Abstract
The c-Myc oncoprotein promotes cell growth by enhancing ribosomal biogenesis. Overexpression of c-Myc and aberrant ribosomal biogenesis lead to deregulated cell growth and tumorigenesis. Hence, c-Myc activity and ribosomal biogenesis must be tightly coordinated during normal homeostasis. We previously found that ribosomal protein L11 inhibits c-Myc activity by blocking the recruitment of its co-activator transformation/transcription domain-associated protein (TRRAP) to the promoter regions of c-Myc target genes that are transcribed by RNA polymerases I and II. In this study, we extended the role of L11 to the regulation of c-Myc-driven transcription of the 5 S rRNA and tRNA genes by RNA polymerase III. L11 co-resided with c-Myc at the 5 S rRNA and tRNA genes and significantly inhibited the binding of TRRAP to these genes. Knocking down endogenous L11 enhanced c-Myc-dependent transcription of these genes. Interestingly, in response to ribosomal stress induced by the treatment of cells with a low dose of actinomycin D or serum starvation, L11 binding to these genes was increased, and inversely TRRAP binding to these genes was decreased. Consistently, knockdown of L11 rescued the reduction of the expression of these genes by the two treatments. These results demonstrate that L11 suppresses c-Myc-dependent and RNA polymerase III-catalyzed transcription of 5 S rRNA and tRNA genes in response to ribosomal stress, ensuring a tight coordination between c-Myc activity and ribosomal biogenesis.
Keywords: RNA/Ribosomal RNA, Transcription/Myc, Cell Cycle, RNA Polymerase III, Transcription Coactivators, RNA Polymerase III, TRAPP, Transcription, c-Myc, Ribosomal Protein L11
Introduction
The c-Myc oncoprotein is a critical transcription factor that is essential for cell growth, proliferation, and development. It regulates the expression of numerous genes involved in cell cycle, apoptosis, differentiation, angiogenesis, metabolism, stem cell renewal, and neoplastic transformation (1, 2). However, deregulated overexpression of c-Myc contributes to ∼70% of human cancers (3). Constitutive or induced expression of the c-myc transgene leads to neoplastic and malignant phenotypes in mice (4–7). Thus, it is essential to maintain the proper physiological level and activity of c-Myc during normal cell homeostasis.
One of the important functions of c-Myc is to regulate ribosomal biogenesis (8). Ribosomal biogenesis is a tightly regulated cellular process that requires coordinated transcription mediated by all three RNA polymerases (Pols)3 in order to ensure efficient and accurate production of ribosomes. c-Myc enhances the transcription of many ribosomal biogenesis-related genes catalyzed by RNA Pol II, such as those encoding ribosome assembly proteins, translation initiation and elongation factors, and ribosomal proteins (9–12). c-Myc also enhances RNA Pol I-catalyzed rRNA synthesis (13–15) and RNA Pol III-mediated 5 S and tRNA transcription (16). Although these are essential functions for c-Myc in regulating cell growth and proliferation, overuse of them would convert this transcriptional factor into an oncogenic factor and favor tumor growth. Indeed, deregulation of c-Myc activity as mentioned above and/or of ribosomal biogenesis has been highly associated with tumorigenesis (17).
In our previous attempt to understand the coordination between ribosomal biogenesis and c-Myc activity, we revealed that ribosomal protein L11 acts as a novel c-Myc inhibitor via a feedback mechanism (18). L11 binds to c-Myc at Myc box II, a critical region required for all known c-Myc functions. This binding leads to inhibition of the recruitment of the TRRAP co-activator and subsequent reduction of histone acetylation at promoters of c-Myc target genes, including RNA Pol II-transcribed nucleolin, E2F2, eIF4E, and RNA Pol I-transcribed pre-rRNA genes (18). Consequently, L11 suppresses the expression of these genes. However, it remains unclear whether the inhibitory effect of L11 on c-Myc activity is ubiquitously true to c-Myc-dependent gene transcription catalyzed by all three RNA Pols, because the promoter organizations for RNA Pol I and II target genes are different from those for RNA Pol III target genes, such as 5 S rRNA and tRNA genes.
Interestingly, regardless of the difference in their target promoter architectures, mechanisms underlying c-Myc-mediated transcription by all RNA Pols appear to be similar. For instance, similar to the case of c-Myc-induced transcription by RNA Pol I and II, the TRRAP co-activator is also required for c-Myc-mediated transcription by RNA Pol III of the 5 S rRNA and tRNA genes, and c-Myc can bind to these genes and promote the association of TRRAP with them, although no typical E-box elements specific for c-Myc-binding were identified in the promoters of these genes (19). Because the 5 S rRNA and tRNA genes are essential for ribosomal biogenesis and translation, the expression of these genes must also be tightly regulated for normal cell growth. Otherwise, overexpression of RNA Pol III-specific transcription factor, Brf1, results in highly specific elevation of tRNA and 5 S rRNA expression and oncogenic transformation in cells (20). Strikingly, overexpression of the RNA Pol III-transcribed tRNAiMet alone is sufficient to drive cell proliferation and induce tumors in mice (20). This study suggests that deregulated overexpression of Pol III activity may contribute to tumorigenesis too.
