Abstract
We identify and characterize two matrix (m)-AAA proteases (AtFtsH3 and AtFtsH10) present in the mitochondria of Arabidopsis thaliana. AtFtsH3 is the predominant protease in leaves of wild type plants. Both proteases assemble with prohibitins (PHBs) into high molecular weight complexes (∼2 MDa), similarly to their yeast counterparts. A smaller PHB complex (∼1 MDa), without the m-AAA proteases, was also detected. Unlike in yeast, stable prohibitin-independent high molecular weight assemblies of m-AAA proteases could not be identified in A. thaliana. AtFtsH3 and AtFtsH10 form at least two types of m-AAA-PHB complexes in wild type plants. The one type contains PHBs and AtFtsH3, and the second one is composed of PHBs and both AtFtsH3 and AtFtsH10. Complexes composed of PHBs and AtFtsH10 were found in an Arabidopsis mutant lacking AtFtsH3 (ftsh3). Thus, both AtFtsH3 and AtFtsH10 may form hetero- and homo-oligomeric complexes with prohibitins. The increased level of AtFtsH10 observed in ftsh3 suggests that functions of the homo- and hetero-oligomeric complexes containing AtFtsH3 can be at least partially substituted by AtFtsH10 homo-oligomers. The steady-state level of the AtFtsH10 transcripts did not change in ftsh3 compared with wild type plants, but we found that almost twice more of the AtFtsH10 transcripts were associated with polysomes in ftsh3. Based on this result, we assume that the AtFtsH10 protein is synthesized at a higher rate in the ftsh3 mutant. Our results provide the first data on the composition of m-AAA and PHB complexes in plant mitochondria and suggest that the abundance of m-AAA proteases is regulated not only at the transcriptional but also at the translational level.
Keywords: Arabidopsis, Gene/Knockout, Proteases/Metalloprotease, Protein/Assembly, Protein/Protein-Protein Interactions, Subcellular Organelles/Mitochondria, m-AAA Proteases, Prohibitins
Introduction
It is well established now that mitochondria have their own system for protein degradation. In the case of membrane proteins, this system is mainly based on AAA proteases (AAA stands for ATPases associated with diverse cellular activities) (1–3). AAA proteases, also called FtsH proteases, belong to a conserved family of ATP-dependent metallopeptidases with members found in bacteria, yeast, plants, and humans. The FtsH proteases are bifunctional enzymes, in which a proteolytic domain is accompanied by an ATPase domain with a chaperone-like activity. The location of FtsH proteases in eukaryotic cells is restricted to mitochondria and chloroplasts. In mitochondria, two types of AAA proteases have been identified. Both of them are anchored in the inner mitochondrial membrane, but their active centers are directed to opposite membrane surfaces as follows: the m-AAA2 proteases face the matrix, whereas the i-AAA proteases are oriented toward the intermembrane space (1, 4).
Mutations in m-AAA proteases cause severe defects in yeast (5), mouse (6), and humans (7, 8). They have been shown to degrade misfolded and unassembled membrane proteins (1, 9–11) and are also responsible for proteolytic activation and maturation of several mitochondrial proteins. Substrates that are cleaved rather than degraded by the m-AAA proteases include mitochondrial maturase responsible for splicing of group II introns (yeast) (5), MrpL32 subunit of mitochondrial ribosome (yeast and mammals) (7), and OPA1, a dynamin-like GTPase of the mitochondrial inner membrane (mammals) (12). Apart from their proteolytic roles, the m-AAA proteases mediate ATP-dependent membrane dislocation of the heme-binding reactive oxygen scavenger protein Ccp1 (13). Thus, diverse mitochondrial processes are under control of m-AAA proteases.
In all organisms investigated to date, the m-AAA proteases form large membrane complexes composed of identical or closely related subunits of molecular masses of 70–80 kDa (1, 4). In yeast, the m-AAA protease exists as a hetero-oligomer of ∼900 kDa composed of two subunits as follows: Yta10 (Afg3p) and Yta12 (Rca1p) present in equimolar amounts (14). Similar hetero-oligomeric complexes have also been identified in human and mouse mitochondria (8, 15), where two (Paraplegin and AFG3L2) or three (Paraplegin, Afg3l1, and Afg3l2) m-AAA proteases are present, respectively. In yeast, when either subunit is lacking, the other forms an inactive homo-oligomer of ∼250 kDa. In humans, mutations in the paraplegin gene cause an autosomal recessive form of hereditary spastic paraplegia, a disabling disorder with a neurological background (8). Analysis of cells from hereditary spastic paraplegia patients revealed a complete absence of Paraplegin. Surprisingly, hereditary spastic paraplegia mitochondria still possessed a 900-kDa m-AAA complex composed of only the AFG3L2 subunits, indicating that this protein is able to homo-oligomerize (15). This was confirmed by reconstitution of the human m-AAA complex in yeast mitochondria, where AFG3L2, but not Paraplegin, was able to form a 900-kDa homo-oligomeric complex. Similarly, murine Afg3l1 and Afg3l2 were able to form homo-oligomeric complexes in yeast mitochondria (15).
Solubilization of yeast mitochondria with a very mild detergent, digitonin, has revealed that m-AAA protease assembles with another multimeric protein complex composed of two highly homologous proteins Phb1p and Phb2p into a supercomplex with a molecular mass of ∼2 MDa (16). Phb1p and Phb2p, belonging to the Prohibitin family, form a large, ring-shaped complex on the outward face of the mitochondrial inner membrane. The functional unit of this complex is the Phb1p-Phb2p heterodimer. Between 10 and 14 Phb1p-Phb2p heterodimers are assembled into a high molecular weight complex with a mass of ∼1 MDa (17). Phb1/2 (or PHB) complex was proposed to be a chaperone/holdase binding and stabilizing subunit of the respiratory chain complexes (16, 18). However, recent data suggest a scaffolding role of PHB complexes (19–20). It is worth mentioning that until now stable m-AAA-PHB assemblies have been detected only in yeast mitochondria.
Sequencing of the Arabidopsis thaliana genome has revealed the presence of 12 homologs of AAA proteases (named AtFtsH1–12) (3) and seven genes encoding homologs of Phb1p and Phb2p (named AtPHB1–7) (21). Recently, it has been shown that AtPHB3 protein co-purifies with other PHB proteins (AtPHB1, AtPHB2, AtPHB4, and AtPHB6), presumably forming with them a hetero-oligomeric high molecular weight complex(es) in mitochondria (22). Several additional proteins interacting with AtPHB3 have been identified, none of them belonging to the AAA protease family. Previously, our group identified the first plant m-AAA protease in Pisum sativum (PsFtsH) (23). The PsFtsH protein is localized to the mitochondrial inner membrane and is able to complement the respiratory deficiency of the yta10 and/or yta12 null yeast mutants, indicating that the plant protein may compensate for loss of at least some functions of m-AAA in yeast mitochondria. Here, we investigated AtFtsH3 and AtFtsH10 proteins, the closest A. thaliana homologs of the pea PsFtsH protease. Our results indicate that the AtFtsH3 and AtFtsH10 proteins are present in an ∼2-MDa complex with prohibitins. AtFtsH3 and AtFtsH10 may form both homo- and hetero-oligomers. A lack of AtFtsH3 protease in A. thaliana mutant is compensated by increased synthesis of the AtFtsH10 protein. To our knowledge, this is the first data on the presence and interactions between m-AAA and prohibitin complexes in plant mitochondria.
