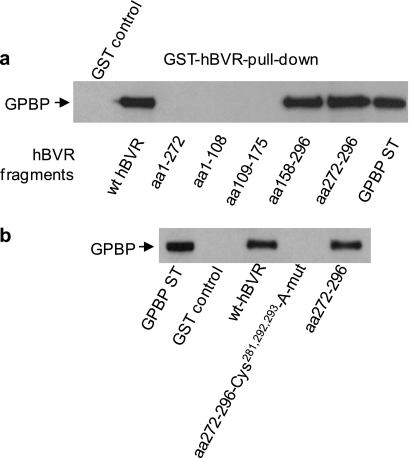
The region(s) of hBVR that binds to GPBP was examined using truncated GST-hBVR expression constructs encoding the fragments aa 1–108, 1–272, 109–175, 158–296, or 272–296 in a GST pulldown assay for their ability to bind to GPBP (Fig. 4a). The results were compared with that of the full-length 296-residue GST-hBVR. Two fragments, aa 158–296 and 272–296, bound GPBP in this assay, whereas no interaction of GPBP with hBVR fragments spanning aa 1–108, 1–272, or 109–175 was detected. The C-terminal 25 residues of hBVR compose the common segment of the two effective active peptides, hence, suggesting the possibility of involvement of this segment of hBVR in interaction with GPBP. The outstanding feature of the C-terminal fragment of hBVR is the presence of three cysteine residues (Cys281, Cys292, and Cys293); cysteine residues are highly interactive and can be involved in protein-protein binding Therefore, the involvement of the cysteine residues in hBVR-GPBP binding was examined using a truncated GST-hBVR fusion expression construct containing aa 272–296 in which all three residues were changed to Ala. Binding of this peptide to GPBP was compared with that of the wt aa 272–296 peptide in the GST-hBVR pulldown assay. As shown in Fig. 4b, although the same binding of GPBP was observed with full-length hBVR and the aa 272–296 fragment, no binding was detected using the cysteine mutant aa 272–296 fragment. The finding is consistent with the likelihood of the key function of cysteine in the protein-protein interaction.
FIGURE 4.
hBVR binds to GPBP through its C terminus. a, GST pulldown of GPBP with hBVR fragments is shown. Lysate was prepared from cells transfected with the FLAG-GPBP expression plasmid. Truncated hBVR proteins were expressed as GST fusions and purified as described under “Experimental Procedures.” These were then used in the GST pulldown assay as described in Fig. 1a; full-length hBVR was used as the control. b, the C-terminal cysteine residues are essential for binding of GPBP to hBVR. Similar GST pulldown experiments to those in a were performed using either the full-length wt hBVR, hBVR aa 272–296 fragment, or a mutant form of the aa 292–296 fragment with Cys281,292,293 changed to Ala. ST, standard.
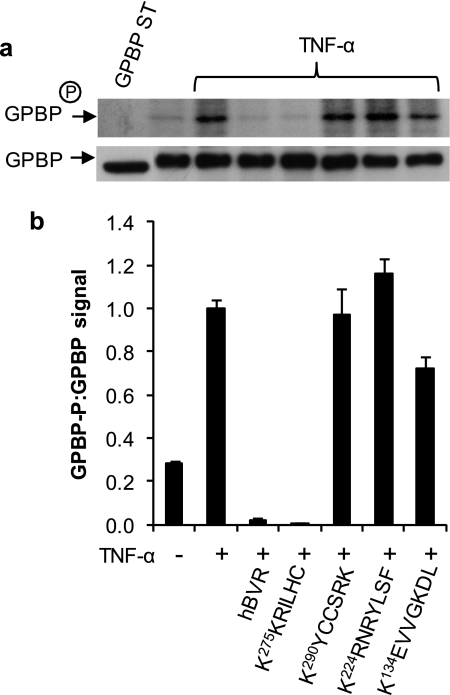
hBVR C-terminus-based 7-Residue Peptide Is as Effective as wt hBVR in Blocking Activation of GPBP by TNF-α
Because both proteins are implicated in the TNF-α signaling cascade, it was of interest to test whether hBVR-derived peptides, in addition to hBVR itself, would affect TNF-α-stimulated phosphorylation of GPBP. To examine this, cells were transfected with FLAG-GPBP and either co-transfected with wt hBVR or treated with myristoylated hBVR-based peptides (KEVVGKDL, KRNRYLSF, KKRILHC, and KYCCSRK) as shown in Fig. 5. Cells were stimulated with TNF-α for 15 min, and the kinase activity of GPBP in cell extracts was tested in an assay system optimized for GPBP autophosphorylation. Again, GPBP phosphorylation was markedly increased in response to TNF-α stimulation. Notably, the increased phosphorylation response was essentially undetectable in the presence of the peptide 275KKRILHC, whereas neither of the peptides 134KEVVGKDL and 290KYCCSRK was effective in suppressing GPBP autophosphorylation. The 224KRNRYLSF peptide caused a modest enhancement of TNF-α-stimulated phosphorylation of GPBP.
