Abstract
DYRK1A (the dual specificity tyrosine phosphorylation-regulated kinase 1A) plays an important role in body growth and brain physiology. Overexpression of this kinase has been associated with the development of Down syndrome in both human and animal models, whereas single copy loss-of-function of DYRK1A leads to increased apoptosis and decreased brain size. Although more than a dozen of DYRK1A targets have been identified, the molecular basis of its involvement in neuronal development remains unclear. Here we show that DYRK1A and another pro-survival member of the DYRK family, DYRK3, promote cell survival through phosphorylation and activation of SIRT1, an NAD+-dependent protein deacetylase that is essential in a variety of physiological processes including stress response and energy metabolism. DYRK1A and DYRK3 directly phosphorylate SIRT1 at Thr522, promoting deacetylation of p53. A SIRT1 phosphorylation mimetic (SIRT1 T522D) displays elevated deacetylase activity, thus inhibiting cell apoptosis. Conversely, a SIRT1 dephosphorylation mimetic (SIRT1 T522V) fails to mediate DYRK-induced deacetylation of p53 and cell survival. We show that knockdown of endogenous DYRK1A and DYRK3 leads to hypophosphorylation of SIRT1, sensitizing cells to DNA damage-induced cell death. We also provide evidence that phosphorylation of Thr522 activates SIRT1 by promoting product release, thereby increasing its enzymatic turnover. Taken together, our findings provide a novel mechanism by which two anti-apoptotic DYRK members promote cell survival through direct modification of SIRT1. These findings may have important implications in understanding the molecular mechanisms of tumorigenesis, Down syndrome, and aging.
Keywords: Apoptosis, Enzyme Catalysis, p53, Protein Kinases, SIRT, DYRK1A, DYRK3, SIRT1, Apoptosis, Phosphorylation
Introduction
The DYRK (dual specificity tyrosine-phosphorylated and regulated kinase) proteins are a highly conserved subfamily of protein kinases that catalyze the autophosphorylation on tyrosine residues and the phosphorylation of serine/threonine residues on exogenous substrates (1). The mammalian DYRK family consists of at least seven members, DYRK1A, DYRK1B (also known as MIRK), DYRK1C, DYRK2, DYRK3, DYRK4A, and DYRK4B (2). Among them, DYRK1A is the best characterized member. DYRK1A is a nuclear protein ubiquitously expressed in all tissues. Previous studies have shown that DYRK1A regulates body growth and normal brain development in a dose-sensitive manner. Single copy loss-of-function of DYRK1A leads to decreased body and brain size (3, 4). On the other hand, modest overexpression of DYRK1A in both humans and mice is associated with the development of Down syndrome (5, 6). Therefore, DYRK1A is also known as Down syndrome-associated kinase.
Accumulating studies have revealed that the DYRK protein kinases play an important role in the regulation of cell proliferation and apoptosis. DYRK1A has been shown to function as a caspase-9 Thr125 kinase that inhibits the intrinsic apoptotic pathways in the developing retina (7) as well as several human cancer cell lines (8). DYRK3 has also been shown to attenuate apoptosis in response to cytokine withdrawal in hematopoietic cells of erythroid lineage (9, 10). Conversely, DYRK2, a family member closely related to DYRK3, is activated by ATM upon genotoxic stress and induces apoptosis by direct phosphorylation of p53 at Ser46 (11).
The Sir2 (silent information regulator 2) family of proteins, also known as sirtuins, are highly conserved NAD+-dependent protein deacetylases and/or ADP ribosyltransferases (12). First identified in yeast as key components in gene silencing complexes (reviewed in Ref. 13), sirtuins have been increasingly recognized as crucial regulators for a variety of cellular processes, ranging from energy metabolism and stress response to tumorigenesis and aging (14). The mammalian genome encodes seven sirtuins, collectively known as SIRT1 to SIRT7 (15). A wealth of data has demonstrated that SIRT1, the mammalian ortholog of yeast Sir2 protein, directly couples NAD+ hydrolysis to the deacetylation of histones and numerous transcription factors and co-factors, such as p53, E2F1, NFκB, FOXO, c-Myc, HIF-2α, and HSF1 (16–24). Therefore, SIRT1 directly links cellular metabolic status to gene regulation, playing an important role in a number of pro-survival activities (14).
We have identified DYRK3 as a SIRT1 interacting partner in a yeast-two hybrid screen (supplemental Fig. S1A). In this report, we show that two anti-apoptotic members of the DYRK family, DYRK1A and DYRK3, directly interact with and phosphorylate SIRT1 on threonine 522. Phosphorylation at this residue activates SIRT1, promoting cell survival upon genotoxic stress in a p53-dependent manner. More importantly, our findings indicate that phosphorylation of Thr522 of SIRT1 is required for DYRK1A- and DYRK3-mediated cell survival. These findings imply that abnormalities in the DYRK/SIRT1 signaling axis may underlie the DYRK gene-dosage imbalance effects observed in conditions such as Down syndrome and cancer.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
Myc-tagged mouse SIRT1 was cloned in pcDNA3, pBabe, and pGEX-4T-3 vectors. HA2-tagged DYRK1A, DYRK2, and DYRK3 were cloned in pcDNA3 vector. The substitution mutants of SIRT1 were constructed by PCR-based site-directed mutagenesis. Anti-human SIRT1 and anti-mouse SIRT1 antibodies were from Sigma, anti-DYRK3 antibody was from Santa Cruz Biotechnology, and anti-DYRK1A antibody was from Abnova. Anti-acetyl-p53 K382 antibody was from Abcam. Rabbit anti-Thr(P)522 antisera were generated against keyhole limpet hemocyanin-conjugated phosphopeptide KKLSELPP-pT-PLHISE.
Yeast Two-hybrid Screen
To identify the potential interacting partners of SIRT1, a bait plasmid was constructed by fusing the full-length mouse SIRT1 in-frame to the C terminus of the GAL4 DNA-binding domain in a modified pAS2-1 vector (Clontech). Because the testis has the highest expression of SIRT1, a murine testis cDNA library fused to the transactivation domain of GAL4 (mouse testis MATCHMARKER cDNA library; Clontech) was screened with the SIRT1 bait plasmid as per the manufacturer's instructions. The candidate interacting clones were then confirmed and sequenced. The interaction between DYRK3 and SIRT1 was reconfirmed by reintroducing purified plasmids into host yeast strain (pJ69-4A) (supplemental Fig. S1A).
Cell Culture
HEK293T cells stably infected with pSuper or pSuper-SIRT1 RNAi have been described (25). HEK293T, MEFs, and H1299 cells were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum (HyClone). U2OS cells were cultured in Dulbecco's modified Eagle's medium/F-12 (1:1) medium with 10% fetal bovine serum.
