Abstract
The Dock180 family of atypical Rho family guanine nucleotide exchange factors (Rho-GEFs) regulate a variety of processes involving cellular or subcellular polarization, including cell migration and phagocytosis. Each contains a Dock homology region-1 (DHR-1) domain that is required to localize its GEF activity to a specific membrane compartment where levels of phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3) are up-regulated by the local activity of PtdIns 3-kinase. Here we define the structural and energetic bases of phosphoinositide specificity by the DHR-1 domain of Dock1 (a GEF for Rac1), and show that DHR-1 utilizes a C2 domain scaffold and surface loops to create a basic pocket on its upper surface for recognition of the PtdIns(3,4,5)P3 head group. The pocket has many of the characteristics of those observed in pleckstrin homology domains. We show that point mutations in the pocket that abolish phospholipid binding in vitro ablate the ability of Dock1 to induce cell polarization, and propose a model that brings together recent mechanistic and structural studies to rationalize the central role of DHR-1 in dynamic membrane targeting of the Rho-GEF activity of Dock180.
Keywords: Calorimetry, Cell Migration, Computer Modeling, Crystal Structure, Phosphatidylinositol Signaling, Plasma Membrane, Protein Targeting, Site-directed Mutagenesis, Thermodynamics, X-ray Crystallography
Introduction
The family of Rho-GTPases, including Rac, Cdc42, and RhoA, localize to specific compartments of the plasma membrane in response to external cues that lead to the modification of membrane phospholipids (1). Rho-GEFs4 co-localize to the same membrane locales, where they catalyze the removal of bound GDP from inactive Rho-GTPases, enabling reloading of GTP and binding of effector molecules that mediate a variety of signaling pathways (2). This localized activation controls the organization and dynamics of the actin cytoskeleton, thereby regulating a large number of processes involving cell morphology, polarity, and migration (3).
Two structurally unrelated families of Rho family GEFs have been characterized: the “classical” Rho-GEFs (2) and the “atypical” Dock180 Rho-GEFs (4–6). Dock1 (also called Dock180) is a GEF for Rac1 (7, 8). It is a large protein (1865 residues) that includes an N-terminal SH3 domain and flanking helical bundle that mediate formation of a functional complex with the “engulfment and cell motility” protein, ELMO1 (9–11); a DHR-1 domain required for targeting to membranes enriched in PtdIns(3,4,5)P3 (12); and a DHR-2 domain that houses the GEF activity (4, 6). The structure and novel GEF mechanism of the Dock9 DHR-2 domain (a GEF for Cdc42) was recently elucidated (13), providing an enzymatic model for the entire Dock family. Long intervening sequences between the recognized domains are predicted to be mostly helical, and may fold as HEAT/ARM repeat domains (2). The C-terminal region includes (in Dock1 and Dock2) a linear motif that augments membrane binding (14, 15), and a proline-rich region that forms a complex with the adaptor protein, CrkII (16).
There are 11 Dock180 members in humans (Dock1–Dock11), at least two in zebrafish (17), and well characterized orthologs in Drosophila (“Myoblast City” or “MBC”) (18) and Caenorhabditis elegans (“ced-5”) (19), which form complexes with orthologous partners and share many related functions (9, 20). Dock180 proteins have been implicated in a variety of cellular processes, including cell migration, axonal polarization, tumor suppression, the engulfment of apoptotic cells, and phagocytosis of pathogens (9, 12, 21–26). During cell migration, for example, elevated levels of PtdIns(3,4,5)P3 are created at the leading edge by the local activity of PtdIns 3-kinase, which promotes membrane attachment by the Dock1-ELMO complex leading to polarized activation of Rac1 (4, 5, 8, 12, 27–32).
In all cases tested, the DHR-1 domain has been found necessary for the cellular function of full-length Dock180 family proteins. Here, we report the crystal structure of the DHR-1 domain of Dock1, which comprises a C2 core with several large insertions, and identify the structural and energetic determinants of phospholipid recognition in vitro and in cells. These findings, when combined with recent advances by others, lead us to propose a model for how full-length Dock1 coordinates membrane specificity conferred by the DHR-1 domain with the GEF activity of the DHR-2 domain, which provides a structural framework for probing the diverse biological functions mediated by the Dock180 family of GEFs.
EXPERIMENTAL PROCEDURES
Protein Production
The domain boundaries of Dock1 DHR-1 were previously defined (12). The Dock1 DHR-1 domain (422–619) was cloned in pET28a and expressed in Escherichia coli as a His6-tagged fusion protein. The protein was purified using HiTrap Ni2+-chelating and Superdex 200 gel filtration columns (GE Healthcare). Protein purity and identity were confirmed by SDS-PAGE, immunoblot, and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Site-directed mutagenesis was performed using the QuikChange method (Stratagene) in plasmid pET-28a, and confirmed by DNA sequencing. Mutant proteins were expressed and purified as for wild-type, and melting temperature profiles were determined using a CSC Nano II Differential Scanning Calorimeter.
Isothermal Titration Calorimetry (ITC)
Experiments were carried out on a Microcal VP ITC200 isothermal titration calorimeter from Microcal (Northampton, MA). Each titration involved 19 injections of 2-μl aliquots of ∼1 mm water-soluble di-C8 phosphoinositide analogs (Echelon Biosciences, Salt Lake City, UT) or InsP4 into cells containing 25–100 μm DHR-1 at 23 °C. Buffers were comprised of 10 mm Tris-HCl, pH 8.0, 2 mm β-mercaptoethanol, and either 14 or 145 mm NaCl. Folding and monodispersity of DHR-1 were confirmed by size exclusion chromatography and differential scanning calorimetry. To account for heat changes associated with dilution of the phosphoinositide analogs, control titrations into buffer were employed. The use of di-C8 phosphoinositide stock solutions at ∼1 mm concentration avoids complications arising from micelle formation (critical micellar concentration ∼5 mm). By lowering the NaCl concentration to 15 mm, the enthalpy change increased by ∼4-fold, and reproducible binding curves that achieved full saturation were obtained for wild-type and mutant proteins. For the competition assays, a 1.5 m excess of either the PtdIns(3,4,5)P3 or PtdIns(4,5)P2 analogs was first incubated with DHR-1 in the ITC cell, prior to titration of the other phospholipid. For Ca2+ binding experiments, aliquots of 4.0 or 8.0 mm CaCl2 were injected into cells containing 200 or 400 μm DHR-1 (which had previously been dialyzed in 2 mm EGTA). No binding was observed (data not shown). Thermodynamic parameters were obtained using the Origin 7 software package (Microcal).
Crystallization and Data Collection
DHR-1 crystals were grown at 293 K using the microbatch method under oil. 1.5 μl of solution containing 5.5 mg/ml of DHR-1 in 12.5 mm Tris-HCl, pH 8.8, 75 mm NaCl, 2.5 mm β-mercaptoethanol, was mixed with 3 μl of reservoir containing 30% (w/v) polyethylene glycol 10K, 0.2 m sodium acetate, 0.1 m Tris-HCl, pH 8.8. Crystals grew as monoclinic plates within 2–3 weeks, and were flash frozen in liquid nitrogen after briefly rinsing in paratone-N oil. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory beamline 9-2 at a wavelength of 0.912 Å, and recorded with a Mar325 CCD detector controlled by BLU-ICE (33). Data were integrated, scaled, and reduced using HKL2000 (34) (see Table 1).
TABLE 1.
Crystallographic analysis and model refinement
| Space group | P21 |
|---|---|
| Unit cell parameters | a = 47.0Å, b = 63.5Å, c = 63.4Å, β = 109.3° |
| Wavelength (Å) | 0.9116 |
| Resolution range (Å) | 50–2.37 |
| Number of observations | 53,672 |
| Completeness (%) | 99.4 (97.5)a |
| Mean I/σ(I) | 22.6 (6.9)a |
| RSYM on Ib | 0.076 (0.216)a |
| Outer resolution shell (Å) | 2.45-2.37 |
| Model and refinement statistics | |
| Resolution range (Å) | 50–2.37 |
| Protein residues (all atoms) | 349 (2,926) |
| Average B-factor (Å2) | 44.0 |
| Unique reflections (test set) | 13,960 (679) |
| Rwork (%)c | 21.6 |
| Rfree (%)d | 26.8 |
| Stereochemical parameters | |
| Observed rms deviations from ideality | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.43 |
| Torsion angles (°) | 25.34 |
| Improper torsion angles (°) | 0.74 |
| Ramachandran outliersb | 0.3% |
| Estimated coordinate error | |
| Low resolution cut-off (Å) | 5.0 |
| Luzzati coordinate error (Å) | 0.29 |
| σ A coordinate error (Å) | 0.24 |
a Outer resolution shell in parentheses.
b RSYM = ΣIi − 〈Ii〉|/Σ|Ii|, where Ii is the scaled intensity of the ith measurement and 〈Ii〉 is the mean intensity for that reflection.
c Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fc and Fo are the calculated and observed structure factor amplitudes, respectively.
d Rfree is the same as Rwork, but calculated on the 5% of reflections that were omitted from refinement.