FIGURE 5.
A 7-residue long peptide corresponding to hBVR aa 275–281 is as effective as the full-length hBVR polypeptide in blocking TNF-α-stimulated GPBP autophosphorylation. a, HEK293A cells were transfected with FLAG-tagged GPBP expression construct. 24 h later cells were starved overnight and treated for 2 h with the 10 μm myristoylated hBVR-based peptides indicated followed by TNF-α (20 ng/ml for 15 min). Cell lysate was obtained, immunoprecipitated with anti-FLAG antibody, and used for analysis of GPBP autophosphorylation. Analysis of kinase activity was carried out as described (6). b, quantification of GPBP phosphorylation signals is shown. The graph represents the ratio of band intensities of phosphorylated GPBP to GPBP protein shown in a, determined by densitometry of autoradiograph and ECL films, respectively.
DISCUSSION
Recent studies have uncovered multiple functions of the hBVR that include kinase, reductase, and intracellular transporter of regulatory factors. These functions are defined by the primary and secondary features of the protein (for review, see Ref. 10). The present study extends the span of hBVR functions by describing its regulation of GPBP Ser/Thr kinase activity and, by extension, a possible role in the pathophysiology of GPS and potentially in other immune-based disorders (3, 44).
A connection between increased GPBP kinase activity and its expression with autoimmune disorders was initially suggested by detection of an increased expression of GPBP in Goodpasture patients (44) and by the finding that NZW mice develop an age-dependent lupus-like autoimmune response and immune complex-mediated glomerulonephritis. These autoimmune responses are characterized by elevated glomerular GPBP, glomerular basement membrane, collagen disorganization and associated deposits of IgA or IgG. GPBP-dependent serine phosphorylation of the N-terminal region of the human α3(IV)NC1 is suspected to regulate type IV collagen assembly and its up-regulation to induce collagen-based glomerular basement membrane dissociation (3).
As revealed by this study, hBVR disrupts the kinase activity of GPBP and also plays a regulatory function in the response of GPBP to TNF-α and its transcriptional regulation by NF-κB. The finding that hBVR has an inhibitory effect on GPBP kinase activity is somewhat surprising given that the binding and interaction of hBVR with other kinases, such as PKC-βII, PKC-ζ, and ERK1/2, result in their activation and, in the case of insulin receptor substrate 1 (IRS1), which is functionally inactivated and serine-phosphorylated by hBVR (13, 19–21). We have no evidence for hBVR phosphorylation of GPBP as a result of their interaction.3
When examining the effect of hBVR GPBP transcription, we focused on a likely regulatory link between the two genes that involves TNF-α and NF-κB, the downstream effector of TNF-α signaling (23, 24). Both hBVR and GPBP are stimulated by the cytokine; however, hBVR is an upstream activator of NF-κB and binds to p65, the major regulatory subunit of NF-κB; GPBP is downstream of the transcription factor (4, 25). What we observed was a time-dependent change in the profile of GPBP mRNA levels in response to TNF-α and a display of an early rapid rise (Fig. 3). Findings with si-hBVR convincingly indicate that the increase is dependent on hBVR, and the observation with si-p65 indicates that NF-κB activation is required for the increase (Figs. 3, a and b); those observations together with findings that hBVR stimulates p65-dependent transcription (Fig. 3c) lead us to reason that the stimulation of hBVR kinase activity by TNF-α is a contributing factor in the initial rise in GPBP transcription. As shown in supplemental Fig. S1a, hBVR is activated within 5 min in response to TNF-α treatment. This proposal is also consistent with previous findings that linked the rapid activation of hBVR kinase activity to the increased transcription of CREB (cAMP-response element-binding protein) and heme oxygenase-1 (40, 41). Moreover the evidence presented here clearly indicates that the increase in the GPBP mRNA levels is not related to a reduction in the rate of its decay (Fig. 3d).