Immunoprecipitation and Immunoblot Analysis
To investigate the interaction between SIRT1 and DYRKs, HEK293T cells transfected with the indicated expressing constructs were lysed 48 h post-transfection in Nonidet P-40 buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40) containing CompleteTM protease inhibitors (Roche Applied Science) and MG-132 (Calbiochem). The whole cell extracts were immunoprecipitated with monoclonal anti-Myc antibody-conjugated agarose beads at 4 °C for 2 h. Immunocomplexes were eluted with SDS sample buffer and resolved by SDS-PAGE. To detect the acetylation status of endogenous p53 proteins, the cells were treated with 50 μm of MG-132 for 20 min to stabilize the p53, followed by treatment with 20 μm of etoposide in the presence of 25 μm of MG-132 for an additional hour to stimulate p53 acetylation. The total cell lysates were then analyzed with anti-acetyl-p53 K382 antibodies (Abcam).
GST Pulldown
To examine the interaction between SIRT1 and DYRKs in vitro, recombinant GST fusion proteins were incubated with in vitro translated HA-DYRKs or Myc-SIRT1 proteins (TnT-coupled reticulocyte lysate system; Promega) in Nonidet P-40 buffer. The protein complexes were then pulled down with GST beads, eluted with SDS sample buffer, and resolved by SDS-PAGE. The levels of the GST proteins were detected by Ponceau S stain.
Immunofluorescence Assay
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS and then permeabilized with 0.2% Triton X-100 in PBS buffer for 10 min. The coverslips were then incubated with primary antibodies (as indicated) in 10% fetal bovine serum/PBS for 45 min at room temperature, followed by 10 min of incubation with secondary antibodies. Finally, the cells were counterstained with 4′,6-diamidino-2-phenylindole to visualize the nuclei.
In Vitro Phosphorylation Assay and in Vitro Acetylation and Deacetylation Assays
To test whether DYRKs directly phosphorylate SIRT1, 1 μg of purified GST-SIRT1, GST-SIRT1T522V, and GST-SIRT1S531A proteins were incubated with 0.5 μg of GST-DYRK1A (Millipore) or 2 μg of GST-DYRK3 in kinase buffer (20 mm HEPES, pH 7.5, 10 mm MgCl2, 0.1 mm Na3VO4, 2 mm dithiothreitol, and 1 mm ATP) at 30 °C for 30 min. The samples were then resolved by SDS-PAGE and analyzed by anti-Ser(P)/Thr(P)-Pro antibodies (MPM-2; Millipore) or anti-phosphoserine antibodies (Invitrogen). To analyze the activity of wild-type SIRT1 and SIRT1 mutants in vitro, the in vitro acetylation and deacetylation of GST-p53 were carried out essentially as described (16).
Cell Viability and Apoptosis Assays
U2OS cells, HEK293T pSuper, or T1RNAi cells transfected with the indicated expression constructs or siRNA duplexes were treated with DMSO or 20 μm etoposide for 30 h or as indicated, and cell viability was determined with cell proliferation reagent WST-1 (Roche Applied Science) according to the manufacturer's protocol. To analyze cell apoptosis in MEFs, SIRT1-deficient MEFs (SIRT1−/−) infected with retroviruses expressing empty vector, wild-type SIRT1, or SIRT1 mutants were treated with DMSO or 20 μm etoposide for 48 h. The cells were then washed with PBS and stained with fluorescein isothiocyanate-annexin V and propidium iodide according to the manufacturer's instructions (BD Bioscience annexin V kit). Apoptotic cells (annexin V-positive, phosphatidylinositol-negative) were determined by flow cytometry analysis. The data were quantified from three independent experiments and presented as the means ± S.E.
RNA Analysis
To analyze the expression of DYRKs and p53 target genes, the cells were treated with DMSO or etoposide (20 μm) for 8 h. Total RNA was then isolated using Qiagen RNeasy mini-kit. For quantitative real time PCR, cDNA was synthesized with the ABI reverse transcriptase kit and analyzed using SYBR Green Supermix (Applied Biosystems). All of the data were normalized to 18 S RNA expression and presented as the means ± S.E.
Statistical Analysis
The values are expressed as the means ± S.E. The significant differences between the means were analyzed by two-tailed, unpaired Student's t test, and the differences were considered significant at p < 0.05.
RESULTS
DYRK1A and DYRK3 Interact with SIRT1 in Vitro and in Vivo
We have identified DYRK3, a member of DYRK family, as a SIRT1 interacting protein in a yeast-two hybrid screen with a bait plasmid encoding the full-length mouse SIRT1 protein and a murine testis cDNA library. The interaction between the full-length DYRK3 protein and SIRT1 was reconfirmed by an independent yeast-two hybrid assay (supplemental Fig. S1A).
The DYRK family of kinases contains a highly conserved catalytic core domain with shared substrate specificity (26, 27). The interaction between DYRK3 and SIRT1 in the yeast two-hybrid assay therefore suggests that other DYRK proteins may also interact with SIRT1. To test this possibility, we chose to investigate the interaction between SIRT1 and three well studied DYRKs: DYRK1A, DYRK2, and DYRK3. As shown in Fig. 1A, GST pulldown between GST-SIRT1 fusion protein and in vitro transcribed and translated HA-DYRKs showed that GST-SIRT1 specifically interacts with DYRK1A and DYRK3, but not DYRK2, indicating that SIRT1 selectively interacts with pro-survival members of DYRK family.
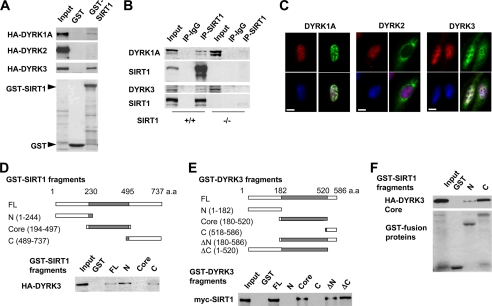
FIGURE 1.
SIRT1 interacts with DYRK1A and DYRK3 in vitro and in cells. A, SIRT1 interacts with DYRK1A and DYRK3, but not DYRK2 in vitro. The GST-SIRT1 fusion protein was incubated with HA-tagged DYRK1A, DYRK2, or DYRK3 in Nonidet P-40 buffer. Top three panels, immunoblots of HA-DYRK1A, HA-DYRK2, or HA-DYRK3. Bottom panel, Ponceau S staining of GST fusion proteins. B, endogenous SIRT1 interacts with endogenous DYRKs in MEFs. Whole cell extracts from wild-type (+/+) and SIRT1-deficient (−/−) MEFs were immunoprecipitated with control IgG or anti-SIRT1 antibodies. C, localization of DYRKs in U2OS cells. U2OS cells were transfected with constructs expressing HA-tagged DYRK as indicated. HA-DYRK (green) and SIRT1 (red) were detected by indirect immunofluorescence, and the nucleus was stained blue with 4′,6-diamidino-2-phenylindole. Bar, 10 μm. D, DYRK3 interacts with the N- and C-terminal domains of SIRT1. GST full-length (FL) SIRT1 or indicated fragments were incubated with HA-tagged DYRK3. E, SIRT1 interacts with the catalytic core domain of DYRK3. GST-full-length DYRK3 or the indicated fragments were incubated with Myc-SIRT1. F, the catalytic core domain of DYRK3 interacts with the N- and C-terminal domains of SIRT1. Top panel, immunoblot of HA-DYRK3 Core domain. Bottom panel, Ponceau S staining of GST-SIRT1 fusion proteins.
