Abstract
Genomic integration of human papillomavirus (HPV) DNA accounts for more than 90% of cervical cancers. High-risk genital HPVs encode E6 proteins that can interact with a cellular ubiquitin ligase E6-associated protein (E6AP) and target the tumor suppressor p53 for ubiquitin-mediated proteolysis. Currently, how this critical event is regulated is largely unknown. Here we report that activating transcription factor 3 (ATF3), a broad DNA damage sensor whose expression is frequently downregulated in cervical cancer, interacted with E6 and prevented p53 from ubiquitination and degradation mediated by the viral protein. Consistent with its role as a potent E6 antagonist, ATF3 expressed enforcedly in HPV-positive SiHa cells activated p53, leading to expression of p53-target genes (e.g. p21 and PUMA), cell cycle arrest and apoptotic cell death. The leucine zipper domain of ATF3 appears indispensable for these effects as an ATF3 mutant lacking this domain failed to interact with E6 and activate p53 in the cervical cancer cells. The prevention of p53 degradation was unlikely caused by binding of ATF3 to the tumor suppressor, but rather was a consequence of disruption of the E6-E6AP interaction by ATF3. These results indicate that ATF3 plays a key role in a mechanism defending against HPV-induced carcinogenesis, and could serve as a novel therapeutic target for HPV-positive cancers.
Keywords: Apoptosis, p53, Protein Degradation, Tumor Suppressor, Ubiquitination, ATF3, E6, E6AP, HPV, Cervical Cancer
Introduction
Human papillomavirus (HPV)3 infection is a major risk factor for cervical cancer (1, 2). Genomic integration of HPV DNA, occurring in more than 90% of cervical cancers, results in expression of a viral protein E6, which in turn inactivates the tumor suppressor p53 by driving its proteolysis (3). The E6 protein can bind to p53 at both the C terminus and the central DNA-binding region (4) and recruit a cellular protein E6AP to p53 (5, 6). E6AP belongs to the HECT family of E3 ubiquitin ligases and can catalyze the addition of ubiquitin moieties to p53, leading to its degradation by the 26 S proteasomes (6). Because the p53 gene is rarely mutated in cervical cancer, the E6-mediated degradation serves as the major mechanism inactivating p53 and promoting cervical carcinogenesis (7).
In addition to p53 and E6AP, the E6 protein interacts with many other cellular proteins including Bak (8), Bax (9), CBP/p300 (10), and BRCA1 (11), presumably through the four CXXC motifs sparsely distributed in the viral protein (12). These interactions account for the oncogenic activities of E6, which include not only promoting transformation but also enhancing cell proliferation and survival (13). Therefore, it is important to dissect the E6 interaction network for a better understanding of the molecular basis for cervical cancer and identification of therapeutic targets for the disease.
ATF3 is a member of the ATF/CREB family of transcription factors and can be rapidly induced by DNA damage and other oncogenic stimuli (14, 15). Whereas consequences of ATF3 induction are unclear, it is often assumed that ATF3 functions as a transcription factor to regulate gene expression thereby contributing to cellular responses to oncogenic stresses (14, 15). However, emerging evidence indicates that this DNA-binding protein can also regulate cellular functions through mechanisms beyond transcriptional regulation (16). ATF3 contains a central leucine zipper domain (Zip) that is well characterized as a mediator of protein-protein interaction (17). ATF3 binds to p53 via this domain, and as a consequence, p53 ubiquitination catalyzed by MDM2, the major ubiquitin ligase in HPV-negative cells (18, 19), is blocked, leading to up-regulation of the p53 tumor suppressor activity independent of the ATF3 transcriptional activity (16). These findings are consistent with recent reports that ATF3 expression is down-regulated in a wide range of human cancers (20–22) including cervical cancer (23), arguing for a link between ATF3 expression and tumor suppression (23). In line with this notion, we report here the identification of ATF3 as a novel E6 repressor that can compete with E6AP for binding to the viral protein thereby activating p53 in HPV-positive cervical cancer cells. These results indicate that ATF3 could contribute to suppression of HPV-induced carcinogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
H1299 were cultured in RPMI1640 medium while SiHa, CaSki, and HT1080 cells were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotics. Transfections were performed with Lipofectamine 2000 (Invitrogen) or Fugene 6.0 (Roche) according to the manufacturers' recommendations. The latter transfection reagent exerted no toxicity to SiHa and CaSki cells, and therefore was used for apoptosis assays. Cells were harvested 48 h after transfections for immunoblotting or luciferase reporter assays.
Protein Purification and in Vitro Translation
Histidine-tagged ATF3 and Δ102 proteins were expressed in Escherichia coli BL21 cells, and purified with Ni+-nitrilotriacetic acid-agarose (Invitrogen) as described previously (16). For in vitro translation, the coding sequences of ATF3, p53, E6, and E6AP were cloned into pcDNA3.1 at downstream of a T7 promoter. In vitro translation was performed using the TNT Quick-coupled Transcription/Translation System (Promega) following the manufacturer's protocol. Briefly, 1 μg of plasmids were incubated with 40 μl of rabbit reticulocytes lysates supplemented with 20 μm methionine or 2 μl of [35S]methionine (1,000 Ci/mmol, PerkinElmer) at 30 °C for 90 min.
GST-pulldown Assays
The coding sequence for HPV16 E6 was PCR amplified using the genomic DNA prepared from SiHa cells as template and cloned into pGEX-3X (Amersham Biosciences). The plasmid expressing the GST-ATF3 fusion protein was described previously (16). These plasmids were transformed into E. coli BL21 strain, and expression of GST (glutathione S-transferase) or GST fusion proteins was induced by isopropyl-1-thio-β-d-galactopyranoside. Bacterial pellets were lysed in TENT buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 2 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40, and 1% Blotto) with mild sonication. For pulldown assays, lysates containing 1 μg of GST or GST fusion proteins were immobilized on glutathione-agarose (Sigma) for 1 h at room temperature. After washes, the agarose was resuspended in Buffer I (20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 2 mm EDTA, 2 mm DTT, 1 mg/ml BSA, 5% glycerol, and 0.1% Nonidet P-40) and then incubated with purified or in vitro-translated proteins at 4 °C for 4 h or overnight. After extensive washes with Buffer II (Buffer I with 100 mm NaCl), bound proteins were eluted, resolved with SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by immunoblotting or autography as described previously (24).