To further consolidate the role of L11 in regulating c-Myc-specific transcriptional activity, we have carried out a series of experiments to test whether L11 also regulates c-Myc-dependent transcription of class III genes by RNA Pol III. Indeed, our study as presented here shows that L11 can inhibit c-Myc-dependent transcription of the 5 S rRNA and tRNA genes. L11 bound to c-Myc at the 5 S rRNA and tRNA genes, and this binding leads to reduction of TRRAP association with these genes. Knockdown of endogenous L11 significantly enhanced c-Myc-mediated transcription of these genes. Interestingly, L11 reduction of TRRAP association with these genes was responsive to ribosomal stress (also called nucleolar stress) induced by the treatment of cells with either a low dose of actinomycin D (Act D) or serum starvation. This reduction was alleviated by knockdown of endogenous L11. Taken together, these results not only validate a general role for L11 in regulating c-Myc transcriptional activity but also demonstrate that L11-mediated negative regulation of c-Myc is responsive to ribosomal stress. This L11-c-Myc feedback regulation may contribute to a tight coordination between c-Myc activity and ribosomal biogenesis.
MATERIALS AND METHODS
Cell Lines, Plasmids, and Antibodies
Human lung non-small cell adenocarcinoma H1299 cells and human osteosarcoma U2OS cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere as described previously (21). pD40-His/V5-c-Myc, FLAG-tagged L11 (FLAG-L11), FLAG-TRRAP, and Myc-tagged L11 plasmids have been described previously (18). Anti-FLAG M2 (Sigma), anti-V5 (Invitrogen), polyclonal anti-c-Myc (N262, Santa Cruz Biotechnology, Inc.), monoclonal anti-c-Myc (9E10, Zymed Laboratories Inc.), and anti-TRRAP (sc-5405, Santa Cruz Biotechnology, Inc.) antibodies were purchased. Rabbit polyclonal anti-L11 was generated as described (22).
Cotransfection, Immunoblot (IB), and Co-immunoprecipitation (co-IP) Analyses
Cells were transfected with plasmids as indicated in the figure legends using TransIT®-LT1 reagents following the manufacturer's protocol (Mirus Bio Corp.). The cells were harvested at 48 h posttransfection and lysed in lysis buffer consisting of 50 mm Tris/HCl (pH 8.0), 0.5% Nonidet P-40, 1 mm EDTA, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 0.25 μg/ml pepstatin A, and 1 mm leupeptin. Co-IP and IB analyses were performed as described (23).
Reverse Transcription and Quantitative Real-time PCR Analysis (RT-qPCR)
Total RNA was isolated from cells using Qiagen RNeasy minikits (Qiagen). Reverse transcriptions were performed using iScript reagents (Bio-Rad) following the manufacturer's protocol. Quantitative PCR was performed on an ABI StepOne real-time PCR system (Applied Biosystems) using SYBR Green mix (Bio-Rad) as described previously (18). All reactions were carried out in triplicate. Relative gene expression was calculated using the ΔCτ method following the manufacturer's instructions. The primers used were as follows: 5′-GATTCCACCCATGGCAAATTC-3′ and 5′-AGCATCGCCCCACTTGATT-3′ (glyceraldehyde-3-phosphate dehydrogenase); 5′-GGCCATACCACCCTGAACGC-3′ and 5′-CAGCACCCGGTATTCCCAGG-3′ (5 S rRNA); 5′-CCTTCGATAGCTCAGCTGGTAGAGCGGAGG-3′ and 5′-CGGAATCGGAACCAGCGACCTAAGGATGTCC-3′ (tRNATyr); 5′-GTCAGGATGGCCGAGTGGTCTAAG-3′ and 5′-CCACGCCTCCATACGGAGAACCAGAAGACCC-3′ (tRNALeu).
RNA Interference
The 21-nucleotide siRNA duplexes with a 3′-dTdT overhang were synthesized by Dharmacon (Lafayette, CO). The target sequences were 5′-GGTGCGGGAGTATGAGTTA-3′ for L11, 5′-CAGAAATGTCCTGAGCAAT-3′ for c-myc, and 5′-AAGTTCCTGAGGAACATGCGC-3′ for L29 as described (18). The scrambled II RNA duplex was used as a control (23). Transfection of siRNA was performed as previously described using siLentFectTM Lipid (Bio-Rad) following the manufacturer's protocol (23). The cells were harvested at 48 h post transfection for IB and RT-qPCR assays.
Chromatin Immunoprecipitation (ChIP)-qPCR
ChIP analysis was performed as described (18) using anti-c-Myc (N262), anti-L11, or anti-TRRAP antibodies. Immunoprecipitated DNA fragments were analyzed by qPCR amplification. The primers were 5′-ACGGCCATACCACCCTGAA-3′ and 5′-GCCAAAGAAAAAGCCTACAGCA-3′ (for 5 S rRNA); 5′-AGGACAACGGGGACAGTAAG-3′ and 5′-TCCACCAGAAAAACTCCAGCC-3′ (for tRNALeu); 5′-GCGGAAAGTCCAGTGATCCA-3′ and 5′-GGAATCGAACCAGCGACCTAAG-3′ (for tRNATyr); and 5′-CTTGGCTGATCCATCTGCCT-3′ and 5′-TCCCATATCCTCGTCCGACT-3′ (for ARPP0). For detection of co-occupancy of both c-Myc and L11 in promoters, H1299 cells were transfected with V5-c-Myc alone or together with FLAG-L11. After cross-linking and sonication, the cell lysates were immunoprecipitated with anti-FLAG antibodies. The immunoprecipitated FLAG-L11 protein-DNA complexes were eluted with 0.1 mg/ml FLAG peptide (Sigma) using methods as described (23). Eluates were then immunoprecipitated with anti-V5 antibody followed by DNA purification and qPCR amplification as described above.