To confirm that the interaction between SIRT1 and DYRKs occurs at endogenous levels, whole cell extracts from MEFs were precipitated with anti-SIRT1 antibodies. As shown in Fig. 1B, anti-SIRT1 antibodies co-immunoprecipitated endogenous DYRK1A (top two panels) and DYRK3 (bottom two panels) from wild-type (+/+) MEFs but not SIRT1-deficient (−/−) MEFs, supporting the specificity of the interaction between SIRT1 and DYRKs. Similar results were observed in human osteosarcoma U2OS cells (supplemental Fig. S1B). Immunofluorescent staining in U2OS cells showed that both HA-DYRK1A and SIRT1 localized in the nucleus (Fig. 1C, left panels). HA-DYRK3 was distributed in both the nucleus and cytoplasm, with a strong nuclear staining that closely resembles the pattern of SIRT1 (Fig. 1C, right panels). HA-DYRK2, on the other hand, was located predominately in the cytoplasm (Fig. 1C, middle panels). These observations suggest that DYRK1A and DYRK3 function as physiological interacting partners of SIRT1.
To identify the regions of SIRT1 and DYRKs that are responsible for their interaction, we generated truncated mutants of SIRT1 (Fig. 1D and supplemental Fig. S1C) and DYRK3 (Fig. 1E and supplemental Fig. S1D) and analyzed their interactions by in vitro GST pulldown. Our data indicate that the catalytic core domain of DYRK3 interacts with both the N- and C-terminal domains of SIRT1 (Fig. 1, D–F). Full-length DYRK1A also interacted with the N- and C-terminal domains of SIRT1 (supplemental Fig. S1C). Taken together, our observations demonstrate that SIRT1 forms protein complexes with both DYRK1A and DYRK3 in vitro and in cells.
DYRK1A and DYRK3 Activate SIRT1 Deacetylase Activity through Phosphorylation at Thr522
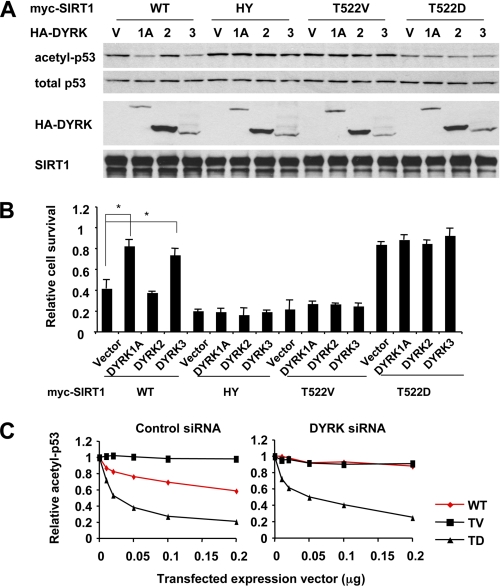
The interaction of DYRK1A and DYRK3 with SIRT1 suggests that these kinases may directly regulate SIRT1 activity. To test this possibility, we transfected control HEK293T cells and stable SIRT1 knockdown cells (HEK293T T1RNAi) with the indicated expression vectors (Fig. 2A) and then treated them with etoposide, a DNA-damaging reagent that induces the acetylation of p53, a well established SIRT1 substrate. DYRK1A and DYRK3, but not DYRK2, promoted deacetylation of p53 following etoposide treatment in control cells but not in HEK293T T1RNAi cells (Fig. 2A). These results indicated that DRYK1A and DYRK3 activate SIRT1, promoting the deacetylation of p53. It is well known that deacetylation of p53 results in inhibition of its activity, which in turn leads to resistance to apoptosis upon DNA damage. Consistent with this notion, DYRK1A and DYRK3 promoted cell survival after etoposide treatment in a SIRT1-dependent manner (Fig. 2B). Similar results were observed in HEK293T T1RNAi cells (supplemental Fig. S2) and U2OS cells (Fig. 2, C and D) overexpressing SIRT1 or the SIRT1HY mutant. Moreover, DYRK1A and DYRK3, but not DYRK2, also promoted the deacetylation of H3K9 and H4K16, two other SIRT1 targets, in a SIRT1 dependent manner (Fig. 2E). Together, our data demonstrate that in independent cell lines, DYRK1A and DYRK3 interact with and activate SIRT1, thereby promoting cell survival upon genotoxic stress.
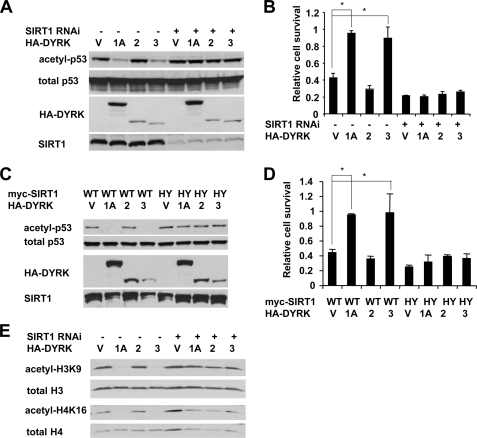
FIGURE 2.
DYRK1A and DYRK3 activate the deacetylase activity of SIRT1. A, DYRK1A and DYRK3, but not DYRK2, promote deacetylation of p53 in HEK293T cells. Control HEK293T and HEK293T T1RNAi cells were transfected with either vector (lane V) or HA-tagged DYRK1A (lane 1A), DYRK2 (lane 2), or DYRK3 (lane 3). The cells were then treated with MG-132/etoposide as described under “Experimental Procedures.” B, DYRK1A and DYRK3, but not DYRK2, increase cell survival after genotoxic stress in a SIRT1-dependent manner. Control and SIRT1 RNAi HEK293T cells were transfected with indicated expressing vectors and treated with etoposide. Cell viability was analyzed as described under “Experimental Procedures” (n = 4; *, p < 0.01). C, DYRK1A and DYRK3, but not DYRK2, promote the deacetylation of endogenous p53 in a SIRT1-dependent manner in U2OS cells. U2OS cells were transfected with either vector (lane V) or HA-tagged DYRK1A (lane 1A), DYRK2 (lane 2), or DYRK3 (lane 3) together with wild type (WT) or H355Y mutant (HY) of SIRT1. The cells were then treated with MG132/etoposide as described under “Experimental Procedures.” D, DYRK1A and DYRK3, but not DYRK2, promote cell survival upon genotoxic stress in U2OS cells. U2OS cells expressing indicated proteins were treated with etoposide and analyzed as described under “Experimental Procedures” (n = 4; *, p < 0.01). E, DYRK1A and DYRK3 promote deacetylation of H3K9 and H4K16 in a SIRT1-dependent manner in HEK293T cells. HEK293T and HEK293T T1RNAi cells were transfected and treated as in A.
