Abstract
Phosphorylation regulates transcription factor activity by influencing dimerization, cellular localization, activation potential, and/or DNA binding. Nevertheless, precisely how this post-translation modification mediates these processes is poorly understood. Here, we examined the role of phosphorylation on the DNA-binding properties of MafA and MafB, closely related transcriptional activators of the basic-leucine zipper (b-Zip) family associated with cell differentiation and oncogenesis. Many common phosphorylation sites were identified by mass spectrometry. However, dephosphorylation only precluded the detection of MafA dimers and consequently dramatically reduced DNA-binding ability. Analysis of MafA/B chimeras revealed that sensitivity to the phosphorylation status of MafA was imparted by sequences spanning the C-terminal dimerization region (amino acids (aa) 279–359), whereas the homologous MafB region (aa 257–323) conveyed phosphorylation-independent DNA binding. Mutational analysis showed that formation of MafA dimers capable of DNA binding required phosphorylation within the distinct N-terminal transactivation domain (aa 1–72) and not the C-terminal b-Zip region. These results demonstrate a novel relationship between the phosphoamino acid-rich transactivation and b-Zip domains in controlling MafA DNA-binding activity.
Keywords: DNA-binding Protein, General Transcription Factors, Insulin Secretion, Protein-DNA Interaction, Protein Phosphorylation, Basic L-zip, MafA, MafB, Protein Dimer
Introduction
Phosphorylation influences the activity of many transcription factors. However, the precise manner by which this post-translational modification impacts regulation has been defined mechanistically in only a few cases (1, 2). Here, we have focused on understanding how phosphorylation affects the DNA-binding potential of MafA and MafB, whose basic-leucine zipper (b-Zip)2 region defines their dimerization and DNA-binding properties. There are two subfamilies of mammalian musculoaponeurotic fibrosarcoma (i.e. Maf) proteins, termed large and small Mafs (3). The small Maf proteins (MafF, MafG, MafK, and MafT) lack a transactivation domain and affect transcription through dimerization with related and distinct proteins (4–7). The large Maf proteins (MafA, MafB, c-Maf, and NRL) contain an N-terminal transactivation domain (8–11), which has considerable identity among MafA, MafB, and c-Maf (12, 13).
Large Mafs are required in promoting many distinct physiological processes by binding as dimers to Maf-responsive elements and activating transcription (14, 15). Among other properties, chicken L-Maf (termed MafA in mammals) is involved in lens development (16), mammalian MafB is required for segmentation of the hindbrain (17), mammalian c-Maf contributes to chondrocyte differentiation (9, 18), and mammalian NRL functions in eye rod formation (10). Moreover, MafA and MafB have recently been shown to be essential within the mammalian pancreas, with islet α and β cell production requiring the actions of MafB during development and adult β islet activity uniquely MafA (19–21). In addition, large Maf proteins mediate cellular transformation in vitro and are overexpressed in human angioimmunoblastic T cell lymphomas and multiple myeloma and contribute directly to cancer progression (22–24).
The activity of MafA is regulated by a variety of post-translational modification mechanisms, including phosphorylation, ubiquitination, and sumoylation (25–29). The best studied MafA modification is phosphorylation, which impacts protein stability (26–28), transactivation (26, 29), and DNA binding (30). For example, a priming phosphorylation at serine 65 in MafA (or Ser70 in MafB) is necessary for both ubiquitin-mediated degradation (26) and glycogen synthase kinase 3-mediated phosphorylation (27, 28), the latter enhancing transactivation and transformation potential (29). In addition, the in vitro DNA-binding capabilities of MafA are reduced by endogenous and exogenous phosphatases (30). Inhibition could entail phosphorylation directly within the basic region of MafA, as found for a variety of different transcription factors, including c-Myb (31), PRH/Hex (31), and HNF4 (31). Alternatively, this modification might influence MafA dimer formation and, as a result, DNA-binding potential. Such a mechanism has been described for STAT1, wherein tyrosine phosphorylation of cytoplasmic STAT1 potentiates dimerization and transcriptional activation (32, 33).