In Vitro p53 Degradation and Ubiquitination Assays
These were performed essentially as described with modifications (25, 26). For degradation assays, E6 was preincubated with in vitro-translated ATF3 (at a ratio of 1:4) or the same amount of reticulocyte lysates programmed with the empty vector for 30 min at room temperature, followed by added in a reaction mixture (15 μl) containing 35S-labeled p53 (in vitro-translated, at a ratio to E6 of 1:1), 3 μl of rabbit reticulocyte lysates and the degradation buffer (25 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 3 mm DTT). 60 min after incubations, the reaction mixtures were mixed with 2× SDS loading buffer, resolved in SDS-PAGE, and subjected to autography. For E6-mediated p53 ubiquitination assays, 35S-labeled E6 was similarly incubated with ATF3 and p53, and 20 μg of methylated ubiquitin (Boston Biochem) was added into the reaction mixtures to accumulate ubiquitinated p53 proteins for easy detection (26). Modified p53 proteins were visualized by autography.
shRNA Knockdown and Retroviral Infections
p53 knockdown was performed using a Lentivector-based shRNA system (pSIH-H1 shRNA Cloning and Lentivector Expression system, System Biosciences) according to the manufacturer's recommendation. The p53-targeted sequence was 5′-GAC TCC AGT GGT AAT CTA C-3′, based on a publication (27). The ATF3-targeted sequences were 5′-GCA AAG TGC CGA AAC AAG A-3′ and 5′-GAG AAA CCT CTT TAT CCA A-3′, based on siRNA used in one of our earlier reports (16). For negative controls, a luciferase-targeted sequence (5′-CTT ACG CTG AGT ACT TCG A-3′) was cloned into the Lentivector. For retroviral-mediated gene transfer, the ATF3-coding sequence was cloned into pBabe-Neo, and transfected into Ampho293 (Clontech) to pack retrovirions. Supernatants were then collected and used to infect SiHa and CaSki cells as described previously (16). The retroviral vector expressing E6 shRNA was kindly provided by T. Kiyono (28).
Immunoblotting
This was performed as described previously (29). Antibodies for p53 (DO-1) and ATF3 (C-19) were purchased from Santa Cruz Biotechnology. Antibodies for p21, c-Myc, and PUMA were from BD Pharmingen, Invitrogen, and ProSci, respectively.
Flow Cytometry
SiHa cells were transfected with GFP or GFP-ATF3 with Fugene 6.0 for 2 days, and fixed with 1% formaldehyde at 4 °C for 1 h. Cells were then permeabilized with cold 70% ethanol overnight, stained with a solution containing 50 μg/ml propidium iodide (Sigma) and 20 μg/ml RNase A at 37 °C for 20 min, and subjected to flow cytometry analysis as described previously (24). The FlowJo software was used to calculate percentages of GFP-positive cells in each cell cycle phase.
TUNEL Staining
Apoptotic cells were labeled with In Situ Cell Death Detection Kit TMR Red (Roche) according to the manufacturer's protocol. Briefly, SiHa or CasKi cells cultured on coverslips were transfected with GFP, GFP-ATF3, or GFP-IRES-ATF3, or infected with ATF3-expressing retroviruses, for 3 days, and fixed with 4% paraformaldehyde for 1 h. After permeabilization, the cells were incubated with 50 μl of reaction mixture containing the labeling enzyme and the TMR red labeled-dUTP at 37 °C for 1 h. After extensive washes, cells were counterstained with DAPI and observed under a fluorescence microscope. For quantitation, at lease 300 GFP-positive cells (for transfections) or infected cells (for retroviral infections) were randomly chosen and the numbers of TUNEL-positive cells were counted.
Colony Formation Assays
SiHa cells were infected with lentiviruses carrying shp53 or shLuc for 3 days, followed by infections with retroviruses expressing ATF3 or the empty vector for 2 days. The cells were then plated in 6-well plates (200 cells/well) and stained with crystal violet after 14 days of incubation as described previously (16).
RESULTS
ATF3 Directly Binds to E6
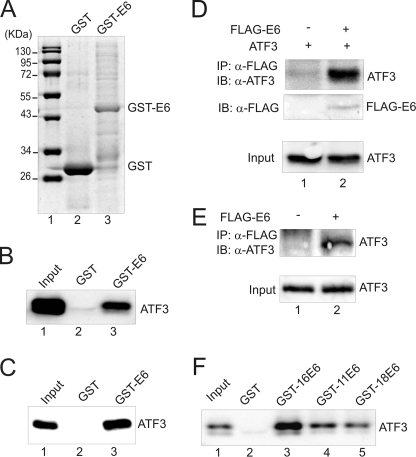
Our recent observation that ATF3 expression is down-regulated in cervical cancers (23) prompted us to explore a possibility that ATF3 could be involved in regulation of cervical carcinogenesis. We therefore sought to determine whether ATF3 regulates the oncogenic activities of HPV proteins. Given that both ATF3 and E6 proteins contain motifs that can mediate protein-protein interaction, we tested whether these two proteins could interact. We thus cloned the E6 gene from the genomic DNA of HPV-positive SiHa cells using PCR, and fused it with a GST-coding sequence. The recombinant DNA was introduced into E. coli and expressed a fusion protein with an anticipated molecular weight (Fig. 1A, lane 3). The recombinant protein immobilized on glutathione-agarose was then incubated with ATF3 prepared by in vitro translation for GST-pulldown assays. The results showed that the GST-E6 protein, but not GST, was able to pull down ATF3 (Fig. 1B, lane 3 versus lane 2), indicating an association between ATF3 and the viral protein. The ATF3-E6 interaction was likely caused by direct protein-protein binding, because the immobilized E6 protein, but not GST, could also pull down a recombinant ATF3 protein (Fig. 1C, lane 3) purified with Ni2+-nitrilotriacetic acid chromatography (supplemental Fig S1B, lane 1). Moreover, ATF3 could be immunoprecipitated by the FLAG-antibody in the presence of a FLAG-tagged E6 protein that was co-expressed with ATF3 in H1299 cells null for both E6 and p53 (Fig. 1D, lane 2), indicating that the ATF3-E6 interaction occurred in vivo as well. The endogenous E6 protein cannot be detected by immunoblotting (10, 30) and thus no assay is available to examine the interaction of the endogenous proteins. However, we found that the anti-FLAG antibody could precipitate the endogenous ATF3 protein when the FLAG-E6 protein was expressed in HCT116 cells that are null for p53 (Fig. 1E, lane 2), suggesting that the endogenous proteins likely interact. Interestingly, in addition to the E6 protein derived from HPV16-positive SiHa cells, ATF3 also bound to the E6 proteins derived from HPV11 and HPV18 (Fig. 1F, lanes 4 and 5), a result which was anticipated given that the primary structures of these E6 proteins are strikingly similar (12). Of note, the difference in the amounts of ATF3 down-pulled by these E6 proteins was more likely due to the variation in the amounts of the viral proteins immobilized on the glutathione-agarose (supplemental Fig S1B, lanes 3–5).
FIGURE 1.