RESULTS
Ablation of Endogenous L11 Enhances c-Myc-dependent Transcription of 5 S rRNA and tRNA Genes
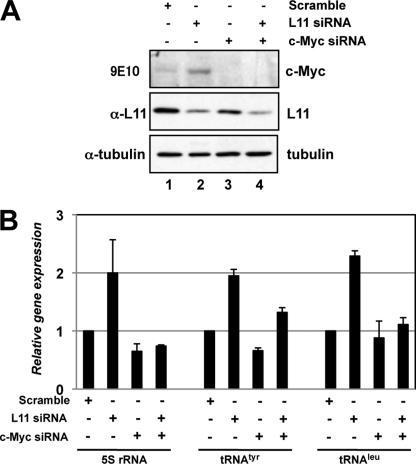
It has been shown that c-Myc directly binds to 5 S rRNA and tRNA genes and enhances their transcription catalyzed by RNA Pol III (16, 19). Because we recently showed that L11 inhibits c-Myc-dependent transcription of pre-rRNA, nucleolin, E2F2, and eIF4E genes, which are transcribed by RNA Pol I and II, respectively (18), we wanted to determine whether L11 plays a ubiquitous role in regulating c-Myc-mediated transcription of all three classes of genes by further testing if it affects c-Myc-dependent transcription of the 5 S rRNA and tRNA genes by RNA Pol III. First, we tested if ablation of endogenous L11 would affect c-Myc-mediated transcription of these genes. Consistent with our previous observations (18), knocking down endogenous L11 markedly induced the level of c-Myc protein (Fig. 1A, compare lane 2 with lane 1). To determine whether the effect of L11 knockdown on gene expression is dependent on c-Myc activity, we also depleted endogenous c-Myc using siRNA as shown in Fig. 1A (lanes 3 and 4). These siRNA-transfected cells were subjected to RT-qPCR analysis to examine the expression of the 5 S rRNA and tRNA genes. As shown in Fig. 1B, knockdown of endogenous L11 enhanced the expression of the 5 S rRNA, tRNATyr, and tRNALeu genes by 2-fold. This enhancement was c-Myc-dependent because further ablation of endogenous c-Myc by siRNA drastically reduced the L11 siRNA-induced expression of the above genes. These results suggest that L11 indeed inhibits c-Myc-induced transcription of the 5 S rRNA and tRNA genes in cells.
FIGURE 1.
Endogenous L11 regulates c-Myc-mediated transcription of the 5 S rRNA and tRNA genes. A, knockdown of endogenous L11 increases the level of endogenous c-Myc. U2OS cells were transfected with siRNAs as indicted followed by IB analysis using antibodies as indicated. B, knockdown of endogenous L11 enhances c-Myc dependent transcription of the 5 S rRNA, tRNATyr, and tRNALeu genes. U2OS cells were transfected with siRNAs as indicated, followed by RT-qPCR assays to determine the relative expression of the 5 S rRNA, tRNATyr, and tRNALeu genes compared with that of glyceraldehyde-3-phosphate dehydrogenase mRNA. Error bars indicate mean ± S.D.
L11 Binds to the 5 S rRNA and tRNA Genes
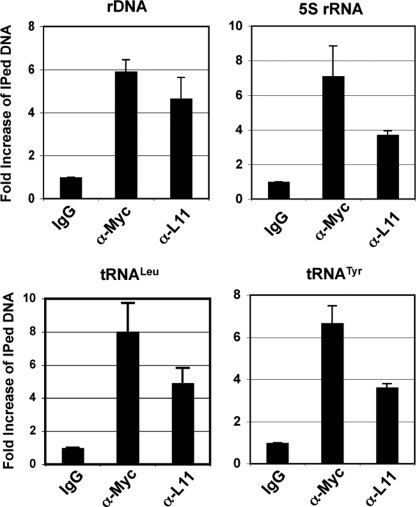
Unlike the rDNA genes, which harbor a number of E-box elements, the 5 S rRNA and tRNA gene promoters do not usually contain typical E-box elements (19). However, c-Myc does bind to these genes, and the binding is important for their c-Myc-dependent expression (16, 19). To elucidate how L11 regulates c-Myc-mediated transcription of these genes, we next tested if L11 associates with c-Myc at these genes using ChIP-qPCR assays because we previously showed that L11 physically associates with c-Myc at the promoters of several RNA Pol I- and II-transcribed genes (18). The binding of L11 to the rDNA genes was used as positive control (18) (Fig. 2). Consistent with the report by another group (19), endogenous c-Myc readily bound to the rDNA genes as well as the 5 S rRNA, tRNALeu, and tRNATyr genes (Fig. 2). Interestingly, endogenous L11 also specifically associated with each of these genes using the anti-L11 antibody, but not the IgG control (Fig. 2). This result confirms that endogenous L11 can associate with the 5 S rRNA and tRNA genes in cells.
FIGURE 2.
L11 binds to the 5 S rRNA and tRNA genes. ChIP-qPCR assays were conducted to detect the binding of endogenous c-Myc and L11 in the 5 S rRNA and tRNA genes in H1299 cells using anti-c-Myc (N262) or anti-L11 antibodies. Error bars indicate mean ± S.D.