EXPERIMENTAL PROCEDURES
Sequence Analysis
The amino acid sequences of i-AAA and m-AAA proteases from Mus musculus, Homo sapiens, and Saccharomyces cerevisiae were collected from the NCBI data base. Plant homologs were identified in A. thaliana, Oryza sativa, Populus trichocarpa, and Pisum sativum using BLAST and PsFtsH from P. sativum (23) and AtFtsH4 and AtFtsH11 from A. thaliana (24) as a query for m-AAA and i-AAA proteases, respectively. The following sequences were used to build a phylogenetic tree (first two letters identify species): AtFtsH3 (NP_850129); AtFtsH10 (NP_172231); AtFtsH4 (NP_565616); AtFtsH11 (NP_568787); OsFtsH3.1 (EEC79350); OsFtsH3.2 (BAB86453); OsFtsH4 (NP_001043385); OsFtsH11 (EEE54999); PtFtsH3.1 (EEE87381); PtFtsH3.2 (EEF05269); PtFtsH4 (EEE92077); PtFtsH11 (EEE95971); PsFtsH3 (AAK77908); PsFtsH4 (CAG25608); PsFtsH11 (CAH10348); MmParaplegin (AAN03852); MmAfg3l1 (BAE39961); MmAfg3l2 (BAE29204); MmYme1l1 (CAM19538); HsParaplegin (AAD28099); HsAFG1L2 (EAX01551); HsYME1L1 (CAC19650); ScYme1 (CAA95020), ScYta10 (CAA56953); and ScYta12 (CAA89236). The sequences were aligned, and the phylogenic tree was constructed using MEGA software. The TMpred tool was used to predict transmembrane domains.
The AGI IDs of Arabidopsis genes used in this work are as follows: At2G29080 (AtFtsH3); At1G07510 (AtFtsH10); At3G18780 (AtAct2, Actin 2); At4G28510 (AtPHB1); At1G03860 (AtPHB2); At5G14300 (AtPHB3); At3G27280 (AtPHB4); and At5G40770 (AtPHB6).
Plant Material and Growth Conditions
Wild type and T-DNA knock-out mutant A. thaliana plants were of the Columbia (Col-0) ecotype. The T-DNA insertion lines ftsh3 (SAIL_767_F02/TAIR) and ftsh3-1 (SALK_037144/TAIR) were from the Salk Institute collection. Plants were grown in soil in a climate-controlled chamber at 22 °C and 100 μmol m−2 s−1 light with 8-h light/16-h dark photoperiod. Mitochondria and chloroplasts were isolated from 11- to 12-week-old plants according to procedures adopted from Urantowka et al. (24) and Aronsson and Jarvis (25), respectively.
Isolation and Characterization of ftsh3-1 Insertion Mutant Line
Homozygous ftsh3-1 mutant plants with insertion in the AtFtsH3 gene were selected from line N537144 by PCR with primers flanking the genomic region of the insertion and a standard LBb1 primer complementary to the T-DNA insertion. Total DNA was extracted using GeneMATRIX plant and fungi DNA purification kit (Eurx, Poland). The sequences of the primers designed by the SIGnAL iSect Primer Design were as follows: LP, CAGAACCACATCTGCAGTTCC; RP, TATTTCCCCCAATCAAAAACC; LBb1, GCGTGGACCGCTTGCTGCAACT. To establish the exact location of the T-DNA insertion, the PCR product obtained using primers RP + LBb1 was sequenced, and the obtained sequence was aligned to the genomic sequence of A. thaliana. The absence of normal AtFtsH3 transcript was tested by reverse transcription-PCR using primers with sequences complementary to the 5′- and 3′-untranslated region of the transcript. Total RNA from wild type plants and ftsh3-1 mutant was extracted using GeneMATRIX Universal RNA purification kit (Eurx). To remove genomic DNA contamination, RNA samples were treated with RNase-free DNase I (Fermentas) according to the manufacturer's instructions. cDNA was synthesized using Omniscript reverse transcription kit (Qiagen) and oligo(dT)18 primer. The sequences of primers used for gene-specific amplification were as follows: AtFtsH3, 5′-TCCTCTTCTAAGCTCGTTTCAG (forward primer) and 5′-GCTAACAAATTCCGTCGTCACT (reverse primer); AtAct2 (actin 2), 5′-ACACTGTGCCAATCTACGAGGG (forward primer) and 5′-CTCTTACAATTTCCCGCTCTGC (reverse primer).
Two-dimensional BN/SDS-PAGE and Western Blot Analysis
Blue native gel electrophoresis was carried out as described elsewhere (24). In short, 100 μg of mitochondrial protein was resuspended in buffer (30 mm Hepes, pH 7.4, 150 mm potassium acetate, 10% glycerol) and solubilized by adding digitonin (Sigma) or dodecyl maltoside (ICN Biomedicals, OH) in the proportion of 5 or 10 g of detergent per 1 g of protein, respectively. After a clarifying centrifugation for 20 min at 24,000 × g, supernatant was supplemented with Coomassie Brilliant Blue G-250 solution and loaded on a 3–12% polyacrylamide gel. For the second dimension, lanes of the BN gel were cut out, incubated for 20 min in solution containing 2% SDS and 1% β-mercaptoethanol, and placed in a casting unit above the SDS gel. After electrophoresis, proteins were transferred to polyvinylidene difluoride membrane (Bio-Rad) and immunostained using appropriate antibodies. Anti-cytochrome f antibodies were obtained from Prof. Andrzej Szczepaniak, University of Wroclaw. The following antibodies were purchased from Agrisera AB: anti-AtVDAC1 (AS07 212); AtFtsH3/10 (AS07 204); and AtFtsH10 (AS07 251). Anti-AtFtsH3 was produced by GenScript. AtFtsh3-specific antibodies were raised against peptide TEKDSAATPTVEPV, and AtFtsH10-specific antibodies were raised against peptide SEKESQKESVPVKP. Detection was performed using ECL reagent. Chemiluminescence was recorded using x-ray film (Eastman Kodak Co.) or chemiluminescence imager (Typhoon 8600, GE Healthcare). Bands were quantified using QuantityOne software (Bio-Rad). Mass spectrometry analysis of PHB complexes was carried out as follows: gels after two-dimensional BN/SDS-PAGE were stained with colloidal Coomassie. Gel fragments corresponding to PHB complexes (coordinates 2 MDa/30 kDa and 1 MDa/30 kDa) were cut out and sent for mass spectrometry analysis to Laboratory of Mass Spectrometry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland.
Production of Anti-prohibitin Antibodies
For generating anti-prohibitin antibodies, a cDNA corresponding to the entire coding region of the Arabidopsis prohibitin homolog, AtPHB3 (At5g40770), was generated by reverse transcription-PCR and cloned into an expression vector pCR T7/NT-TOPO (Invitrogen). AtPHB3 was expressed as the His tag fusion protein in Escherichia coli strain BL21 and purified as described previously (2). The purified His-AtPHB3 protein was used to commercially raise rabbit antibodies.