ATF3 directly binds to E6. A, GST-E6 and GST proteins eluted from glutathione-agarose were subjected to SDS-PAGE and stained with Coomassie Blue. B, immobilized GST-E6 or GST protein was incubated with in vitro-translated ATF3 protein at 4 °C overnight. After extensive washes, bound proteins were eluted and subjected to immunoblotting with an ATF3 antibody. Lane 1 represents 10% of total input protein. C, immobilized GST-E6 or GST was incubated with 200 ng of purified ATF3 protein followed by immunoblotting to visualize bound proteins. The input lane represents 10% of total input protein. D, ATF3 was expressed in H1299 cells with or without FLAG-E6 by transfections. Cell lysates were immunoprecipitated by agarose conjugated with an anti-FLAG antibody, and bound proteins were eluted for immunoblotting as indicated. E, HCT116 p53−/− cells were transfected with the plasmid encoding FLAG-E6. After treated with 25 μm MG132, cells were subjected to immunoprecipitations as in D to examine the interaction of FLAG-E6 with the endogenous ATF3 protein. F, GST-E6 proteins derived from HPV11 (GST-11E6), HPV16 (GST-16E6), and HPV18 (GST-18E6) were immobilized on glutathione-agarose, and incubated with in vitro-translated ATF3 proteins for GST-pulldown assays as in B.
ATF3 Blocks E6-mediated p53 Degradation and Ubiquitination
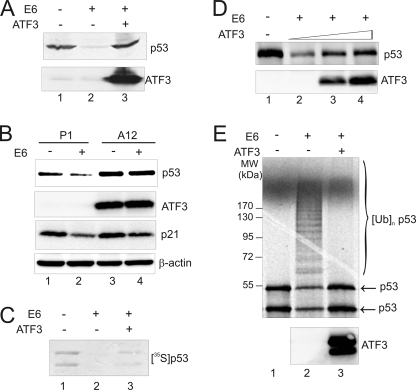
One major oncogenic activity of E6 is to drive ubiquitin-dependent degradation of p53. Intrigued by the finding that ATF3 directly bound to E6, we determined whether ATF3 could affect E6-mediated p53 degradation. We thus co-expressed p53, E6 and/or ATF3 in H1299 cells, and measured p53 levels using immunoblotting. Whereas E6 largely reduced the p53 level as expected (3) (Fig. 2A, lane 2), the presence of ATF3 abolished this effect (Fig. 2A, lane 3). Because expression of both p53 and E6 was directed by a constitutively active CMV promoter that is refractory to regulation by ATF3 (16), these results strongly suggest that ATF3 suppressed E6-mediated p53 degradation. Indeed, while enforced E6 expression decreased the endogenous p53 level in a cell line (HT1080) carrying a functional p53 protein (Fig. 2B, lane 2), it failed to do so in the A12 clone stably expressing ATF3 and developed in our previous studies (24) (Fig. 2B, lane 4). Of note, the observation that the ATF3-expressing clone expressed high level of p53 (Fig. 2B, lane 3 versus lane 1) was consistent with our previous report that ATF3 can stabilize p53 in HPV-negative cells (16). To corroborate these important findings, we took use of a well-established in vitro p53 degradation assay (25) to further study the effects of ATF3 on E6-mediated p53 degradation. This assay utilizes rabbit reticulocyte lysates that contain components required for p53 ubiquitination and degradation, and the presence of E6 in the assay system effectively promoted p53 degradation (Fig. 2C, lane 2) as expected (25). Consistent with Fig. 2A, ATF3 strongly suppressed E6-mediated decrease of the p53 level (Fig. 2C, lane 3 versus lane 2), and this effect was in a dose-dependent manner (Fig. 2D). Measurements of p53 degradation rates by immunoblotting confirmed that ATF3 indeed decrease the degradation rate of p53 (supplemental Fig. S2). Because E6-mediated p53 degradation is mainly caused by ubiquitination (12), we determined effects of ATF3 on p53 ubiquitination using an in vitro assay that is similar to the above-mentioned degradation assay (Fig. 2C) but employs methylated ubiquitin to block degradation and accumulate mono-ubiquitinated proteins for ready detection (26). Indeed, while E6 promoted p53 ubiquitination evident by the appearance of an array of slowly migratory bands (Fig. 2E, lane 2), the presence of ATF3 in the assay system almost completely abolished the modifications of p53 (Fig. 2E, lane 3). These results argue for a notion that ATF3 blocks E6-mediated p53 ubiquitination thereby preventing degradation of the tumor suppressor.
FIGURE 2.
ATF3 prevents p53 from E6-mediated degradation and ubiquitination. A, H1299 cells were co-transfected with p53, E6, RSV-Luc, and ATF3 as indicated. 48 h after transfections, cell lysates were normalized against luciferase activity (as transfection efficiency control) and subjected to immunoblotting for p53 and ATF3 expression. B, HT1080 cells stably expressing ATF3 (A12) or its empty vector (P1) (24) were transfected with E6 for 2 days, and lysed for immunoblotting analysis. C, E6 protein was preincubated with (lane 3) or without ATF3 (lane 2) at 4 °C for 30 min, and then added into a reaction mixture containing 35S-labeled p53. The reactions were terminated after 60 min, and p53 was visualized by autography. D, E6 was preincubated with increasing amounts of ATF3, and mixed with in vitro-translated p53 for 60 min. The p53 levels were measured by immunoblotting using the DO-1 antibody. E, E6 protein preincubated with (lane 3) or without ATF3 (lane 2) was incubated with 35S-labeled p53 in the presence of 20 μg of methylated ubiquitin for 60 min followed by SDS-PAGE and autography.
ATF3 Activates p53 in HPV-positive Cells
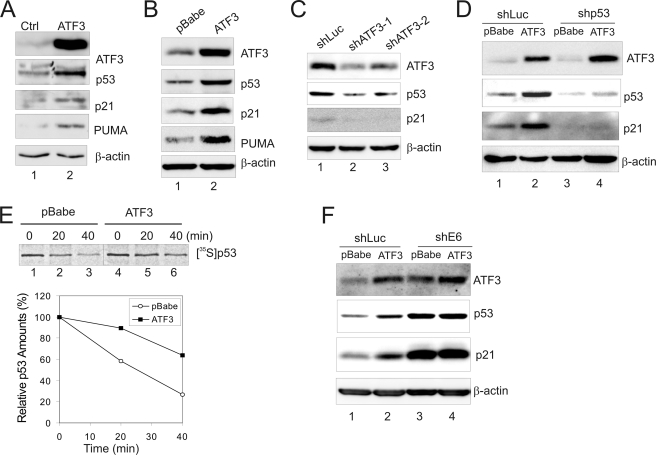
Given that p53 inactivation in HPV-positive cervical cancer cells is completely dependent on E6 (7), we next determined whether ATF3 could restore p53 activity in these cells. We thus expressed ATF3 bicistronically with GFP in HPV-positive SiHa cells, and sorted GFP-expressing cells (i.e. transfected cells) for immunoblotting to determine expression of p53 and its target genes. The results show that ectopic ATF3 expression increased the p53 level in SiHa cells (Fig. 3A). Importantly, expression of p21 and PUMA, two representative p53-target genes, was largely induced by ATF3 (Fig. 3A). Similar results were obtained when ATF3 was expressed in SiHa and CaSki cells through retrovirus-mediated gene transfer (Fig. 3B, supplemental Fig. S3A). Consistent with these results, two independent short-hairpin RNA (shRNA) that could down-regulate ATF3 expression by more than 60% significantly decreased the levels of p53 and p21 in SiHa cells (Fig. 3C). Interestingly, knockdown of p53 expression by shRNA expressed via a lentiviral vector almost completely abolished p21 expression induced by ATF3 in the cells (Fig. 3D, lanes 3–4 versus lanes 1–2), suggesting that the induction of p53-target gene expression was a direct consequence of p53 activation by ATF3. These results thus demonstrate that ATF3 could activate p53 in HPV-positive cells, an effect likely achieved through a mechanism that prevents E6-mediated degradation of the tumor suppressor. Indeed, by measuring the p53 half-life using pulse-chase assays, we confirmed that the half-life of p53 was significantly increased by ectopic expression of ATF3 in SiHa cells (Fig. 3E). Moreover, knockdown of E6 expression using E6 shRNA (28) diminished ATF3-induced stabilization and activation of p53 in HPV-positive SiHa cells (Fig. 3F).