Structure Determination
Initial molecular replacement efforts were unsuccessful, as were efforts to incorporate conventional heavy atoms or selenomethionine. Consequently, a more extensive molecular replacement search strategy was pursued using the search pipeline (35) based on MOLREP (36), with automated parameter-space screening. The search employed models derived from the distant homology sequence-recognition algorithm (37). The top solution was the RIM2 C2A domain (PDB code 2BWQ), which has 10% sequence identity over 50% of the DHR-1 domain. This solution was independently confirmed using PHASER (38), and further optimized in a second round of parallelized searches with a large set of trimmed RIM2 C2A polyalanine models generated by combinatorial removal of loops, regions of neighboring gaps in the model, and regions of lowest sequence similarity. Initial phases allowed a partial model to be built. Phase combination, iterative model building, and refinement with Coot (39), REFMAC5 (40), and CNS (41), using σ A-weighted composite omit maps and 2Fo − Fc maps, allowed most of the model to be built. The asymmetric unit includes two DHR-1 domains, 81 water molecules, and a modified residue (the β-mercaptoethanol adduct of C595) in each domain. Electron density was not observed for the His tag, the N terminus (422–424 in both copies), or the C terminus (610/612–619), or for a surface loop within the β7–β8 subdomain (580/581–587). The L1 and L3 loops have relatively weak density, but distinct conformations are evident for molecules A and B (supplemental Fig. S1). The root mean square deviation between backbone atoms of the two molecules in the asymmetric unit is 0.54 Å (for 161 equivalent residues). Stereochemical quality of the final model was assessed using AutoDepInputTool (42), MolProbity (43), SFcheck 4.0 (44), and WHATIF 5.0 (45). 97.2 and 99.8% of the main chain torsion angles are in the favored and allowed regions, respectively. Data collection and refinement statistics are summarized in Table 1.
Molecular Docking and Modeling of Full-length Dock1
Docking simulations were performed with Molegro Virtual Docker (46) using di-C2 PtdIns(3,4,5)P3 or its head group, inositol 1,3,4,5-tetraphosphate (InsP4). The docking runs included a maximum of 1500 iterations with a population size of 50 and a minimum of 10 runs. Binding modes were analyzed using the “re-ranking score.” Docking solutions from the initial runs, with no search space constraints and default parameters, were clustered at two sites. In the subsequent docking simulations, side chain flexibility was introduced, and the search space radius was set to 20 or 15 Å from the centers of the binding cavities. Distance restraints penalized the interaction between the aliphatic carbon atoms di-C2 PtdIns(3,4,5)P3 and the protein. The “MolDock [GRID]” scoring function and “Tabu pose” clustering algorithms were used for side chain flexible docking simulations. Side chain torsional angles allowed to vary during simulation were: on the upper surface, Lys439, Ser441, Lys442, Thr443, Thr444, Lys446, Asn447, Tyr484, Gln485, His515, Gln519, Asp523, Lys524, Phe529, and Lys555; and in the β-groove, Glu449, Thr451, Ser453, Tyr455, Arg461, Glu477, Lys479, Arg510, Arg514, Arg516, Lys527, Ile528, Glu562, and Asp600.
For modeling full-length Dock1, dimeric DHR-2/Rac1 and ARM-repeat Dock1 models were based on the crystal structures of Dock9 DHR-2/Cdc42 (PDB code 2WM9) (13) and importin-β (PDB code 2QNA) (47). Homology models were built using SCWRL4 (48) and Phyre (49). The ARM-DHR-2 interaction interface was determined using the RosettaDock docking algorithm (50).
Cell Culture, Transfection, and Cell Spreading Assays
CHO LR73 cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and penicillin-streptomycin antibiotics, and assayed as previously described (11, 12). Briefly, cells were transfected with the indicated plasmids. Cells were starved overnight in 0.1% serum medium, then allowed to spread on fibronectin, and stained for Dock1, actin, and cell nuclei. Anti-Dock1 (H-4 and H-70) and anti-Myc (9E10) antibodies were obtained from Santa Cruz Biotechnologies.
RESULTS
Crystal Structure of the Dock1 DHR-1 Domain
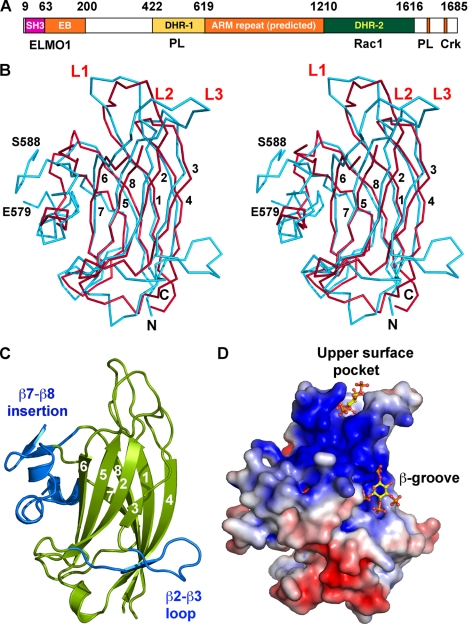
We expressed a construct of Dock1 in E. coli comprising residues 422–619, which is the minimal fragment that maintains specific PtdIns(3,4,5)P3 binding activity (12) (Fig. 1A). We determined its crystal structure at 2.37-Å resolution (Table 1, Figs. 1 and 2 using automated molecular replacement searches and phase recombination (see “Experimental Procedures” and Table 1). The central core of the structure adopts an anti-parallel β-sandwich with the “type II” C2 domain fold (a circular permutation of the more common “type I” topology (51, 52)), in which two 4-stranded sheets with strand orders 6-5-2-3 and 7-8-1-4 create convex- and concave-exposed faces, respectively. Structural comparisons using DALI (53) show that the β-sandwich core is similar to at least 38 C2 domains (of either topology) in the PDB with root mean square deviations between 1.7 and 3.6 Å for 101–143 Cα atoms, but very low identities in all cases: 7–16% (supplemental Fig. S2). Elaborations to the core include three loops (β1–β2 = L1; β3–β4 = L2; and β5–β6 = L3) on the upper surface, as well as two further large insertions (see below) that may explain why sequence alignment algorithms had difficulty in recognizing the C2-fold (12). The most extensive structural similarity occurs with the C2 domain of PtdIns 3-kinase (supplemental Fig. S3; root mean square difference = 2.8 Å for 143 Cαs; 10% identity), which, intriguingly, is the enzyme that generates PtdIns(3,4,5)P3 at the leading edge of migrating cells.
FIGURE 1.
Structure of the Dock1 DHR-1 domain. A, domain organization of Dock1, with residue limits and major binding partners indicated. ELMO1 binds to a module comprising the SH3 domains and “EB” helical segment, as well as segments at the C terminus (not shown) (11, 85). ARM, Armadillo/HEAT repeat; EB, ELMO-binding; PL, phospholipid; Crk, CrkII. B, stereo Cα plot of DHR-1 (cyan) overlaid with the C2 domain of PtdIns 3-kinase-γ (red). The latter is disordered at loop L3 and has shorter β2–β3 and β7–β8 insertions. DHR-1 is disordered between residues 579 and 588. Strands and loops are numbered, and termini labeled. C, schematic of DHR-1 in the same view as in B. The β-sandwich core (with strands numbered) is in green, insertions in blue. D, surface charge potential (83) on DHR-1. The view is rotated ∼90° about a vertical axis to show the basic surfaces, with computational docking solutions (see text) for the PtdIns(3,4,5)P3 head group in the upper surface pocket and β-groove (note that there is no experimental evidence for binding in the β-groove).