Large Maf proteins appear to be heavily phosphorylated (27, 28). Here, we first used mass spectrometry to identify the phosphoamino acids in MafA and MafB. A high phosphorylation state was shown to be uniquely important for the production of dimers capable of DNA binding in purified, full-length MafA, as phosphatase treatment induced multimerization and eliminated DNA binding. Exchanging the C-terminal b-Zip-spanning sequences between MafA and MafB conferred phosphorylation-independent binding to MafA and sensitivity to MafB. Significantly, phosphorylation-dependent DNA binding was controlled by both the phosphorylation-rich N-terminal transactivation domain (amino acids (aa) 1–72) and the leucine zipper region. These results demonstrate that phosphorylation within the N-terminal region is not only critical to transactivation by enabling recruitment of co-activators like P/CAF (27), but also plays a novel role in DNA binding by mediating intramolecular interactions with the b-Zip domain.
EXPERIMENTAL PROCEDURES
DNA Constructs
S14A, S65A, S67A, S72A, S290A/S297A/S343A (MafA-C3A), and S14A/S65A/S67A/S72A (MafA-N4A) were prepared in a cytomegalovirus (CMV)-driven myc-MafA expression plasmid (pCMV4-myc) using the QuikChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA). MafA S14A/S49A/S51A/S52A/S56A/S60A/S61A/S64A/S65A/S67A/S72A/T53A/T57A (N13A) and S14E/S49E/S51E/S52E/S56E/S60E/S61E/S64E/S65E/S67E/S72E/T53E/T57E (N13E) were constructed in a similar manner in the CMV-driven pcDNA3.1(+) neo plasmid. Mouse MafA/MafB chimeras were produced by two-step PCR and cloned into the pCMV4-myc vector as described previously (34). MafA(73–359), MafA(151–359), MafA(279–359), and MafBBA(78–359) were prepared by PCR and cloned into pCMV4-myc. Primer sequence information is available upon request. DNA sequencing analysis confirmed the fidelity of each construct.
Cell Culture Transfection and Nuclear Extract Preparation
HeLa monolayer cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% calf serum. Each expression plasmid (4 μg) was transfected using Lipofectamine (Invitrogen) in cells plated on 100-mm dishes. Nuclear extracts were prepared 48 h after transfection in the absence of sodium orthovanadate (Na3VO4), as described previously (30). Each experiment was repeated at least three times using independently isolated plasmid preparations.
Purification of MafA and MafB
Adenovirus-driven MafA- or MafB-infected HeLa cells were cultured for 18 h (10 plaque-forming units/cell, 10 × 150-mm dishes). Cells were washed twice with phosphate-buffered saline and suspended in lysis buffer containing 100 mm Tris-HCl (pH 8.0) and 0.5% Nonidet P-40 at 4°C for 5 min. After centrifugation at 10,000 × g at 4°C for 5 min, the nuclear pellet was dissolved for 30 min in 8 m urea (in 20 mm Tris-HCl (pH 8.0), 100 mm NaCl). The supernatant obtained after a 15,000 × g centrifugation was loaded onto a 1-ml nickel-agarose (Qiagen) column, which was washed with 10 ml of 8 m urea (in 20 mm Tris-HCl (pH 8.0), 100 mm NaCl), 10 ml of 50 mm imidazole (in 20 mm Tris-HCl (pH 8.0), 100 mm NaCl), and then MafA or MafB was eluted with 500 mm imidazole (in 20 mm Tris-HCl (pH 8.0), 100 mm NaCl). The proteins were dialyzed at 4 °C overnight against a buffer containing 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm dithiothreitol, and 5% glycerol, and then stored at −80 °C.