FIGURE 3.
ATF3 activates p53 in HPV-positive cancer cells. A, SiHa cells were transfected with GFP (lane 1) or GFP-IRES-ATF3 (lane 2) for 2 days. The sorted GFP-positive cells were lysed for immunoblotting. B, SiHa cells were infected with retroviruses expressing ATF3 (lane 2) or its vector pBabe (lane 1) for 2 days followed by immunoblotting. C, SiHa cells were infected with lentiviruses expressing two independent shRNA targeting ATF3 (lanes 2 and 3) or luciferase (lane 1) for 3 days followed by immunoblotting. D, SiHa cells were sequentially infected with lentiviruses expressing shRNA specific to p53 (shp53) or luciferase (shLuc), and retroviruses expressing ATF3 or its vector as indicated. Infected cells were lysed and subjected to immunoblotting. E, SiHa cells infected with retroviruses expressing ATF3 or pBabe were labeled with Tran[35S] for 1 h and lysed at indicated time. p53 was immunoprecipitated with the DO-1 antibody and visualized by autography. The graph shows results of densitometric analysis. F, SiHa cells were infected sequentially with retroviruses expressing ATF3 and E6-targeted shRNA, and then lysed for immunoblotting as indicated.
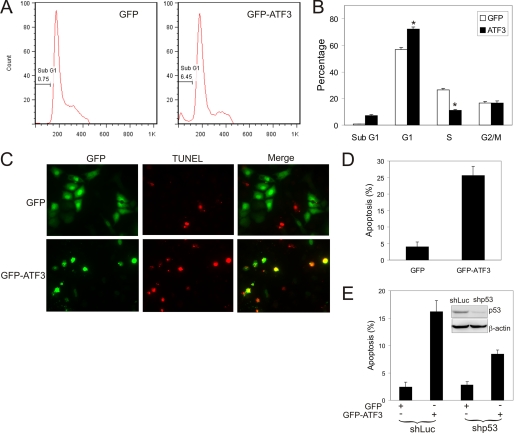
ATF3 Promotes p53-mediated Cell Cycle Arrest and Apoptosis in Cervical Cancer Cells
The major consequences of p53 activation include cell cycle arrest and apoptotic cell death due to activation of expression of cell cycle checkpoint proteins (e.g. p21) and pro-apoptotic genes (e.g. PUMA) (31). To determine the consequences of ATF3-induced p53 activation, we expressed a GFP-ATF3 fusion protein or GFP (as a control) in SiHa cells for 2 days, and subjected transfected (GFP-positive) cells to flow cytometry for cell cycle analysis. While ectopic expression of ATF3 significantly increased the number of cells in the G0/G1 phase, there were fewer ATF3-expressing cells staying in the S phase as compared with GFP-expressing cells (Fig. 4, A and B), suggesting that ATF3 activated p53 leading to inhibition of cell cycle progression. Most importantly, we found that the number of ATF3-expressing sub-G0/G1 cells that were mostly constituted by late-stage apoptotic cells was largely increased (Fig. 4, A and B), suggesting that ATF3 expression could result in apoptotic cell death. Because induction of apoptosis is one of the major mechanisms underlying the p53 tumor suppressor function in vivo (32), we further characterized the effects of ATF3 on apoptosis of HPV-positive cells. We expressed GFP-ATF3 or GFP in SiHa cells for 3 days, and stained apoptotic cells using TUNEL assays. In contrast to flow cytometry, TUNEL assays detect both early and late apoptotic cells and thus would more accurately measure apoptosis rates. Consistent with Fig. 4, A and B, while very few GFP-expressing cells were stained by TUNEL, GFP-ATF3 expression increased the percentage of apoptotic (TUNEL-positive) cells to more than 25% (Fig. 5, C and D), demonstrating that ATF3 indeed induced the HPV-positive cells to undergo apoptosis. This effect was not merely a consequence of the fusion protein, because a native ATF3 protein bicistronically expressed with GFP also increased the number of TUNEL-positive cells (supplemental Fig. S4). Moreover, ectopic expression of ATF3 via a retroviral vector also promoted SiHa cells to apoptosis (Fig. 6E), but with a reduced efficacy likely due to inefficient transduction of SiHa cells by the retroviruses. Interestingly, retrovirus-mediated ATF3 expression also induced apoptosis in CaSki cells (supplemental Fig. S3, B and C), which contains more than 600 copies of HPV DNA and presumably expresses a high level of E6, indicating that ATF3 could actively promote apoptosis in other HPV-positive cells. To confirm that the ATF3-induced apoptosis was due to p53 activation, we knocked down p53 expression in SiHa cells using shRNA (Fig. 4E, inset) and subjected the cells to TUNEL assays. Knockdown of p53 expression alone had no effect on apoptosis, likely due to the fact that the cells have adapted to a growth condition where p53 expresses at the basal level. However, p53 down-regulation significantly impaired the activity of ATF3 to induce apoptosis (Fig. 4E), suggesting that the ATF3-induced apoptosis was mediated, at least in part, by activation of the p53 pro-apoptotic activity. The incomplete suppression of apoptosis by the p53 shRNA might be due to the residue p53, effects of ATF3 on p53-independent E6 functions (e.g. repression of pro-apoptotic Bak and Bax) (8, 9), or ATF3-mediated activation of other p53 family members such as p73 (33). We therefore concluded that ATF3 can activate p53 and promote p53-mediated cell cycle arrest and apoptosis in HPV-positive cells.
FIGURE 4.
ATF3 induces SiHa cells to cell cycle arrest and apoptosis as a consequence of p53 activation. A and B, SiHa cells were transfected with GFP or GFP-ATF3 for 2 days, and stained with PI. GFP-positive cells were subjected to cell cycle analysis using the FlowJo software. *, p < 0.001. C and D, SiHa cells were transfected with GFP or GFP-ATF3 for 3 days, and subjected to TUNEL staining. Quantitation was performed by counting at least 300 GFP-positive cells. E, SiHa cells were infected with Lentiviruses expressing shp53 or shLuc for 3 days, followed by transfections with GFP or GFP-ATF3 for 3 days. The cells were stained with TUNEL and counted as C. The inset shows an immunoblot for p53 expression.