The interaction between SIRT1 and the kinase core domain of DYRK3 (Fig. 1F) raises the possibility that SIRT1 may be a bona fide DYRK substrate. To test this hypothesis, we purified Myc-tagged full-length murine SIRT1 from HEK293T cells in the absence or presence of overexpressed DYRK3 and subjected them to mass spectrometry analysis. Interestingly, a doubly phosphorylated form of the peptide corresponding to amino acids 512–535 of SIRT1 (Fig. 3A and supplemental Fig. S3A) was identified in the DYRK3 co-expressed sample. Further mass spectrometry analyses suggested that among six serine/threonine sites in this peptide, four (Thr522, Ser530, Ser531, and Ser532) are putative phosphorylation sites of SIRT1. To investigate whether the phosphorylation of SIRT1 by DYRK3 plays a role in SIRT1 activity and to locate the exact phosphorylation sites, we generated four single phosphorylation site mutants by site-directed mutagenesis and then analyzed their deacetylase activity. Although none of these mutants displayed obvious defects in their deacetylase activities in vitro on Fluor-de lys, a fluorogenic 4-amino acid peptide substrate based on the p53 K382 motif (Biomol International) (supplemental Fig. S3B), the T522V mutant showed dramatically lower deacetylase activity on endogenous p53 protein in response to etoposide treatment in HEK293T T1RNAi cells (Fig. 3B). Importantly, a SIRT1 mutant that mimics the phosphorylation of the Thr522 residue, T522D, displayed increased deacetylase activity not only in cells (Fig. 3C and supplemental Fig. S4) but also in vitro (Fig. 3D). These results suggest that DYRK3-mediated phosphorylation of the Thr522 residue is responsible for the direct activation of SIRT1 deacetylase activity on native protein substrates. Consistent with these observations, the Thr522 residue lies within a conserved amino acid motif (PPTP; supplemental Fig. S3C) that resembles the consensus target sequence for DYRK family (RPX(T/S)P) (26, 27). To further confirm that DYRKs indeed phosphorylate Thr522 residue of SIRT1, we carried out in vitro phosphorylation assays with recombinant GST fusion proteins. As shown in Fig. 3E, GST-DYRK1A and GST-DYRK3, but not control GST protein, phosphorylated wild-type SIRT1, which was detectable by the MPM-2 antibody that recognizes Ser(P)/Thr(P) followed by a proline (P(S/T)P) and by an antibody specific to Thr(P)522 (supplemental Fig. S8A). Further confirming the specificity of DYRKs for the Thr522 of SIRT1, the T522V mutant lost the majority of its phosphorylation signal. Additionally, DYRK1A and DYRK3, but not DYRK2, were found to induce the phosphorylation of endogenous SIRT1 at Thr522 in cells (Fig. 3F). Taken together, our results indicate that DYRK1A and DYRK3 phosphorylate Thr522 of SIRT1 and activate its deacetylase activity in cells and in vitro.
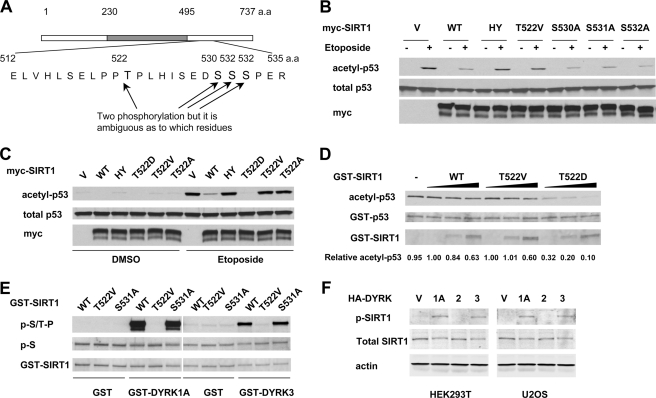
FIGURE 3.
DYRK1A and DYRK3 phosphorylate SIRT1 at Thr522. A, schematic representation of mouse SIRT1 depicting the putative phosphorylation sites induced by DYRK3. B, the SIRT1 T522V mutant displays defective deacetylase activity in cells. HEK293T T1RNAi cells expressing the indicated mouse SIRT1 proteins were treated, and the acetylation of p53 was analyzed as described under “Experimental Procedures.” C, the SIRT1 T522D mutant displays increased deacetylase activity in HEK293T T1 RNAi cells. D, the in vitro deacetylase activity of wild-type (WT) SIRT1 and T522D and T522V mutants. Purified wild-type, T522V, and T522D GST fusion proteins (0.1, 0.5, or 1.0 μg) were incubated with in vitro acetylated GST-p53 fusion proteins (2 μg) in the presence of 50 μm of NAD+ and 200 nm of trichostatin A. E, DYRK1A and DYRK3 phosphorylate the SIRT1 Thr522 residue in vitro. Purified GST-SIRT1 proteins were incubated with GST-DYRK1A (left panels) or GST-DYRK3 (right panels) and analyzed as described under “Experimental Procedures.” F, DYRK1A and DYRK3, but not DYRK2, phosphorylates SIRT1 in vivo. The phosphorylation levels of Thr522 of endogenous SIRT1 in HEK293T and U2OS cells transfected with the indicated vector were analyzed by anti-Thr(P)522 antibody.
RNAi of DYRK1A and DYRK3 Induces Cell Death in Response to DNA Damage through p53
To further examine the role of DYRK1A and DYRK3 in the activation of SIRT1 deacetylase, we investigated whether knockdown of these two DYRKs would decrease the phosphorylation of SIRT1 while inducing the acetylation of p53. Because both DYRK1A and DYRK3 are highly expressed in U2OS cells and function redundantly, knockdown of one DYRK appears to induce a compensative increase of the others (supplemental Fig. S5A). Therefore, we chose to knock down both proteins by simultaneously transfecting siRNAs against DYRK1A and DYRK3 (abbreviated as DYRK siRNA) (Fig. 4 and supplemental Fig. S5B). As shown in Fig. 4A, knockdown of endogenous DYRK1A and DYRK3 significantly decreased the phosphorylation of endogenous SIRT1. As a consequence, the acetylation levels of endogenous p53 were significantly increased following etoposide treatment (Fig. 4B, lane 9). Notably, knockdown of both SIRT1 and DYRK dramatically elevated the acetylation levels of p53 (lane 10), suggesting that SIRT1 and DYRK1A/DYRK3 coordinately repress p53. As expected, RNAi knockdown of DYRK1A and DYRK3 in U2OS cells resulted in increased expression of two major p53 downstream targets, p21 and PUMA (Fig. 4C). These changes were decreased in U2OS cells with RNAi of p53 and were completely abolished in H1299 cells, a p53-null cell line (Fig. 4C). These results suggest that DYRK1A and DYRK3 regulate apoptotic gene expression through p53. Finally, knockdown of DYRK1A and DYRK3 sensitized U2OS cells to DNA damage-induced cell death, whereas RNAi of p53 completely rescued cells from death (Fig. 4D). Taken together, our observations demonstrate that DYRK1A and DYRK3 are critically involved in regulating the p53-mediated apoptotic response by phosphorylating and activating SIRT1.