In Vitro Phosphatase Treatment and the Dimerization/DNA Binding Assays
MafA- or MafB-transfected nuclear extract (25 μg, 20-μl total reaction volume) was incubated at 37 °C in 1× phosphatase buffer (20 mm Tris-HCl (pH 7.4), 1 mm dithiothreitol, 0.1 mm EGTA, 5 mm MgCl2, 1× protease inhibitor mixture (Roche Applied Science)) with either 10 mm NaCl or 10 mm Na3VO4. (The endogenous large Maf phosphatases are inhibited by 10 mm Na3VO4) (35). MafA or MafB protein levels were determined from 2 μl of the reaction sample by Western blotting, and DNA-binding ability was analyzed from 9 μl in the electrophoretic mobility shift assay with 32P-labeled rat insulin II C1 element (5′-GGAAACTGCAGCTTCAGCCCCTC-3′) (30). MafA and MafB dimerization capabilities were determined with the remaining 9 μl, which was mixed with DNA-binding buffer (30 μl) and 1 μl of cold 20 fmol/μl C1 element. After 10 min at room temperature, the reaction mixture was separated on a 5% native-PAGE, and MafA or MafB protein levels were examined by Western blotting. These same assays were performed under similar conditions with 1.2 μg of purified MafA and MafB treated with 6 units of calf intestine alkaline phosphatase (CIAP; Promega) in the presence or absence of phosphatase inhibitors (1× general phosphatase inhibitor (Roche Applied Science), 10 mm sodium pyrophosphate (NaPPi), or 10 mm Na3VO4) for 1 h at 37 °C. The following antibodies were used in these studies: rabbit α-mouse MafA (Bethyl Laboratories, 1225 (raised against an N-terminal peptide) and 1069 (raised against a C-terminal peptide)), rabbit α-mouse MafB (Bethyl Laboratories, 658 (raised against an N-terminal peptide)), mouse α-c-myc (9E10; Santa Cruz Biotechnology), α-rabbit IgG horseradish peroxidase conjugate (W401B; Promega), and α-mouse IgG horseradish peroxidase conjugate (W402B; Promega).
Separation of MafA and MafB by Sucrose Gradient Ultracentrifugation
Each phosphatase-treated sample (300 μl) was loaded onto a 30% to 5% sucrose gradient prepared in Beckman 50 Ultra-Clear™ tubes (30% 0.25 ml; 25% 0.5 ml; 20% 0.75 ml; 15% 1.0 ml; 10% 1.0 ml; 5% 1.0 ml), and centrifuged at 48,000 rpm for 18 h at 4 °C. Three-hundred μl/fractions were collected after removing 1.3 ml from the top of the tube. Western blotting analysis was performed to detect MafA or MafB in the fractions.
RESULTS
Phosphorylation Is Required for MafA Dimerization, but not MafB
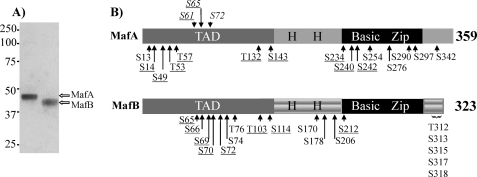
Because a wide range of MafA and MafB activities is controlled by phosphorylation, the extent of modification with purified MafA and MafB (Fig. 1A) was determined using an unbiased proteomics approach. Mass spectrometry analysis revealed that the phosphorylated serines and threonines within these highly charged proteins were concentrated principally within the N- and C-terminal regions (Fig. 1B and supplemental Tables 1 and 2). The number of phosphorylated amino acids detected was similar to the estimate obtained from radiolabeled inorganic phosphate incorporation into endogenous MafA (26).
FIGURE 1.