FIGURE 5.
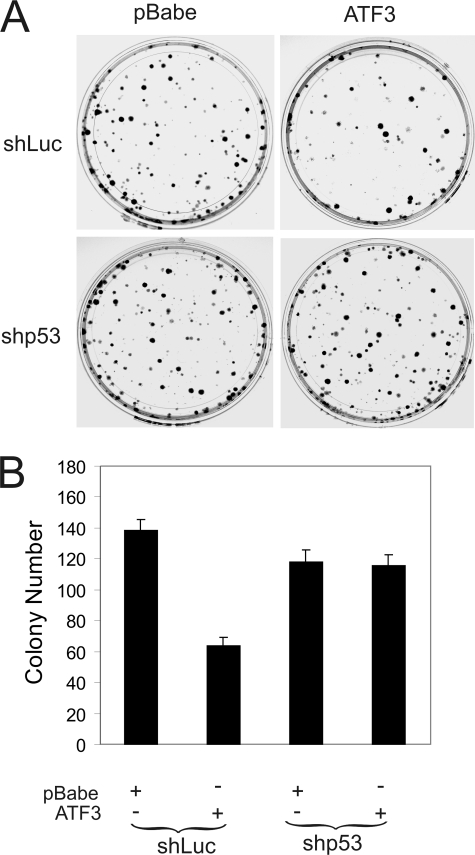
ATF3 decreases the growth potential of HPV-positive cells. SiHa cells infected with shp53- or shLuc-expressing Lentiviruses were further infected with retroviruses expressing ATF3 or its vector pBabe for 2 days. The infected cells (200/well) were plated into 6-well plates, and cultured for 14 days. The colonies were stained with crystal violet (A) and counted (B).
FIGURE 6.
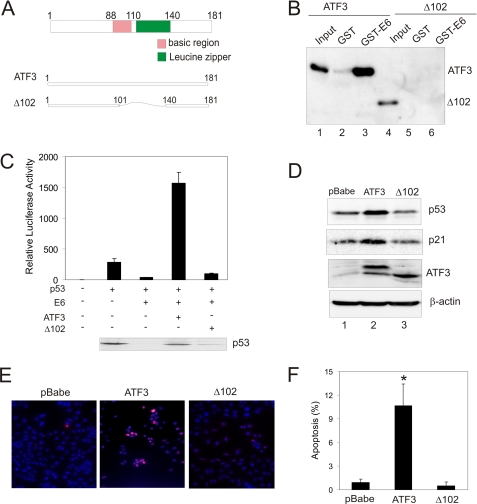
The ATF3 Zip domain is required for E6 binding and p53 activation in HPV-positive cells. A, schematic representation of the full-length ATF3 protein and the mutant protein lacking the Zip domain (Δ102) purified from bacterial culture using Ni2+-nitrilotriacetic acid-agarose. The recombinant proteins contain a c-Myc tag that can be used for detection of the mutant protein by immunoblotting. B, purified ATF3 and Δ102 proteins were incubated with immobilized GST-E6 or E6, and subjected to GST-pulldown assays as in Fig. 1C. The bound proteins were detected with an anti-c-Myc antibody. The input lane represents 10% of total input protein. C, H1299 cells were co-transfected with a p53 luciferase reporter, pRL-TK, p53, E6, ATF3, or Δ102 as indicated. Dual luciferase assays were performed to measure the p53 transcription activity. The immunoblot shows p53 expression in the transfected cells. D, SiHa cells were infected with retroviruses expressing ATF3, Δ102, or pBabe, and subjected to immunoblotting for detection of p53 and p21 expression. E and F, SiHa cells were infected with pBabe, ATF3, or Δ102, and then subjected to TUNEL staining followed by counterstaining with DAPI. Representative, merge images were shown in E. At least 300 cells were counted to calculate percentages of apoptotic cells (F). *, p < 0.001 compared with the pBabe group.
ATF3 Decreases the Growth Potentials of HPV-positive Cells
Having shown that ATF3 antagonizes E6 and promote the tumor suppressor activity of p53, a question was surfaced as to whether ATF3 affects the growth of HPV-positive cells. We thus expressed ATF3 in SiHa cells through retroviral infections, and plated the infected cells for colony formation assays. The results show that ecotopic expression of ATF3 decreased the number of colonies by more than 1-fold (Fig. 5, A and B), in support of a notion that targeting the E6 oncogenic activity could be a promising strategy to treat HPV-positive cancers (34). To determine whether this effect was dependent on p53 activation, we knocked down p53 expression by shRNA prior to retroviral infections. Consistent with Fig. 4E, down-regulation of p53 expression alone had little effect on SiHa cell growth. However, reduced p53 expression diminished the activity of ATF3 to decrease the growth potential of the HPV-positive cells (Fig. 5, A and B). Therefore, ATF3 likely inhibited HPV-positive cancer cell growth through antagonizing E6 for p53 activation.
The ATF3 Zip Domain Is Required for Its Interaction with E6 and Its Effects on p53 Activation
ATF3 contains a leucine zipper domain (Zip) (Fig. 6A) that is known to mediate its interactions with other proteins such as p53 (16). To determine whether this domain is also indispensable for its interaction with E6, we purified a mutant ATF3 protein (Δ102) lacking the Zip domain (amino acids 102–139) (supplemental Fig. S1A, lane 2) and performed GST-pulldown experiments. As same as Fig. 1C, the purified, full-length ATF3 protein was associated strongly with the immobilized E6 protein (Fig. 6B, lane 3). In striking contrast, deletion of the Zip domain (Δ102), which unlikely results in a misfolded protein as demonstrated in our previous report (16), completely abolished the ATF3-E6 binding (Fig. 6B, lane 6), suggesting that ATF3 likely bound to the HPV protein via the Zip domain. Interestingly, consistent with its inability to interact with E6, the ATF3 mutant failed to prevent p53 from E6-mediated degradation (Fig. 6C, see the immunoblot), and consequently, the p53 transcriptional activity down-regulated by the viral protein, as shown by a dramatic reduction in the activity of a p53 reporter (16), was not restored (Fig. 6C). It is worth to note that the full-length ATF3 protein not only restored E6-mediated down-regulation, but also largely elevated the reporter activity, suggesting that ATF3 might also counteract E6-mediated repression of the p53 trans-activation activity (10, 35) (see “Discussion”). In line with these observations, Δ102 expressed via retroviral infections lost the activity to increase the p53 and p21 levels in SiHa cells (Fig. 6D), and also failed to induce apoptosis in both SiHa and CaSki cells evident by TUNEL staining (Fig. 6, E and F, supplemental Fig. S3, B and C). Of note, Δ102 was expressed at a level comparable to the full-length protein measured by immunoblotting (Fig. 4D) and was also co-localized in the nuclei with p53 like the full-length protein (16). Therefore, the ATF3 Zip domain is required for its interaction with E6 and its E6-antagonizing activity.