Co-immunoprecipitation and Immunodepletion
Mitochondria (100 μg) were solubilized in IP buffer (1% digitonin, 150 mm NaCl, 100 mm Tris-HCl, 4 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, pH 7.5) and centrifuged at 24,000 × g for 15 min. Supernatants were incubated for 2 h at 4 °C with gentle shaking with serum (4.5 μl of anti-AtFtsH10 serum or preserum) coupled to protein A-Sepharose beads (Pierce). Beads were washed three times with IP buffer and once with IP buffer without the detergent. Finally, beads were boiled with 2× Laemmli buffer and subjected to standard SDS-PAGE and Western blot analysis. Immunoprecipitation for mass spectrometry analysis was scaled up to 3 mg of mitochondria. Immunoprecipitates separated in SDS-PAGE were stained with Coomassie Brilliant Blue G-250. The band corresponding to prohibitins was cut out and subjected to mass spectrometry analysis.
Immunodepletion experiments were performed using saturating amounts of anti-AtFtsH10 antibodies. Mitochondria (100 μg) were solubilized in ID buffer (1% digitonin, 150 mm NaCl, 100 mm Tris-HCl, pH 7.5) supplemented with protease inhibitor mixture (Roche Applied Science) and centrifuged at 24,000 × g for 15 min. Supernatants were incubated with serum (30 μl of anti-AtFtsH10 serum or preserum) coupled to protein A-Sepharose beads. However, this time the antibodies were chemically cross-linked to protein A using disuccinimidyl suberate (Pierce) according to manufacturer's instructions. After incubation (7 h at 4 °C) with gentle shaking, the beads were removed by centrifugation (6,000 × g for 2 min), and supernatants were subjected to trichloroacetic acid precipitation, SDS-PAGE, and immunoblotting.
Isolation of Polysomal RNA
Polysomes were fractionated from homogenized leaf tissue as described previously by Kahlau and Bock (26) with slight modifications. 200 mg of leaf tissue was ground in 1 ml of freshly prepared extraction buffer (200 mm Tris-HCl, pH 9.0, 200 mm KCl, 35 mm MgCl2, 25 mm EGTA, 200 mm sucrose, 100 mm β-mercaptoethanol, 1% (v/v) Triton X-100, 2% (v/v) polyoxyethylene-10-tridecyl ether, 6% (w/v) digitonin, 1 mg/ml heparin, 100 μg/ml chloramphenicol, and 25 μg/ml cycloheximide) and then centrifuged at 13,200 × g to remove cell debris. Leaf extracts were supplemented with 1/20 volume of 10% (w/v) sodium deoxycholate and separated in sucrose density gradient (56:40:30:15% (w/v) sucrose in 40 mm Tris-HCl, pH 8.5, 20 mm KCl, 10 mm MgCl2, 100 μg/ml chloramphenicol, 500 μg/ml heparin) by ultracentrifugation (4 °C, 200,000 × g, 80 min). Ten aliquots of ∼410 μl each obtained by fractionation of gradients were subjected to RNA extraction using phenol/chloroform/isoamyl alcohol (25:24:1).
Real Time PCR Analysis
DNA-free RNA prepared as described above was subjected to reverse transcription using high capacity cDNA reverse transcription kit (Applied Biosystems). Real time PCR analysis was performed on a LightCycler (Roche Applied Science) using real time 2× PCR master mix SYBR (A & A Biotechnology) and the following primers: 5′-CGCTGAGCTCTTGCTAGAGAAG (forward) and 5′-ATTCCATCATCCTCCACAGGCT (reverse) for AtFtsH10 transcript; CGCCGAGCTTTTACTAGAGAAA (forward) and AGCTCCATCATCCACCACAGGC (reverse) for AtFtsH3 transcript; and 5′-ACACTGTGCCAATCTACGAGGG (forward) and 5′-CTCTTACAATTTCCCGCTCTGC (reverse) for AtAct2 transcript. The AtFtsH3/AtFtsH10 transcript ratio was determined in RNA samples isolated from leaves of wild type plants using an external standard. The standard was obtained by serial dilutions of a mixture prepared by combining in a 1:1 ratio AtFtsH3 and AtFtsH10 cDNAs cloned into the pTZ57R vector. cDNAs were amplified using TCCTCTTCTAAGCTCGTTTCAG (forward) and GCTAACAAATTCCGTCGTCACT (reverse) or CCAAAGCTGCCATCTTTCTCCT (forward) and GCGTGTTTTGTTATTGCCTTGG (reverse) primers for AtFtsH3 and AtFtsH10, respectively.
Functional Complementation in Yeast
DNA sequences encoding AtFtsH3 or AtFtsH10 were PCR-amplified using pTZ57R-AtFtsH3 or pTZ57R-AtFtsH10 constructs (see above) and pairs of primers as follows: CATCGGGATCCATGACAATGATCTTCTTCTC and CGCTTGCGGCCGCTTACGTCGGAACAACCTGGG for AtFtsH3 or CATCGGGATCCATGATATTCTCCAAGCTTGG and CGCTTGCGGCCGCCTACGTCGGAACTACCTGTG for AtFtsH10 (the start and stop codons are in italic, restriction enzyme sites are underlined). PCR products were digested with NotI and BamHI and ligated into BamHI-NotI sites of yeast shuttle vector pCM185. Yeast strains were transformed by the lithium acetate method (27). W303 (WT), YHA101 (Δyta10), YHA201 (Δyta12) and YHA301 (Δyta10/Δyta12) yeast strains used in this study and complementation analysis have been described previously (23).
RESULTS
Identification of AtFtsH3 and AtFtsH10 Proteins as Functional m-AAA Proteases
In our previous work, we identified and partially characterized the first plant homolog of yeast m-AAA protease from P. sativum (PsFtsH) (23). To investigate plant m-AAA proteases more closely, we turned our efforts to the model plant A. thaliana. Among the 12 FtsH proteases identified in A. thaliana, two, AtFtsH3 and AtFtsH10, show the highest homology to PsFtsH and consequently were proposed to be Arabidopsis m-AAA proteases (28).
A phylogenetic analysis (supplemental Fig. S1A) of amino acid sequences of known and predicted mitochondrial AAA proteases from several organisms (see “Experimental Procedures” for details) revealed that the selected sequences form two distinct branches. One of them contains all experimentally confirmed mitochondrial i-AAA proteases, whereas sequences of all identified m-AAA proteases are located in the second branch. The position of AtFtsH3 and AtFtsH10 on this phylogentic tree clearly indicates that they are members of the m-AAA subfamily.
As expected, cell fractionation confirms that both AtFtsH3 and AtFtsH10 are present in mitochondria, but not in chloroplasts (supplemental Fig. S1B). Mitochondrial localization of these two proteins was also reported by Sakamoto et al. (2). Yeast m-AAA proteases span the membrane twice, and TMpred predicted two transmembrane domains both in FtsH3 (amino acid positions 136–152 and 263–281) and in FtsH10 (amino acid positions 133–156 and 267–285).