Findings in this study point to the likelihood that the hBVR-mediated inhibition of TNF-α-dependent GPBP kinase activation requires protein-protein interaction. It is reasonable to suggest that hBVR-bound GPBP is shielded from activation by the cytokine, and/or its inactivation/dephosphorylation is accelerated as a consequence of the interaction.
The hBVR GPBP binding/interacting segment of hBVR was traced to the C terminus of the reductase (Fig. 4). Based on the solved crystal structure of rat BVR (15, 18) and the recently solved crystal structure of the human enzyme (PDB 2H63) much of the 150-residue long C-terminal half of hBVR folds into a large flat six-stranded β-sheet, which is a characteristic of a protein-protein interaction site. Furthermore, the cysteine residues in the aa 272–296 segment proved to be essential to hBVR binding to GPBP. This segment encompasses the 281CX10C motif, and the primary structure of GPBP also predicts the presence of this configuration in its C terminus (544CX10C). Therefore, it is conceivable that the hBVR-GPBP interaction may involve disulfide bond formation or coordination of a divalent sulfhydryl-reactive metal ion. It is noted that the C terminus cysteine residues of hBVR are high affinity Zn2+ binding sites (45). Additionally, the same region of hBVR is required for interaction with PKC-ζ (20). The atypical PKC-ζ cysteine-rich domain binds two Zn2+ and contains a CX10C motif (47, 48). Admittedly, at this time it is not clear whether all three cysteine residues are involved in the protein-protein interaction. The finding that 275KKRILHC281 peptide, which is within the identified GPBP binding segment of hBVR, aa 272–296, inhibits GPBP phosphorylation lends strong support for the involvement, minimally, of this specific cysteine residue in protein-protein binding. Regarding the blockade of GPBP response in the cell to TNF-α, we reason that upon binding of the inhibitory peptide or the intact hBVR, GPBP is blocked from interaction with the docking protein(s) required for its translocation to the cell membrane and points of assembly for signaling molecules that include upstream kinases (49).
It is noteworthy that this hBVR-derived peptide also interacts with and inhibits PKC-ζ activation by TNF-α (20). Furthermore, this peptide, which is the core of hBVR D-Box (275KKRILHCLGLA285), has been characterized as the anchor for MAPK kinase substrates (17, 50). This segment of hBVR was recently identified as the site of ERK-hBVR interaction. This identification was essential to uncovering the function of hBVR as a scaffold protein for MEK/ERK interaction in the cytoplasm and for ERK/Elk interaction in the nucleus (21).
We propose the idea that GPS pathophysiology may be in part related to a suppressed hBVR activity and/or expression in the affected tissues that would result both in an attenuated ability to temper GPBP kinase activity and to generate the quencher of oxygen radicals, bilirubin (30). ROS have been shown to expose cryptic epitopes associated with autoimmune GPS (35) Furthermore, GPBP has been identified as a product of a human-specific TNF-α-responsive transcriptional unit (4) and has been implicated in apoptotic pro-autoimmune signaling (44). In contrast, hBVR has been shown to protect against cell death by apoptosis (41). The co-expression of hBVR and GPBP in various organs together with the observed inhibitory action of the 7-residue hBVR-derived peptide leads us to envision the potential utility of hBVR or its fragments in therapeutic settings to suppress autoimmune responses. hBVR is expressed at high levels in the kidney (51), and expression has also been reported in the lung (46), the two organs that display the symptoms of GPS, and also display significant expression of GPBP (3, 6).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants ES04066 and ES12187 (to M. D. M.). This work was also supported by SAF 2006-12520-C02-01 from the Plan Nacional I+D of the Ministerio de Educacion y Ciencia and Prometeo/2009/065 from Generalitat Valenciana, of Spain (to J. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
T. Miralem, P. E. M. Gibbs, F. Revert, J. Saus, and M. D. Maines, unpublished observations.
- GPS
- Goodpasture syndrome
- GPA
- Goodpasture antigen
- NC1
- noncollagenous-1
- GPBP
- Goodpasture antigen-binding protein
- BVR
- biliverdin reductase
- hBVR
- human BVR
- HEK
- human embryonic kidney
- ROS
- reactive oxygen species
- siRNA
- small interfering (si-) RNA
- TNF-α
- tumor necrosis factor α
- MAPK
- mitogen-activated protein kinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- IP
- immunoprecipitation
- aa
- amino acids
- wt
- wild type
- GST
- glutathione S-transferase
- RT
- reverse transcription.
