Abstract
Tandem repeats of various trinucleotide motifs are present in the human transcriptome, but the functions of these regular sequences, which likely depend on the structures they form, are still poorly understood. To gain new insight into the structural and functional properties of triplet repeats in RNA, we have performed a biochemical structural analysis of the complete set of triplet repeat transcripts, each composed of a single sequence repeated 17 times. We show that these transcripts fall into four structural classes. The repeated CAA, UUG, AAG, CUU, CCU, CCA, and UAA motifs did not form any higher order structure under any analyzed conditions. The CAU, CUA, UUA, AUG, and UAG repeats are ordered according to their increasing tendency to form semistable hairpins. The repeated CGA, CGU, and all CNG motifs form fairly stable hairpins, whereas AGG and UGG repeats fold into stable G-quadruplexes. The triplet repeats that formed the most stable structures were characterized further by biophysical methods. UV-monitored structure melting revealed that CGG and CCG repeats form, respectively, the most and least stable hairpins of all CNG repeats. Circular dichroism spectra showed that the AGG and UGG repeat quadruplexes are formed by parallel RNA strands. Furthermore, we demonstrated that the different susceptibility of various triplet repeat transcripts to serum nucleases can be explained by the sequence and structural features of the tested RNAs. The results of this study provide a comprehensive structural foundation for the functional analysis of triplet repeats in transcripts.
Keywords: RNA/Metabolism, Gene Expression, RNA Folding, RNA-Protein Interaction, RNA Structure, G-quadruplex, G-tetraplex, RNA Structure Probing, Triplet Repeat Hairpin
Introduction
Trinucleotide repeats (TNRs)4 belong to a family of microsatellite sequences also known as short tandem repeats or simple sequence repeats. TNRs are present in both prokaryotic and eukaryotic genomes and, similar to other microsatellites, show high mutation rates resulting in frequent length variability. Polymorphic TNRs are better tolerated than dinucleotide and tetranucleotide repeats in translated sequences because their length variation does not change the open reading frame.
A recent survey of the human genome reference sequence showed that it harbors more than 32,000 tracts of uninterrupted TNR sequences composed of six or more repeated units. In annotated human exons, which account for less than 3% of the genomic sequence, there are as many as 1,030 TNR tracts (61). Some AT-rich TNR types, such as CTT, AAC, and AAT, are particularly underrepresented in exons, whereas GC-rich repeats (CGG, CAG, and CCG) are highly overrepresented, implying that these sequences have a functional significance. The AT-rich and GC-rich TNRs tend to localize preferentially in the 3′-UTR and 5′-UTR, respectively (1). About 60% of exonic TNRs are localized in the open reading frame. These coding repeats are primarily translated to the poly-Gln, poly-Ala, poly-Glu, and poly-Leu tracts. However, the amino acid coding property of TNRs is not the only feature for which these sequences are selected in exons. The other properties of TNR sequences that manifest themselves on the levels of DNA, RNA, or both may also contribute to the functional importance of these sequences and their prevalence in exons. These properties of TNRs may include their ability to form higher order structures in single-stranded DNA and transcripts. TNR structures and their unstructured sequences may play important regulatory roles in numerous cellular processes, such as DNA replication repair, and at various steps of gene expression, ranging from transcription to mRNA decay (1–4).
Little is known about the structures and physiological functions of normal TNR tracts in genes and transcripts. Existing knowledge has been gathered mainly on a subset of TNRs known to undergo pathogenic expansions that lead to human neurological diseases (1–4). This fragmentary knowledge does not allow for either a deeper insight into the possible roles played by TNRs in transcripts or a more comprehensive characterization of structural, functional, and evolutionary relationships within the TNR family.
In this study, we performed a survey of RNA structures formed by all types of TNRs, using biochemical and biophysical methods. To obtain more information about the detailed features of these sequences, we carried out a structural analysis in various solutions and under different temperature conditions. We classified TNRs into four groups according to their ability to form higher order structures. TNRs showing the highest structure-forming potential were further subjected to more detailed structure characterization using biophysical methods such as UV-monitored structure melting and circular dichroism (CD). We also investigated the biochemical properties of various TNR RNA sequences by analyzing their susceptibility to endogenous nucleases present in serum and HeLa cell extracts. Taken together, our results provide new information regarding the structures and properties of triplet repeat RNAs, which may shed new light on the functions of these abundant sequences in cellular transcripts.
EXPERIMENTAL PROCEDURES
RNA Preparation
The RNAs used in this study were prepared by in vitro transcription of synthetic DNA templates with T7 RNA polymerase as described earlier (5). For transcripts forming tetraplex structure as well as transcripts synthesized from tetraplex-forming DNA templates, the in vitro transcription was significantly less efficient. Therefore, some RNAs, especially those which were used for biophysical analysis, were chemically synthesized (Metabion). All transcripts were purified by electrophoresis in a denaturing 10% polyacrylamide gel.
Nuclease Digestions and Lead Ion Cleavages
Prior to structure-probing reactions, 32P-labeled RNAs were subjected to a denaturation and renaturation procedure in a buffer containing either 2 mm (buffer A) or 20 mm MgCl2 (buffer B) and 80 mm NaCl, 20 mm Tris-HCl, pH 7.2, or in a buffer containing 2 mm MgCl2, 270 mm KCl, 30 mm NaCl, 20 mm Tris-HCl, pH 7.2 (buffer C) by heating the sample at 80 °C for 1 min and then slowly cooling to the reaction temperature. Limited RNA digestion was initiated by mixing 5 μl of RNA sample (20–40 nm; 50,000 cpm) with 5 μl of a probe solution containing lead ions, nuclease S1, mung bean, or ribonucleases T1, T2, Cl3, and V1. Final probe concentrations were as follows: S1 nuclease (0.25, 0.5, and 1 units/μl; 1 mm ZnCl2 was present in each reaction); RNase T1 (0.05, 0.075, 0.1 unit/μl); RNase T2 (0.025, 0.05, 0.1 units/μl); nuclease V1 (0.025, 0.05, 0.1 units/ml); nuclease mung bean (0.0625, 0.125, 0.25 units/μl); RNase Cl3 (0.025, 0.05, 0.075 units/μl); and lead ions (0.25, 0.5, 1 mm). For all transcripts, the reactions were performed at three different temperatures, 4 °C (for 60 min), 25 °C (for 15 min), and 37 °C (for 10 min), and stopped by adding an equal volume of stop solution (7.5 m urea and 20 mm EDTA with dyes) and freezing the samples. To determine cleavage sites, the products of lead-induced hydrolysis and nuclease digestion were separated on 15% polyacrylamide gels containing 7.5 m urea along with an alkaline hydrolysis ladder and a C- or G-specific ladder obtained with Cl3 or T1 ribonuclease, as described earlier (5).