L11 Associates with c-Myc at the 5 S rRNA and tRNA Genes
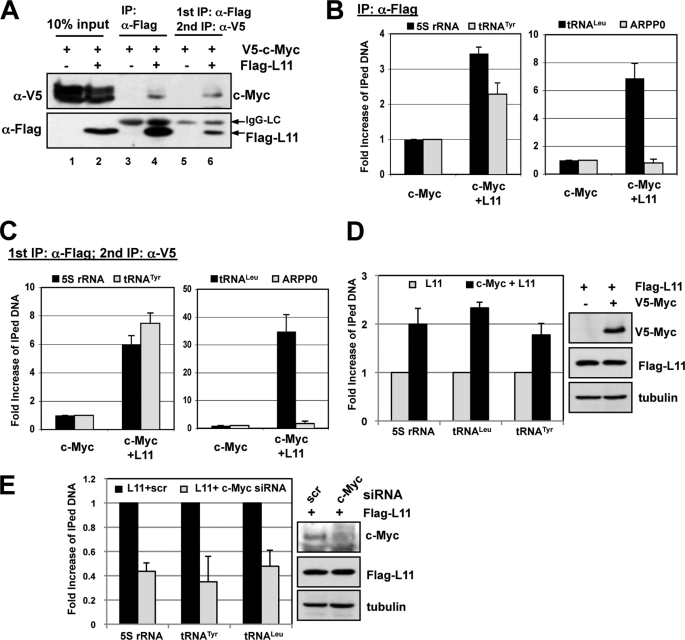
To further test if L11 associates with c-Myc at the 5 S rRNA and tRNA genes, we introduced V5-c-Myc alone or V5-c-Myc with the FLAG-L11 plasmid into H1299 cells through transfection followed by sequential ChIP-qPCR assays as described previously (18). The sequential co-IP of the L11-c-Myc complex using anti-FLAG antibodies followed by anti-V5 antibodies is shown in Fig. 3A. The second co-IP using the anti-V5 antibody immunoprecipitated a significant amount of FLAG-L11, suggesting that this complex is indeed a core-L11-c-Myc complex and therefore suitable for the following sequential ChIP assays. After cross-linking and sonication of the transfected cells, cell lysates were first immunoprecipitated with the anti-FLAG antibody, followed by elution using FLAG peptides to affinity-purify the FLAG-L11-associated protein-DNA complexes (23). Eluted protein-DNA complexes were then immunoprecipitated with the anti-V5 antibody. As shown in Fig. 3B, the anti-FLAG antibody readily immunoprecipitated the genomic regions of the 5 S rRNA, tRNALeu, and tRNATyr, but not ARPP0 (as a control), genes. Importantly, subsequent anti-V5 IP of the eluted FLAG-L11-associated protein-DNA complexes showed the co-occupancy of L11 and c-Myc at the aforementioned genes (Fig. 3C), as compared with control IP without FLAG-L11 expression. Of note, the -fold increase of immunoprecipitated DNA in the secondary anti-V5 IP was higher than that in the first anti-FLAG IP compared with IgG control, indicating the increased purity of the L11-c-Myc-DNA complex in the secondary co-IP. These results indicate that L11 binds to c-Myc at the 5 S rRNA and tRNA genes.
FIGURE 3.
L11 co-resides with c-Myc in the 5 S rRNA and tRNA genes. A, co-IP between ectopic L11 and c-Myc. H1299 cells were transfected with FLAG-L11 with or without V5-c-Myc. The cell lysates were first immunoprecipitated with anti-FLAG antibodies (lanes 3 and 4). The FLAG-L11-associated proteins were eluted with FLAG peptide, and the elution was then immunoprecipitated with anti-V5 antibodies (lanes 5 and 6), followed by an IB assay. B and C, sequential ChIP-qPCR assays were conducted to detect the binding of L11 to c-Myc at the 5 S rRNA and tRNA genes. H1299 cells were transfected with FLAG-L11 with or without V5-c-Myc. The cells were cross-linked, and the cell lysates were immunoprecipitated with anti-FLAG antibody. The FLAG-L11 protein-DNA complexes were eluted with FLAG peptide, and 10% of eluates were used for DNA purification and qPCR amplification (B). The rest of the eluates were subjected to a second IP using anti-V5 antibody, followed by qPCR amplification (C) to detect the 5 S rRNA and tRNA genes. D, overexpression of c-Myc enhances L11 binding to the 5 S rRNA, tRNATyr, and tRNALeu genes. H1299 cells were transfected with FLAG-L11 alone or together with c-Myc. The cells were subjected to ChIP-qPCR assays to detect the binding of L11 to the aforementioned genes. Protein expression of the transfected genes is shown on the right. E, knockdown of endogenous c-Myc decreases L11 binding to the 5 S rRNA, tRNATyr, and tRNALeu genes. U2OS cells were transfected with scrambled (scr) or L11 siRNA, followed by ChIP-qPCR assays to detect the binding of L11 to the aforementioned genes. Protein expression of the transfected genes is shown on the right. The expression of c-Myc and L11 is also shown on the right. Error bars indicate mean ± S.D.
To further test if L11 binding to above genes requires c-Myc, we examined the L11 binding to aforementioned genes upon either overexpression or knockdown of c-Myc. H1299 cells were transfected with FLAG-L11 in the absence or presence of V5-c-Myc. The cells were subjected to ChIP-qPCR assays using anti-FLAG antibodies. As shown in Fig. 3D, co-expression of c-Myc enhanced the binding of L11 to the 5 S rRNA, tRNALeu, and tRNATyr genes approximately 2-fold in all cases. Conversely, when endogenous c-Myc was knocked down, the binding of L11 to these genes was decreased by 2–3-fold (Fig. 3E). Taken together, these results further support the statement that L11 associates with c-Myc at the aforementioned gene promoters.