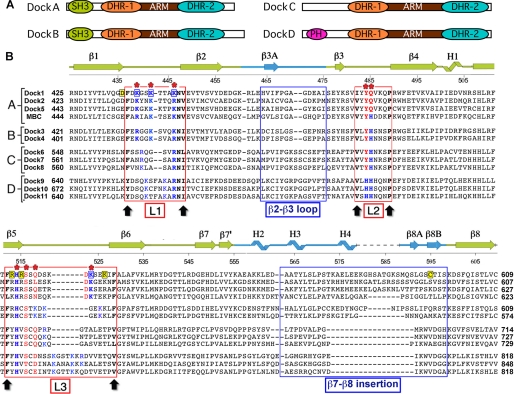
FIGURE 2.
Sequences of Dock180 family DHR-1 domains. A, domain organization of the 4 groups of human full-length proteins. B, structure-based alignment of the 11 human DHR-1 sequences and Drosophila myoblast city, which belongs to the DockA group. The secondary structure is indicated. The 3 surface loops, L1–L3, are indicated by red boxes; they are delimited by conserved residues marked by black arrows. Residues predicted to contact phospholipid in Dock1 DHR-1 are marked with a red asterisk; those supported by mutagenesis are boxed in blue. Contact residues in other family members predicted by homology are shown in blue (H/K/R) or red (others). Conserved structural residues are in black (bold). Residues mutated in this study that do not affect phospholipid binding are highlighted in yellow; 3 of these are on L3 but point into the β-groove. The 2 large insertions in the C2 core are also indicated.
The crystals contain two independent copies of the DHR-1 domain (molecules “A” and “B”), which are very similar except for the conformations of the large loops (L1 and L3) on the upper surface of the domain (supplemental Fig. S3). In Molecule A, which is free from lattice contacts in the loop region, a cluster of lysines points inward, whereas acidic residues point outward, creating a positively charged pocket at the top of the molecule. In Molecule B, a major lattice contact appears to trigger or stabilize a distinct organization of the loops in which the charged residues create a network of salt bridges, and the positively charged pocket seen in Molecule A is not observed. The absence of crystal contacts, as well as mutagenesis and modeling studies (see below), are fully consistent with Molecule A representing the functional conformation in solution. Moreover, we found that a point mutant (D436Y) that disrupts a salt bridge observed only in Molecule B had no functional effect (see below). Molecule A was therefore used for all modeling and structural analysis.
Insertions into the C2 Core
In addition to the 3 surface loops, there are 2 major elaborations of the C2 core: a 20-residue insertion (457–476) between β2 and β3 on one side of the sandwich; and a 42-residue (557–598) subdomain inserted between β7 and β8 that packs against the opposite side (Figs. 1 and 2). The β2–β3 insertion first forms a ridge along the bottom of the 6-5-2-3 sheet; it then crosses over to the 7-8-1-4 sheet, forming a short strand (β3A) that augments the β4 end of the sheet and provides two hydrophobic side chains (Val465 and Ile466) that insert into and seal this edge of the sandwich, before returning to the C2 core via a well defined loop that is unique to DHR-1. β3 is irregular at its C-terminal end, where it adopts cross-sheet interactions with the side chains of Glu493 and the buried Trp491.
The β7–β8 insertion forms a subdomain at the opposite edge of the β-sandwich, beginning with an extension of the β7 strand (β7′) that crosses over to the other sheet, forming main chain hydrogen bonds with the beginning of β6, and inserting two side chains (Leu551 and Val553) between the sheets, sealing this edge of the sandwich (the two cross-over strands, β7′ and β3A, effectively create a β-barrel). The subdomain next adopts two short helical elements (H2/H3) that form a ridge along the left side of the concave face, which connects with the β2–β3 insertion at the bottom, and a bulge formed by the start and end of L3, to create a continuous ridge and a distinctive pocket on the concave face, which is highly basic (Fig. 1D). Many C2 domains have a pocket here, which is often called the “β-groove” (54). A third helical segment (H4) extends away from the body of the domain, followed by a disordered loop (residues 581 and 587) and a β-hairpin, before finally returning to the C2 core via the β8 strand. The hairpin includes a partly buried cysteine (Cys595) that forms a stable adduct with β-mercaptoethanol in the crystals; however, we found no evidence that the cysteine plays a functional role, because a C595A mutant behaved like wild-type in phospholipid binding and cell-based assays (see below).
DHR-1 Binds Short Chain Analogs of PtdIns(3,4,5)P3 and PtdIns(4,5)P2 at Overlapping Sites
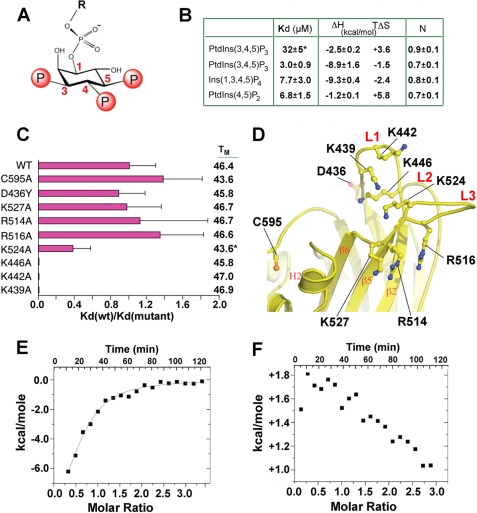
We explored the specificity of phospholipid binding in solution by using monodisperse phosphoinositide analogs and ITC, which allows precise determination of binding constants and thermodynamic parameters (Figs. 3 and supplemental S4). We first defined conditions for the reproducible binding of DHR-1 to di-C8-PtdIns(3,4,5)P3. Although we could observe 1:1 binding in 145 mm NaCl, the affinity (Kd = 32 ± 6 μm) was close to the technical limits of detection, given the need to limit the lipid concentration to avoid micelle formation. To enhance detection, we shifted to a lower ionic strength (∼20 mm), which increased the binding affinity 10-fold (Kd = 3.0 ± 0.9 μm) without changing the stoichiometry (Fig. 3B). These conditions allowed us to study the effects of mutations and compare the binding of other phospholipids. We found that Ins(1,3,4,5)P4 (the PtdIns(3,4,5)P3 head group) bound with a comparable affinity (Kd = 7.7 ± 3.0 μm) and stoichiometry, consistent with a dominant role for ionic interactions between a positively charged protein surface and the negatively charged phosphate groups. In both cases, binding was largely enthalpy-driven.
FIGURE 3.
Phosphoinositide binding to the DHR-1 domain. A, the head group of PtdIns(3,4,5)P3 with phosphate positions numbered. B, thermodynamic parameters for phosphoinositide and head group binding to wild-type DHR-1 (Kd, dissociation constant; ΔH, enthalpy change; TΔS, temperature (K) × entropy change; ΔG (the free energy change) = ΔH-TΔS = −RT ln Kd); N is the apparent stoichiometry. No binding was observed for PtdIns(3,5)P2, PtdIns(3,4)P2, or PtdIns(3)P1. * indicates the experiment was performed in 145 mm NaCl. All other experiments were carried out in low salt (see “Experimental Procedures”). Representative ITC profiles are provided in supplemental Fig. S4. C, effect of point mutations on PtdIns(3,4,5)P3 binding, defined as Kd(wt)/Kd(mutant). Error estimates are from fitting of ITC titration curves. At right are melting temperatures, Tm, for each mutant, in °C. *, the Tm value given for K524A is actually for the triple mutant, K446A/K524A/K555A. D, mutation sites mapped onto the DHR-1 structure. E and F, ITC competition titrations (negative values of energy indicate an exothermic reaction; positive values, endothermic). E, DHR-1 equilibrated with a 1.5 m excess of PtdIns(4,5)P2 titrated with PtdIns(3,4,5)P3. F, the converse experiment: DHR-1/PtdIns(3,4,5)P3 titrated with PtdIns(4,5)P2.