REFERENCES
- 1.Hudson B. G., Tryggvason K., Sundaramoorthy M., Neilson E. G. (2003) N. Engl. J. Med. 348, 2543–2556 [DOI] [PubMed] [Google Scholar]
- 2.Saus J. (1998) in Encyclopedia of Immunology, 2nd Ed., pp. 1005–1011, Academic Press, Ltd., London [Google Scholar]
- 3.Revert F., Merino R., Monteagudo C., Macias J., Peydró A., Alcácer J., Muniesa P., Marquina R., Blanco M., Iglesias M., Revert-Ros F., Merino J., Saus J. (2007) Am. J. Pathol. 171, 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Granero F., Revert F., Revert-Ros F., Lainez S., Martínez-Martínez P., Saus J. (2005) FEBS J. 272, 5291–5305 [DOI] [PubMed] [Google Scholar]
- 5.Quinones S., Bernal D., García-Sogo M., Elena S. F., Saus J. (1992) J. Biol. Chem. 267, 19780–19784 [PubMed] [Google Scholar]
- 6.Raya A., Revert F., Navarro S., Saus J. (1999) J. Biol. Chem. 274, 12642–12649 [DOI] [PubMed] [Google Scholar]
- 7.Revert F., Ventura I., Martínez-Martínez P., Granero-Moltó F., Revert-Ros F., Macías J., Saus J. (2008) J. Biol. Chem. 283, 30246–30255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litersky J. M., Johnson G. V. (1992) J. Biol. Chem. 267, 1563–1568 [PubMed] [Google Scholar]
- 9.Brown K., Gerstberger S., Carlson L., Franzoso G., Siebenlist U. (1995) Science 267, 1485–1488 [DOI] [PubMed] [Google Scholar]
- 10.Kapitulnik J., Maines M. D. (2009) Trends Pharmacol. Sci. 30, 129–137 [DOI] [PubMed] [Google Scholar]
- 11.Maines M. D. (2005) Physiology 20, 382–389 [DOI] [PubMed] [Google Scholar]
- 12.Maines M. D., Trakshel G. M. (1993) Arch. Biochem. Biophys. 300, 320–326 [DOI] [PubMed] [Google Scholar]
- 13.Lerner-Marmarosh N., Shen J., Torno M. D., Kravets A., Hu Z., Maines M. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7109–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter T. (1998) Harvey Lect. 94, 81–119 [PubMed] [Google Scholar]
- 15.Whitby F. G., Phillips J. D., Hill C. P., McCoubrey W., Maines M. D. (2002) J. Mol. Biol. 319, 1199–1210 [DOI] [PubMed] [Google Scholar]
- 16.Jacobs J. M., Marek D., Walton H. S., Sinclair P. R., Sinclair J. F. (1999) Arch. Biochem. Biophys. 371, 8–14 [DOI] [PubMed] [Google Scholar]
- 17.Minden A., Karin M. (1997) Biochim. Biophys. Acta 1333, F85–F104 [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi A., Park S. Y., Miyatake H., Sun D., Sato M., Yoshida T., Shiro Y. (2001) Nat. Struct. Biol. 8, 221–225 [DOI] [PubMed] [Google Scholar]
- 19.Maines M. D., Miralem T., Lerner-Marmarosh N., Shen J., Gibbs P. E. (2007) J. Biol. Chem. 282, 8110–8122 [DOI] [PubMed] [Google Scholar]
- 20.Lerner-Marmarosh N., Miralem T., Gibbs P. E., Maines M. D. (2007) FASEB J. 21, 3949–3962 [DOI] [PubMed] [Google Scholar]
- 21.Lerner-Marmarosh N., Miralem T., Gibbs P. E., Maines M. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tudor C., Lerner-Marmarosh N., Engelborghs Y., Gibbs P. E., Maines M. D. (2008) Biochem. J. 413, 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duran A., Diaz-Meco M. T., Moscat J. (2003) EMBO J. 22, 3910–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karin M., Greten F. R. (2005) Nat. Rev. Immunol. 5, 749–759 [DOI] [PubMed] [Google Scholar]
- 25.Gibbs P. E., Maines M. D. (2007) Int. J. Cancer 121, 2567–2574 [DOI] [PubMed] [Google Scholar]