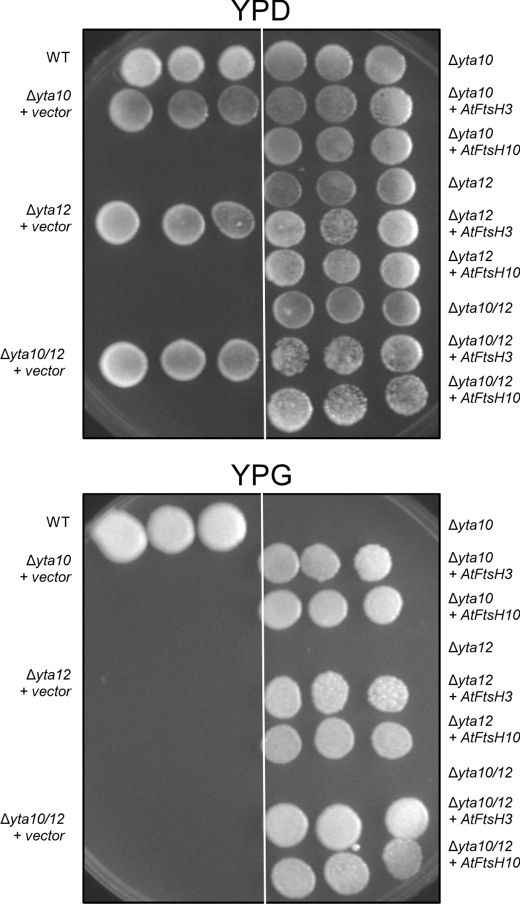
To confirm that AtFtsH3 and AtFtsH10 are functional homologs of yeast m-AAA proteases, we placed both sequences under the control of the CYC1 promoter in the yeast centromeric vector pCM185 and introduced them into yeast lacking Yta10p, Yta12p, or both Yta10p and Yta12p. Because of impaired mitochondrial function, yeast cells with disrupted expression of YTA10 or YTA12 fail to grow on nonfermentable carbon sources. Single yta10 or yta12 disruptants as well as double yta10/yta12 knock-out strain transformed with AtFtsH3 or AtFtsH10 were able to grow on glycerol at 30 °C (Fig. 1). This result strongly indicates that both plant proteins can substitute for the function of the yeast m-AAA proteases.
FIGURE 1.
AtFtsH3 and AtFtsH10 restore growth of the yta10 and/or yta12 null mutants on glycerol. Yeast strain W303 or Δyta10 and/or Δyta12 mutants were transformed with recombinant plasmid pCM185 carrying AtFtsH3 (+ AtFtsH3) or AtFtsH10 (+ AtFtsH10) genes cloned into pCM185 vector or empty vector (+ vector). 10-Fold serial dilutions of cells were replica-plated on YP plates containing 2% glucose (YPD) or 4% glycerol (YPG) and incubated at 30 °C for 3 days. WT, wild type.
Accumulation of AtFtsH10 Protein Differs in ftsh3 and Wild Type Plants
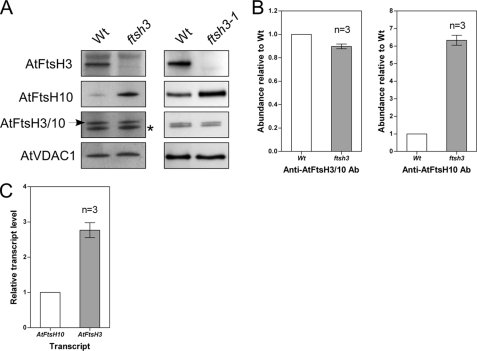
To characterize the Arabidopsis m-AAA proteases, we took advantage of two T-DNA insertion mutants ftsh3 and ftsh3-1 lacking the AtFtsH3 protease. The ftsh3 mutant was characterized in our previous publication (29), although here we describe the ftsh3-1 mutant selected from line N537144. The T-DNA insert in ftsh3-1 was localized in exon 2 upstream of nucleotide 218 counting from the translation start codon (supplemental Fig. S2, A and B). The absence of FtsH3 transcript was confirmed by reverse transcription-PCR with primers encompassing the complete open reading frame (supplemental Fig. S2C). Here, we also show a lack of the FtsH3 protein in the mitochondria of ftsh3 and ftsh3-1 using AtFtsh3-specific antibodies (Fig. 2A). Interestingly, we noticed that the level of the AtFtsH10 protein in both mutants was substantially higher than that in wild type plants. Despite this, using antibodies raised against a peptide common for both AtFtsH3 and AtFtsH10, we found that the overall level of m-AAA proteases (AtFtsH3 + AtFtsH10) in both mutants was only slightly affected. Densitometric analysis of Western blots revealed that the abundance of the AtFtsH10 protein was over 6-fold higher in ftsh3 than in the wild type, whereas the level of m-AAA proteases (AtFtsH3 + AtFtsH10) measured with anti-FtsH3/10 antibodies was only ∼10% lower in ftsh3 than in wild type plants (Fig. 2B). Thus, the absence of the AtFtsH3 protein in the ftsh3 and ftsh3-1 mutants is almost completely compensated by an increased abundance of the AtFtsH10 protein. The more than 6-fold increase for the AtFtsH10 protein required to compensate quantitatively for the lack of AtFtsH3 indicates that in the wild type mitochondria AtFtsH3 is at least five times more abundant than the AtFtsH10 protein. Analysis of the relative transcript abundance using real time PCR indicates that the ratio of AtFtsH3/AtFtsH10 transcripts is 2.77 ± 0.21 (Fig. 2C). Thus, AtFtsH3 is the predominant m-AAA protease in rosette leaves of wild type A. thaliana plants.
FIGURE 2.
Increased abundance of AtFtsH10 in ftsh3 and ftsh3-1 mutants. A, mitochondria isolated from wild type (Wt) plants and ftsh3 and ftsh3-1 mutants were separated by 10% SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with antibodies raised against epitopes specific to AtFtsH3 and AtFtsH10 or an epitope common to AtFtsH3 and AtFtsH10 (anti-AtFtsH3/10 antibodies). AtVDAC1 (porin1) was used as a loading control. B, levels of AtFtsH3 + AtFtsH10 proteins (anti-AtFtsH3/10 antibody (Ab)) or AtFtsH10 protein (anti-AtFtsH10 antibody) in ftsh3 mutant relative to wild type (Wt) plants. C, steady-state level of the AtFtsH3 mRNAs relative to the AtFtsH10 mRNAs in leaves of wild type plants. Relative quantities of the mRNAs were counted using real time PCR and external standards (see “Experimental Procedures” for details). Data are means ± S.E. from three independent experiments. *, nonspecific band.