L11 Inhibits the Recruitment of the c-Myc Coactivator TRRAP at the 5 S rRNA and tRNA Genes
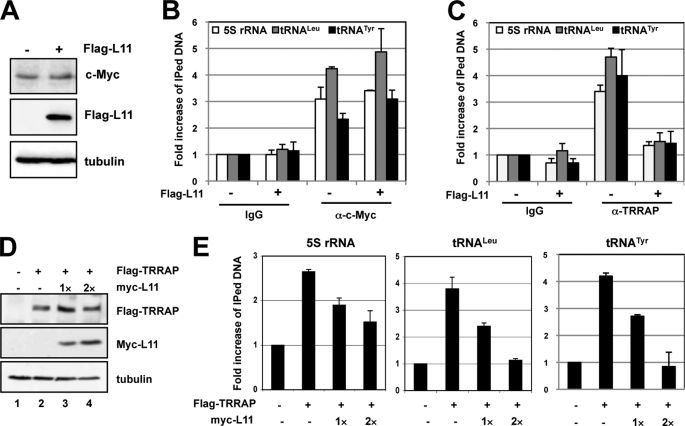
To test whether L11 affects the recruitment of the c-Myc co-activator TRRAP to the 5 S rRNA and tRNA genes, we conducted ChIP-qPCR assays in H1299 cells transfected with control or FLAG-L11 vector. Indeed, the binding of TRRAP to the 5 S rRNA, tRNALeu, and tRNATyr genes was reduced by 3–4-fold when L11 was overexpressed (Fig. 4C). By contrast, the c-Myc level (Fig. 4A) and the binding of c-Myc to the above genes (Fig. 4B) were not changed significantly, suggesting that L11 competes with TRRAP for binding to c-Myc at these genes. To further confirm this result, we co-transfected the cells with TRRAP with increasing amounts of L11, followed by ChIP-qPCR assays. As shown in Fig. 4D, ectopic L11 did not significantly change the level of ectopic TRRAP. However, the binding of ectopic TRRAP to the 5 S rRNA, tRNALeu, and tRNATyr genes was drastically reduced by overexpression of L11 in a dose-dependent manner (Fig. 4E). These results further confirm that L11 inhibits the recruitment of TRRAP to the 5 S rRNA, tRNALeu, and tRNATyr genes by competing with it for binding to c-Myc.
FIGURE 4.
L11 reduces the binding of TRRAP to the 5 S rRNA and tRNA genes. A, H1299 cells transfected with control or FLAG-L11 plasmid were subjected to IB with anti-c-Myc or anti-FLAG antibodies. B and C, L11 reduces the binding of TRRAP to the 5 S rRNA and tRNALeu genes. The transfected cells in A were subjected to ChIP-qPCR assays using goat anti-TRRAP, control goat IgG, rabbit polyclonal anti-c-Myc (N262), or control rabbit IgG, followed by detection of the 5 S rRNA, tRNALeu, and tRNATyr genes. D and E, L11 competes with TRRAP for binding to the 5 S rRNA and tRNALeu genes. H1299 cells transfected with FLAG-TRRAP in the presence of increasing amounts of Myc-tagged L11 were immunoblotted with antibodies as indicated (D). The transfected cells were also subjected to ChIP assays using anti-FLAG antibody, followed by detection of the TRRAP binding to the 5 S rRNA, tRNALeu, and tRNATyr using qPCR assays (E). Error bars indicate mean ± S.D.
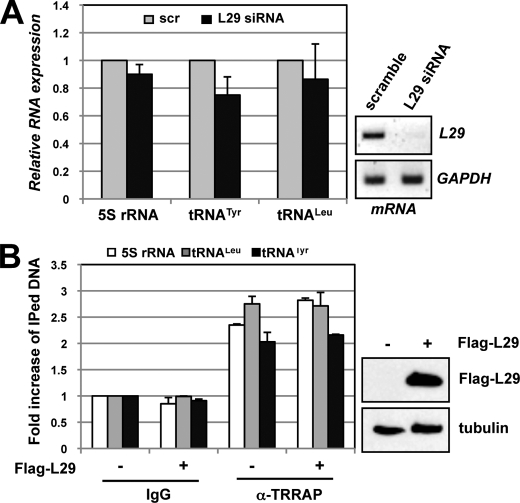
L29 Does Not Regulate c-Myc-mediated Transcription of the 5 S rRNA and tRNA Genes
To test if L11 regulation of c-Myc-induced transcription of the 5 S rRNA and tRNA genes is specific, we examined the role of L29, which does not bind to c-Myc and does not regulate c-Myc-driven transcription of the nucleolin gene (18), on the transcription of the 5 S rRNA and tRNA genes. As shown in Fig. 5A, knockdown of L29 did not significantly affect the levels of the 5 S rRNA, tRNATyr, and tRNALeu genes. Also, unlike L11, overexpression of L29 did not inhibit the TRRAP binding to the 5 S rRNA, tRNATyr, and tRNALeu genes (Fig. 5B). Together, these results suggest that L11 inhibition of c-Myc-driven transcription of the 5 S rRNA, tRNATyr, and tRNALeu genes is specific to L11 but not a general effect of all individual ribosomal proteins.