Because many C2 domains have a broad specificity for anionic lipids, we explored the binding of PtdIns mono- and bisphosphates. No binding was observed by titrating with PtdIns(3)P1, PtdIns(3,4)P2, or PtdIns(3,5)P2. However, we found that PtdIns(4,5)P2 bound reproducibly, with a Kd similar to that of PtdIns(3,4,5)P3 and a stoichiometry approaching 1:1. However, in this case, binding was largely entropy-driven, with only a small enthalpic contribution (Fig. 3B). To determine whether PtdIns(3,4,5)P3 and PtdIns(4,5)P2 bound at overlapping or distinct sites, we used a competition assay in which DHR-1 was first preincubated with an excess of one lipid and titrated with the other. Given the similar binding free energies but different enthalpic contributions, if binding is reversible and competitive, then titration of PtdIns(3,4,5)P3 into DHR-1/PtdIns(4,5)P2 should displace PtdIns(4,5)P2 in an exothermic reaction, whereas titration of PtdIns(4,5)P2 into DHR-1/PtdIns(3,4,5)P3 should be endothermic; and this was indeed the case (Fig. 3, E and F).
Phosphoinositide Binding Is Mediated by the Upper Loops of DHR-1, Not the β-Groove
Most C2 domains that engage membrane do so in a Ca2+-mediated fashion via a cluster of acidic residues on their surface loops (supplemental Fig. S5) (52, 54, 55), whereas a subset utilize a positively charged β-groove (56–58) (see Figs. 1D and supplemental S6). Ca2+-independent binding via the upper loops is far less common, and those that do bind phosphoinositide in this way typically show little specificity (59). The DHR-1 C2 domain displays regions of broad positive potential both in the β-groove and on the surface loops (see Figs. 1D and supplemental S6), and we confirmed that DHR-1 does not bind Ca2+ by calorimetry (data not shown). Both regions are thus candidates for binding negatively charged lipids in a Ca2+-independent fashion. As an objective first step, we employed computational docking using the Molegro Virtual Docker (46), which allows for ligand and protein side chain flexibility, to search for phospholipid binding sites. Using Ins(1,3,4,5)P4 as the ligand, this procedure generated two clusters of docking solutions in the expected regions (see Figs. 1D and supplemental S6). The first cluster, on the upper surface, occurs where a pocket is formed by basic and polar residues from all three loops, and includes a histidine, tyrosine, and four lysines. The second cluster is in the β-groove, which contains a similar arrangement of basic and aromatic side chains.
In a previous study, DHR-1 with six lysines simultaneously mutated to alanine failed to bind PtdIns(3,4,5)P3 in a bead pulldown assay (12). Our crystal structure shows that 4 of these lysines (Lys439, Lys442, Lys446, and Lys524) line the pocket on the upper surface, one (Lys527) forms part of the β-groove, and one (Lys522) is in neither pocket (Fig. 3D). Although this mutant is consistent with our docking studies, it does not delineate the roles of the two pockets; moreover, its fold-integrity was not demonstrated. We therefore generated single site mutants, and determined their melting temperatures using differential scanning calorimetry, which provides a sensitive measure of tertiary fold-integrity. We first made Lys → Ala mutants of each of the 4 lysines in the upper pocket (Lys439, Lys442, Lys446 (L1), and Lys524 (L3)), none of which form salt bridges or hydrogen bonds in the wild-type structure. All 4 mutants folded well, as judged by melting temperatures, Tm (Fig. 3C), and reproducible binding data were collected by ITC in each case. We found that the K524A mutation reduced PtdIns(3,4,5)P3 binding 2-fold, whereas mutation of any one of the 3 L1 lysines completely abolished binding (Fig. 3C).
We next made point mutants in the β-groove, and were able to collect reliable data from 3 of them (R514A, R516A, and K527A). Note that these residues lie on L3, but that their side chains point away from the surface pocket. In contrast to the surface pocket mutants, none of these mutants significantly affected PtdIns(3,4,5)P3 binding (Fig. 3C). Other mutants in this region, including K479A and K479Q, folded either with reduced stability (ΔTm ≥ 5 °C) or, in the case of R510A, not at all, presumably because these residues play important structural roles. Thus, Arg510, which lies at the heart of the β-groove, is partly buried, making several intramolecular hydrogen bonds; whereas Lys479 makes inter-strand salt bridges and hydrogen bonds.
We also tested binding of the L1 mutants, K442A and K446A, to PtdIns(4,5)P2, and found, as for PtdIns(3,4,5)P3, that binding was completely abolished in both cases (data not shown). Taken together, our studies suggest that DHR-1 has a high level of specificity for PtdIns(3,4,5)P3 and PtdIns(4,5)P2, which bind competitively to the same or overlapping sites within the pocket on the upper surface. Moreover, we conclude that the β-groove plays no role in binding to these or any of the phospholipids tested.
Point Mutants That Ablate Phospholipid Binding in Vitro Inhibit Cellular Polarization
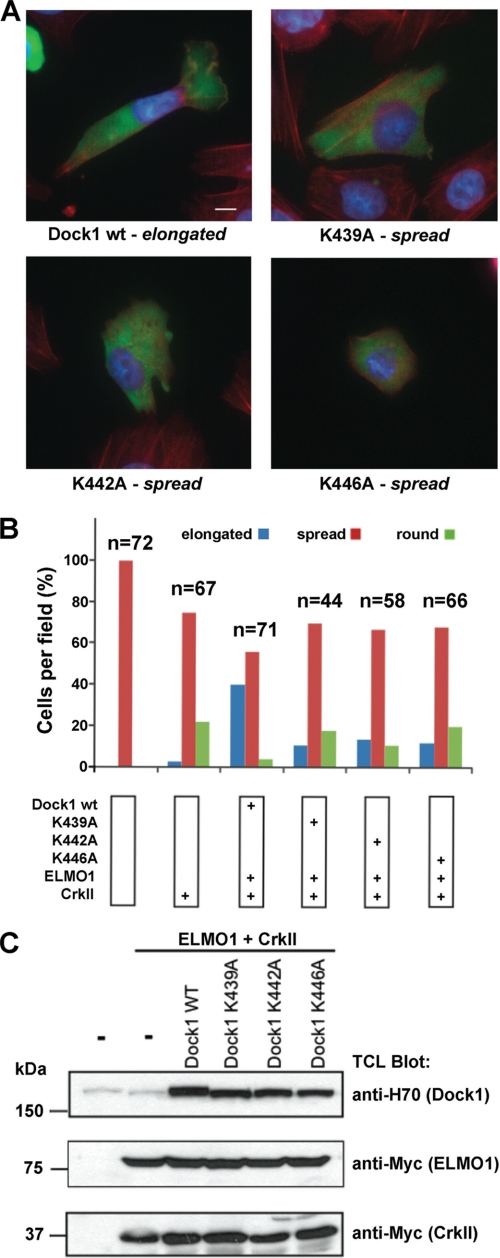
We next constructed 3 full-length Dock1 point mutants with Lys → Ala mutations in the L1 lysines, and tested their function in a cell-based assay. We used the assay previously established (11, 12) in which co-transfection of Dock1/ELMO1/CrkII into LR73 cells triggers cell elongation in a PI 3-kinase-dependent fashion when cells are replated on fibronectin (a measure of the polarizability of spreading cells, which correlates with cell migration). We found that all 3 mutants caused a consistent reduction in cell elongation, as well as an increase in cell rounding (Fig. 4), indistinguishable from the effects seen previously for Dock1 with the entire DHR-1 domain deleted (12). Thus, point mutations in DHR-1 that ablate the binding of PtdIns(3,4,5)P3 in vitro manifest as a loss-of-function of the full-length molecule in this cell-based assay.
FIGURE 4.
Point mutations in the DHR-1 domain inhibit cell elongation by Dock1. A, cells transfected with plasmids expressing wild-type or mutant Dock1, together with plasmids for ELMO1 and CrkII, were detached and plated on fibronectin-coated chambers for 2 h, and stained with anti-Dock1 antibody, rhodamine, phalloidin, and 4,6-diamidino-2-phenylindole (photographed at ×100 magnification: scale bar, 10 μm). B, quantification of cell behavior. Independent fields were photographed at lower magnification, and scored for three phenotypes: round, spread, and elongated. Data in A and B are representative of three independent experiments. C, expression levels of transfected proteins analyzed by immunoblotting cell lysates.