More Efficient Translation Is Responsible for Increased Abundance of AtFtsH10 Protein
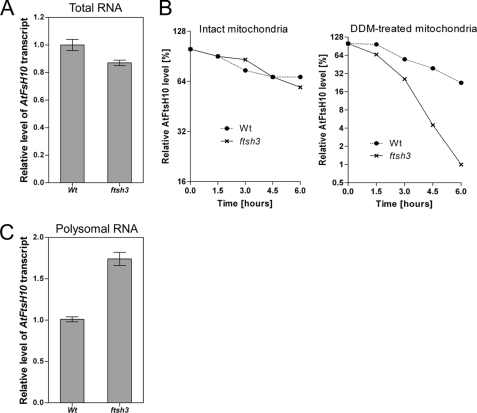
Next, we performed experiments to find out which step of the AtFtsH10 gene expression was responsible for the compensatory effect observed in the ftsh3 and ftsh3-1 mutants. First, we investigated the steady-state level of the AtFtsH10 transcript. Results presented in Fig. 3A indicate that the level of this transcript is not elevated in ftsh3 compared with wild type plants, and it is even slightly lower. Thus, the increased abundance of the AtFtsH10 protein is not due to an increased amount of the AtFtsH10 transcript and hence must be obtained at a post-transcriptional level. Therefore, we then investigated the stability of the AtFtsH10 protein using two experimental approaches. In the first experiment, we examined the stability of the AtFtsH10 protein in intact isolated mitochondria. In the second experiment, we treated mitochondria with the detergent dodecyl maltoside (DDM) to release complexes from mitochondrial membranes. The incubation period in both experiments was 6 h. The results are presented in Fig. 3B. The rates of AtFtsH10 degradation are comparable in intact mitochondria isolated from wild type plants and the ftsh3 mutant, which means that the elevated abundance of AtFtsH10 in ftsh3 does not result from its increased stability. Surprisingly, we observed that degradation of AtFtsH10 in the presence of DDM was even faster in ftsh3 mitochondria than in the wild type. This finding indicates that release of AtFtsH10 from the membrane results in its increased susceptibility to degradation in the ftsh3 mutant. Taken together, the analysis of the AtFtsH10 stability suggested that the observed compensatory effect in the ftsh3 mutant is most probably achieved at the level of protein synthesis.
FIGURE 3.
Expression of AtFtsH10 gene in ftsh3 mutant. A, level of AtFtsH10 transcript in ftsh3 mutant and wild type (Wt) plants. RNA samples were isolated from whole 6-week-old plants. Data are means ± S.E. from three independent experiments. B, turnover of AtFtsH10 protein in wild type (Wt) and ftsh3 mitochondria. Mitochondria were incubated for 6 h. Steady-state levels of AtFtsH10 protein were analyzed in 1.5-h intervals by Western blotting. Left, results obtained for intact mitochondria. Right, results obtained for mitochondria treated with DDM at time 0. Data are representative of two independent experiments. C, abundance of AtFtsH10 transcript in polysomal RNA from wild type plants and ftsh3 mutant. RNA was isolated from adult leaves harvested from 10- to 11-week-old plants. The AtFtsH10 transcript was standardized relative to the AtAct2 transcript. The difference in abundance of the AtFtsH10 transcript in polysomal RNA from ftsh3 mutant and from wild type plants is statistically significant (Student's t test, p value = 0.01). The relative abundance of the AtFtsH10 transcript in total RNA was set as 1. Data presented as mean ± S.E. are representative of two biological repeats, each performed in three technical repeats.
To address this possibility, ftsh3 and wild type plants were analyzed for association of the AtFtsH10 transcript with cytoplasmic polysomes. A quantitative analysis of polysomal mRNA revealed that the abundance of the AtFtsH10 transcript in polysomal RNA from the ftsh3 mutant is almost twice (1.74 ± 0.08) as high as in wild type plants (Fig. 3C). This substantial enrichment of the AtFtsH10 transcript in polysomal RNA suggests its enhanced translation in the ftsh3 mutant compared with wild type plants. Based on this result, we assume that, despite an unchanged overall level of its mRNA, the AtFtsH10 protein is synthesized at a higher rate in the ftsh3 mutant than in wild type plants.
It has been reported that the percentage of particular mRNAs bound to polysomes in A. thaliana varies typically from 35 to 85% (30). For the AtFtsH10 transcript, this value was 59% (30). Taking into consideration the above and the fact that the steady-state level of the FtsH10 transcript is similar in ftsh3 and wild type plants, but almost twice as many molecules of this transcript are bound to polysomes in ftsh3, we conclude that practically all AtFtsH10 transcripts are associated with polysomes in the ftsh3 mutant.
AtFtsH3, AtFtsH10, and Prohibitins Are Present in High Molecular Weight Complexes
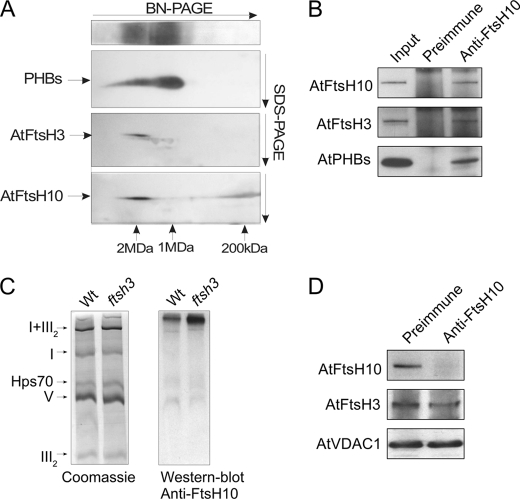
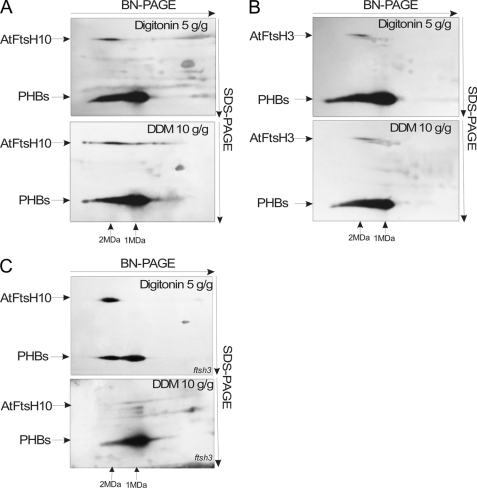
To explore whether the A. thaliana m-AAA proteases form multimeric complexes, like their homologs in yeasts and mammals do, we applied two-dimensional BN/SDS-PAGE linked with immunoblotting. The obtained results (Fig. 4A) indicate that AtFtsH3 and AtFtsH10 form high molecular weight complex(es), with a molecular mass of ∼2 MDa. Using anti-AtFtsH10 antibodies, we observe an additional signal in the range of 150–250 kDa.
FIGURE 4.