ATF3 Prevents E6AP from Binding to E6
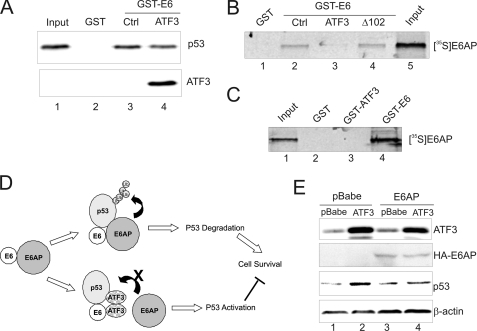
E6-mediated ubiquitination and degradation of p53 requires the binding of the viral protein to both p53 and E6AP. Because we have shown that ATF3 directly binds to both p53 and E6 (Fig. 1) (16) and because E6-mediated ubiquitination occurs at regions other than the p53 C terminus (36) where ATF3 binds (16), we reasoned that the ATF3 binding may not directly cover the lysine residues for ubiquitination, but rather interfere with the p53-E6-E6AP interaction. To explore this possibility, we first determine whether ATF3 interacting with p53 prevents the latter protein from binding to E6. We thus performed GST-pulldown assays in which the immobilized E6 protein precipitated p53 from rabbit reticulocyte lysates (Fig. 7A, lane 3) as expected (3). The purified ATF3 protein (supplemental Fig. S1A, lane 1) only slightly decreased the amount of p53 down-pulled by the E6 protein (Fig. 7A, lane 4 versus lane 3). Thus, it is unlikely that ATF3 antagonized the E6 activity through disrupting the E6-p53 interaction. We therefore explore an alterative possibility that ATF3 interferes with the interaction between E6 and E6AP by binding to the viral protein. Toward this end, we preincubated the purified ATF3 protein with the immobilized GST-E6 protein, and assayed the association of 35S-labeled E6AP protein with E6. Interestingly, while GST-E6, but not GST, pulled down E6AP (Fig. 7B, lane 2), preincubation of E6 with ATF3 dramatically decreased the amount of E6AP down-pulled by E6 (Fig. 6B, lane 3 versus lane 2). Importantly, the ATF3 mutant deficient in E6 binding (Δ102) (Fig. 6B) and deficient in p53 activation (Fig. 6D) lost the capability to block the binding of E6AP to E6 (Fig. 7B, lane 4), indicating that the ATF3 binding to E6 can prevent E6AP from binding to the viral protein. Interestingly, unlike E6 (Fig. 7C, lane 4), ATF3 did not bind to E6AP (Fig. 7C, lane 3). These results thus argue for a model that ATF3 competes with E6AP for forming a complex with E6 and p53 (Fig. 7D). This model predicts that overexpression of E6AP in HPV-positive cells could compromise the effects of ATF3 on p53 activation. Indeed, when SiHa cells were enforced to overexpress E6AP via retroviral infections, the capability of ATF3 to increase the p53 level was dramatically diminished (Fig. 7E, lane 3 versus lane 4). The failure of ectopically expressed E6AP to decrease the p53 level (Fig. 7E, lane 3 versus lane 1) might be due to the fact that SiHa cells contain one copy of the HPV DNA (34) and thus the endogenous E6AP protein is excess for E6 binding. Of note, the same ectopic E6AP protein could promote p53 degradation in HeLa cells (supplemental Fig. S5) as previously reported (37). We concluded that ATF3 can disrupt the E6-E6AP interaction by binding to E6, leading to diminished ubiquitination and degradation of p53 thereby activating the tumor suppressor in HPV-positive cancer cells.
FIGURE 7.
ATF3 prevents E6AP from binding to E6. A, in vitro-translated p53 protein was preincubated with BSA (lane 3) or ATF3 (lane 4) before incubated with immobilized GST-E6 or GST. Bound proteins were visualized by immunoblotting. The input lane represents 10% of total input protein. B, immobilized E6 protein was preincubated with BSA (lane 2), ATF3 (lane 3), or Δ102 (lane 4), and then incubated with 3 μl of 35S-labeled E6AP for 4 h. Bound proteins were visualized by autography. C, 3 μl of 35S-labeled E6AP was incubated with immobilized GST, GST-ATF3, or GST-E6, followed by autography to visualize bound proteins. D, schematic representation of a model in which ATF3 competes with E6AP for binding to E6 thereby blocking p53 ubiquitination catalyzed by the E3 ubiquitin ligase. E, SiHa cells were sequentially infected with retroviruses expressing HA-E6AP and ATF3 for 2 days and lysed for immunoblotting to determine the p53 level.
DISCUSSION
Expression of HPV proteins such as E6 is a driving force for cervical carcinogenesis (1, 2). The HPV proteins promote malignant transformation by intervening in functions of various cellular proteins through protein-protein interactions (38). While cellular proteins regulated by the viral proteins have been well documented (38), little is known as to how the oncogenic activities of the HPV proteins are regulated. In this study, we identified ATF3 as a novel repressor of the HPV E6 protein. ATF3 bound to E6 and prevented p53 degradation mediated by the viral protein. Because it is widely induced by oncogenic stimuli (e.g. DNA damage) (14, 15), ATF3 could serve as a key activator of p53 in HPV-infected cervix epithelium thereby defending against oncogenic transformation, a notion which is supported by our recent observation that ATF3 expression is frequently down-regulated in cervical cancers (23). Whereas the mechanism(s) for ATF3 down-regulation in cervical cancer remains elusive, decreased ATF3 expression allows E6 to drive p53 degradation thereby contributing to cervical carcinogenesis. Moreover, the demonstration of ATF3 as a direct binding partner of E6 suggests that ATF3 could regulate other oncogenic activities of E6 such as activation of telomerase and the Src family kinase Blk (39, 40). Activation of the latter proteins promotes cell proliferation, which acts in concert with p53 inactivation to promote the genesis and progression of cervical cancer (13). Therefore, this study has identified a novel mechanism by which the oncogenic activities of HPV proteins are antagonized. Because enforced expression of ATF3 can restore p53 activity and induce apoptosis of HPV-positive cancer cells, targeting ATF3 expression through chemical interventions or gene therapy could be a promising strategy to treat cervical cancer. Interestingly, recent studies have revealed a causal role of HPV infection in the genesis of head and neck cancer (41, 42). Therefore, the significance of the current findings may extend beyond the understanding of the etiology of cervical cancer. Indeed, we recently reported that ATF3 promotes anti-cancer effects of curcumin (43), an agent currently in clinical trials, in a p53-wild-type head and neck cancer cell line (44). It is very likely that the HPV-antagonizing activity of ATF3 described here contributes to such an effect.