Phosphorylation sites in mouse MafA and MafB identified by mass spectrometry. A, purified MafA and MafB isolated from adenovirus-infected HeLa cells. Protein purity was determined by Coomassie Blue staining after SDS-PAGE and Western blotting (data not shown). B, phosphorylation sites in MafA and MafB identified by mass spectrometry. The phosphoamino acids site identified in more than two spectra are denoted, whereas those described previously in vitro and not found here are italicized (27, 28). The underlined amino acids are conserved in MafA and MafB. TAD, transcription activation domain; H H, histidine-rich region.
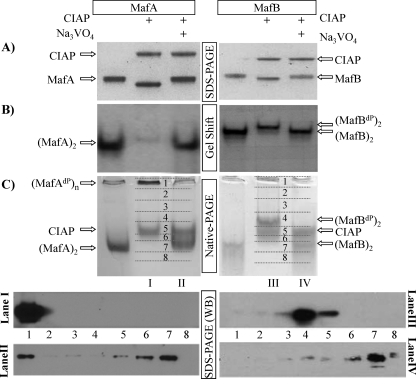
We next examined how phosphorylation influenced the biochemical properties of MafA and MafB, specifically in regard to SDS-PAGE mobility, DNA binding, and homodimerization. Purified MafA and MafB were incubated with CIAP in the presence and absence of Na3VO4, a general phosphatase inhibitor under the conditions of analysis. (Notably, NaPPi and a commercially available mixture of phosphatase inhibitors behaved similarly to Na3VO4 (supplemental Fig. 1).) CIAP treatment increased the mobility of both MafA and MafB (Fig. 2A), a result expected from earlier studies (29). Dephosphorylation of MafA in β cell line nuclear extracts profoundly reduced insulin C1 element DNA-binding potency (30), as also observed here with purified MafA (Fig. 2B). In contrast, there was little to no effect on the DNA-binding ability of MafB under these conditions (Fig. 2B). The same DNA-binding properties were obtained with CIAP-treated MafA and MafB using the Maf-responsive element probes from the human β-globin and mouse crystalline genes (supplemental Fig. 2). These data demonstrated that phosphorylation only influenced the DNA-binding capabilities of MafA and not closely related and highly phosphorylated MafB.
FIGURE 2.
Phosphorylation affects (MafA)2 dimer formation and DNA binding, but not MafB. Purified MafA and MafB were treated with CIAP in the presence of Na3VO4 or NaCl for 60 min at 37 °C. A, the location of CIAP and the large Maf proteins was determined by Coomassie Blue staining after SDS-PAGE. Note that MafA and MafB have a much faster mobility after treatment with CIAP alone. B, MafA DNA binding was specifically reduced by CIAP treatment. In contrast, both unphosphorylated and phosphorylated MafB effectively binds the insulin gene C1 element probe. MafA and MafB dimers were labeled as (MafA)2 and (MafB)2, respectively. (MafBdP)2 denotes the dephosphorylated MafB dimer. C, (MafA)2 dimers were no longer observed following CIAP treatment. Upper panels, treated proteins were visualized by Coomassie Blue staining after native-PAGE. The location of the (MafA)2 and (MafB)2 dimers was determined in cross-linking studies with tetranitromethane (data not shown). (MafAdP)n is the dephosphorylated MafA multimer. Lower panels, lanes I, II, III, and IV were dissected into eight pieces and the eluted proteins analyzed by Western blotting using α-MafA or α-MafB antibody.