A Model for Phospholipid Binding to the Dock1 DHR1 Domain
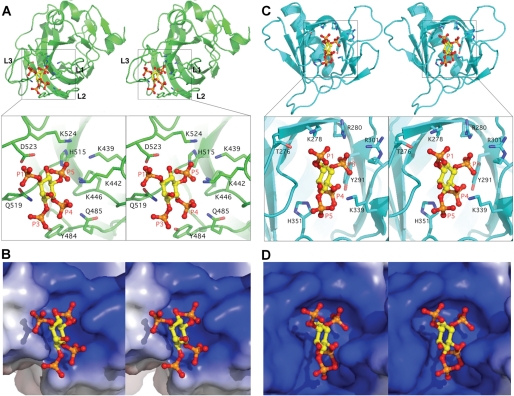
We were not able to co-crystallize the DHR-1 domain with PtdIns(3,4,5)P3 or Ins(1,3,4,5)P3, and indeed no experimental structures of inositol phosphates bound to Type II C2 domains have been determined, presumably because of their weak affinities and/or nonspecific binding. By contrast, many signaling molecules contain PH domains that bind inositol phosphates with high affinity, and many structures have been determined. For example, the PH domain of a splice variant of ARNO (60) displays many characteristics of the DHR-1 binding site: thus, the pockets have a strikingly similar shape and character, including a tyrosine and histidine in addition to a cluster of Arg/Lys; and both accommodate 2 of the 3 phosphate moieties of PtdIns(3,4,5)P3 buried deep within the pocket, with the 3rd phosphate more surface exposed (Fig. 5).
FIGURE 5.
The PtdIns(3,4,5)P3 binding pocket in DHR-1 and comparison with ARNO. A, stereo image of the PtdIns(3,4,5)P3 head group docked into the upper surface pocket of the DHR-1 domain, with a zoom-in to show residues predicted to form the pocket and/or bind phospholipid. B, same view as the zoom-in with the binding pocket shown as electrostatic surface. C and D, ARNO PH domain and its complex with Ins(1,3,4,5)P4 (PDB code 1U27) (60), with the pocket oriented to show surface similarity.
Although atomic details must remain speculative in the absence of direct structural data, the model presented in Fig. 5A is fully consistent with the phospholipid binding preferences we observed. Thus, the 4- and 5-phosphate moieties are buried in two basic subpockets, forming strong ionic interactions. The 3-phosphate can make hydrogen bonds to the polar residues, Tyr484 and Gln519, but these surface-exposed bonds are likely to contribute much less energetically. The lack of a 3-phosphate on PtdIns(4,5)P2, assuming it binds in the same orientation, would therefore rationalize the similar affinities of PtdIns(4,5)P2 and PtdIns(3,4,5)P3. And in the case of PtdIns(3,4)P2, PtdIns(3,5)P2, or PtdIns(3)P1, only one of the high affinity subsites could be occupied, consistent with the lack of binding observed in these cases.
Implications for the Dock180 Family
The 11 human DOCK180 paralogs have been divided into four classes, A–D, based on overall sequence identity and domain organization (4–6) (see Fig. 2). Classes A/B and C/D form distinct groups with higher levels of structural and functional similarity. Sequence alignments and secondary structure predictions, as well as three-dimensional homology modeling (not shown), are consistent with the DHR1 domains of all Dock180 family members adopting a similar fold with a C2 core. The most obvious differences between them are the lengths of two of the phospholipid-binding loops (L1 and L3) and the β7–β8 subdomain, which is mostly conserved in the A/B group, but significantly shorter in classes C and D.
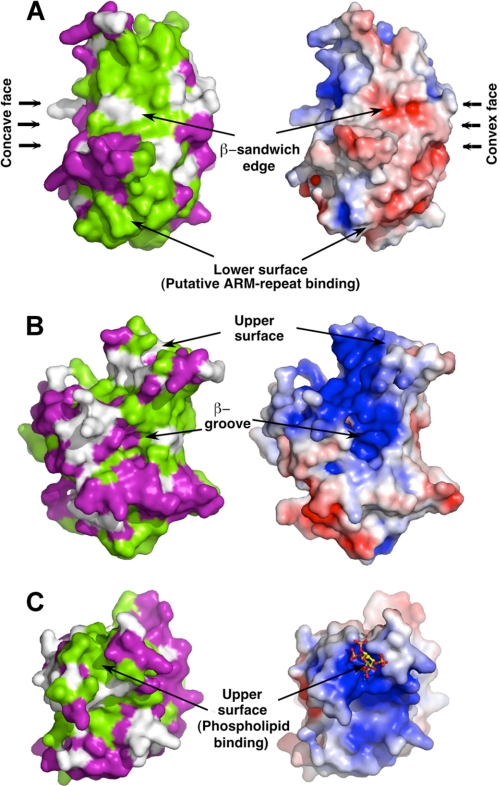
Mapping sequence conservation among the A/B group onto the surface of the domain reveals, in addition to the phospholipid binding region, two further surfaces with a high level of conservation, raising the possibility that they are also involved in intra- or intermolecular interactions (Fig. 6). One conserved surface occurs along an edge (β3–β4) of the β-sandwich, and includes the β2–β3 inserted loop (Fig. 6A). There is a precedent for a C2 domain utilizing this edge of the sheet for protein-protein interactions: via recognition of a phosphotyrosine in the target protein that inserts between the sheets (61). One possibility, which is consistent with our model of full-length Dock1 (see below) and supported by experiments with Dock9 (62), is that this surface interacts with either (or both) DHR2 and Rac1. The second conserved region encompasses the lower surface of the DHR-1 domain (Fig. 6C). In the structure of PtdIns 3-kinase, the lower surface of its C2 domain packs against an ARM-repeat scaffold; given the extended structural similarity in their C2 domains and the prediction of ARM-repeats in Dock1 (see below), it is possible that DHR-1 makes a similar interaction (supplemental Fig. S3). For classes C and D, surface conservation is class-specific, and the features conserved throughout the family are elements of the phospholipid binding site and the edge of the β-sandwich (not shown).
FIGURE 6.
Surface conservation of Dock180 proteins. Sequence conservation within the DockA/B group (green, conserved; purple, variable) of DHR-1 mapped onto Dock1 DHR-1. 3 views are shown and compared with the electrostatic surface potential. A is a view along the edge of the β-sandwich; B is a side view showing the β-groove; and C is a top view looking down onto the upper surface, with InsP4 docked into the upper surface binding pocket.
Phopholipid Binding by the Dock180 Family
The 6 residues delimiting loops L1–L3 pack against each other as well as their counterparts on the adjacent loop(s), forming an aromatic/hydrophobic “quilt” that seals the top of the β-sandwich, and which is highly conserved throughout the Dock180 family. These residues provide reference points for discussing phospholipid binding (see Figs. 2 and supplemental S7).
In group A/B, the L1 loop comprises the motif 437FXBXXBXXXBNV448 (where B = K/R; phospholipid-binding residues in bold; Dock1 numbering). Classes C and D diverge in length and sequence of L1, although they all contain the BNV motif (Asn446 plays a structural role, cross-linking the bases of the three loops) at the end of L1, as well as 1 or 2 further basic residues that can be modeled to interact with phospholipid. The L2 loop is invariant in length in all 4 classes, with a conserved sequence 463VXYQ/HXXXP470 in which the YQ/H pair (HH in class D) are predicted to form the “3-phosphate” end of the binding pocket. The L3 loops are the most divergent in terms of length and sequence, and are class-specific. They are delimited by Phe513 and Phe/Val529, and include the invariant His515, which lies at the base of the binding pocket. An alternating pattern of basic residues, consistent with an extended loop, is also discernible in each case (supplemental Fig. S7), and can be modeled to interact with the phospholipid head group.
It therefore seems very likely that all DHR-1 domains will recognize phospholipids using similar motifs, and the available experimental data support this conjecture. Thus, Dock1, Dock2 and Drosophila myoblast city (Class A), and Dock4 (Class B), bind PtdIns(3,4,5)P3 in vitro and function in a PI 3-kinase-dependent fashion (12, 28, 29, 32). For Class C, although direct binding in vitro has not been tested, the function of at least one member, Dock7, is mediated by Rac activation and dependent on PtdIns 3-kinase activity (63).
For Class D, an additional factor is the presence of a PH domain that may also contribute to membrane targeting. The only lipid binding study, on Dock9, showed that the PH domain bound phosphatidylinositol monophosphates and PtdIns(3,5)P2, as well as PtdSer in vitro (64). The solution structure of the Dock9 PH domain (PDB code 1WG7) is consistent with these binding data, because its canonical phospholipid binding site displays a basic but shallow pocket suitable for engaging a single phosphate (11) (supplemental Fig. S8); and a tyrosine, conserved in PH domains that engage a 4-phosphate, is replaced by a phenylalanine in Dock9 (65). Although the isolated DHR-1 domain was not studied, full-length Dock9 showed additional preferences for PtdIns(3,4)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3, which are at least consistent with a DHR-1 specificity similar to other Dock180 family members; however, further experiments are clearly needed here.