Electrophoresis under Non-denaturing Conditions
RNA structures formed by each transcript were analyzed by electrophoresis of radiolabeled RNA samples in 10% non-denaturing polyacrylamide gels (acrylamide/bisacrylamide, 29/1) buffered with 45 mm Tris borate (0.5× Tris borate) at a fixed temperature of 4 or 25 °C. In some experiments, either 1 mm or 10 mm MgCl2, as well as 40 mm NaCl, was present in the electrophoretic buffer. Prior to gel electrophoresis, ∼1 pmol of the 32P-labeled transcripts was subjected to a denaturation and renaturation procedure in a buffer containing either 1 mm or 10 mm MgCl2 and 40 mm NaCl, 10 mm Tris-HCl, pH 7.2. The sample was mixed with an equal volume of 7% sucrose with dyes before gel loading. Electrophoresis was performed at 15 watts.
Thermodynamic Measurements
Oligoribonucleotides were melted in standard buffers containing either 100 mm NaCl, 20 mm sodium cacodylate, 0.5 mm Na2EDTA (pH 7.0) or 100 mm KCl, 20 mm sodium cacodylate, 0.5 mm Na2EDTA (pH 7.0). Absorbance versus temperature melting curves were measured simultaneously at 260 and 295 nm with a heating rate of 1 °C/min from 0 to 90 °C on a Beckman DU 640 spectrometer with a water-cooled Peltier thermoprogrammer. Melting curves were analyzed, and thermodynamic parameters were calculated using the MeltWin 3.5 program (6). To calculate thermodynamic parameters for melting recorded at 295 nm for (AGG)17 and (UGG)17, the spectra were inverted.
UV and CD Spectra
Experiments were performed on Beckman DU 640 and Jasco J-810 spectrophotometers for UV and CD spectra, respectively, in the same buffers as for UV melting measurements. For UV experiments, the RNA samples were either heated at 90 °C or cooled at 4 °C for 3 min, and duplicate spectra were recorded. The UV spectra were corrected for buffer and adjusted at 330 nm. The CD spectra were recorded in quadruplicate in both buffers at room temperature.
Stability of RNA in Serum and Cell Extract
Approximately 20 pmol of non-labeled and 45,000 cpm of 32P-labeled transcript were added to RPMI 1640 medium containing 5% thermally inactivated fetal bovine serum or HeLa whole-cell extract (1 μg/μl protein) (7). Samples were incubated at 37 °C for different time intervals (from 1 to 135 min) and frozen on a dry ice bath after the stop solution was added. Each sample was analyzed using 8% denaturing polyacrylamide gel-electrophoresis. Quantitative analysis was performed using FLA-5100 (Fujifilm) and Multi Gauge 3.0 software.
RESULTS
A total of 20 transcripts composed from each of the 20 different types of trinucleotide motifs reiterated 17 times were assayed to determine what structures they form in solutions at different temperatures and salt conditions. We utilized a structure-probing method involving limited digestion of transcripts with nucleases and lead ions at low (1 mm) and high (10 mm) magnesium ion concentration in the presence of sodium and potassium ions at temperatures of 4, 25, and 37 °C. The probes used included single strand-specific ribonucleases T1 (G-specificity); Cl3 (C-specificity), T2 (A-preference), and S1 and mung bean nucleases (known to be sequence-nonspecific); the ribonuclease V1 (shows a preference for double-stranded RNA) (8); and lead ions that cleave single-stranded RNA and relaxed double-stranded RNA structures (9). The structural features of the transcripts were also analyzed by (i) electrophoresis in non-denaturing polyacrylamide gels, (ii) UV absorbance-based melting measurements, and (iii) UV and CD spectra.
RNAs Composed of Triplet Repeats Fall into Four Structural Classes According to Structure Probing and Migration in Native Gel
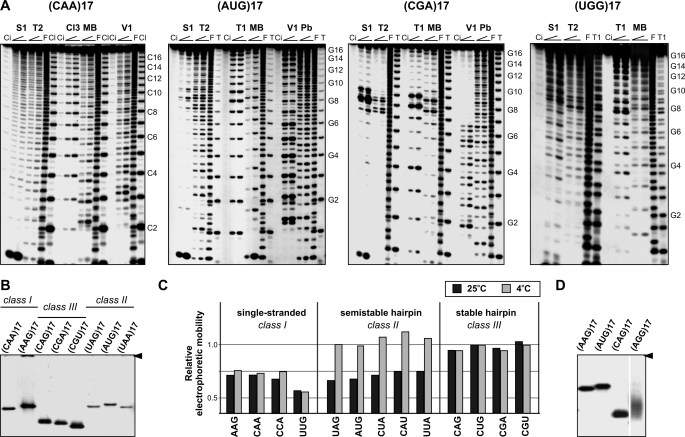
Four different types of cleavage patterns could be distinguished for the 20 transcripts (Table 1). Examples of these patterns are shown in Fig. 1A for (CAA)17, (AUG)17, (CGA)17, and (UGG)17. In (CAA)17, all probes generated cuts of similar intensity along the entire sequence. This class of unstructured transcripts (class I) also includes RNAs composed of UUG, AAG, CUU, CCU, CCA, and UAA motifs. Five transcripts composed of AUG, UAG, UUA, CAU, and CUA motifs fall into the second class. At 25 °C, cleavages at central repeats were 2–10-times stronger than at the side repeats. V1 cleavages occurred throughout the entire transcript, but cuts at the 5′ end were stronger. When structure probing was performed at 37 °C, cleavage intensities were similar along the entire RNA sequence. These five transcripts appeared to form a semistable hairpin structure (class II). The cleavage pattern characteristic for the third class of transcripts represented by the (CGA)17 in Fig. 1A is different. (CGU)17 and all (CNG)17 transcripts also belong to this class. Single strand-specific nucleases showed 10–100 times stronger activity at central repeats than at repeats located at the 5′ and 3′ ends, indicative of hairpin structures. Ribonuclease V1 cleaved only the 5′ end of the hairpin stem formed by these six types of repeats. A similar cleavage pattern was also obtained at 37 °C. The fourth class includes transcripts composed of UGG and AGG repeats. The cleavage patterns obtained at 25 °C resemble those observed for transcripts forming semistable hairpins. The stability of their structures is as high as that of transcripts forming stable hairpins (class III) despite their inability to form W-C base pairs. At 60 °C, enhanced nuclease cuts were still clearly visible in the central repeats of these transcripts (supplemental Fig. 1). The intensities of the nuclease cuts were different at both ends of the highly reactive region, being relatively stronger at its 3′ end (Fig. 1A).
TABLE 1.
Structural classes of triplet repeat RNAs
| Class I (unstructured RNAs) | Class II (semistable hairpins) | Class III (stable hairpins) | Class IV (stable tetraplexes) |
|---|---|---|---|
| (CAA)17 | (UAG)17 | (CAG)17 | (AGG)17 |
| (UUG)17 | (AUG)17 | (CUG)17 | (UGG)17 |
| (AAG)17 | (UUA)17 | (CCG)17 | |
| (CCA)17 | (CUA)17 | (CGG)17 | |
| (CUU)17 | (CAU)17 | (CGA)17 | |
| (CCU)17 | (CGU)17 | ||
| (UAA)17 |
FIGURE 1.