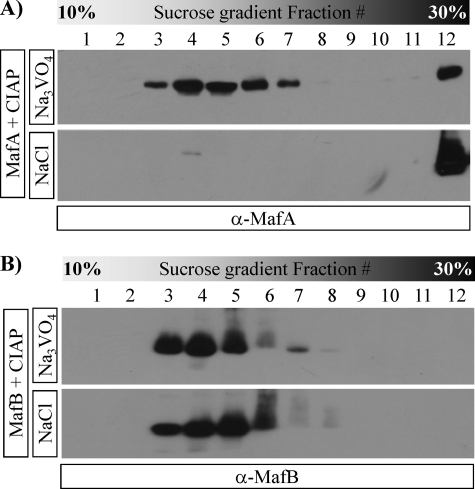
CIAP-treated MafA and MafB were analyzed by native-PAGE to test whether the loss in MafA dimer formation caused the change in DNA-binding activity. The resulting MafA and MafB products were visualized by Coomassie Blue staining and by dividing the gel lanes into eight fractions and Western blotting the eluted proteins (Fig. 2C). MafA treated with only CIAP did not enter the gel, whereas MafA and MafB treated with Na3VO4 + CIAP migrated similarly to the control, untreated proteins. Dephosphorylation also shifted the mobility of MafB, but to a much lesser extent than the MafA protein. Because protein charge and size affect mobility in this assay, we further evaluated these proteins by sucrose gradient ultracentrifugation. MafA treated with CIAP alone was found in samples containing high molecular weight proteins, here represented by the 30% sucrose fraction (Fig. 3), suggesting that dephosphorylation induced the multimerization of MafA. However, MafA protected by Na3VO4 co-sedimented with phosphorylated and dephosphorylated MafB in lower sucrose fractions (Fig. 3). Significantly, only MafA dimers (i.e. (MafA)2) and not the high molecular weight form (MafAdP)n was active in gel shift assays. These data showed that a phosphorylation event(s) only controlled the stable formation of the dimer species required for DNA binding in MafA.
FIGURE 3.
Dephosphorylated MafA has a profoundly different sucrose gradient sedimentation rate than phosphorylated MafA or phosphorylated/unphosphorylated MafB. Purified MafA or MafB was treated with CIAP in the presence of Na3VO4 or NaCl and separated by sucrose gradient ultracentrifugation (see “Experimental Procedures”). The MafA (A) or MafB (B) proteins in the collected fractions were detected by Western blotting. Notably, dephosphorylated MafA (i.e. treated with CIAP/NaCl) is shifted to the 30% sucrose high molecular weight protein fraction, whereas the sedimentation rates of phosphorylated MafA and phosphorylated/unphosphorylated MafB are similar.
The MafA C-terminal Region Conveys Phosphatase Sensitivity
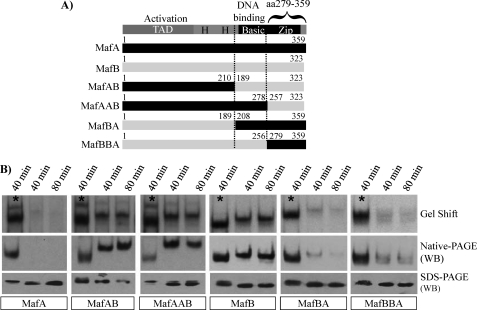
MafA/MafB chimeras were constructed to localize the region(s) controlling phosphorylation-responsive DNA binding in MafA (Fig. 4A). Nuclear extracts prepared from transfected cells were incubated with or without Na3VO4, conditions that allow or prevent (plus Na3VO4) an endogenous phosphatase(s) in HeLa or islet β cell lines from inhibiting MafA DNA-binding activity (30). Strikingly, phosphorylation-independent dimer formation and DNA-binding activity were obtained upon exchanging the C-terminal region of MafA with that of MafB (compare MafAB and MafAAB with MafA and MafB in Fig. 4B). Significantly, the SDS-PAGE mobility of these dephosphorylated protein chimeras was expectedly faster (also see Fig. 2A), whereas there was no reduction in steady-state protein levels (Fig. 4B). Furthermore, the MafA C terminus also imparted phosphorylation dependence to MafB in these assays (compare MafBA and MafBBA with MafA and MafB in Fig. 4B). These data illustrated the importance of sequences within the leucine zipper-spanning region of MafA and MafB in regulating phosphorylation-dependent DNA binding (i.e. MafA, aa 279–359; MafB, aa 257–323).
FIGURE 4.