ATF3 is well recognized as a stress responsive protein (14). Although ATF3 induction occurs frequently in the early stage of the DNA damage response, the role of ATF3 in cancer development remains undecided. Whereas earlier studies indicate that ATF3 may promote growth and dissemination of malignant cells (45, 46), results from many other studies strongly argue for a detrimental role that ATF3 plays in cancer development (47, 48). Indeed, ATF3 deficiency leads to cellular transformation induced by the Ras oncogene (16, 49). Our current findings that ATF3 antagonizes the oncogenic activity of a HPV viral protein further argues for the notion that ATF3 induction can function as an anticancer barrier to defend against oncogenic transformation (16, 23).
In addition to suppression of E6-mediated p53 degradation, we previously demonstrated that ATF3 stabilizes and activates p53 in HPV-negative cells (16) where the p53 level is mainly controlled by MDM2 (50). Interestingly, it appears that ATF3 regulates p53 degradation mediated by MDM2 and E6 through distinct mechanisms. While ATF3 blocks MDM2-mediated ubiquitination by binding to p53 and altering its conformation (16), we have shown here that the ATF3-p53 interaction was dispensable for suppression of the E6-mediated degradation. Indeed, although ATF3 binding may cover the lysine residues clustered at the p53 C terminus (16), p53 ubiquitination mediated by E6 does not occur in the C terminus (36, 51). Moreover, because it can also bind to p53 at the core DNA-binding domain (4), E6 might not compete with ATF3 for binding to the C terminus of the tumor suppressor, a consideration supported by the results that ATF3 failed to disrupt the E6-p53 interaction (Fig. 7A). Our results suggest that ATF3 rather suppresses E6-mediated p53 degradation through direct binding to the viral protein. Such an interaction prevents E6AP from binding to E6 (Fig. 7B), and thus blocks the recruitment of the ubiquitin ligase to p53, leading to diminished ubiquitination and proteolysis of the latter protein (Fig. 7D). Therefore, these results reiterate the importance of ATF3 in activating p53 in response to distinct oncogenic challenges.
Whereas the binding of E6 to the p53 C terminus does not require E6AP, the presence of the E3 ubiquitin ligase is likely indispensable for E6 binding to the p53 DNA binding domain (4). If the ATF3 binding disrupts the E6-E6AP interaction (Fig. 7B), it raises the questions as to how this achieves and how the E6-p53 interaction occurs in the absence of E6AP. E6 is a small protein containing four CXXC motifs that form two conserved internal sequence repeats (52). It might be that ATF3 competes with E6AP for binding to the same region(s) of the viral protein while the p53 conformation changes conferred by ATF3 interaction with its C terminus (16) allow E6 bind to p53 via the central DNA binding domain (Fig. 7D). Alternatively, the p53 C terminus might be preferably bound by E6 despite the presence of ATF3. Interestingly, although the leucine-zipper domain of ATF3 does not contain the charged leucine motif (LXXLL) that commonly exists in E6-associated proteins (53), it enriches leucine residues and can form a α-helix, which is one of the common structural characteristics of E6-binding proteins (54). Thus, it would be of interest to investigate whether other leucine zipper-containing proteins also interact with E6 and regulate the oncogenic activities of the viral protein.
Independent of its activity to drive p53 degradation, E6 also represses p53 trans-activation activity by inhibiting p300-mediated acetylation of p53 and histones (10, 35). The results from our reporter assays indicate that ATF3 not only restored the p53 activity decreased by E6, but also increased the p53 trans-activation activity by several folds (Fig. 6C). These results argue for a possibility that ATF3 regulates the p53 transcriptional activity independent of regulating its stability. The ATF3 binding might alter p53 conformation to a state favoring its binding to DNA, or its interaction with transcription activators/co-activators (e.g. p300, TFIIH, mediators) (55–57). Alternatively, ATF3 might directly recruit these transcription activators/co-activators to p53-target promoters. It might also be that ATF3 competes with transcription co-repressors for binding to p53 and its target promoters (58). Regardless of the mechanism(s), the capability of ATF3 to promote p53 trans-activation provides the cell with an additional means of activating p53 in response to a large variety of oncogenic stresses.
In summary, we have identified ATF3 as a novel repressor of the HPV E6 protein. ATF3 activates p53 by dissociating E6AP from the viral protein thereby preventing p53 degradation. Because E6 is the major protein that inactivates p53 in HPV-positive cells, these findings argue for a model in which ATF3 induction by DNA damage antagonizes the oncogenic activity of the E6 protein and thereby serves as an anticancer barrier to prevent carcinogenesis upon HPV infections (Fig. 7D).
Supplementary Material
Acknowledgments
We thank Drs. Tsonwin Hai, Peter Howley, Bert Vogelstein, Tohru Kiyono, Christine Blattner, and Zhi-Ming Zheng for the materials. We are grateful to Dr. Suzanne Camus for discussions and comments.
This work was supported in part by Department of Defense Grant PC061106 (to C. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- HPV
- human papillomavirus
- ATF
- activating transcription factor
- E6AP
- E6-associated protein
- GST
- glutathione S-transferase
- shRNA
- short-hairpin RNA
- TUNEL
- terminal dUTP nick-end labeling
- DTT
- dithiothreitol
- GFP
- green fluorescent protein
- BSA
- bovine serum albumin.