Triplet repeat transcripts differ in their ability to form higher order structures. A, cleavage patterns obtained for the 5′-labeled transcripts (CAA)17, (AUG)17, (CGA)17, and (UGG)17 were denatured and renatured in buffer B and treated with two concentrations of nuclease S1, V1, mung bean (MB), RNase T1, T2, Cl3, and lead ions (Pb). Lane Ci, incubation control (no probe); lane F, formamide ladder; lane T, guanine-specific ladder; lane Cl, cytosine specific ladder. Electrophoresis was performed on 15% polyacrylamide gels under denaturing conditions. Positions of selected G-residues (or C-residues) of repeated sequences are indicated. B, non-denaturing 10% polyacrylamide gel electrophoresis of eight transcripts representing three structural classes in structure-probing buffer containing 1 mm magnesium ion at 25 °C. An arrowhead indicates the position of the wells. C, electrophoretic mobilities of different triplet repeat transcripts relative to the migration rate of (CUG)17 transcript in non-denaturating gels and two different electrophoresis buffers (black bars and gray bars represent the results obtained in the absence or presence of magnesium ions, respectively) and temperature conditions (at 25 or 4 °C). D, non-denaturing gel electrophoresis of (AGG)17 transcripts forming a tetraplex structure and three control transcripts; (CAG)17 represents hairpin-forming transcripts, (AAG)17 represents single-stranded transcripts, and (AUG)17 represents semistable hairpin-forming transcripts. Experiments were performed at 25 °C.
All 20 transcripts were also analyzed in non-denaturing gels at different temperatures and salt conditions. The electrophoretic mobility of transcripts representing the first three structural classes in 0.5× Tris borate buffer at 25 °C is shown in Fig. 1B. Transcripts forming stable hairpin structures migrated the fastest, whereas all transcripts belonging to classes I and II showed slower electrophoretic mobility. When analyzed at 4 °C in the presence of 10 mm MgCl2 and 40 mm NaCl, the migration rates of all transcripts belonging to class II, including (UAG)17, (AUG)17, (CAU)17, (CUA)17, and (UUA)17, were similar to those of stable hairpin-forming transcripts (Fig. 1C). This means that at a low temperature, class II transcripts form hairpin structures. (UGG)17 and (AGG)17 exhibited electrophoretic migration different from all other transcripts as shown for (UGG)17 in Fig. 1D. They migrated faster than single strands and more slowly than hairpins and gave rise to diffuse bands. These two G-rich transcripts appear to form a tetraplex structure (more data addressing this issue are described below).
UAN Repeats Form More Stable Structures than AUN Repeats
To determine the influence of base arrangement within the stem on the stability of hairpins formed by AU-rich sequences, we compared two pairs of transcripts, (UAG)17 and (AUG)17, as well as (CUA)17 and (CAU)17 (mismatched bases are underlined). Quantitative analysis of cleavages generated by nucleases revealed that UAN17 transcripts formed more stable structures than AUN17 (supplemental Figs. 2 and 3). For example, T1 cuts at 25 °C were only two times stronger in the loop of (AUG)17 when compared with the stem. The same difference, but in favor of stem cleavages, was observed for V1 nuclease (supplemental Fig. 2). Conversely, the relative cleavage intensities in (UAG)17 were nearly identical at 4 and 25 °C. In the case of (UAG)17, cleavages at central repeats (loop region) were enhanced at 37 °C, whereas this effect was not observed for (AUG)17 (Table 2).
TABLE 2.
Stability of hairpin structure of AU-rich triplet repeat-containing transcripts
Symbols +, +/−, and − indicate that cleavages generated in the terminal loop were more efficient than those generated in the stem region at different temperatures and buffer conditions. + indicates double efficiency, +/− indicates 1.5–2× efficiency, and − denotes no change.
| Conditions | Transcripts |
|||||
|---|---|---|---|---|---|---|
| (UAG)17 | (AUG)17 | (UUA)17 | (CUA)17 | (CAU)17 | (UAA)17 | |
| 37 °C; 1 mm Mg2+ | − | − | − | − | − | − |
| 37 °C; 10 mm Mg2+ | +/− | − | − | − | − | − |
| 25 °C; 1 mm Mg2+ | + | + | +/− | − | − | − |
| 25 °C; 10 mm Mg2+ | + | + | + | + | − | − |
| 4 °C; 1 mm Mg2+ | + | + | + | + | +/− | − |
| 4 °C; 10 mm Mg2+ | + | + | + | + | + | − |
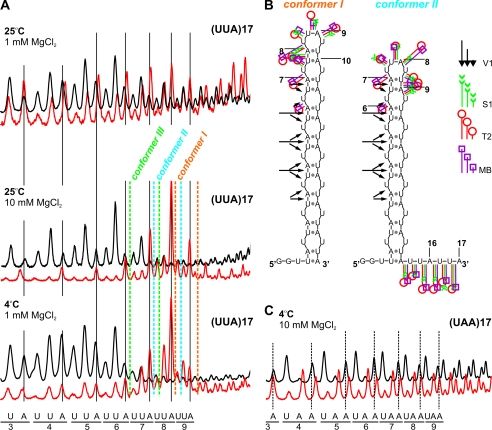
Two transcripts, (UUA)17 and (UAA)17, could potentially form hairpins containing two alternative base-pairing systems in their stems, 5′-UA/5′-UA and 5′-AU/5′-AU. Our results indicated that only (UUA)17 folded into hairpin structures (Fig. 2), whereas (UAA)17 did not form any higher order structure, even at 4 °C and in 10 mm MgCl2. Thus, putative A/A mismatches in the context of AU base pairs were prohibited. As revealed by structure-probing results, other mismatches were ordered (G/G > U/U > C/C) according to their structure-stabilizing effect (Table 2). The number of cuts occurring at central repeats that correspond to a hairpin terminal loop of five transcripts belonging to this structural group was higher than predicted for the 4-nucleotide loops (Fig. 2 and supplemental Figs. 2 and 3). This result was interpreted to mean that several slipped conformers of the AU-rich repeat hairpin co-exist (Fig. 2B), as shown previously for all CNG repeat hairpins (5).
FIGURE 2.
Ability of AU-rich transcripts to form hairpin structures depends on mismatch type. A, cleavage patterns obtained with RNase T2 (red line) and V1 (black line) for the (UUA)17 transcript in the terminal loop region are shown as PhosphorImager peaks. Enzymatic digestions were performed at 25 °C in buffer A or B containing 1 mm or 10 mm magnesium ion, as well as at 4 °C in buffer A. The positions of individual nucleotides are shown below the peaks. Peaks corresponding to terminal loop cuts in conformers I–III are indicated with different colors. B, two alternative structures of (UUA)17 transcripts (conformer I and conformer II) are presented. Cleavage sites and intensities are specified for each probe according to the symbols in the figure legend. MB, mung bean. C, cleavage patterns obtained for the (UAA)17 transcript central region are shown as PhosphorImager peaks. Enzymatic digestions were performed at 4 °C in buffer containing 10 mm magnesium. The positions of individual nucleotides are shown below the peaks.