The C-terminal region of MafA and MafB is involved in phosphosensitive dimer formation. A, schematics illustrating the transactivation, histidine-rich, and basic-dimerization (b-Zip) domains common to MafA, MafB, and the MafA/B chimeras are shown. B, the large Maf construct in HeLa nuclear extracts was incubated at 37 °C for 40 or 80 min in the presence of Na3VO4 (denoted by asterisks) or NaCl. Each of the treated samples was analyzed for DNA-binding activity (i.e. Gel Shift), dimerization ability (Native-PAGE), and mobility (SDS-PAGE). The α-MafA (BL1225) antibody was used in the Western blot (WB) analysis to locate the MafAB and MafAAB, whereas α-MafB (BL628) antibody was utilized for MafBA and MafBBA.
Phosphatase Sensitivity Is Not Due to Phosphorylation within the MafA C Terminus
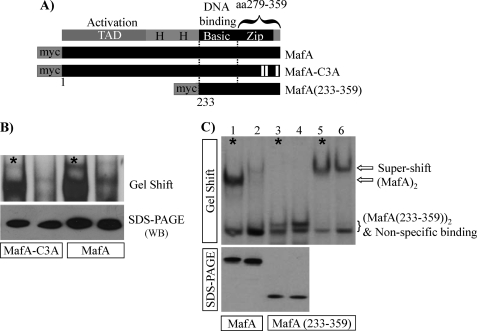
To determine whether the unique C-terminal phosphoamino acids at Ser290, Ser297, and Ser342 (Fig. 1) directly influenced DNA binding, a compound alanine mutant was constructed and transfected into HeLa cells (MafA-C3A in Fig. 5A). S290A/S297A/S342A and wild type MafA had similar DNA-binding ability, and both were sensitive to phosphatase treatment (Fig. 5B). Moreover, the cellular phosphatase(s) did not reduce the inherent DNA-binding activity of MafA(233–359), a myc-tagged construct expressing only the C-terminal b-Zip-spanning region of MafA (dephosphorylation did not affect (MafA(233–359))2 supershifted levels in lanes 5 and 6 of Fig. 5C as well). These data strongly indicated that the phosphorylated amino acid(s) regulating MafA-binding activity resided outside the MafA C-terminal b-ZIP region.
FIGURE 5.
The unique phosphorylated amino acids in the MafA C-terminal region do not control phosphatase(s)-sensitive DNA binding. A, schematic diagrams of the MafA-C3A (S290A/S297A/S342A) and MafA(233–359) mutants are shown. B, MafA-C3A and wild type MafA activity in HeLa nuclear extracts was sensitive to endogenous phosphatase(s)-mediated loss in DNA binding and SDS-PAGE mobility. WB, Western blotting. C, MafA(233–359) gel shift activity was unaffected by endogenous phosphatase(s) treatment. An α-myc antibody supershift is shown in lanes 5 and 6 due to a nonspecific binding complex co-migrating with (or near) MafA(233–359). The asterisks denote the samples containing with Na3VO4.
Multiple Phosphorylation Events within the MafA N-terminal Activation Domain Sustain Dimer/DNA Binding
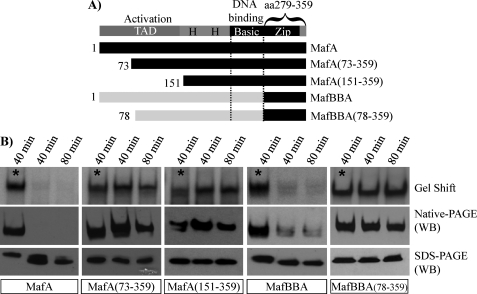
To localize the phosphorylated amino acid(s) regulating (MafA)2 dimer formation, N-terminal deletion mutants of MafA were constructed (Fig. 6A). Interestingly, the HeLa endogenous phosphatase(s) did not inhibit (MafA)2 production or DNA-binding activity in MafA(73–359) or MafA(151–359) (Fig. 6B). Furthermore, sensitivity was also lost upon removing aa 1–77 in MafBBA (Fig. 6B), a highly conserved region with MafA. In addition, the sucrose gradient sedimentation rate of MafA(73–359) was unaffected by phosphatase(s) treatment, with properties now indistinguishable from MafAAB and phosphorylated MafA (supplemental Fig. 3). These results revealed the significance of the MafA N-terminal region in forming stable (MafA)2 dimers.