FIGURE 5.
c-Myc transcription of the 5 S rRNA and tRNA genes is not affected by L29. A, knockdown of L29 does not affect the expression of the 5 S rRNA, tRNALeu, and tRNATyr genes. U2OS cells were transfected with scrambled or L29 siRNA. The levels of the 5 S rRNA, tRNALeu, and tRNATyr in the cells were analyzed by using RT-qPCR. The knockdown efficiency of L29 mRNA is shown on the right. B, overexpression of L29 does not affect the binding of TRRAP to the 5 S rRNA, tRNALeu, and tRNATyr genes. H1299 cells transfected with control or FLAG-L29 plasmid were subjected to ChIP-qPCR assays using goat anti-TRRAP or control goat IgG, followed by detection of the 5 S rRNA, tRNALeu, and tRNATyr genes. The expression of FLAG-L29 is shown on the right using IB with anti-FLAG antibodies. Error bars indicate mean ± S.D.
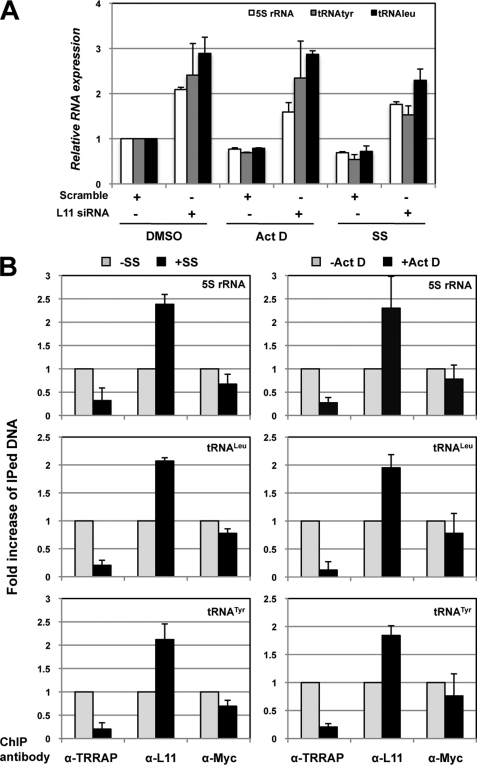
L11 Is Required for Ribosomal Stress-induced Reduction of the 5 S rRNA and tRNA Genes
To analyze the physiological significance of L11 inhibition of c-Myc-dependent transcription of the 5 S rRNA and tRNA genes, we examined whether L11 plays a role in the expression of these genes in response to ribosomal (nucleolar) stress, as L11 has been shown to mediate p53 activation in response to such stress induced by a low dose of Act D and serum starvation (24, 25). Initially, we found that treatment of cells with Act D and serum starvation reduced the expression of the 5 S rRNA, tRNALeu, and tRNATyr genes by ∼25–40% (statistically significant), as determined by RT-qPCR assays (the columns with scrambled siRNAs in Fig. 6A). Interestingly, this reduction was dependent on L11 because knockdown of L11 abolished the reduction of these genes by Act D and serum starvation (the columns with L11 siRNA in Fig. 6A). These results suggest that L11 regulation of c-Myc-mediated expression of the 5 S rRNA and tRNA genes is responsive to ribosomal stress.
FIGURE 6.
L11 is required for the regulation of the 5 S rRNA and tRNA genes in response to ribosomal stress. A, L11 is required for the reduction of the 5 S rRNA, tRNALeu, and tRNATyr genes in response to the treatment of cells with Act D or serum starvation. U2OS cells were transfected with scrambled or L11 siRNA followed by treatment with Act D (5 nm) for 12 h or serum starvation (SS) for 24 h before harvesting. The cells were subjected to RT-qPCR assays to detect the relative expression of the 5 S rRNA and tRNATyr genes. B, the binding of L11 to the 5 S rRNA, tRNALeu, and tRNATyr genes inversely correlates with that of TRRAP in response to Act D treatment or serum starvation. U2OS cells were cultured in 0.2% fetal calf serum-containing medium for 24 h or treated with 5 nm Act D for 12 h. The cells were subjected to ChIP assays using anti-L11, anti-c-Myc, or anti-TRRAP antibodies followed by qPCR detection of the 5 S rRNA, tRNALeu, and tRNATyr genes. Error bars indicate mean ± S.D.
L11 Binding to the 5 S rRNA and tRNA Genes Displays a Reverse Profile to That of TRRAP Binding to These Genes in Response to Ribosomal Stress
To determine how L11 plays a role in c-Myc regulation of the 5 S rRNA and tRNA gene expression in response to ribosomal stress, we performed ChIP-qPCR assays to test the occupancy of L11 and TRRAP at these genes following Act D treatment or serum starvation. As shown in Fig. 6B, TRRAP binding to the 5 S rRNA, tRNALeu, and tRNATyr genes was all significantly reduced in response to Act D treatment for 12 h or serum starvation for 24 h. By contrast, the association of L11 with these genes was increased (Fig. 6B). Of note, Act D treatment did not significantly affect c-Myc binding to the aforementioned genes, whereas serum starvation slightly reduced (<2-fold) the binding of c-Myc to these genes. In contrast, TRRAP binding to these genes was dramatically decreased (up to 5-fold) (Fig. 6B), suggesting that the reduction of TRRAP recruitment to above RNA Pol III-catalyzed genes was not merely due to the reduction of c-Myc binding in the serum-starved cells. Taken together, these results indicate that L11 competes with TRRAP for binding to the 5 S rRNA and tRNA genes in response to ribosomal stress, further supporting the role of L11 in inhibition of c-Myc-driven transactivational activity toward the RNA Pol III-catalyzed transcription of the 5 S rRNA and tRNA genes by RNA Pol III.