Magnesium Ion Concentration Affects the Distribution of Slipped Conformers
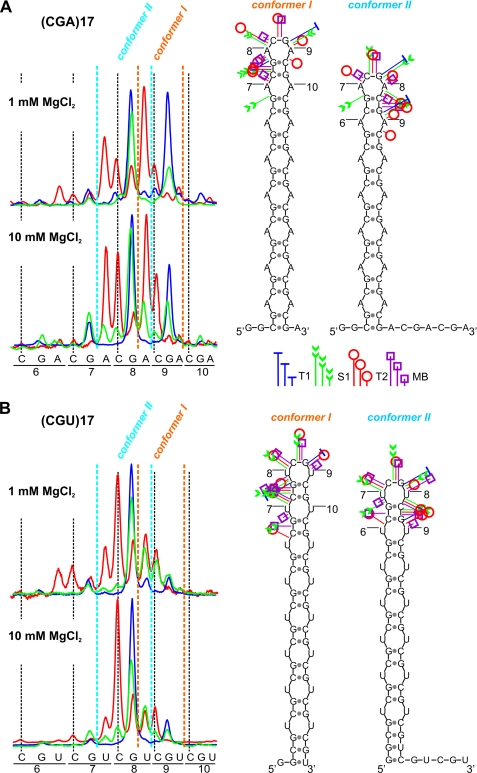
The structures formed by four transcripts, (CAG)17, (CUG)17, (CGA)17, and (CGU)17, were probed at 37 °C at low and high MgCl2 concentrations. Only quantitative differences in relative cleavage intensities within transcripts were evident (Fig. 3). The number of cleavages generated by nucleases occurring at central repeats was higher than predicted for the single 4-nucleotide loop. This means that several slipped conformers of the repeat hairpin co-exist for these transcripts. Two dominant conformers of (CGA)17 and (CGU)17 (conformers I and II) are shown in Fig. 3. Conformer I predominated in (CGA)17, whereas conformer II predominated in (CGU)17. Differences in the distribution of slipped conformers observed for (CGA)17 and (CGU)17 at low and high MgCl2 concentrations (Fig. 3) were observed for (CAG)17 and (CUG)17, as well as for all AU-rich transcripts forming semistable hairpins.
FIGURE 3.
Effect of magnesium concentration on the distribution of slipped conformers in (CGA)17 and (CGU)17 hairpin structures. A, cleavages generated in the terminal loop of the (CGA)17 transcript are shown as PhosphorImager peaks. Enzymatic digestions were performed at 37 °C in two buffers containing either 1 mm or 10 mm MgCl2. Positions of individual nucleotides are shown below the peaks. Peaks corresponding to terminal loop cuts in conformers I and II are indicated. Two alternative structures of the (CGA)17 transcript (conformer I and conformer II) are presented. Cleavage sites and intensities are specified for each probe according to the symbols shown in the figure. The colors of the peaks representing cleavages are indicated in the legend. B, as in A but for the (CGU)17 transcript.
(CGG)20 Forms the Most Stable Hairpins, whereas (CCG)20 Forms the Least Stable CNG Repeat Hairpins
The thermodynamic stabilities of four CNG trinucleotide repeat RNAs were obtained from UV-melting experiments at 260 nm (Table 3). All of the investigated RNAs formed hairpins with similar architectures (5) and showed differences in stability, arising from the presence of different single-nucleotide N/N mismatches (10). The thermodynamic stabilities (ΔG°37) of (CCG)20, (CUG)20, (CAG)20, and (CGG)20 in 100 mm NaCl were −6.68, −6.18, −4.57, and −2.57 kcal/mol, respectively. When the thermodynamic stability of the same RNAs was measured in 100 mm KCl, the free energies were −4.32, −6.00, −3.88, and −2.38 kcal/mol, respectively. Shorter CGG repeat RNA was previously reported to form a G-tetraplex structure (11–13), as described earlier for DNA oligonucleotides of the same sequence (14, 15). Comparison of both free energy values and melting temperatures (Tm) of (CCG)20, (CUG)20, (CAG)20, and (CGG)20 in the presence of sodium and potassium salt demonstrated that none of these RNAs formed a tetraplex structure. However, the properties of (CGG)20 differed slightly from those of other CNG repeat RNAs. In particular, a large decrease of free energy (ΔΔG°37) equal to 2.36 kcal/mol was observed for (CGG)20 melting in the presence of NaCl and KCl (Table 3). In the case of (CCG)20, (CUG)20, and (CAG)20, the changes in free energies were much smaller (0.18–0.69 kcal/mol). Also, the lack of a melting transition for (CGG)20 at 295 nm argues against G-quadruplex formation.
TABLE 3.
Thermodynamic parameters of triplet repeat transcripts
| Transcript | Wavelength | 100 mm NaCla |
100 mm KClb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| −ΔH° | −ΔS° | −ΔG°37 | Tmc | −ΔH° | −ΔS° | −ΔG°37 | Tmc | ΔΔG°37 | ΔTm | ||
| nm | kcal/mol | eud | kcal/mol | °C | kcal/mol | eud | kcal/mol | °C | kcal/mol | °C | |
| (CGG)20 | 260 | 60.9 ± 5.0 | 175.1 ± 14.7 | 6.68 ± 0.47 | 75.2 | 38.8 ± 1.6 | 111.3 ± 4.9 | 4.32 ± 0.14 | 75.8 | 2.36 | 0.6 |
| (CAG)20 | 260 | 72.9 ± 1.9 | 215.3 ± 5.5 | 6.18 ± 0.35 | 65.7 | 73.3 ± 6.8 | 217.0 ± 20.1 | 6.00 ± 0.61 | 64.6 | 0.18 | −1.1 |
| (CUG)20 | 260 | 59.5 ± 1.8 | 177.2 ± 5.4 | 4.57 ± 0.14 | 62.8 | 53.2 ± 3.2 | 159.1 ± 9.8 | 3.88 ± 0.21 | 61.4 | 0.69 | −1.4 |
| (CCG)20 | 260 | 39.7 ± 5.3 | 119.9 ± 16.6 | 2.57 ± 0.25 | 58.4 | 39.9 ± 2.4 | 121.2 ± 7.4 | 2.38 ± 0.13 | 56.7 | 0.19 | −1.7 |
| (AGG)17 | 260 | 52.5 ± 4.5 | 156.9 ± 14.1 | 3.87 ± 0.23 | 61.7 | 75.8 ± 8.8 | 212.7 ± 25.1 | 9.80 ± 1.11 | 83.1 | −5.93 | 21.4 |
| (AGG)17 | 295 | 57.5 ± 2.9 | 171.9 ± 8.7 | 4.17 ± 0.18 | 61.3 | 73.5 ± 6.1 | 205.7 ± 17.7 | 9.71 ± 0.65 | 84.2 | −5.54 | 22.9 |
| (UGG)17 | 260 | 62.0 ± 3.5 | 186.9 ± 10.6 | 4.04 ± 0.21 | 58.6 | 81.1 ± 8.5 | 227.2 ± 24.3 | 10.65 ± 0.99 | 83.9 | −6.61 | 25.3 |
| (UGG)17 | 295 | 62.0 ± 1.4 | 187.1 ± 4.3 | 4.02 ± 0.13 | 58.5 | 74.1 ± 2.9 | 207.2 ± 8.2 | 9.82 ± 0.45 | 84.4 | −5.80 | 25.9 |
a Average of curve fits 100 mm NaCl, 20 mm sodium cacodylate, and 0.5 mm Na2EDTA, pH 7.0.
b Average of curve fits 100 mm KCl, 20 mm sodium cacodylate, and 0.5 mm Na2EDTA, pH 7.0.
c Calculated for 10−4 m RNA concentration.
d eu, entropy units.