DHR-1 Mutations and Disease
Two mutations in the DHR-1 domain have been linked to disease. A spontaneous 9-residue deletion in Dock5, Asp488–Asp496 (Dock1 numbering), causes “rupture of lens cataract” in mice (86). Our crystal structure shows that this deletion removes most of strand β4, which immediately follows the L2 residues forming the back of the phospholipid binding pocket. The deletion is therefore predicted to disrupt phospholipid binding (and very likely compromises folding of the entire domain). Our experiments therefore point to this mutant losing its membrane targeting ability and thus, whereas it may retain GEF activity in vitro, will be dysfunctional in cells.
Several Dock180 family members have been linked to tumorigenesis (66), and screening to identify somatically acquired epigenetic mutations in human cancers identified the point mutation, D436Y, within Dock1 DHR-1. Asp436 is conserved in the DockA/B subfamily, surface-exposed, and adjacent to the PtdIns(3,4,5)P3-binding pocket (Fig. 2D). We constructed the DHR-1 D436Y mutant and determined that its fold-integrity was not compromised, and that it bound PtdIns(3,4,5)P3 with wild-type affinity (Kd = 3.4 μm). We conclude that if the mutation is linked to tumorigenesis, then it must affect a distinct inter- or intra-molecular interaction.
DISCUSSION
We have shown that a basic pocket on the upper surface of the DHR-1 domain mediates binding to the head group of PtdIns(3,4,5)P3 in vitro, and that point mutations in any one of three lysines lining this pocket ablate the ability of full-length Dock1 to induce cell elongation (in the context of its binding partners, ELMO and CrkII). Our observation of specific binding of DHR-1 to PtdIns(3,4,5)P3 is consistent with previous qualitative studies in vitro using different methodologies, as well as pulldown of Dock1 from cell lysates, and the PI 3-kinase dependence of Dock1 function in cells (12). This level of phospholipid specificity is unusual for a Type II C2 domain (54), but our structural and modeling studies suggest how this is achieved, through the creation of a pocket that closely resembles those found in PH domains, albeit in a distinct structural context (Fig. 4, B and D).
Our observation that DHR-1 also binds to (short chain) PtdIns(4,5)P2 was more surprising, but consistent with our modeling studies as well as a recent proteomics study, in which Dock1 was isolated from cytosolic extracts by interacting specifically with liposomes containing PtdIns(4,5)P2 (but not PtdIns(3,5)P2) (67). However, in another study, employing phospholipid-coated beads, stronger binding to DHR-1 was observed with PtdIns(3,5)P2 than with PtdIns(4,5)P2 (12). Differences of this kind are not unexpected (68), and presumably arise from the distinct experimental approaches, each of which presents the phospholipid in a different context (and none of which is likely to recapitulate conditions in vivo, for which the components and dynamics of intracellular lipid microdomains have not been defined).
Membrane Localization by Dock1
How does the DHR-1 domain localize Dock1 to PtdIns(3,4,5)P3-enriched membranes, and how does it control the GEF activity of DHR-2? First, it should be noted that overexpression of Dock1 lacking the DHR-1 domain leads to efficient GTP loading of Rac1 (12), and does not require ELMO1 (11). However, both elements are required for cellular processes that require local, polarized activation of Rac1, such as cell migration, where Rac1 activation at the leading edge, and rapid assembly and disassembly of activation complexes, drive dynamic cycles of elongation, spreading, and retraction.
The low membrane affinity of the DHR-1 domain is a common feature of signaling molecules, one that may allow them to rapidly sample the membrane until they reach their optimal location (69, 70). In the case of DHR-1, a significant affinity for the abundant lipid, PtdIns(4,5)P2, as we observed in vitro, could provide a pathway to more rapidly locate the much rarer PtdIns(3,4,5)P3 (71). Once located, it has been shown in the case of another signaling protein, GAP1 (which also binds PtdIns(4,5)P2 and PtdIns(3,4,5)P3 with a similar affinity in vitro), that a subtle (2-fold) increase in retention time at the membrane (caused by a reduced off-rate) is sufficient for an effective functional response to the activation of PtdIns 3-kinase (70).
Given the relatively weak affinity of DHR-1 for phospholipids, it may seem remarkable that point mutations in the DHR-1 domain (which do not compromise fold integrity) can have such a profound functional effect. However, specific lipid recognition is typically only one element of membrane targeting. In a process of combinatorial signal integration that has also been called “coincidence detection” (72, 73), there is abundant evidence that simultaneous engagement of a second ligand (e.g. a protein or another lipid) that independently localizes to the same site provides a combinatorial signal that defines (and refines) the optimal membrane location. In the context of such a combinatorial binding/spatial coincidence event, weak but specific membrane binding of the DHR-1 domain (or any one of the contributors) would be sufficient to tip the balance between assembly and disassembly of the membrane-targeted GEF complex. We previously proposed a related model for the combinatorial/coincidence activation of vinculin at cell-matrix adhesions (74), which was corroborated by subsequent experiments (75).
Thus, in the case of Dock2, a linear C-terminal motif that recognizes phosphatidic acid has been shown to be necessary for correct membrane localization in leukocytes (15); and an analogous motif in Dock1 (14), or the PH domain of Dock9 (64), could provide a similar “coincidence” signal. Moreover, Dock1 forms a near-constitutive complex with ELMO1, which binds to several proteins, including RhoG, Bai1, and ERM, that independently localize to PtdIns(3,4,5)P3-enriched membranes (76–80). Finally, the target of Dock1, Rac1, localizes to the same sites via a basic motif at its C terminus (81), and the DHR-2 domain has significant affinity for its target (the GDP-bound form).
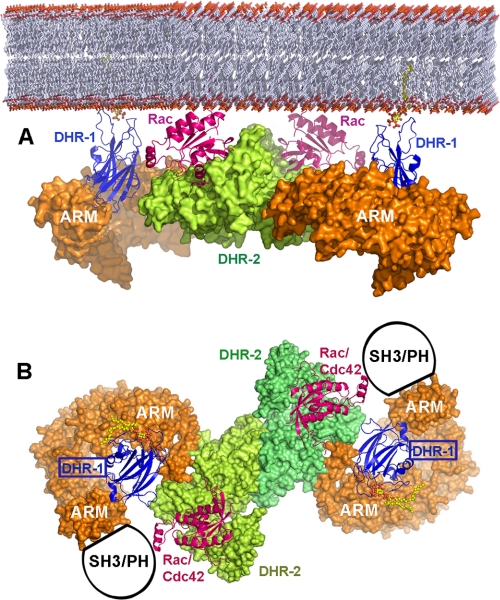
A Three-dimensional Model for Dock180
The above considerations suggest that the system is finely poised, energetically and dynamically, which would require that there is a close structural and functional linkage between the DHR-1 and DHR-2 domains, despite being separated by ∼600–800 residues in the primary sequence. Evidence for a functional linkage comes from studies of Dock9, where N-terminal fragments including either the DHR-1 or PH domain bound to DHR-2 in vitro and affected intrinsic GEF activity (62); and in Dock1, the N-terminal SH3 domain-autoinhibited GEF activity (82). In addition, Dock9 forms dimers mediated by its DHR-2 domain in solution (64) and in crystals (13), and homology modeling of other Dock family members suggests that they will all form dimers in the same way (13), consistent with experiments on Dock1 (64). Moreover, electron micrographs of Dock9 suggest a compact, dimeric organization for the full-length molecule (64), indicative of a molecule stabilized by intramolecular and interdimer interactions.
The FFAS fold-recognition server (87), among others, predicts significant sequence similarity (confidence level >95%; supplemental Figs. S9 and S10) for the entire interdomain region (residues 620–1210) of Dock1 with β-importin (PDB code 2QNA) and other proteins that adopt a HEAT/ARM-repeat tertiary fold, a suprahelical structure providing a semicircular scaffold. By aligning helices at the N terminus of the DHR-2 domain crystal structure with the HEAT/ARM repeats, and positioning DHR-1 on top of the repeats by analogy with the organization of PtdIns 3-kinase, the DHR-1 domain is brought into close apposition with DHR-2 and Rac1 (Fig. 7), consistent with a structural and functional linkage between membrane specificity and GEF activity. The model further predicts that the membrane-binding elements of DHR-1 and Rac-1 (and their symmetry mates across the dimer axis) achieve coplanarity, which would enable simultaneous engagement of the plasma membrane by four components, enhancing affinity through avidity effects. A continuation of the ARM-repeat curvature would also bring the SH3 (DockA/B) or PH (DockD) domains into close apposition with DHR-2, consistent with the functional studies.