Characteristics of high molecular weight complexes of PHBs and m-AAA proteases. A, high molecular weight complexes of AtPHBs, AtFtsH3, and AtFtsH10 separated on two-dimensional BN/SDS-polyacrylamide gels. Wild type (Wt) mitochondria solubilized with digitonin were separated in 3–12% BN-PAGE in the first dimension and in 10% SDS-PAGE in the second dimension, transferred onto polyvinylidene difluoride membrane, and probed with anti-PHBs, anti-AtFtsH3, and anti-AtFtsH10 antibodies. For PHBs, a lane from the first dimension is also presented in order better to visualize the existence of two complexes of PHBs. The molecular weight of the m-AAA and PHB complexes was estimated in relation to the positions of respiratory chain complexes. B, co-immunoprecipitation of AtFtsH10 protein. Wild type mitochondria (100 μg) were solubilized with digitonin. Co-immunoprecipitation was carried out using anti-AtFtsH10 antibodies or preimmune antiserum (negative control). Immunoprecipitants were analyzed by SDS-PAGE and immunoblotting. Input, mitochondria used for immunoprecipitation. C, high molecular weight complex of AtFtsH10 in ftsh3 mutant. Mitochondria solubilized with digitonin were separated by 5% nongradient BN-PAGE in order better to visualize the AtFtsH10 complex. Left, Coomassie staining of respiratory chain complexes. Right, immunoblotting with anti-AtFtsH10 antibodies. D, immunodepletion of AtFtsH10. Wild type mitochondria (100 μg) were solubilized with digitonin and incubated for 7 h with protein A-Sepharose beads with cross-linked anti-AtFtsH10 antibodies or preimmune serum (negative control). After centrifugation, supernatant was taken for immunoblotting with anti-AtFtsH3 and anti-AtFtsH10 antibodies. AtVDAC1 (porin1) was used as a loading control.
In the next step, we examined the complexes formed by prohibitins. Our results indicate that PHBs form two multimeric complexes in Arabidopsis mitochondria (Fig. 4A). Their molecular masses were estimated to be ∼1 and 2 MDa. Mass spectrometry analysis demonstrated that both complexes comprise five prohibitins AtPHB1, AtPHB2, AtPHB3, AtPHB4, and AtPHB6. Based on the alignment presented in Fig. 4A, the 2-MDa PHB complex migrates identically as a complex detected by anti-AtFtsH3 and anti-AtFtsH10 antibodies. This suggests that the m-AAA proteases and PHBs could be subunits of the same high molecular weight complex, as they are in yeast. To confirm this conjecture, we applied co-immunoprecipitation using the AtFtsH10 protein as the bait. Western blot analysis of the immunoprecipitate revealed the presence of AtFtsH3, AtFtsH10, and PHBs (Fig. 4B). Mass spectrometry analysis of the band corresponding to PHBs indicated the presence of peptides specific to four prohibitins as follows: AtPHB1, AtPHB2, AtPHB3, and AtPHB6. These results confirm a direct physical interaction of plant m-AAA and prohibitins in Arabidopsis mitochondria and demonstrate that AtFtsH3, AtFtsH10, and PHBs are all part of a high molecular weight complex of about 2 MDa.
AtFtsH3 and AtFtsH10 Form Both Hetero- and Homo-oligomeric Complexes
The results of immunoprecipitation indicated that in the wild type A. thaliana mitochondria, AtFtsH3 and AtFtsH10 form hetero-oligomeric complexes. On the other hand, the presence of only one m-AAA protease, AtFtsH10, in the ftsh3 mutant suggested that this protein may form homo-oligomeric complexes as well. By homo-oligomeric complexes, we understand complexes comprising only one kind of m-AAA protease, AtFtsH3 or AtFtsH10, but we do not exclude the possibility that they contain other proteins as well. To confirm the suggestion that m-AAA proteases can form homo-oligomeric complexes in Arabidopsis mitochondria, we checked whether an FtsH10 complex is present in the mitochondria of the ftsh3 mutant. Fig. 4C shows also that in the absence of AtFtsH3 protein AtFtsH10 is present in a complex whose molecular mass is the same as that of the m-AAA-PHB complex present in wild type mitochondria.
Using a mutant lacking AtFtsH10 would be the easiest way to also examine whether AtFtsH3 is able to form homo-oligomers. However, not having plants without the AtFtsH10 protein, we took advantage of the earlier observation that in wild type plants AtFtsH3 is much more abundant that AtFtsH10. It was conceivable that the excess of the AtFtsH3 protein forms homo-oligomers. If so, then complete depletion of the AtFtsH10 protein from a mitochondrial protein extract containing intact supramolecular complexes should result in only partial depletion of the AtFtsH3 protein. Thus, we applied anti-AtFtsH10 antibodies to immunodeplete AtFtsH10 protein from a protein extract obtained by treatment of mitochondria with digitonin. As expected, complete immunodepletion of AtFtsH10 resulted in only partial depletion of AtFtsH3. Therefore, only a fraction of AtFtsH3 exists as the AtFtsH3/AtFtsH10 hetero-oligomer. The remaining probably forms AtFtsH3 homo-oligomers. These results indicate that both A. thaliana m-AAA proteases are able to form homo-oligomeric complexes, but in wild type mitochondria the majority of m-AAA complexes are homo-oligomers of AtFtsH3 and hetero-oligomeric complexes of AtFtsH3 and AtFtsH10. Complementation of the yeast yta10/yta12 null mutant by AtFtsH3 or AtFtsH10 indicates that homo-oligomers formed by both proteins are functional.
Stability of m-AAA Protease Complexes Depends on Their Subunit Composition
In yeast, the m-AAA-PHB supercomplex is observed only when mitochondria are solubilized with a mild detergent such as digitonin (16). Usage of stronger detergents results in dissociation of the supercomplex into two independent complexes of m-AAA proteases and prohibitins, respectively, with molecular masses of ∼1 MDa each. Application of digitonin in the experiments described above showed that in mitochondria of A. thaliana m-AAA proteases exist exclusively in a complex with PHBs, whereas prohibitins also form a second complex not containing the m-AAA proteases. To assess whether AtFtsH3 and AtFtsH10 may form a complex independent of prohibitins, we replaced digitonin with a stronger DDM. The results obtained for wild type mitochondria compared with the data obtained with digitonin are presented in Fig. 5, A–C. Western blot with anti-PHB antibodies indicates that the 2-MDa complex is still present, but it seems to be less abundant when digitonin is replaced with DDM (Fig. 5, A and B). Anti-AtFtsH3 antibodies indicate that the 2-MDa complex formed with AtFtsH3 can still be observed in the presence of DDM (Fig. 5B), although after inspection, a light smear down to 1 MDa can be perceived. In the case of AtFtsH10, the detection signal corresponding to a molecular mass of ∼2 MDa was shifted down in the presence of DDM to a somewhat lower molecular mass, with a distinctive smear down to ∼150 kDa. These results indicate that the 2-MDa complex composed of PHBs and AtFtsH3 homo-oligomers is fairly stable in DDM, although the 2-MDa complex composed of PHBs and AtFtsH3-AtFtsH10 hetero-oligomers undergoes partial but substantial dissociation. However, we did not observe dissociation of the AtFtsH3-AtFtsH10-PHB complex into separate AtFtsH3-AtFtsH10 and PHB complexes but rather its gradual disintegration with release of the AtFtsH10 subunits.
FIGURE 5.
Stability of m-AAA and PHB complexes in DDM. Mitochondria solubilized with either digitonin or DDM were separated by two-dimensional BN/SDS-PAGE, transferred on polyvinylidene difluoride membrane, and immunoblotted for PHBs and AtFtsH10 (A and C) or PHBs and AtFtsH3 (B). A and B, results for wild type mitochondria. C, results for mitochondria isolated from ftsh3 mutant.