FIGURE 6.
Phosphorylation within the N-terminal transactivation domain is important to MafA DNA-binding activity. A, schematics of the N-terminally deleted MafA and MafBBA constructs are shown. B, HeLa nuclear extracts containing the transfected constructs were incubated at 37 °C for 40 or 80 min in the presence of Na3VO4 (denoted by asterisks) or NaCl. Each of the treated samples was analyzed for DNA-binding activity (i.e. Gel Shift), dimer formation (Native-PAGE), and mobility (SDS-PAGE). The α-MafA (BL1069) antibody was used in the Western blot (WB) analysis. Notably, the endogenous phosphatase(s) does not influence MafA(73–359), MafA(151–359), or BBA(78–359) dimer formation, DNA binding, or SDS-PAGE mobility.
Phosphoamino acid mutants within the MafA N terminus were made to define their role in (MafA)2 formation more precisely. It is noteworthy that many more phosphosites were found within this region of MafB by mass spectrometry than MafA (Fig. 1). We believe our inability to detect phosphorylation at conserved Ser60, Ser61, Ser64, Ser65, and Ser67 is due a technical limitation(s) in our analysis, as even the corresponding unphosphorylated peptides were not found (supplemental Tables 1 and 2). Additionally, the phosphorylation-independent-binding properties of MafA(73–359) and MafBBA(77–359) indicated that phosphosite usage within the N-terminal region of MafA and MafB was conserved (Fig. 6B), and mutants were constructed accordingly.
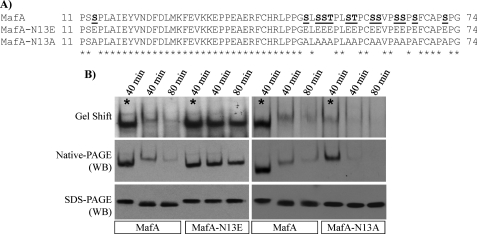
Because single and limited combination phosphosite mutants had little or no significant effect on MafA DNA binding (supplemental Fig. 4), comprehensive Ser/Thr to Ala or Glu mutants were constructed to block or mimic the phosphorylation state of the N-terminal region (Fig. 7A). Meaningfully, MafA-N13E migrated to the same position as Na3VO4-protected MafA after SDS-PAGE (Fig. 7B). The ability of MafA-N13E to form stable dimers and bind DNA was also unaffected by the endogenous phosphatase(s), with the level and mobility indistinguishable from phosphorylated MafA (Fig. 7B). In contrast, MafA-N13A behaved like unphosphorylated MafA. Thus, MafA-N13A-binding activity was comparatively poor, and it migrated like dephosphorylated MafA after SDS-PAGE (Fig. 7B). These results demonstrate that multiple phosphorylation events within the N-terminal region are fundamental to the detection of (MafA)2 or (MafBBA)2 dimers and consequently DNA binding.
FIGURE 7.
The high negative charge density in the N-terminal regions appears significant to MafA activity. A, location of the phosphosite mutants within the MafA N-terminal region. The S/T mutant sites in N13A and N13E are highlighted and underlined in the wild type. B, the DNA-binding activity (Gel Shift) and dimer formation ability (Native-PAGE) of MafA-N13E were irresponsive to the endogenous phosphatase(s) and migrated like Na3VO4-treated MafA after SDS-PAGE. In contrast, the DNA binding of the N13A mutant was very low and ran like dephosphorylated MafA. WB, Western blotting; *, with Na3VO4.