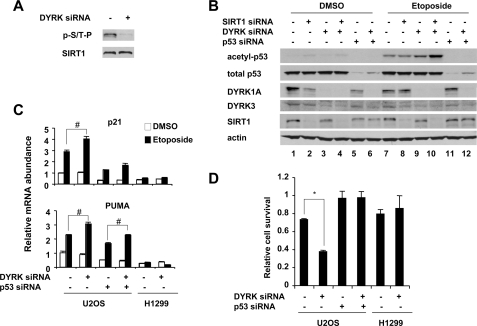
FIGURE 4.
Knockdown of DYRK1A and DYRK3 leads to hyperacetylation of p53 and hypersensitivity to genotoxic stress. A, RNAi of DYRK1A and DYRK3 decreases the phosphorylation of SIRT1. Endogenous SIRT1 was immunopurified from U2OS cells transfected with control siRNA or siRNA specific to DYRK1A and DYRK3 and then analyzed with anti-Ser(P)/Thr(P)-Pro (p-S/T-P) antibodies. B, RNAi of DYRK1A and DYRK3 induces hyperacetylation of p53. U2OS cells transfected with indicated siRNA were treated with DMSO or etoposide as described, and acetylation of p53 was analyzed. C, RNAi of DYRK1A and DYRK3 induces overexpression of p53 target genes in a p53-dependent manner. U2OS cells and H1299 cells were transfected with indicated siRNA and treated as described. The expression of p53 target genes was analyzed by quantitative real time PCR (n = 3; *, p < 0.01; #, p < 0.05). D, RNAi of DYRK1A and DYRK3 sensitizes cells to genotoxic stress induced cell death. U2OS cells and H1299 cells were transfected with indicated siRNA. 30 h later, the cells were treated with etoposide for additional 24 h, and cell viability was analyzed as described under “Experimental Procedures” (n = 3; *, p < 0.01).
Phosphorylation of SIRT1 Thr522 Is Required for DYRK1A- and DYRK3-mediated Cell Survival
The observations that DYRK1A and DYRK3 promote cell survival in a SIRT1-dependent manner (Fig. 2, B and D, and supplemental Fig. S2) suggest that these two kinases promote cell survival directly through phosphorylation of the Thr522 residue of SIRT1. To test this hypothesis, we investigated whether their anti-apoptotic activities require an intact Thr522 residue. As shown in supplemental Fig. S6A, the SIRT1 T522V mutant displayed normal affinity to DYRK1A and DYRK3. It also localized with DYRK1A and DYRK3 in the nucleus of U2OS cells (supplemental Fig. S6B). However, the SIRT1 T522V mutant was unable to mediate DYRK1A and DYRK3-stimulated p53 deacetylation (Fig. 5A) and cell survival (Fig. 5B) in U2OS cells. In contrast, the SIRT1 T522D mutant was able to promote the deacetylation of p53 (Fig. 5A) and cell survival (Fig. 5B) independent of cellular DYRK levels. Moreover, the SIRT1 T522V mutant displayed decreased deacetylase activity, whereas the T522D mutant was constitutively active after knockdown of endogenous DYRK1A and DYRK3 proteins (Fig. 5C and supplemental Fig. S7). These results further highlight the importance of DYRK-mediated phosphorylation in the regulation of SIRT1 activity. Collectively, these data verify that Thr522 phosphorylation of SIRT1 is an essential element in the anti-apoptotic functions of DYRK proteins.
FIGURE 5.
DYRK1A and DYRK3 promote cell survival through phosphorylation of Thr522 residue of SIRT1. A, the SIRT1 T522V mutant fails to mediate DYRK1A and DYRK3-induced deacetylation of p53, whereas the SIRT1 T522D mutant is constitutively active. U2OS cells expressing the indicated proteins were treated with etoposide, and the acetylation of endogenous p53 was analyzed. B, DYRK1A and DYRK3 fail to significantly promote cell survival in SIRT1 T522V-expressing U2OS cells, whereas the SIRT1 T522D mutant constitutively promotes cell survival. U2OS cells transfected with the indicated constructs were treated with etoposide, and 30 h later cell viability was analyzed as described under “Experimental Procedures” (n = 6; *, p < 0.01). C, the SIRT1 T522A mutant is inactive, whereas the SIRT1 T522D mutant is constitutively active regardless of cellular DYRK1A and DYRK3 levels. U2OS cells transfected with control siRNA or siRNA to DYRK1A/3 were transfected with increased doses of vectors for wild-type SIRT1, T522V, or T522D mutants. The acetylation levels of p53 in supplemental Fig. S7 were quantified with the ImageJ program and normalized to the total p53 levels. WT, wild type; HY, H355Y mutant.
Phosphorylation of Thr522 Is Required for Pro-survival Activity of SIRT1 in Cells
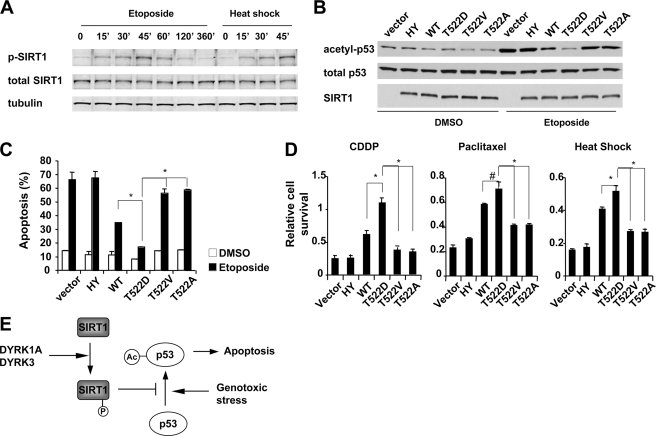
To further explore the connection between SIRT1 phosphorylation and its deacetylase activity in vivo, we generated an antibody specific to Thr(P)522 (supplemental Fig. S8A). As shown in Fig. 6A, the Thr(P)522 levels of endogenous SIRT1 quickly increased upon etoposide treatment and peaked at ∼45 min. The Thr(P)522 levels were also elevated as early as 15 min following heat shock, a stress condition known to activate SIRT1 (24) (Fig. 6A). These observations suggest that phosphorylation of Thr522 is an important modification to activate SIRT1 in response to various stresses. To further explore the connection between SIRT1 phosphorylation and its deacetylase activity, we infected SIRT1-deficient MEFs with retroviruses expressing either empty vector, wild-type, or selected SIRT1 mutants (supplemental Fig. S8B). We then analyzed their ability to rescue SIRT1 deficiency-induced hypersensitivity to genotoxic stress. Compared with wild-type SIRT1 (Fig. 6B), the T522D mutant displayed increased activity to deacetylate p53 upon etoposide treatment, whereas the T522V SIRT1 mutant failed to rescue defective deacetylase activity of SIRT1. The T522D mutant also increased its ability to suppress the induction of p21 and PUMA upon genotoxic stress, whereas the SIRT1 T522V mutant failed to significantly suppress the expression of these targets (supplemental Fig. S8C). This suggests that phosphorylation of SIRT1 directly influences its ability to regulate p53 transcriptional activity. More importantly, phosphorylation of SIRT1 at Thr522 appears essential for SIRT1-mediated stress resistance. As shown in Fig. 6C, 48 h of etoposide treatment induced 66.3% of apoptotic cells in SIRT1-deficient MEFs. Expression of wild-type SIRT1 but not the HY mutant reduced levels of apoptosis to 34.9%. Compared with cells expressing wild-type SIRT1, cells expressing the T522D phosphorylation mimic were more resistant to genotoxic stress. On the other hand, the T522V and T522A phosphorylation mutants of SIRT1 displayed only a partial resistance to genotoxic stress. Similar results were observed when cells were treated with another genotoxic stress inducer, CDDP (cisplatin), a mitotic inhibitor, paclitaxel, or heat shock (Fig. 6D). Collectively, these data indicate that phosphorylation of Thr522 is an important post-translational modification of SIRT1 involved in regulating stress resistance.