Finally, we assessed the stability of the 2-MDa complex present in the ftsh3 mutant (Fig. 5C). This complex is composed of PHBs and AtFtsH10 homo-oligomers. Western blot analysis with anti-PHBs and anti-AtFtsH10 antibodies indicated that DDM treatment of mitochondria isolated from the ftsh3 mutant results in a complete absence of the 2-MDa complex. Only a 1-MDa PHB complex can be easily detected, although the AtFtsH10 protein is observed as a slight smear below 2 MDa. We conclude from this experiment that the AtFtsH10-PHB complex is unstable, and the stronger detergent results in its disintegration. Moreover, in the presence of this detergent, AtFtsH10 is unable to form an independent homo-oligomeric complex free of prohibitins.
Taken together, our results indicate that in A. thaliana mitochondria an intact m-AAA complex cannot be sequestered from the m-AAA-PHB complex. Moreover, the different m-AAA-PHB complexes differ in their stability. Complexes containing only AtFtsH3 and prohibitins are relatively stable, whereas those containing only AtFtH10 and prohibitins are highly unstable. The complexes comprising prohibitins and both AtFtsH3 and AtFtsH10 show intermediate stability.
DISCUSSION
Our results demonstrate that Arabidopsis m-AAA proteases (AtFtsH3 and AtFtsH10), like their yeast counterparts, assemble with prohibitins into high molecular weight complexes of ∼2 MDa. A smaller PHB complex (∼1 MDa) not containing m-AAA proteases was also detected in Arabidopsis mitochondria. In contrast, the m-AAA proteases were found exclusively in complexes with prohibitins. The results presented here also offer an insight into subunit composition of the high molecular weight complexes of m-AAA proteases with prohibitins. Homo-oligomeric AtFtsH3 and AtFtsH10 complexes and hetero-oligomeric assembles of both these proteases were identified in leaf mitochondria.
Based on literature data, it seems that the ability of m-AAA protease subunits to form homo-oligomers is different in various taxons, despite their relatively high overall sequence homology. Yeast m-AAA proteases (Yta10 and Yta12) (14) and Paraplegin in mammals (15) cannot form functional homo-oligomeric complexes, whereas AFG3l1 and AFG3l2 in mouse and hAFG3L2 in human are capable of oligomerization in the absence of the respective partner (15). Thus, at least one m-AAA protease present in the organisms examined was not able to homo-oligomerize. Our results indicate that both Arabidopsis m-AAA proteases, AtFtsH3 and AtFtsH10, can homo-oligomerize. The evidence for the co-existence of homo-oligomeric complexes composed of AtFtsH3 and hetero-oligomeric complexes formed by AtFtsH3 and AtFtsH10 in Arabidopsis mitochondria isolated from leaves of wild type plants was obtained by co-immunoprecipitation, immunodepletion, and BN/SDS-PAGE analysis. Furthermore, using the BN-PAGE technique, we found that in the absence of AtFtsH3, homo-oligomeric complexes composed of AtFtsH10 assemble in the ftsh3 mutant. A lack of an easily detectable phenotype for ftsh3 (data not shown) suggests that the homo-oligomeric AtFtsH10 complexes at least partially substitute for the loss of the native homo- and hetero-oligomeric complexes containing AtFtsH3. Moreover, successful complementation of yeast yta12/yta12 null mutant by both m-AAA proteases indicates that AtFtsH3 or AtFtsH10 homo-oligomers are functional and can substitute the endogenous Yta10p-Yta12p complex. Taken together our results are consistent with the view (15) that the housekeeping functions of the m-AAA proteases can be carried out by complexes with variable subunit composition, whereas individual m-AAA proteases/subunits may possess additional specific functions or substrates.
Of the two m-AAA proteases present in A. thaliana leaves of wild type plants, AtFtsH3 seems to be the predominant one. Our results indicate that the AtFtsH3 transcript is almost three times more abundant than the AtFtsH10 transcript, whereas the AtFtsH3 protein is at least five times more abundant than AtFtsH10 in this plant organ. Analysis of publicly available expression data accessible through the Genevestigator data base indicates that the steady-state level of the AtFtsH3 transcripts is extremely low in pollen (31). Therefore, the high abundance of AtFtsH10 homo-oligomers may be expected at least in this plant tissue. Variable, tissue-specific composition of m-AAA complexes has already been shown in mammals (15). It has also been proposed that different subunit composition leads to altered substrate specificity to meet the demands of the various cell types (12). It is therefore of interest to evaluate the extent of cooperation and redundancy between AtFtsH3 and AtFtsH10 as well as to determine the distinct functions of these isoenzymes to understand the role of m-AAA proteases in growth and development of A. thaliana.
The results presented here suggest that relative abundance of the two isoenzymes of Arabidopsis m-AAA in different plant tissues is not only regulated at the transcriptional level but also at the translational level. We found that the significantly increased abundance of the AtFtsH10 protein in ftsh3 relative to wild type plants was not due to an increased level of the respective transcript or an elevated stability of the AtFtsH10 protein. Rather, a more efficient allocation of AtFtsH10 mRNA to polysomes seemed to be (at least partially) responsible for the increased accumulation of AtFtsH10. The mechanism leading to the increased efficiency of synthesis of the AtFtsH10 protein in ftsh3 remains unknown; however, this finding also suggests that in wild type plants translational regulation may modulate the level of the m-AAA subunits.
However, complementation studies indicate that functions of plant and yeast m-AAA proteases are at least partially overlapping. It has been shown that control of proteolytic maturation of a mitoribosomal subunit Mrpl32 is the key function of yeast m-AAA proteases (7). The observation that AtFtsH3 and AtFtsH10 can compensate for the loss of Yta10-Yta12 complex suggests that both plant proteases are able to process yeast MrpL32. It is conceivable that AtFtsH3 and AtFtsH10 play the same role in Arabidopsis mitochondria. However, the plant homolog of Mrpl32 has not been identified so far.
It has been reported that the PHB complex appears to act as a chaperone, stabilizing various respiratory chain subunits (16, 18). However, it is currently suggested that the PHB complexes act rather as protein and lipid scaffolds controlling inner membrane organization and integrity (19–20, 32). In this context, we present results that indicate that the complexes containing AtFtsH10, hetero-oligomeric in wild type plants and homo-oligomeric in ftsh3, are unstable unless assembled together with prohibitins to form the supercomplexes of 2 MDa. Under conditions similar to those successfully used to pry apart the yeast m-AAA complex from the m-AAA-PHB supercomplex, we could not identify prohibitin-independent complexes of AtFtsH10. We believe that the failure to detect stable AtFtsH10 complexes without PHBs can be explained by their rapid breakdown, because we did observe disassembly intermediates. Moreover, the homo-oligomeric complexes composed of AtFtsH10 are definitely less stable than the AtFtsH3/AtFtsH10 hetero-oligomers. This conclusion based on BN-PAGE/SDS-PAGE analysis is in agreement with our results showing that the degradation rate of the AtFtsH10 protein is accelerated in the ftsh3 mutant (where AtFtsH10 only forms homo-oligomers) compared with wild type plants (AtFtsH3/AtFtsH10 hetero-oligomers are present). The highest resistance to detergent treatment was exhibited by the homo-oligomers of AtFtsH3 present in mitochondria of wild type plants. Thus, the stability of the Arabidopsis m-AAA complexes depends on their subunit composition. Interestingly, the same conclusion can be reached for mammalian m-AAA complexes after analysis of the data published by Atorino et al. (8). The homo-oligomers composed of AFG3L2 are much more sensitive to detergent treatment than the hetero-oligomers composed of both AFG3L2 and Paraplegin.