FIGURE 7.
Hypothetical model of the dimeric Dock1-Rac1 membrane complex. A, the predicted interdomain HEAT/ARM repeats (see also supplemental Figs. S9 and S10) are shown as orange molecular surfaces, onto which DHR-1 (blue ribbon) is modeled by analogy with the structure of PtdIns 3-kinase (84). The DHR2 dimer (green/lime surface) bound to Rac1 (magenta ribbon) is modeled on the Dock9-Cdc42 complex (13). Membrane attachment sites for DHR-1 (PtdIns(3,4,5)P3 head group) and Rac-1 (prenylated C terminus) are indicated. B, the same as A but rotated by 90° about a horizontal axis, providing a “membrane view” of the complex. The region N-terminal to the DHR-1 domain (shown schematically) is predicted to lie adjacent to the DHR-2 domain; it includes the SH3 domain in DockA/B or the PH domain in DockD.
We therefore propose the following scenario of Dock180 function: (i) localization of the Dock180-ELMO complex to the leading edge is initially promoted by a combination of the recognition by ELMO of one of its partners and the avidity of dimeric Dock180 for phospholipids, mediated by its DHR-1 domain, with secondary contributions from its PH domain (DockD) or C-terminal linear motifs (DockA). (ii) The dimeric organization and ARM-repeat linkage between the DHR-1 and DHR-2 domains creates a rather rigid complex that can “land” on (and “take off” from) the membrane, presenting DHR-2 at an optimal height and orientation to engage Rac1. (iii) By binding with relatively low affinity, DHR1 can search for an optimal site by repeatedly sampling the membrane and/or promoting diffusion of PtdIns(3,4,5)P3 through the membrane toward Dock180. The probability of encountering Rac1 is enhanced when DHR-1 finds an elevated concentration of PtdIns(3,4,5)P3, by virtue of an increased membrane retention time and/or the independent co-localization of Rac1 to the same site. (iv) Once DHR-2 engages GDP-bound Rac1, it remains bound until GDP is released and GTP is loaded. (v) The dynamic DHR-1/membrane interaction then promotes removal of Dock180 from the vicinity of GTP-loaded Rac1, enabling its effectors to bind, and the search for the next GDP-bound Rac1 to begin.
Supplementary Material
Acknowledgments
We thank the outstanding beam-line support team at the SSRL for data collection facilities and Drs. Pellecchia, Crowell, and Pascual for helpful discussions. The SSRL is a national synchrotron user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences, for synchrotron access.
This work was supported, in whole or in part, by National Institutes of Health NIGMS Cell Migration Consortium Grant U54 GM06346 (to R. C. L.) and the Canadian Institute of Health Research (to J.-F. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
The atomic coordinates and structure factors (code 3L4C) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GEF
- guanine exchange factor
- PtdIns
- phosphatidylinositol
- P3
- Tris phosphate
- P2
- bisphosphate
- P1
- monophosphate
- DHR
- Dock homology region
- PH domain
- pleckstrin homology domain
- ELMO
- engulfment and cell motility protein
- ITC
- isothermal titration calorimetry
- InsP4
- inositol 1,3,4,5-tetraphosphate
- PDB
- Protein Data Bank
- SH3
- Src homology domain 3.
REFERENCES
- 1.Jaffe A. B., Hall A. (2005) Annu. Rev. Cell Dev. Biol. 21, 247–269 [DOI] [PubMed] [Google Scholar]
- 2.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 3.Raftopoulou M., Hall A. (2004) Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 4.Côté J. F., Vuori K. (2002) J. Cell Sci. 115, 4901–4913 [DOI] [PubMed] [Google Scholar]
- 5.Meller N., Irani-Tehrani M., Kiosses W. B., Del Pozo M. A., Schwartz M. A. (2002) Nat. Cell Biol. 4, 639–647 [DOI] [PubMed] [Google Scholar]
- 6.Meller N., Merlot S., Guda C. (2005) J. Cell Sci. 118, 4937–4946 [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa H., Kiyokawa E., Tanaka S., Nagashima K., Gotoh N., Shibuya M., Kurata T., Matsuda M. (1996) Mol. Cell. Biol. 16, 1770–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiyokawa E., Hashimoto Y., Kobayashi S., Sugimura H., Kurata T., Matsuda M. (1998) Genes Dev. 12, 3331–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., Nishiwaki K., Walk S. F., Nemergut M. E., Macara I. G., Francis R., Schedl T., Qin Y., Van Aelst L., Hengartner M. O., Ravichandran K. S. (2001) Cell 107, 27–41 [DOI] [PubMed] [Google Scholar]
- 10.Grimsley C. M., Kinchen J. M., Tosello-Trampont A. C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O., Ravichandran K. S. (2004) J. Biol. Chem. 279, 6087–6097 [DOI] [PubMed] [Google Scholar]
- 11.Komander D., Patel M., Laurin M., Fradet N., Pelletier A., Barford D., Côté J. F. (2008) Mol. Biol. Cell 19, 4837–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Côté J. F., Motoyama A. B., Bush J. A., Vuori K. (2005) Nat. Cell Biol. 7, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., Zhang Z., Roe S. M., Marshall C. J., Barford D. (2009) Science 325, 1398–1402 [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi S., Shirai T., Kiyokawa E., Mochizuki N., Matsuda M., Fukui Y. (2001) Biochem. J. 354, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikimi A., Fukuhara H., Su W., Hongu T., Takasuga S., Mihara H., Cao Q., Sanematsu F., Kanai M., Hasegawa H., Tanaka Y., Shibasaki M., Kanaho Y., Sasaki T., Frohman M. A., Fukui Y. (2009) Science 324, 384–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda M., Ota S., Tanimura R., Nakamura H., Matuoka K., Takenawa T., Nagashima K., Kurata T. (1996) J. Biol. Chem. 271, 14468–14472 [DOI] [PubMed] [Google Scholar]
- 17.Moore C. A., Parkin C. A., Bidet Y., Ingham P. W. (2007) Development 134, 3145–3153 [DOI] [PubMed] [Google Scholar]
- 18.Erickson M. R., Galletta B. J., Abmayr S. M. (1997) J. Cell Biol. 138, 589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y. C., Horvitz H. R. (1998) Nature 392, 501–504 [DOI] [PubMed] [Google Scholar]
- 20.Nolan K. M., Barrett K., Lu Y., Hu K. Q., Vincent S., Settleman J. (1998) Genes Dev. 12, 3337–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert M. L., Kim J. I., Birge R. B. (2000) Nat. Cell Biol. 2, 899–905 [DOI] [PubMed] [Google Scholar]
- 22.Wu Y. C., Tsai M. C., Cheng L. C., Chou C. J., Weng N. Y. (2001) Dev. Cell 1, 491–502 [DOI] [PubMed] [Google Scholar]
- 23.Fukui Y., Hashimoto O., Sanui T., Oono T., Koga H., Abe M., Inayoshi A., Noda M., Oike M., Shirai T., Sasazuki T. (2001) Nature 412, 826–831 [DOI] [PubMed] [Google Scholar]
- 24.Handa Y., Suzuki M., Ohya K., Iwai H., Ishijima N., Koleske A. J., Fukui Y., Sasakawa C. (2007) Nat. Cell Biol. 9, 121–128 [DOI] [PubMed] [Google Scholar]
- 25.Janardhan A., Swigut T., Hill B., Myers M. P., Skowronski J. (2004) PLoS Biol. 2, E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yajnik V., Paulding C., Sordella R., McClatchey A. I., Saito M., Wahrer D. C., Reynolds P., Bell D. W., Lake R., van den Heuvel S., Settleman J., Haber D. A. (2003) Cell 112, 673–684 [DOI] [PubMed] [Google Scholar]
- 27.Brugnera E., Haney L., Grimsley C., Lu M., Walk S. F., Tosello-Trampont A. C., Macara I. G., Madhani H., Fink G. R., Ravichandran K. S. (2002) Nat. Cell Biol. 4, 574–582 [DOI] [PubMed] [Google Scholar]