REFERENCES
- 1.zur Hausen H. (2000) J. Natl. Cancer Inst. 92, 690–698 [DOI] [PubMed] [Google Scholar]
- 2.Schiffman M., Castle P. E., Jeronimo J., Rodriguez A. C., Wacholder S. (2007) Lancet 370, 890–907 [DOI] [PubMed] [Google Scholar]
- 3.Scheffner M., Werness B., Huibregtse J. M., Levine A. J., Howley P. M. (1990) Cell 63, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 4.Li X., Coffino P. (1996) J. Virol. 70, 4509–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huibregtse J. M., Scheffner M., Howley P. M. (1991) EMBO J. 10, 4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. (1993) Cell 75, 495–505 [DOI] [PubMed] [Google Scholar]
- 7.Hengstermann A., Linares L. K., Ciechanover A., Whitaker N. J., Scheffner M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas M., Banks L. (1998) Oncogene 17, 2943–2954 [DOI] [PubMed] [Google Scholar]
- 9.Vogt M., Butz K., Dymalla S., Semzow J., Hoppe-Seyler F. (2006) Oncogene 25, 4009–4015 [DOI] [PubMed] [Google Scholar]
- 10.Patel D., Huang S. M., Bagila L. A., McCane D. J. (1999) EMBO J. 18, 5061–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Fan S., Meng Q., Ma Y., Katiyar P., Schlegel R., Rosen E. M. (2005) J. Biol. Chem. 280, 33165–33177 [DOI] [PubMed] [Google Scholar]
- 12.Huibregtse J. M., Beaudenon S. L. (1996) Semin. Cancer Biol. 7, 317–326 [DOI] [PubMed] [Google Scholar]
- 13.zur Hausen H. (2002) Nat. Rev. Cancer 2, 342–350 [DOI] [PubMed] [Google Scholar]
- 14.Hai T., Wolfgang C. D., Marsee D. K., Allen A. E., Sivaprasad U. (1999) Gene Expression 7, 321–335 [PMC free article] [PubMed] [Google Scholar]
- 15.Hai T., Hartman M. G. (2001) Gene 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 16.Yan C., Lu D., Hai T., Boyd D. D. (2005) EMBO J. 24, 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phizicky E. M., Fields S. (1995) Microbiol. Rev. 59, 94–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 19.Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 20.Beer D. G., Kardia S. L., Huang C. C., Giordano T. J., Levin A. M., Misek D. E., Lin L., Chen G., Gharib T. G., Thomas D. G., Lizyness M. L., Kuick R., Hayasaka S., Taylor J. M., Iannettoni M. D., Orringer M. B., Hanash S. (2002) Nat. Med. 8, 816–824 [DOI] [PubMed] [Google Scholar]
- 21.Chen X., Cheung S. T., So S., Fan S. T., Barry C., Higgins J., Lai K. M., Ji J., Dudoit S., Ng I. O., Van De Rijn M., Botstein D., Brown P. O. (2002) Mol. Biol. Cell 13, 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapointe J., Li C., Higgins J. P., Van De Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Ekman P., DeMarzo A. M., Tibshirani R., Botstein D., Brown P. O., Brooks J. D., Pollack J. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan C., Boyd D. D. (2006) Cell Cycle 5, 926–929 [DOI] [PubMed] [Google Scholar]
- 24.Yan C., Wang H., Boyd D. D. (2002) J. Biol. Chem. 277, 10804–10812 [DOI] [PubMed] [Google Scholar]
- 25.Turnell A. S., Grand R. J., Gorbea C., Zhang X., Wang W., Mynryk J. S., Gallimore P. H. (2000) EMBO J. 19, 4759–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camus S., Menéndez S., Cheok C. F., Stevenson L. F., Laín S., Lane D. P. (2007) Oncogene 26, 4059–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brummelkamp T. R., Bernards R., Agami R. (2002) Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 28.Narisawa-Saito M., Handa K., Yugawa T., Ohno S., Fujita M., Kiyono T. (2007) Oncogene 26, 2988–2996 [DOI] [PubMed] [Google Scholar]
- 29.Yan C., Wang H., Toh Y., Boyd D. D. (2003) J. Biol. Chem. 278, 2309–2316 [DOI] [PubMed] [Google Scholar]
- 30.An J., Mo D., Liu H., Veena M. S., Srivastsan E. S., Massoumi R., Rettig M. B. (2008) Cancer Cell 14, 394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogelstein B., Lane D., Levine A. J. (2000) Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 32.Hemann M. T., Zilfou J. T., Zhao Z., Burgess D. J., Hannon G. J., Lowe S. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9333–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh Y. K., Lee H. J., Jeong M. H., Rhee M., Mo J. W., Song E. H., Lim J. Y., Choi K. H., Jo I., Park S. I., Gao B., Kwon Y., Kim W. H. (2008) Mol. Cancer Res. 6, 1232–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butz K., Denk C., Ullmann A., Scheffner M., Hoppe-Seyler F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6693–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas M. C., Chiang C. M. (2005) Mol. Cell 17, 251–264 [DOI] [PubMed] [Google Scholar]
- 36.Camus S., Higgins M., Lane D. P., Lain S. (2003) FEBS Lett. 536, 220–224 [DOI] [PubMed] [Google Scholar]
- 37.Talis A. L., Huibregtse J. M., Howley P. M. (1998) J. Biol. Chem. 273, 6439–6445 [DOI] [PubMed] [Google Scholar]
- 38.Boulet G., Horvath C., Vanden Broeck D., Sahebali S., Bogers J. (2007) Int. J. Biochem. Cell Biol. 39, 2006–2011 [DOI] [PubMed] [Google Scholar]
- 39.Veldman T., Horikawa I., Barrett J. C., Schlegel R. (2001) J. Virol. 75, 4467–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oda H., Kumar S., Howley P. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9557–9562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran N., Rose B. R., O'Brien C. J. (2007) Head Neck 29, 64–70 [DOI] [PubMed] [Google Scholar]
- 42.Chung C. H., Gillison M. L. (2009) Clin. Cancer Res. 15, 6758–6762 [DOI] [PubMed] [Google Scholar]
- 43.Yan C., Jamaluddin M. S., Aggarwal B., Myers J., Boyd D. D. (2005) Mol. Cancer Ther. 4, 233–241 [PubMed] [Google Scholar]
- 44.Xu L., Davidson B. J., Murty V. V., Li R. G., Sacks P. G., Garinchesa P., Schantz S. P., Chaganti R. S. (1994) Int. J. Cancer 59, 383–387 [DOI] [PubMed] [Google Scholar]
- 45.Ishiguro T., Nakajima M., Naito M., Muto T., Tsuruo T. (1996) Cancer Res. 56, 875–879 [PubMed] [Google Scholar]
- 46.Perez S., Vial E., van Dam H., Castellazzi M. (2001) Oncogene 20, 1135–1141 [DOI] [PubMed] [Google Scholar]
- 47.Bottone F. G., Jr., Moon Y., Kim J. S., Alston-Mills B., Ishibashi M., Eling T. E. (2005) Mol. Cancer Ther. 4, 693–703 [DOI] [PubMed] [Google Scholar]
- 48.Kang Y., Chen C. R., Massagué J. (2003) Mol. Cell 11, 915–926 [DOI] [PubMed] [Google Scholar]
- 49.Lu D., Wolfgang C. D., Hai T. (2006) J. Biol. Chem. 281, 10473–10481 [DOI] [PubMed] [Google Scholar]
- 50.Michael D., Oren M. (2003) Semin. Cancer Biol. 13, 49–58 [DOI] [PubMed] [Google Scholar]
- 51.Nakamura S., Roth J. A., Mukhopadhyay T. (2002) Oncogene 21, 2605–2610 [DOI] [PubMed] [Google Scholar]
- 52.Liu Y., Baleja J. D. (2008) Front. Biosci. 13, 121–134 [DOI] [PubMed] [Google Scholar]
- 53.Bohl J., Das K., Dasgupta B., Vande Pol S. B. (2000) Virology 271, 163–170 [DOI] [PubMed] [Google Scholar]
- 54.Chen J. J., Hong Y., Rustamzadeh E., Baleja J. D., Andorphy E. J. (1998) J. Biol. Chem. 273, 13537–13544 [DOI] [PubMed] [Google Scholar]
- 55.Teufel D. P., Fruend S. M., Bycroft M., Fersht A. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7009–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao H., Pearson A., Coulombe B., Truant R., Zhang S., Regier J. L., Triezenberg S. J., Reinberg D., Flores O., Ingles C. J. (1994) Mol. Cell Biol. 14, 7013–7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donner A. J., Szostek S., Hoover J. M., Espinosa J. M. (2007) Mol. Cell 27, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitagawa M., Lee S. H., McCormick F. (2008) Mol. Cell 29, 217–231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.