FIGURE 6.
Phosphorylation of Thr522 residue of SIRT1 is required for the pro-survival activity of SIRT1. A, the levels of phosphorylation at Thr522 are elevated in endogenous SIRT1 in response to stress. Wild-type MEFs were treated with 20 μm of etoposide or heat-shocked at 45 °C. The cells were then harvested at the indicated time points, and the phosphorylation levels of Thr522 were analyzed by anti-Thr(P)522 antibody. B, the SIRT1 T522D mutant increases its ability to inhibit etoposide-induced p53 acetylation in SIRT1-deficient MEFs. SIRT1 knock-out MEFs expressing indicated SIRT1 protein were treated and analyzed as described. C, the SIRT1 T522D mutant inhibits etoposide-induced apoptosis in MEFs. SIRT1-deficient MEFs expressing indicated SIRT1 protein were treated with DMSO or etoposide for 48 h. The apoptotic cells were measured by phosphatidylinositol and annexin V double staining as described under “Experimental Procedures” (n = 4; *, p < 0.01). D, the SIRT1 T522D mutant displays increased pro-survival activity in response to various stresses. SIRT1-deficient MEFs expressing indicated SIRT1 protein were treated with 25 nm CDDP (cisplatin) or 25 nm paclitaxel for 48 h or were treated at 45 °C for 45 min followed by 24 h recovery. Cell viability was measured as described (n = 4; *, p < 0.01; #, p < 0.05). E, a working model showing that DYRK1A and DYRK3 promote cell survival through phosphorylation of SIRT1 and deacetylation of p53. WT, wild type; HY, H355Y mutant.
Phosphorylation of Thr522 Activates SIRT1 by Decreasing Its Association with Deacetylated Protein Products
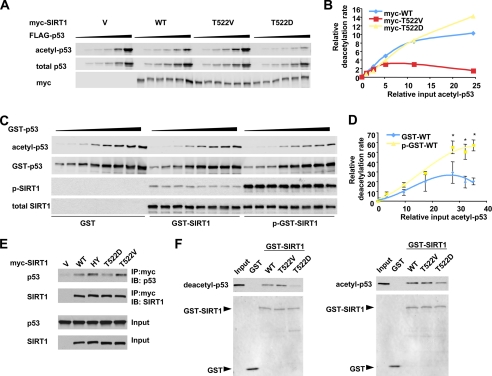
The Thr522 residue of murine SIRT1 is localized within a conserved hinge region linking the core domain and the C-terminal domain (supplemental Fig. S3C). Our observations suggest that Thr522 phosphorylation at this hinge region probably induces conformational changes that reposition the previously reported inhibitory C-terminal extension (28, 29). Consequently, SIRT1 activity is induced, possibly at multiple stages including protein substrate recognition, protein product release, or NAD+ binding. To answer these mechanistic questions, we characterized the enzymatic behavior of wild-type SIRT1, SIRT1 T522V, and SIRT1 T522D mutants at different substrate levels. As shown in Fig. 7(A and B), all three SIRT1 proteins displayed comparable activities when acetyl-p53 levels were low in the H1299 cells. However, the reaction rate of the T522V mutant leveled off (or slightly decreased) with increasing doses of p53, whereas the wild-type SIRT1 and particularly the T522D mutant were able to deacetylate p53 at a higher rate when p53 concentrations were increased. The recombinant wild-type GST-SIRT1 protein is largely unphosphorylated at Thr522 (Fig. 3D and supplemental Fig. S8A). Further in vitro enzymatic analyses of recombinant wild-type GST-SIRT1 (GST-WT) and DYRK1A-phosphorylated GST-SIRT1 (p-GST-WT) confirmed that phosphorylation of Thr522 increases the activity of SIRT1, particularly at high substrate concentrations (Fig. 7, C and D).
FIGURE 7.
Mechanistic characterization of the effect of Thr522 phosphorylation on the enzymatic behaviors of SIRT1 in vivo and in vitro. A and B, a representative in vivo kinetic analysis of Myc-SIRT1 and phosphorylation mutants. p53-null H1299 cells were co-transfected with 0.125 μg of vectors for Myc-SIRT1 or phosphorylation mutants with increased doses of a construct expressing FLAG-p53 (0, 0.125, 0.25, 0.5, 1, and 2 μg). The cells were then treated with etoposide/MG132 for 1 h, and the acetylated FLAG-p53 and total FLAG-p53 was quantified with the Odyssey Infrared Imaging System (Li-Cor Biosciences) using anti-acetyl-p53 K382 antibodies. C and D, representative in vitro enzymatic analysis of GST-SIRT1 and DYRK1A-phosphorylated GST-SIRT1 proteins. 0.05 μg of purified wild-type SIRT1 (GST-WT) or DYRK1A-phosphorylated wild-type SIRT1 (p-GST-WT) proteins were incubated with 0.15, 0.3, 0.6, 1.2, 1.8, 2.4, 3.0, and 3.6 μg of in vitro acetylated GST-p53 fusion proteins in the presence of 3 mm of NAD+ and 200 nm of trichostatin A and incubated at 30 °C for 1 h. The acetylated GST-p53 was analyzed as described for A (n = 3; *, p < 0.01). E, the SIRT1 T522V mutant displays increased affinity to p53 in cells. HEK293T SIRT1 RNAi cells transfected with indicated expression vectors and then treated with etoposide as described. The cell lysates were then immunoprecipitated (IP) with anti-Myc antibodies. F, the SIRT1 T522D mutant displays decreased affinity to deacetylated and acetylated p53 in vitro. Purified GST-SIRT1 and mutants were incubated with purified deacetylated or acetylated FLAG-p53, and SIRT1-associated FLAG-p53 was analyzed by anti-p53 antibodies. WT, wild type; HY, H355Y mutant; IB, immunoblot.