- 28.Kunisaki Y., Nishikimi A., Tanaka Y., Takii R., Noda M., Inayoshi A., Watanabe K., Sanematsu F., Sasazuki T., Sasaki T., Fukui Y. (2006) J. Cell Biol. 174, 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanai A., Ihara S., Ohdaira T., Shinohara-Kanda A., Iwamatsu A., Fukui Y. (2008) IUBMB Life 60, 467–472 [DOI] [PubMed] [Google Scholar]
- 30.Kuramoto K., Negishi M., Katoh H. (2009) J. Neurosci. Res. 87, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 31.Para A., Krischke M., Merlot S., Shen Z., Oberholzer M., Lee S., Briggs S., Firtel R. A. (2009) Mol. Biol. Cell 20, 699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balagopalan L., Chen M. H., Geisbrecht E. R., Abmayr S. M. (2006) Mol. Cell. Biol. 26, 9442–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPhillips T. M., McPhillips S. E., Chiu H. J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., Kuhn P. (2002) J. Synchrotron Radiat. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z., Minor W. (1997) in Methods in Enzymology (Carter C. W., Sweet R. M. eds) Vol. 276A, pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 35.Schwarzenbacher R., Godzik A., Jaroszewski L. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagin A., Teplyakov A. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 1622–1624 [DOI] [PubMed] [Google Scholar]
- 37.Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. (2005) Nucleic Acids Res. 33, W284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 39.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 41.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 42.Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., Westbrook J. D. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 [DOI] [PubMed] [Google Scholar]
- 43.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 44.Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 45.Vriend G. (1990) J. Mol. Graph. 8, 52–56 [DOI] [PubMed] [Google Scholar]
- 46.Thomsen R., Christensen M. H. (2006) J. Med. Chem. 49, 3315–3321 [DOI] [PubMed] [Google Scholar]
- 47.Wohlwend D., Strasser A., Dickmanns A., Ficner R. (2007) J. Mol. Biol. 374, 1129–1138 [DOI] [PubMed] [Google Scholar]
- 48.Krivov G. G., Shapovalov M. V., Dunbrack R. L., Jr. (2009) Proteins 77, 778–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 50.Lyskov S., Gray J. J. (2008) Nucleic Acids Res. 36, W233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grobler J. A., Essen L. O., Williams R. L., Hurley J. H. (1996) Nat. Struct. Biol. 3, 788–795 [DOI] [PubMed] [Google Scholar]
- 52.Hurley J. H., Misra S. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 49–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho W., Stahelin R. V. (2006) Biochim. Biophys. Acta 1761, 838–849 [DOI] [PubMed] [Google Scholar]
- 55.Rizo J., Südhof T. C. (1998) J. Biol. Chem. 273, 15879–15882 [DOI] [PubMed] [Google Scholar]
- 56.Sánchez-Bautista S., Marín-Vicente C., Gómez-Fernández J. C., Corbalán-García S. (2006) J. Mol. Biol. 362, 901–914 [DOI] [PubMed] [Google Scholar]
- 57.Manna D., Bhardwaj N., Vora M. S., Stahelin R. V., Lu H., Cho W. (2008) J. Biol. Chem. 283, 26047–26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guerrero-Valero M., Ferrer-Orta C., Querol-Audí J., Marin-Vicente C., Fita I., Gómez-Fernández J. C., Verdaguer N., Corbalán-García S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunn R., Klos D. A., Adler A. S., Hicke L. (2004) J. Cell Biol. 165, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cronin T. C., DiNitto J. P., Czech M. P., Lambright D. G. (2004) EMBO J. 23, 3711–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benes C. H., Wu N., Elia A. E., Dharia T., Cantley L. C., Soltoff S. P. (2005) Cell 121, 271–280 [DOI] [PubMed] [Google Scholar]
- 62.Meller N., Irani-Tehrani M., Ratnikov B. I., Paschal B. M., Schwartz M. A. (2004) J. Biol. Chem. 279, 37470–37476 [DOI] [PubMed] [Google Scholar]
- 63.Watabe-Uchida M., John K. A., Janas J. A., Newey S. E., Van Aelst L. (2006) Neuron 51, 727–739 [DOI] [PubMed] [Google Scholar]
- 64.Meller N., Westbrook M. J., Shannon J. D., Guda C., Schwartz M. A. (2008) Biochem. J. 409, 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 66.Smith H. W., Marra P., Marshall C. J. (2008) J. Cell Biol. 182, 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Catimel B., Schieber C., Condron M., Patsiouras H., Connolly L., Catimel J., Nice E. C., Burgess A. W., Holmes A. B. (2008) J. Proteome Res. 7, 5295–5313 [DOI] [PubMed] [Google Scholar]
- 68.Narayan K., Lemmon M. A. (2006) Methods 39, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cullen P. J., Cozier G. E., Banting G., Mellor H. (2001) Curr. Biol. 11, R882–893 [DOI] [PubMed] [Google Scholar]
- 70.Hammond G. R., Sim Y., Lagnado L., Irvine R. F. (2009) J. Cell Biol. 184, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stephens L. R., Jackson T. R., Hawkins P. T. (1993) Biochim. Biophys. Acta 1179, 27–75 [DOI] [PubMed] [Google Scholar]
- 72.Balla T. (2005) J. Cell Sci. 118, 2093–2104 [DOI] [PubMed] [Google Scholar]
- 73.Carlton J. G., Cullen P. J. (2005) Trends Cell Biol. 15, 540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., Liddington R. C. (2004) Nature 430, 583–586 [DOI] [PubMed] [Google Scholar]
- 75.Chen H., Choudhury D. M., Craig S. W. (2006) J. Biol. Chem. 281, 40389–40398 [DOI] [PubMed] [Google Scholar]
- 76.deBakker C. D., Haney L. B., Kinchen J. M., Grimsley C., Lu M., Klingele D., Hsu P. K., Chou B. K., Cheng L. C., Blangy A., Sondek J., Hengartner M. O., Wu Y. C., Ravichandran K. S. (2004) Curr. Biol. 14, 2208–2216 [DOI] [PubMed] [Google Scholar]
- 77.Skowronek K. R., Guo F., Zheng Y., Nassar N. (2004) J. Biol. Chem. 279, 37895–37907 [DOI] [PubMed] [Google Scholar]
- 78.Santy L. C., Ravichandran K. S., Casanova J. E. (2005) Curr. Biol. 15, 1749–1754 [DOI] [PubMed] [Google Scholar]
- 79.Grimsley C. M., Lu M., Haney L. B., Kinchen J. M., Ravichandran K. S. (2006) J. Biol. Chem. 281, 5928–5937 [DOI] [PubMed] [Google Scholar]
- 80.Park D., Tosello-Trampont A. C., Elliott M. R., Lu M., Haney L. B., Ma Z., Klibanov A. L., Mandell J. W., Ravichandran K. S. (2007) Nature 450, 430–434 [DOI] [PubMed] [Google Scholar]
- 81.Del Pozo M. A., Kiosses W. B., Alderson N. B., Meller N., Hahn K. M., Schwartz M. A. (2002) Nat. Cell Biol. 4, 232–239 [DOI] [PubMed] [Google Scholar]
- 82.Lu M., Kinchen J. M., Rossman K. L., Grimsley C., Hall M., Sondek J., Hengartner M. O., Yajnik V., Ravichandran K. S. (2005) Curr. Biol. 15, 371–377 [DOI] [PubMed] [Google Scholar]
- 83.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walker E. H., Perisic O., Ried C., Stephens L., Williams R. L. (1999) Nature 402, 313–320 [DOI] [PubMed] [Google Scholar]
- 85.Lu M., Kinchen J. M., Rossman K. L., Grimsley C., deBakker C., Brugnera E., Tosello-Trampont A. C., Haney L. B., Klingele D., Sondek J., Hengartner M. O., Ravichandran K. S. (2004) Nat. Struct. Mol. Biol. 11, 756–762 [DOI] [PubMed] [Google Scholar]
- 86.Omi N., Kiyokawa E., Matsuda M., Kinoshita K., Yamada S., Yamada K., Matsushima Y., Wang Y., Kawai J., Suzuki M., Hayashizaki Y., Hiai H. (2008) Exp. Eye Res. 86, 828–834 [DOI] [PubMed] [Google Scholar]
- 87.Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. (2005) Nucleic Acids Res. 33, W284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.