The experimentally determined thermodynamic stabilities for CNG hairpins are much lower than those predicted by RNA folding algorithms (Mfold and RNAstructure), which use the nearest neighbor thermodynamic parameters. For example, the experimentally determined free energy values are 3–9-times lower. Moreover, the stability order for the CNG repeat hairpins is different for the predicted and experimentally analyzed structures. The structure prediction shows that the CAG repeat hairpin has the lowest stability, whereas CCG repeat hairpin is the least stable according to structure melting experiments. This issue was widely discussed by Broda et al. (16).
AGG and UGG Repeats Form G-quadruplex Structures
Two G-rich transcripts, (UGG)17 and (AGG)17, showed a clear difference in thermodynamic stability (ΔG°37) as calculated from UV-melting analysis in 100 mm NaCl and 100 mm KCl. UV melting at 260 nm in sodium and potassium salts exhibited a typical melting transition with hyperchromism of 10–15%. However, (UGG)17 and (AGG)17 melting recorded at 295 nm also showed hypochromism (25–30%). This feature was reported earlier to be characteristic of a G-quadruplex structure (17, 18). On the contrary, (CCG)20 and (CUG)20 did not show any transition at 295 nm. In the case of (AGG)17 and (UGG)17, the free energies (ΔG°37) of the melting transitions recorded at both 260 and 295 nm in 100 mm NaCl were similar, about −4 kcal/mol (Table 3). Measurements performed in 100 mm KCl showed strongly enhanced thermodynamic stability of both transcripts. The free energies calculated from melting transitions recorded at either 260 nm or 295 nm were about −10 kcal/mol for both (AGG)17 and (UGG)17. In the presence of potassium ions, the Tm also increased significantly, by about 25 °C (Table 3). Higher thermodynamic stabilities in the presence of potassium as well as descending absorbance during UV melting recorded at 295 nm are typical for G-quadruplex structures (17).
The UV spectra of (UGG)17, (AGG)17, (CCG)20, (CUG)20, and (CGG)20 in the presence of sodium or potassium salts were recorded at 5 and 90 °C. Differential spectra were similar for (UGG)17 and (AGG)17 under both conditions (supplemental Fig. 4). Negative peaks were observed at 297 and 265 nm, whereas positive peaks were seen at 274 and 235 nm. This characteristic of UV differential spectra is also typical of G-quadruplexes (19). On the contrary, differential UV spectra for (CCG)20, (CUG)20, and (CGG)20 in the presence of sodium and potassium did not show a negative peak near 295 nm.
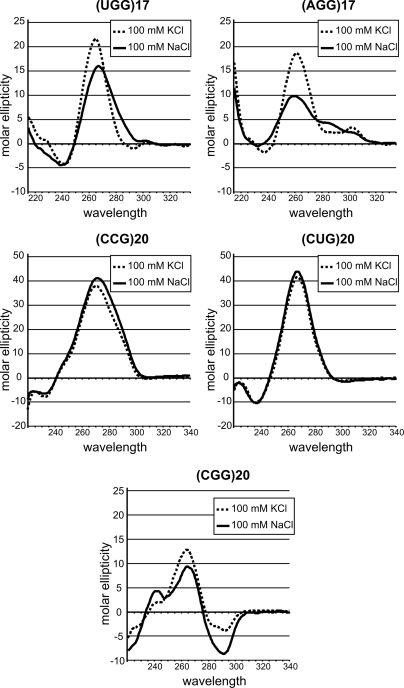
Finally, CD spectra were recorded at room temperature for (UGG)17 and (AGG)17, as well as for (CCG)20, (CUG)20, and (CGG)20 RNAs, in a buffer containing either 100 mm NaCl or 100 mm KCl. The CD spectra were measured in the same buffers used earlier (18, 19) to demonstrate G-quartet structure formation. A difference depending on ionic conditions was observed for (UGG)17 and (AGG)17 (Fig. 4). Both RNAs showed large positive peaks in the 260–265 nm range and small peaks at 300–305 nm. Significant differences in the shape and ellipticity of their CD spectra were also observed. On the other hand, differences in CD spectra recorded in the presence of NaCl and KCl were negligible for (CCG)20 and (CUG)20 hairpins. Positive peaks at 265–270 nm typical for A-RNA structures were observed for these transcripts. In the case of (CGG)20, the CD spectra were more complex, and some differences in ellipticity and peak wavelengths were observed for buffers containing KCl and NaCl. Besides the intense positive peak at 265 nm, an intense negative peak occurred at around 290 nm under both ionic conditions.
FIGURE 4.
Circular dichroism spectra confirm the quadruplex structure of two transcripts, (AGG)17 and (UGG)17. A comparison of CD spectra for five transcripts, (CUG)20, (CCG)20, (CGG)20, (UGG)17, and (AGG)17, incubated in two different buffers containing either 100 mm NaCl (continuous line) or 100 mm KCl (dashed line) is shown.
Because G-quadruplexes may adopt a wide variety of structural variants, including tetra-, bi-, and unimolecular structures, we analyzed these RNAs at several different concentrations ranging from 50 nm to 100 μm. No changes in melting temperature and free energy (ΔG°37) were observed (data not shown). This suggests that transcripts composed of AGG and UGG repeats form unimolecular tetraplex structures.
Sequences and Structures of Triplet Repeat RNAs Explain Their Different Susceptibility to Serum Nucleases
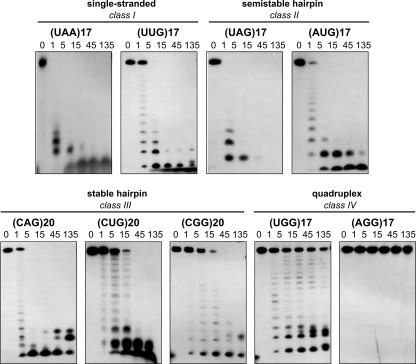
To determine how triplet repeat RNAs differ in their resistance to serum nucleases, we incubated 5′-labeled transcripts representing all of the RNA structural classes in bovine fetal serum and monitored substrate disappearance and formation of products over time (Fig. 5). We found that transcripts that did not form secondary structures (class I) and those transcripts that formed semistable hairpins (class II) had half-lives (t½) shorter than 1 min. Similarly, rapid degradation was observed for the CAG repeat transcript. (CAG)20 was capable of forming a stable hairpin, but it contains reiterations of fragile CpA phosphodiester bonds, making this transcript highly unstable in serum. Two other transcripts that also formed stable hairpins, (CUG)20 and (CGG)20 (class III), had half-lives of 7.3 (±2.1) and 12.5 (±1.9) min, respectively. The serum stability of G-tetraplex-forming (UGG)17 and (AGG)17 transcripts (class IV) was much higher. The t½ for (UGG)17 was 52.4 (±3.9) min, but (AGG)17 transcripts remained intact even after 135 min in serum. A similar relationship between structure and stability was observed for the same transcripts incubated in HeLa whole-cell extract (data not shown). These results are in line with earlier observations suggesting that the formation of higher order structure strongly protects RNAs from attack by nucleases and that pyrimidine-specific pancreatic-type ribonuclease activities are most prevalent in animal cells and sera (20–23).