- 26.Sedlak T. W., Saleh M., Higginson D. S., Paul B. D., Juluri K. R., Snyder S. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5171–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maines M. D. (1997) Annu. Rev. Pharmacol. Toxicol. 37, 517–554 [DOI] [PubMed] [Google Scholar]
- 28.Mancuso C., Pani G., Calabrese V. (2006) Redox Rep. 11, 207–213 [DOI] [PubMed] [Google Scholar]
- 29.Stocker R., Yamamoto Y., McDonagh A. F., Glazer A. N., Ames B. N. (1987) Science 235, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 30.Maghzal G. J., Leck M. C., Collinson E., Li C., Stocker R. (2009) J. Biol. Chem. 284, 29251–29259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halliwell B. (2007) Biochem. Soc. Trans. 35, 1147–1150 [DOI] [PubMed] [Google Scholar]
- 32.Immenschuh S., Fahimi H. D., Baumgart-Vogt E. (2005) Cell. Mol. Biol. 51, 471–477 [PubMed] [Google Scholar]
- 33.Ryter S. W., Alam J., Choi A. M. (2006) Physiol. Rev. 86, 583–650 [DOI] [PubMed] [Google Scholar]
- 34.Ryter S. W., Morse D., Choi A. M. (2007) Am. J. Respir. Cell Mol. Biol. 36, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R., Cantley L. G., Kerjaschki D., Neilson E. G. (2000) J. Biol. Chem. 275, 20027–20032 [DOI] [PubMed] [Google Scholar]
- 36.Nath K. A., Vercellotti G. M., Grande J. P., Miyoshi H., Paya C. V., Manivel J. C., Haggard J. J., Croatt A. J., Payne W. D., Alam J. (2001) Kidney Int. 59, 106–117 [DOI] [PubMed] [Google Scholar]
- 37.Rao R. P., Yuan C., Allegood J. C., Rawat S. S., Edwards M. B., Wang X., Merrill A. H., Jr., Acharya U., Acharya J. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juan S. H., Cheng T. H., Lin H. C., Chu Y. L., Lee W. S. (2005) Biochem. Pharmacol. 69, 41–48 [DOI] [PubMed] [Google Scholar]
- 39.Naidu S., Wijayanti N., Santoso S., Kietzmann T., Immenschuh S. (2008) J. Immunol. 181, 4113–4123 [DOI] [PubMed] [Google Scholar]
- 40.Kravets A., Hu Z., Miralem T., Torno M. D., Maines M. D. (2004) J. Biol. Chem. 279, 19916–19923 [DOI] [PubMed] [Google Scholar]
- 41.Miralem T., Hu Z., Torno M. D., Lelli K. M., Maines M. D. (2005) J. Biol. Chem. 280, 17084–17092 [DOI] [PubMed] [Google Scholar]
- 42.Dolcet X., Llobet D., Pallares J., Matias-Guiu X. (2005) Virchows Arch. 446, 475–482 [DOI] [PubMed] [Google Scholar]
- 43.Graham B., Gibson S. B. (2005) Cell Cycle 4, 1342–1345 [DOI] [PubMed] [Google Scholar]
- 44.Raya A., Revert-Ros F., Martinez-Martinez P., Navarro S., Rosello E., Vieites B., Granero F., Forteza J., Saus J. (2000) J. Biol. Chem. 275, 40392–40399 [DOI] [PubMed] [Google Scholar]
- 45.Maines M. D., Polevoda B. V., Huang T. J., McCoubrey W. K., Jr. (1996) Eur. J. Biochem. 235, 372–381 [DOI] [PubMed] [Google Scholar]
- 46.Baglole C. J., Sime P. J., Phipps R. P. (2008) Am. J. Physiol. Lung Cell. Mol. Physiol. 295, L624–L636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbard S. R., Bishop W. R., Kirschmeier P., George S. J., Cramer S. P., Hendrickson W. A. (1991) Science 254, 1776–1779 [DOI] [PubMed] [Google Scholar]
- 48.Newton A. C. (1995) J. Biol. Chem. 270, 28495–28498 [DOI] [PubMed] [Google Scholar]
- 49.Pawson T., Nash P. (2003) Science 300, 445–452 [DOI] [PubMed] [Google Scholar]
- 50.Kasza A., O'Donnell A., Gascoigne K., Zeef L. A., Hayes A., Sharrocks A. D. (2005) J. Biol. Chem. 280, 1149–1155 [DOI] [PubMed] [Google Scholar]
- 51.McCoubrey W. K., Jr., Eke B., Maines M. D. (1995) Biol. Reprod. 53, 1330–1338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.