DISCUSSION
We have previously shown that L11 regulates c-Myc-mediated transcription of the pre-rRNA and several other genes by RNA Pol I and II, respectively (18). In this study, we show that L11 also regulates c-Myc-mediated transcription of the 5 S rRNA and tRNA genes by RNA Pol III. L11 binds to c-Myc and inhibits the recruitment of TRRAP at the 5 S rRNA and tRNA genes (Figs. 2–4). Knocking down L11 enhances c-Myc-dependent transcription of these genes (Fig. 1). Together with our previous findings (18), these results suggest that L11 can regulate c-Myc-mediated ribosomal biogenesis, forming a tight coordination between c-Myc and ribosomal biogenesis because the L11 gene itself is a transcriptional target of c-Myc (18).
L11 competes with TRRAP for binding to c-Myc at the 5 S rRNA and tRNA genes, a mechanism utilized by L11 to inhibit c-Myc-mediated transcription by Pol I and Pol II of genes that contain E-box elements at their promoters (18). Because no E-box elements exist in the 5 S rRNA and tRNA gene promoters, it remains unclear how c-Myc associates with these genes. One possibility could be that c-Myc may associate with these genes through transcriptional machineries, such as TFIIIB (16), rather than the E-box elements and then recruit its co-activator(s), such as TRRAP, to enhance the transcription of the target genes (19). Our current results support the notion that c-Myc associates with the 5 S rRNA and tRNA genes. Also, our study shows that L11 binding to c-Myc results in marked decrease of TRRAP binding to these genes and consequent reduction of their expression. Moreover, L11 regulates c-Myc levels because knockdown of endogenous L11 results in drastic increase of c-Myc protein (Fig. 1) and mRNA levels (26), although the underlying mechanisms still remain unclear. Thus, it appears that L11 regulates c-Myc-mediated transcription of the 5 S rRNA and tRNA genes via regulation of both the activity and the level of c-Myc (Fig. 7). By contrast, L29, another ribosomal protein from the 60 S ribosome subunit, does not bind to c-Myc (18). Knockdown of L29 neither affects the levels of c-Myc (18) nor induces the expression of target genes. Also, L29 does not compete with TRRAP for binding to these target gene promoters (18) (this study). Together, these results suggest that the regulation of c-Myc by L11 is specific.
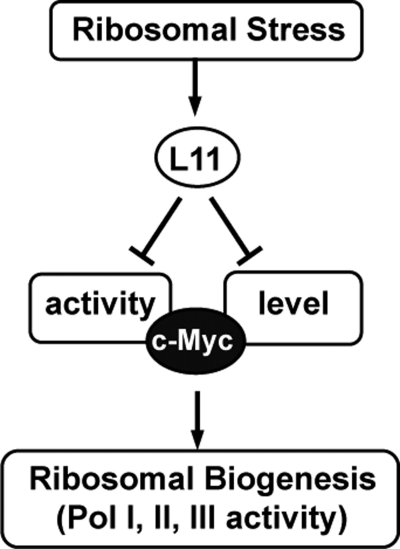
FIGURE 7.
A schematic diagram for the action of L11 in coordinating c-Myc activity with ribosomal biogenesis. In normal growth conditions, c-Myc enhances ribosomal biogenesis through up-regulating Pol I, II, and III activity. In response to ribosomal stress, L11 binds to c-Myc and inhibits TRRAP binding to target gene promoters. Meanwhile, L11 also negatively regulates the level of c-Myc, leading to the inhibition of c-Myc-mediated transcription and ribosomal biogenesis.
Because up-regulation of the 5 S rRNA and tRNA gene transcription by overexpression of Pol III-specific transcription factor Brf1 or even overexpression of Pol III-transcribed tRNAiMet alone can induce oncogenic transformation in cells and induce tumors in mice (20), our results showing the inhibition of c-Myc-mediated 5 S rRNA and tRNA expression by L11 also imply that L11 may function to suppress cell transformation and tumorigenesis mediated by aberrant overexpression of c-Myc. This would be a tempting topic for future exploration.
Interestingly, we also observe that L11 suppresses c-Myc activity in response to ribosomal stress. We show that ribosomal stress caused by treatment of cells with a low dose of Act D or serum deprivation reproducibly decreased the level of the 5 S rRNA and tRNA transcription, and this reduction is dependent on L11, because knockdown of L11 abolished ribosomal stress-induced reduction of the expression of these genes. Furthermore, the elevation of L11 binding to the 5 S rRNA and tRNA genes was well correlated with the decline of TRRAP binding to these genes in response to ribosomal stress (Fig. 6). It has been proposed that ribosomal stress can increase the free form of L11 that is dissociated from the large ribosome complex and then released to the nucleoplasm, where it can target c-Myc (24), leading to a decrease in TRRAP binding at target gene promoters. Also, in response to ribosomal stress, c-Myc binding to the aforementioned genes was slightly decreased, particularly in serum-starved cells (Fig. 6B), and the level of c-Myc was decreased (data not shown), suggesting that regulation of c-Myc level by L11 may also contribute to the c-Myc inhibition by this ribosomal protein in response to ribosomal stress (Fig. 7). Future studies are necessary to determine whether L11 can suppress c-Myc activity in response to other types of stress or physiological signals and to determine how L11 is activated in these conditions.