FIGURE 5.
Triplet repeat-containing transcripts show varying susceptibility to serum nucleases and do not activate the cellular sensors of foreign RNA. Electrophoretic separation of 5′-labeled transcripts incubated in 5% bovine serum at 37 °C for the indicated period of time (from 1 to 135 min) is shown.
DISCUSSION
Ribonucleic acids employ a diverse repertoire of structural motifs to compose their native folded structures. The questions we asked in this study included the following. How does the variety of triplet repeat sequences contribute to RNA structural diversity? What are the thermodynamic properties of triplet repeats forming stable structures? What could be the putative structure-dependent functions of triplet repeats in transcripts? Finally, how do the various triplet repeats differ in their susceptibility to cellular ribonucleases? A survey of triplet repeats, which are present in about 4% of human mRNAs, revealed that some of these repeats are highly prevalent, whereas others are either rare or absent (61). CNG repeats, associated with human neurological diseases, are among the most frequent repeat types and have attracted the highest interest to date. Only AGG repeats occur at a similarly high frequency in human transcripts. These five repeated motifs constitute 77% of the 1,030 total repeat tracts composed of six or more uninterrupted repeats found in 878 human mRNAs. At the other end of the frequency scale are the CGU, CUA, UAG, CAU, CUU, CGA, and UGG repeats (61).
To gain insight into the structural features of all types of triplet repeat RNAs, we used native gel migration tests, biochemical structure probing, and biophysical characterization. The data obtained from this structure survey indicate that triplet repeat RNAs exist in three basic structural forms: single-stranded, semistable hairpins, stable hairpins, and very stable G-quadruplexes. CUU, CAA, AAG, UAA, UUG, CCA, and CCU repeats, which cannot form any higher order structure under any conditions tested, occur in 2, 12, 18, 18, 34, 45, and 50 human mRNAs, respectively (61). Unless these sequences provide suitable sites for some single-stranded RNA-binding proteins, purine-rich repeats may form rigid spacers, and pyrimidine-rich repeats may form flexible adapters between neighboring RNA structure domains in mRNAs. For example, the (GAA)6 was shown by SELEX experiments to be a binding site for SC35 and SF2/ASF (alternative splicing factor) splicing proteins (24). Pyrimidine-rich TNR sequences that are present close to the 3′ splice site may form binding sites for the polypyrimidine tract-binding protein and influence splicing efficiency. Moreover, in some genes, such TNRs are polymorphic in length in population and could play the role of modulating factors in gene expression.
All CNG repeats of sufficient length form imperfect stem and loop structures in transcripts in which the stems are composed of reiterated C-G and G-C base pairs and N/N mismatches (5, 7, 16, 25–30). However, the CGG repeats that form the most stable hairpins (Tm = 75–76 °C) (Table 3) differ considerably from CUG, CCG, and CAG repeats in their tendency to form slipped-hairpin conformers (5). In a more recent NMR study, G/G mismatches were shown to exist in a dynamic equilibrium between the anti- and syn-conformations required to form two symmetric hydrogen bonds between the N1 and carbonyl oxygen atoms of two guanines (31). The CGG repeats were also reported to form G-quadruplexes in both RNA (11–13) and DNA (14, 15). In the current study, we analyzed the structural features of CGG and other types of CNG repeats under identical conditions using biochemical and biophysical methods. Data from structure-probing and gel migration tests indicated that the properties of CGG repeats do not deviate from those of CUG, CCG, and CAG repeats. The Tm of the CGG repeat transcript is not sensitive to potassium ions, which also argues against G-quartet structure formation. Only the CD spectra of CGG repeats do not closely resemble those of CUG and CCG repeats, suggesting a slightly different hairpin architecture. No evidence for tetraplex structure formation by CGG repeats was found by the authors of the NMR study in which RNA was analyzed at millimolar concentrations (31).
By comparison, in the crystal structure of the CUG repeats (32), the U/U mismatches form stretched wobble interactions with only one hydrogen bond between the carbonyl O4 atom of one U-base and the N3 imino group of the second uracil residue. CUG repeats are predicted to assume one of two different conformations depending on whether the uracil residue is inclined toward the minor groove of the CUG repeat RNA-A helix (32). The authors of the first reported crystal structure of CUG repeats (33) suggested that CCG repeats may form a similar structure as pyrimidine-pyrimidine mismatches could well accommodate within the RNA-A helix. They anticipated stronger structure distortions by purine-purine mismatches, which have higher hydrogen bond-forming potential. Whatever the structural differences at the mismatched bases are, our study shows that they do not result in high stability differences between the CUG, CCG, and CAG repeat hairpins.
Two repeated motifs, UGG and AGG, form G-quartet structures known also as G-quadruplexes or G-tetraplexes, which are characteristic of both DNA and RNA sequences rich in guanine. These four-stranded structures are formed by Hoogsteen hydrogen bonding between a tetrad of guanine bases and are strongly stabilized by centrally located potassium ions. The DNA (34–36) and RNA (37, 38) structures formed by AGG repeats have been investigated by NMR and CD. The d(GGA)4 intramolecular parallel quadruplex is composed of G:G:G:G tetrads and G(:A):G(:A):G(:A):G heptads (34, 36). The same structure was formed by d(GGA)8 (35). Similarly, but not identically, the r(GGA)4 intramolecular parallel quadruplex is formed by one G:G:G:G tetrad plane and one G(:A):G:G(:A):G hexad plane (38). The structures of d(GGA)4, d(GGA)8, and r(GGA)4 correlated well with CD spectra characteristics typical for parallel quadruplexes. The (AGG)17 and (UGG)17 transcripts investigated in the current study are much longer, but they seem to form similar types of quadruplex structures as their shorter counterparts based on the similarity of their CD spectra.
The results of the biophysical and biochemical structural studies on the CNG and AGG repeat transcripts obtained by us and others are gathered in supplemental Table 1 and summarized in supplemental Fig. 5. The fact that different structures were often reported for the same repeat types by authors that used different methods reflects the atypical character of these sequences where the structures are highly sensitive to the length of the repeated sequence, RNA concentration, temperature, and ionic conditions. Nevertheless, the x-ray structures of short CUG repeat duplexes (32, 33) may well represent the stem portions of hairpin structures formed by longer CUG repeats (5, 16, 26, 29, 39). Similarly, the NMR structure of short CGG repeats, which were forced to form hairpins (31), may well correspond to the stem structure of hairpins formed spontaneously by long CGG repeat tracts (5, 25).