Recently, Van Aken et al. (22) using an approach based on tandem affinity purification showed that all five expressed prohibitins of type I and type II are present within a multimeric complex in Arabidopsis mitochondria isolated from cell suspension culture. Our data on mitochondria isolated from Arabidopsis leaves support this result. However, in sharp contrast to our results, no m-AAA proteases were found by Van Aken et al. (22) in the PHB complexes purified by tandem affinity purification. The low stability and rapid disintegration of Arabidopsis m-AAA complexes in the presence of detergent observed here could be one of the reasons for this discrepancy, another being the different biological material studied. In conclusion, our findings based on BN-PAGE analysis and co-immunoprecipitation demonstrate a direct physical interaction between prohibitins and m-AAA proteases in Arabidopsis mitochondria. The results imply that Arabidopsis m-AAA proteases cannot form stable high molecular weight assemblies without prohibitins, although prohibitins, conversely, are able to form m-AAA-independent complexes. Contrary to yeast and humans, the formation of the m-AAA complexes in Arabidopsis is strictly dependent on the PHB complex. Thus, in plants prohibitins may be considered as a kind of scaffold for m-AAA proteases.
Supplementary Material
Acknowledgment
We thank Prof. T. Langer, University of Cologne, who provided yeast strains W303, YHA101, YHA201, and YHA301.
This work was supported by Grant N301 119 32/4116 from the Ministry of Science and Higher Education.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- m-AAA
- matrix AAA protease
- i-AAA
- intermembrane AAA protease
- PHB
- prohibitin
- DDM
- detergent dodecyl maltoside
- BN
- blue native.
REFERENCES
- 1.Arnold I., Langer T. (2002) Biochim. Biophys. Acta 1592, 89–96 [DOI] [PubMed] [Google Scholar]
- 2.Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003) Plant Cell 15, 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu F., Park S., Rodermel S. R. (2004) Plant J. 37, 864–876 [DOI] [PubMed] [Google Scholar]
- 4.Langer T. (2000) Trends Biochem. Sci. 25, 247–251 [DOI] [PubMed] [Google Scholar]
- 5.Arlt H., Steglich G., Perryman R., Guiard B., Neupert W., Langer T. (1998) EMBO J. 17, 4837–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreirinha F., Quattrini A., Pirozzi M., Valsecchi V., Dina G., Broccoli V., Auricchio A., Piemonte F., Tozzi G., Gaeta L., Casari G., Ballabio A., Rugarli E. I. (2004) J. Clin. Invest. 113, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., Langer T. (2005) Cell 123, 277–289 [DOI] [PubMed] [Google Scholar]
- 8.Atorino L., Silvestri L., Koppen M., Cassina L., Ballabio A., Marconi R., Langer T., Casari G. (2003) J. Cell Biol. 163, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korbel D., Wurth S., Käser M., Langer T. (2004) EMBO Rep. 5, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaser M., Kambacheld M., Kisters-Woike B., Langer T. (2003) J. Biol. Chem. 278, 46414–46423 [DOI] [PubMed] [Google Scholar]
- 11.Leonhard K., Guiard B., Pellecchia G., Tzagoloff A., Neupert W., Langer T. (2000) Mol. Cell 5, 629–638 [DOI] [PubMed] [Google Scholar]
- 12.Duvezin-Caubet S., Koppen M., Wagener J., Zick M., Israel L., Bernacchia A., Jagasia R., Rugarli E. I., Imhof A., Neupert W., Langer T., Reichert A. S. (2007) Mol. Biol. Cell 18, 3582–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. (2007) EMBO J. 26, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arlt H., Tauer R., Feldmann H., Neupert W., Langer T. (1996) Cell 85, 875–885 [DOI] [PubMed] [Google Scholar]
- 15.Koppen M., Metodiev M. D., Casari G., Rugarli E. I., Langer T. (2007) Mol. Cell. Biol. 27, 758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steglich G., Neupert W., Langer T. (1999) Mol. Cell. Biol. 19, 3435–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Back J. W., Sanz M. A., De Jong L., De Koning L. J., Nijtmans L. G., De Koster C. G., Grivell L. A., Van Der Spek H., Muijsers A. O. (2002) Protein Sci. 11, 2471–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijtmans L. G., de Jong L., Artal Sanz M., Coates P. J., Berden J. A., Back J. W., Muijsers A. O., van der Spek H., Grivell L. A. (2000) EMBO J. 19, 2444–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F. T., von Kleist-Retzow J. C., Waisman A., Westermann B., Langer T. (2008) Genes Dev. 22, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merkwirth C., Langer T. (2009) Biochim. Biophys. Acta 1793, 27–32 [DOI] [PubMed] [Google Scholar]
- 21.Chen J. C., Jiang C. Z., Reid M. S. (2005) Plant J. 44, 16–24 [DOI] [PubMed] [Google Scholar]
- 22.Van Aken O., Pecenková T., van de Cotte B., De Rycke R., Eeckhout D., Fromm H., De Jaeger G., Witters E., Beemster G. T., Inzé D., Van Breusegem F. (2007) Plant J. 52, 850–864 [DOI] [PubMed] [Google Scholar]
- 23.Kolodziejczak M., Kolaczkowska A., Szczesny B., Urantowka A., Knorpp C., Kieleczawa J., Janska H. (2002) J. Biol. Chem. 277, 43792–43798 [DOI] [PubMed] [Google Scholar]
- 24.Urantowka A., Knorpp C., Olczak T., Kolodziejczak M., Janska H. (2005) Plant Mol. Biol. 59, 239–252 [DOI] [PubMed] [Google Scholar]
- 25.Aronsson H., Jarvis P. (2002) FEBS Lett. 529, 215–220 [DOI] [PubMed] [Google Scholar]
- 26.Kahlau S., Bock R. (2008) Plant Cell 20, 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gietz R. D., Woods R. A. (1994) in Molecular Genetics of Yeast: Practical Approaches (Johnston J. A. ed) pp. 121–134, Oxford University Press, New York [Google Scholar]
- 28.Janska H. (2005) Physiol. Plant. 123, 399–405 [Google Scholar]
- 29.Kolodziejczak M., Gibala M., Urantowka A., Janska H. (2007) Physiol. Plant. 129, 135–142 [Google Scholar]
- 30.Zanetti M. E., Chang I. F., Gong F., Galbraith D. W., Bailey-Serres J. (2005) Plant Physiol. 138, 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004) Plant Physiol. 136, 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., Langer T. (2009) J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.