In summary, our study supports a critical and general role for L11 in the feedback regulation of c-Myc activity that is involved in transcription catalyzed by all three RNA Pols. By doing so, L11 may serve as a molecule coordinator between c-Myc and ribosomal biogenesis specifically (Fig. 7) and/or between cell growth and ribosomal biogenesis in general. This feedback regulation may also act as an intrinsic mechanism or surveillance for controlling cell transformation and tumorigenesis because defects of this pathway could lead to deregulated c-Myc activity and tumorigenesis. Recently, it was shown that haploinsufficiency of L11 and other ribosomal proteins, due to loss of function gene mutations or deletions, occurs in patients with Diamond-Blackfan anemia (27, 28), an inherited disorder with bone marrow failure and increased incidence of cancer formation (29). It will be interesting to test if c-Myc activity is enhanced and contributes to cancer susceptibility in this disorder in the near future. Finally, because L11 has been also shown to activate p53 by repressing MDM2 activity (22, 24, 25, 30), it is thus certain that L11 is crucial for monitoring cell growth and proliferation by up-regulating the p53 activity and down-regulating the c-Myc activity. Dissecting how a cell may coordinate the L11 regulation of these two independent pathways in response to ribosomal stress would be another enticing project to pursue.
This work was supported, in whole or in part, by National Institutes of Health Grant K99-CA124137 (to M.-S. D.) and Grants CA93614, CA095441, CA079721, and CA129828 (to H. L.). This work was also supported by startup funds from Oregon Health and Science University (to M.-S. D.).
- Pol
- polymerase
- Act D
- actinomycin D
- IB
- immunoblot
- IP
- immunoprecipitation
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription
- qPCR
- quantitative PCR
- siRNA
- small interfering RNA
- TRRAP
- transformation/transcription domain-associated protein.
REFERENCES
- 1.Eilers M., Eisenman R. N. (2008) Genes Dev. 22, 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer N., Penn L. Z. (2008) Nat. Rev. Cancer 8, 976–990 [DOI] [PubMed] [Google Scholar]
- 3.Nesbit C. E., Tersak J. M., Prochownik E. V. (1999) Oncogene 18, 3004–3016 [DOI] [PubMed] [Google Scholar]
- 4.Felsher D. W., Bishop J. M. (1999) Mol. Cell 4, 199–207 [DOI] [PubMed] [Google Scholar]
- 5.Pelengaris S., Khan M., Evan G. I. (2002) Cell 109, 321–334 [DOI] [PubMed] [Google Scholar]
- 6.Pelengaris S., Littlewood T., Khan M., Elia G., Evan G. (1999) Mol. Cell 3, 565–577 [DOI] [PubMed] [Google Scholar]
- 7.Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. (1985) Nature 318, 533–538 [DOI] [PubMed] [Google Scholar]
- 8.Oskarsson T., Trumpp A. (2005) Nat. Cell Biol. 7, 215–217 [DOI] [PubMed] [Google Scholar]
- 9.Menssen A., Hermeking H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6274–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coller H. A., Grandori C., Tamayo P., Colbert T., Lander E. S., Eisenman R. N., Golub T. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3260–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boon K., Caron H. N., van Asperen R., Valentijn L., Hermus M. C., van Sluis P., Roobeek I., Weis I., Voûte P. A., Schwab M., Versteeg R. (2001) EMBO J. 20, 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Q. M., Malek R. L., Kim S., Chiao C., He M., Ruffy M., Sanka K., Lee N. H., Dang C. V., Liu E. T. (2000) Cancer Res. 60, 5922–5928 [PubMed] [Google Scholar]
- 13.Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. (2005) Nat. Cell Biol. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 14.Grewal S. S., Li L., Orian A., Eisenman R. N., Edgar B. A. (2005) Nat. Cell Biol. 7, 295–302 [DOI] [PubMed] [Google Scholar]
- 15.Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., Larsson L. G., Wright A. P. (2005) Nat. Cell Biol. 7, 303–310 [DOI] [PubMed] [Google Scholar]
- 16.Gomez-Roman N., Grandori C., Eisenman R. N., White R. J. (2003) Nature 421, 290–294 [DOI] [PubMed] [Google Scholar]
- 17.Ruggero D., Pandolfi P. P. (2003) Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 18.Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007) EMBO J. 26, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenneth N. S., Ramsbottom B. A., Gomez-Roman N., Marshall L., Cole P. A., White R. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14917–14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall L., Kenneth N. S., White R. J. (2008) Cell 133, 78–89 [DOI] [PubMed] [Google Scholar]
- 21.Dai M. S., Sun X. X., Lu H. (2008) Mol. Cell. Biol. 28, 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai M. S., Shi D., Jin Y., Sun X. X., Zhang Y., Grossman S. R., Lu H. (2006) J. Biol. Chem. 281, 24304–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Mol. Cell. Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat K. P., Itahana K., Jin A., Zhang Y. (2004) EMBO J. 23, 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Mol. Cell. Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai M. S., Sears R., Lu H. (2007) Cell Cycle 6, 2735–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cmejla R., Cmejlova J., Handrkova H., Petrak J., Petrtylova K., Mihal V., Stary J., Cerna Z., Jabali Y., Pospisilova D. (2009) Hum. Mutat. 30, 321–327 [DOI] [PubMed] [Google Scholar]
- 28.Gazda H. T., Sheen M. R., Vlachos A., Choesmel V., O'Donohue M. F., Schneider H., Darras N., Hasman C., Sieff C. A., Newburger P. E., Ball S. E., Niewiadomska E., Matysiak M., Zaucha J. M., Glader B., Niemeyer C., Meerpohl J. J., Atsidaftos E., Lipton J. M., Gleizes P. E., Beggs A. H. (2008) Am. J. Hum. Genet. 83, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai M. S., Lu H. (2008) J. Cell. Biochem. 105, 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]