Expansions of some triplet repeats in genes, mostly CNG repeats, cause a number of inherited neurological and neuromuscular diseases. Several lines of evidence argue that the pathogenesis of some of these diseases is RNA-mediated. Expanded CUG repeats play a crucial role in the pathogenesis of myotonic dystrophy DM1, and long CGG repeats are associated with the fragile X-associated tremor ataxia syndrome (40, 41). The RNA gain of function mutations have also been implicated in a subgroup of these diseases (so-called poly(Q)-induced diseases), which are caused by CAG repeat expansions (42). The CUG, CAG, as well as CGG repeats form similar hairpin structures. Each of these repeat hairpins is known to be a site of interaction with some double-stranded RNA-binding proteins. For example, the muscleblind-like protein (MBNL1) was found in nuclear inclusions formed by expanded CUG and CAG (43) as well as CGG repeats (44). The affinity of MBNL1 is almost the same for the CUG and CAG repeats (39). It is also known that MBNL1 interacts mainly with the GC element of the repeated sequence (45). The structural similarity of all CNG repeat hairpins was also observed in the in vitro assay with ribonuclease Dicer (46). On the other hand, these hairpin structures are different targets for the binding of small molecule ligands (47, 48). For example, Hoechst 33258 binds to CUG repeats with nanomolar affinity, and KD values are 3–4 times higher for CCG and CAG repeats. This difference could be explained by the different recognition of unique repeating electrostatic patches in the minor groove of the RNA helix.
Sequences capable of forming G-quartet structures are highly prevalent in the human genome (49). The G-rich triplet repeats are, however, rather atypical sequences of this kind. They contain only two consecutive guanine residues instead of the three or four usually present in sequences forming tetraplexes; however, the trinucleotide motif is reiterated many times, which increases the G-quartet structure stability. Analysis of the RNA aptamer confirmed that four GGA repeats are sufficient to form tetraplex conformation. Such a structure is a binding site for bovine prion protein PrP, and adenosine residues are important for this interaction (38). Of the tetraplex-forming repeats analyzed here, AGG is highly prevalent in human mRNAs (122 occurrences), whereas UGG repeats are present only in six transcripts (61). The physiological importance of RNA G-tetraplexes, which are more stable and are more easily formed due to the single-stranded character of RNA than their corresponding DNA structures, was postulated in a number of reports (49–52). Such structures have been implicated in specific protein binding and modulation of gene expression by regulating splicing, polyadenylation, translation, and transcript turnover (18, 53–58). For example, short tetraplex-forming sequences from 5′-UTR of the NRAS and ZIC-1 genes decreased the translation of reporter genes several times when inserted into their 5′-UTRs (18, 57).
The CNG and NGG repeats that are present in numerous genes are often polymorphic in length in a normal population (59). They may form binding sites for specific proteins. Different lengths and the localizations of these sequences may differently regulate gene expression. For example long CGG repeats located in 5′-UTR of FMR1 gene down-regulate fragile X mental retardation protein (FMRP) synthesis (60). Both CNG and AGG repeats predominantly occur in the 5′-UTR regions of hundreds of human transcripts.
In conclusion, the present study combines the structural surveys of triplet repeat RNAs by biochemical methods with detailed thermodynamic characterization by biophysical methods. Novel structural information gathered for repeats that are not known to be involved in the pathogenesis of human diseases, as well as the deeper structural characterization of those previously implicated in human diseases, may help to understand the biological roles of these sequences. Because numerous triplet repeat motifs of various types are present in the transcriptomes of many organisms, our results may also be considered the first step toward RNA structural genomic characterization of the “microsatellite repeatome.”
Acknowledgments
We thank Jacek Gawronski and Pawel Skowronek for help on the CD spectra.
This work was supported by grants from the Sixth Research Framework Programme of the European Union, Project RIGHT (Grant LSHB-CT-2004-005276) and the Ministry of Science and Higher Education, Grants PBZ-MNiI-2/1/2005, N301-112-32/3910, N302 278937, and N N301 284837, and Operational Programme “Innovative economy” Grant POIG.01.03.01-00-098/08.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table 1.
- TNR
- trinucleotide repeat
- UTR
- untranslated region.
REFERENCES
- 1.Jasinska A., Michlewski G., de Mezer M., Sobczak K., Kozlowski P., Napierala M., Krzyzosiak W. J. (2003) Nucleic Acids Res. 31, 5463–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson C. E., Nichol Edamura K., Cleary J. D. (2005) Nat. Rev. Genet. 6, 729–742 [DOI] [PubMed] [Google Scholar]
- 3.Mirkin S. M. (2007) Nature 447, 932–940 [DOI] [PubMed] [Google Scholar]
- 4.Usdin K. (2008) Genome Res. 18, 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobczak K., de Mezer M., Michlewski G., Krol J., Krzyzosiak W. J. (2003) Nucleic Acids Res. 31, 5469–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDowell J. A., Turner D. H. (1996) Biochemistry 35, 14077–14089 [DOI] [PubMed] [Google Scholar]
- 7.Sobczak K., Krzyzosiak W. J. (2004) J. Biol. Chem. 279, 41563–41572 [DOI] [PubMed] [Google Scholar]
- 8.Giege R., Helm M., Florentz C. (2001) in RNA (Soll D., Nishimura S., Moore P. B. eds) pp. 71–89, Elsevier Science Ltd., Amsterdam [Google Scholar]
- 9.Krzyzosiak W. J., Marciniec T., Wiewiorowski M., Romby P., Ebel J. P., Giegé R. (1988) Biochemistry 27, 5771–5777 [DOI] [PubMed] [Google Scholar]
- 10.Kierzek R., Burkard M. E., Turner D. H. (1999) Biochemistry 38, 14214–14223 [DOI] [PubMed] [Google Scholar]
- 11.Khateb S., Weisman-Shomer P., Hershco I., Loeb L. A., Fry M. (2004) Nucleic Acids Res. 32, 4145–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khateb S., Weisman-Shomer P., Hershco-Shani I., Ludwig A. L., Fry M. (2007) Nucleic Acids Res. 35, 5775–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ofer N., Weisman-Shomer P., Shklover J., Fry M. (2009) Nucleic Acids Res. 37, 2712–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry M., Loeb L. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4950–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kettani A., Kumar R. A., Patel D. J. (1995) J. Mol. Biol. 254, 638–656 [DOI] [PubMed] [Google Scholar]