The sirtuin-mediated deacetylation reaction involves the formation of an enzyme:ADP-ribose:acetyl-substrate intermediate and a final product release step that is rate-limiting in the recombinant wild-type enzyme (30–32). We found that the SIRT1 T522V mutant protein (and unphosphorylated wild-type recombinant SIRT1) showed significantly decreased deacetylase activity at high doses of native acetyl-p53 substrates (Fig. 7, A–D), suggesting that the release of the final deacetylated protein products from the intermediate becomes rate-limiting for this unphosphorylated protein. To confirm this model, we examined the interaction between wild-type SIRT1, SIRT1 T522V, or SIRT1 T522D mutants with p53 in cells and in vitro. Consistent with the previous observation that the catalytically inactive SIRT1 H355Y mutant is stalled at the ternary intermediate stage (31), the interaction between SIRT1 and p53 was increased when the SIRT1 HY mutant was expressed in HEK293T SIRT1 RNAi cells (Fig. 7E). Similarly, the T522V mutant displayed an increased association with p53 (Fig. 7E), suggesting that product release may be blunted in this mutant. Conversely, the T522D mutant appeared to have decreased ability to bind p53 compared with the T522V mutant (Fig. 7E). In line with the above observations, the in vitro GST pulldown analyses indicated that the association of purified deacetylated FLAG-p53 with the SIRT1 T522D mutant was significantly decreased compared with its association with unphosphorylated GST-SIRT1 proteins (wild type and T522V mutant) (Fig. 7F). In addition, the SIRT1 T522D mutant also appeared to have weaker association with acetylated FLAG-p53 proteins than the unphosphorylated GST-SIRT1 proteins (Fig. 7F). Taken together, our data suggest that increased release of protein products, and possibly increased accessibility of protein substrates, underlies DYRK-mediated activation of SIRT1.
DISCUSSION
Members of the DYRK family of protein kinases have been increasingly recognized as key regulators of cell proliferation and apoptosis. Although closely related, DYRK1A and DYRK3 have been shown to promote cell survival in various cell types (7–9), whereas DYRK2 induces apoptosis by direct phosphorylation of p53 at Ser46 (11). Our study shows that SIRT1 is selectively activated by the pro-survival DYRK family members through phosphorylation at Thr522 (Fig. 2). Importantly, phosphorylation of SIRT1 is essential for DYRK-mediated cell survival (Figs. 5 and 6E). Although the basis of this selectivity remains unclear, our observations are consistent with the notion that activation of SIRT1 promotes cell survival under nutrient depletion and other stress-associated conditions.
As the best understood member of the sirtuins family, SIRT1 is rising as an important therapeutic target for metabolic diseases, cancer, neurodegenerative disorders, and possibly aging. Although much attention has been focused on the identification of cellular targets of this sirtuin, the regulatory network governing SIRT1 has just begun to emerge (33, 34). Our present study identifies DYRK1A and DYRK3 as key regulators of SIRT1. DYRK1A and DYRK3 phosphorylate and thus activate SIRT1, which in turn deacetylates p53 and promotes cell survival. On the other hand, knockdown of these kinases activates p53, resulting in increased cell death in response to genotoxic stress. Our findings add a new layer to the regulatory network that controls SIRT1 activity and underscore the importance of SIRT1 in the coordination of environmental stimuli and cellular signals that control cell fate.
The observation that SIRT1 is activated by phosphorylation at a single amino acid located at its noncatalytic C-terminal domain is intriguing, but not altogether surprising. Previous structural studies have suggested that the N- and C-terminal extensions of sirtuins may function as inhibitory domains to regulate NAD+ binding and protein substrate recognition (28, 29). Therefore, any protein-protein interactions or modifications that perturb the resting intramolecular orientation of SIRT1 N- and C-terminal domains would increase its activity. In support of this notion, two previously identified protein activators of SIRT1, AROS, and SENP1 both interact with and/or modify the noncatalytic domains (35, 36). In fact, all of the reported phosphorylation sites of SIRT1 that activate its activity are located within the N- or C-terminal domains (37–39). Our study suggests that introduction of a negatively charged phosphate group at Thr522, a site that localizes within a conserved hinge region linking the core and C-terminal domains, likely induces an open conformation. This conformational change does not affect the intrinsic catalytic ability of SIRT1 (supplemental Fig. S3B). Instead, it repositions the inhibitory C-terminal domain, increasing the release of deacetylated products after the reaction is complete, thereby enhancing catalytic turnover and subsequent rounds of activity (Fig. 7). The open conformation of SIRT1 is also likely to increase the accessibility of substrates to the catalytic core domain. Further characterization of conformations of SIRT1 and its phosphorylation mimetics will be helpful to elucidate the molecular mechanisms underlying the phosphorylation regulation of SIRT1.
The phosphorylation of Thr530 of human SIRT1 (equivalent to Thr522 of mouse SIRT1) has been recently reported as a cyclinB/cdk1 target and c-Jun N-terminal kinase 1 (JNK1) kinase (37, 39). Despite claiming that mutation of this site together with Ser540 together affects cell proliferation and cell cycle progression, the study of Sasaki et al. (37) did not find effects of the T530A/S540A double mutant on deacetylase activity of SIRT1 using Fluor-de lys as a substrate. In fact, we also failed to detect any defect of deacetylase activities of the T522V mutant using Fluor-de lys as substrate (supplemental Fig. S3B). The defective SIRT1 activity was only detectable using the native p53 protein. Therefore, the apparent discrepancy between our study and the work from Sasaki et al. lies in the difference of substrates. Although small acetyl-peptide substrates have proven important for analyzing the enzymatic mechanism of sirtuins, a previous study by Blander et al. (40) demonstrated that SIRT1 displays no substrate specificity on these small peptides. This observation suggests that small peptide substrates can freely access the core domain of SIRT1 and subsequently release from the enzyme without steric constrains from the N- and C-terminal domains. Therefore, the activity measured with these small peptides most likely reflects the intrinsic catalytic ability of the core domain, which is distant from the Thr522 site. Nevertheless, it is possible that the phosphorylation at Thr522 will only affect SIRT1 activity on a subset of substrates, not all substrates. Further analyses of substrate specificity of phosphorylated and dephosphorylated SIRT1 proteins will provide insights into the molecular mechanisms underlying the regulation of SIRT1 by phosphorylation.
Several members of the DYRK family, including DYRK1A, Mirk/DYRK1B, and DYRK3, function as survival kinases in cancer cell lines, including cervical cancer (41), certain types of pancreatic cancers and ovarian cancers (42), and possibly anaplastic large cell lymphoma (43). The activation of SIRT1 by these kinases suggests that hyperactive SIRT1 may be part of a tumorigenic program in these cells. However, whether SIRT1 is a tumor promoter or tumor suppressor is currently under intense debate (44, 45). Therefore, it remains difficult to predict whether therapeutic modulation of SIRT1 activity will prove useful for cancer therapy. It has been proposed that the oncogenic and tumor-suppressive effects of SIRT1 are dependent on the status of p53 (45). In cancer cells expressing wild-type p53, SIRT1 promotes cellular senescence and limits cell proliferation. However, in cells containing mutant p53, overexpression of SIRT1 promotes cell survival through p53-independent mechanisms. Therefore, it will be of great interest to determine whether the pro-survival effect of DYRKs requires p53 in cancers.