- 16.Broda M., Kierzek E., Gdaniec Z., Kulinski T., Kierzek R. (2005) Biochemistry 44, 10873–10882 [DOI] [PubMed] [Google Scholar]
- 17.Mergny J. L., Phan A. T., Lacroix L. (1998) FEBS Lett. 435, 74–78 [DOI] [PubMed] [Google Scholar]
- 18.Arora A., Dutkiewicz M., Scaria V., Hariharan M., Maiti S., Kurreck J. (2008) RNA 14, 1290–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vieregg J., Cheng W., Bustamante C., Tinoco I., Jr. (2007) J. Am. Chem. Soc. 129, 14966–14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorrentino S., Libonati M. (1997) FEBS Lett. 404, 1–5 [DOI] [PubMed] [Google Scholar]
- 21.Sorrentino S. (1998) Cell Mol. Life Sci. 54, 785–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shida T., Suda M., Sekiguchi J. (1997) Nucleic Acids Symp. Ser. 37, 129–130 [PubMed] [Google Scholar]
- 23.Qiu L., Moreira A., Kaplan G., Levitz R., Wang J. Y., Xu C., Drlica K. (1998) Mol. Gen. Genet. 258, 352–362 [DOI] [PubMed] [Google Scholar]
- 24.Cavaloc Y., Bourgeois C. F., Kister L., Stévenin J. (1999) RNA 5, 468–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Napierala M., Michalowski D., de Mezer M., Krzyzosiak W. J. (2005) Nucleic Acids Res. 33, 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Napierała M., Krzyzosiak W. J. (1997) J. Biol. Chem. 272, 31079–31085 [DOI] [PubMed] [Google Scholar]
- 27.Michlewski G., Krzyzosiak W. J. (2004) J. Mol. Biol. 340, 665–679 [DOI] [PubMed] [Google Scholar]
- 28.Sobczak K., Krzyzosiak W. J. (2005) J. Biol. Chem. 280, 3898–3910 [DOI] [PubMed] [Google Scholar]
- 29.Michalowski S., Miller J. W., Urbinati C. R., Paliouras M., Swanson M. S., Griffith J. (1999) Nucleic Acids Res. 27, 3534–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzyzosiak W. J., Sobczak K., Napierala M. (2006) in Genetic Instabilities and Neuronal Diseases (Wells R. D., Ashizawa T. eds) pp. 705–713, Elsevier Science Ltd., London [Google Scholar]
- 31.Zumwalt M., Ludwig A., Hagerman P. J., Dieckmann T. (2007) RNA Biol. 4, 93–100 [DOI] [PubMed] [Google Scholar]
- 32.Kiliszek A., Kierzek R., Krzyzosiak W. J., Rypniewski W. (2009) Nucleic Acids Res. 37, 4149–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mooers B. H., Logue J. S., Berglund J. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16626–16631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsugami A., Okuizumi T., Uesugi S., Katahira M. (2003) J. Biol. Chem. 278, 28147–28153 [DOI] [PubMed] [Google Scholar]
- 35.Matsugami A., Ouhashi K., Ikeda T., Okuizumi T., Sotoya H., Uesugi S., Katahira M. (2002) Nucleic Acids Res. Suppl. 2, 49–50 [DOI] [PubMed] [Google Scholar]
- 36.Matsugami A., Ouhashi K., Kanagawa M., Liu H., Kanagawa S., Uesugi S., Katahira M. (2001) J. Mol. Biol. 313, 255–269 [DOI] [PubMed] [Google Scholar]
- 37.Matsugami A., Mashima T., Nishikawa F., Murakami K., Nishikawa S., Noda K., Yokoyama T., Katahira M. (2008) Nucleic Acids Symp. Ser. (Oxf.) 52, 179–180 [DOI] [PubMed] [Google Scholar]
- 38.Nishikawa F., Murakami K., Matsugami A., Katahira M., Nishikawa S. (2009) Oligonucleotides 19, 179–190 [DOI] [PubMed] [Google Scholar]
- 39.Yuan Y., Compton S. A., Sobczak K., Stenberg M. G., Thornton C. A., Griffith J. D., Swanson M. S. (2007) Nucleic Acids Res. 35, 5474–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Rourke J. R., Swanson M. S. (2009) J. Biol. Chem. 284, 7419–7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin J., Charizanis K., Swanson M. S. (2009) Neurosci. Lett. 466, 99–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L. B., Yu Z., Teng X., Bonini N. M. (2008) Nature 453, 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho T. H., Savkur R. S., Poulos M. G., Mancini M. A., Swanson M. S., Cooper T. A. (2005) J. Cell Sci. 118, 2923–2933 [DOI] [PubMed] [Google Scholar]
- 44.Iwahashi C. K., Yasui D. H., An H. J., Greco C. M., Tassone F., Nannen K., Babineau B., Lebrilla C. B., Hagerman R. J., Hagerman P. J. (2006) Brain 129, 256–271 [DOI] [PubMed] [Google Scholar]
- 45.Teplova M., Patel D. J. (2008) Nat. Struct. Mol. Biol. 15, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krol J., Fiszer A., Mykowska A., Sobczak K., de Mezer M., Krzyzosiak W. J. (2007) Mol. Cell 25, 575–586 [DOI] [PubMed] [Google Scholar]
- 47.Pushechnikov A., Lee M. M., Childs-Disney J. L., Sobczak K., French J. M., Thornton C. A., Disney M. D. (2009) J. Am. Chem. Soc. 131, 9767–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee M. M., Childs-Disney J. L., Pushechnikov A., French J. M., Sobczak K., Thornton C. A., Disney M. D. (2009) J. Am. Chem. Soc. 131, 17464–17472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huppert J. L. (2008) Biochimie 90, 1140–1148 [DOI] [PubMed] [Google Scholar]
- 50.Mergny J. L., De Cian A., Ghelab A., Saccà B., Lacroix L. (2005) Nucleic Acids Res. 33, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel D. J., Phan A. T., Kuryavyi V. (2007) Nucleic Acids Res. 35, 7429–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huppert J. L. (2008) Chem. Soc. Rev. 37, 1375–1384 [DOI] [PubMed] [Google Scholar]
- 53.Darnell J. C., Jensen K. B., Jin P., Brown V., Warren S. T., Darnell R. B. (2001) Cell 107, 489–499 [DOI] [PubMed] [Google Scholar]
- 54.Bonnal S., Schaeffer C., Créancier L., Clamens S., Moine H., Prats A. C., Vagner S. (2003) J. Biol. Chem. 278, 39330–39336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez D., Lemarteleur T., Lacroix L., Mailliet P., Mergny J. L., Riou J. F. (2004) Nucleic Acids Res. 32, 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zanotti K. J., Lackey P. E., Evans G. L., Mihailescu M. R. (2006) Biochemistry 45, 8319–8330 [DOI] [PubMed] [Google Scholar]
- 57.Kumari S., Bugaut A., Huppert J. L., Balasubramanian S. (2007) Nat. Chem. Biol. 3, 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wieland M., Hartig J. S. (2007) Chem. Biol. 14, 757–763 [DOI] [PubMed] [Google Scholar]
- 59.Rozanska M., Sobczak K., Jasinska A., Napierala M., Kaczynska D., Czerny A., Koziel M., Kozlowski P., Olejniczak M., Krzyzosiak W. J. (2007) Hum. Mutat. 28, 451–458 [DOI] [PubMed] [Google Scholar]
- 60.Chen L. S., Tassone F., Sahota P., Hagerman P. J. (2003) Hum. Mol. Genet. 12, 3067–3074 [DOI] [PubMed] [Google Scholar]
- 61.Kozlowski P., de Mezer M., Krzyzosiak W. J. (March9, 2010) Nucleic Acids Res. 10.1093/nar/gkq127 [DOI] [PMC free article] [PubMed] [Google Scholar]