Although all of our current data point toward the regulation of SIRT1 by the DYRK family of kinases, it is also possible that DYRK kinases are acetylated proteins, and SIRT1 functions to regulate their activities in response to different cellular signals. Further studies are needed to dissect the interplay between these two families of vital protein modification enzymes.
Supplementary Material
Acknowledgments
We thank Drs. Paul Wade, Amy Galliher-Beckley, John Cidlowski, and Lutz Birnbaumer and members of the Li laboratory for critical reading of the manuscript and the NIEHS flow cytometry facility for apoptosis assays.
This work was supported, in whole or in part, by National Institutes of Health Grant Z01 ES102205 (to X. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
- HA
- hemagglutinin
- RNAi
- RNA interference
- MEF
- mouse embryonic fibroblast
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1.Becker W., Joost H. G. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 1–17 [DOI] [PubMed] [Google Scholar]
- 2.Becker W., Weber Y., Wetzel K., Eirmbter K., Tejedor F. J., Joost H. G. (1998) J. Biol. Chem. 273, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 3.Tejedor F., Zhu X. R., Kaltenbach E., Ackermann A., Baumann A., Canal I., Heisenberg M., Fischbach K. F., Pongs O. (1995) Neuron 14, 287–301 [DOI] [PubMed] [Google Scholar]
- 4.Fotaki V., Dierssen M., Alcántara S., Martínez S., Martí E., Casas C., Visa J., Soriano E., Estivill X., Arbonés M. L. (2002) Mol. Cell. Biol. 22, 6636–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guimera J., Casas C., Estivill X., Pritchard M. (1999) Genomics 57, 407–418 [DOI] [PubMed] [Google Scholar]
- 6.Delabar J. M., Aflalo-Rattenbac R., Creau N. (2006) Sci. World J. 6, 1945–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laguna A., Aranda S., Barallobre M. J., Barhoum R., Fernández E., Fotaki V., Delabar J. M., de la Luna S., de la Villa P., Arbonés M. L. (2008) Dev. Cell 15, 841–853 [DOI] [PubMed] [Google Scholar]
- 8.Seifert A., Allan L. A., Clarke P. R. (2008) FEBS J. 275, 6268–6280 [DOI] [PubMed] [Google Scholar]
- 9.Geiger J. N., Knudsen G. T., Panek L., Pandit A. K., Yoder M. D., Lord K. A., Creasy C. L., Burns B. M., Gaines P., Dillon S. B., Wojchowski D. M. (2001) Blood 97, 901–910 [DOI] [PubMed] [Google Scholar]
- 10.Lord K. A., Creasy C. L., King A. G., King C., Burns B. M., Lee J. C., Dillon S. B. (2000) Blood 95, 2838–2846 [PubMed] [Google Scholar]
- 11.Taira N., Nihira K., Yamaguchi T., Miki Y., Yoshida K. (2007) Mol. Cell 25, 725–738 [DOI] [PubMed] [Google Scholar]
- 12.Blander G., Guarente L. (2004) Annu. Rev. Biochem. 73, 417–435 [DOI] [PubMed] [Google Scholar]
- 13.Guarente L. (2000) Genes Dev. 14, 1021–1026 [PubMed] [Google Scholar]
- 14.Bishop N. A., Guarente L. (2007) Nat. Rev. 8, 835–844 [DOI] [PubMed] [Google Scholar]
- 15.Frye R. A. (2000) Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 16.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 17.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 18.Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., Lowe S. W. (2002) Nat. Cell Biol. 4, 859–864 [DOI] [PubMed] [Google Scholar]
- 19.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 21.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 22.Yuan J., Minter-Dykhouse K., Lou Z. (2009) J. Cell Biol. 185, 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dioum E. M., Chen R., Alexander M. S., Zhang Q., Hogg R. T., Gerard R. D., Garcia J. A. (2009) Science 324, 1289–1293 [DOI] [PubMed] [Google Scholar]
- 24.Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007) Mol. Cell 28, 91–106 [DOI] [PubMed] [Google Scholar]
- 26.Himpel S., Tegge W., Frank R., Leder S., Joost H. G., Becker W. (2000) J. Biol. Chem. 275, 2431–2438 [DOI] [PubMed] [Google Scholar]
- 27.Campbell L. E., Proud C. G. (2002) FEBS Lett. 510, 31–36 [DOI] [PubMed] [Google Scholar]
- 28.Zhao K., Chai X., Clements A., Marmorstein R. (2003) Nat. Struct. Biol. 10, 864–871 [DOI] [PubMed] [Google Scholar]
- 29.Schwer B., North B. J., Frye R. A., Ott M., Verdin E. (2002) J. Cell Biol. 158, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borra M. T., Langer M. R., Slama J. T., Denu J. M. (2004) Biochemistry 43, 9877–9887 [DOI] [PubMed] [Google Scholar]
- 31.Smith B. C., Denu J. M. (2006) Biochemistry 45, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao K., Chai X., Marmorstein R. (2003) Structure 11, 1403–1411 [DOI] [PubMed] [Google Scholar]
- 33.Kwon H. S., Ott M. (2008) Trends Biochem. Sci. 33, 517–525 [DOI] [PubMed] [Google Scholar]
- 34.Zschoernig B., Mahlknecht U. (2008) Biochem. Biophys. Res. Commun. 376, 251–255 [DOI] [PubMed] [Google Scholar]
- 35.Kim E. J., Kho J. H., Kang M. R., Um S. J. (2007) Mol. Cell 28, 277–290 [DOI] [PubMed] [Google Scholar]
- 36.Yang Y., Fu W., Chen J., Olashaw N., Zhang X., Nicosia S. V., Bhalla K., Bai W. (2007) Nat. Cell Biol. 9, 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki T., Maier B., Koclega K. D., Chruszcz M., Gluba W., Stukenberg P. T., Minor W., Scrable H. (2008) PLoS ONE 3, e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang H., Jung J. W., Kim M. K., Chung J. H. (2009) PLoS ONE 4, e6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009) PLoS ONE 4, e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blander G., Olejnik J., Krzymanska-Olejnik E., McDonagh T., Haigis M., Yaffe M. B., Guarente L. (2005) J. Biol. Chem. 280, 9780–9785 [DOI] [PubMed] [Google Scholar]
- 41.Chang H. S., Lin C. H., Yang C. H., Yen M. S., Lai C. R., Chen Y. R., Liang Y. J., Yu W. C. (2007) Int. J. Cancer 120, 2377–2385 [DOI] [PubMed] [Google Scholar]
- 42.Friedman E. (2007) J. Cell. Biochem. 102, 274–279 [DOI] [PubMed] [Google Scholar]
- 43.Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 44.Deng C. X. (2009) Int. J. Biol. Sci. 5, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooks C. L., Gu W. (2009) Nat. Rev. Cancer 9, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.