DISCUSSION
Crystal structure analysis of the fos-jun heterodimer, GCN4 homodimer, and MafG homodimer demonstrated that b-Zip proteins dimerize and bind DNA through a continuous α-helix (36–38), with the basic region principally influencing cis-element binding and the coiled-coil leucine-zipper interface controlling dimerization. This highly structured region can independently mediate dimerization and DNA binding in vitro (39–42). Although the MafA b-Zip region was capable of DNA binding by itself (Fig. 5), we found that phosphorylation within the distinct N-terminal activation domain was also required in the wild type protein. In contrast, this activity in MafB was unaffected by the state of phosphorylation, although analysis of the MafBBA chimera showed that its highly modified N-terminal region conveyed phosphosensitivity. The C-terminal region of MafA (aa 279–359) and MafB (aa 257–323) was shown to respond distinctively to N-terminal domain phosphorylation, apparently by their ability to mediate stable dimer formation. These data illustrate a novel mechanism controlling transcription factor activity, involving phosphoregulated interactions between two distinct functional domains of MafA.
Mass spectrometry analysis of purified MafA and MafB identified many common and unique sites within these highly phosphorylated proteins (Fig. 1). Unfortunately, we were unable to detect MafA phosphorylation at Ser61, described for glycogen synthase kinase 3 in vitro or at Ser65, with the latter required for both glycogen synthase kinase 3 priming and ubiquitin-mediated degradation (26–28). Our underestimation of the phosphorylation events is likely a reflection of poor peptide generation due to the high charge density of the region. The ability to detect Ser49, Thr53, and Thr57 phosphorylation in MafA only after limited CIAP treatment supports this idea (supplemental Table 1). Further evidence for phosphorylation within the N-terminal region results from the ability to impart instability to MafA in the S65E phosphomimetic mutant (26). Moreover, MafA-N13E migrated like the heavily phosphorylated wild type protein after SDS-PAGE, whereas the behavior of N13A was similar to the unphosphorylated protein (Fig. 7B). The ability of the MafB N-terminal region to mediate the formation of stable (MafBBA)2 dimers (Fig. 6) also implies that this area in both proteins is highly phosphorylated, with detection of the MafA Ser65 equivalent phosphorylation at Ser70 in MafB indicating that the overall pattern is possibly conserved (Fig. 1).
Because phosphorylation by glycogen synthase kinase 3 at Ser61, Thr57, Thr53, and Ser49 potentiates MafA transactivation (27), we examined how the N13A and N13E mutants influenced the transcriptional effectiveness of chimeras between MafA aa 1–233 and the Saccharomyces cerevisiae transcription factor Gal4 DNA-binding domain. N13E activity was not significantly different from the wild type, whereas N13A was compromised severely (supplemental Fig. 5). We conclude that a high level of N-terminal region phosphorylation is not only necessary for detection of (MafA)2, but also transactivation domain strength.
Although many transcriptional regulators have been shown to be phosphoproteins, in only a few cases has phosphorylation been shown to affect DNA-binding activity directly. For example, phosphorylation within the basic region can be either a positive (e.g. Gal4) (43) or negative (PRH/Hex) (31) influence. It has also been proposed that dimerization of STAT1 is regulated by phosphorylation (32, 33), although others suggest that STATs exist as inactive dimers in the absence of phosphorylation (44, 45). We believe our data represent the first example of a distinct phosphorylation-rich domain in a transcription factor-stabilizing dimer formation and thereby DNA-binding domain activity. On the other hand, it is unclear whether the dephosphorylated high molecular weight multimers of MafA are due to monomer and/or dimer oligomerization (see Figs. 2 and 3). The phosphoregulatory mechanism described here is not unique to MafA, for at least c-Maf appears to be controlled in a similar manner (supplemental Fig. 6). It was striking that phosphatase(s)-sensitive DNA binding was imparted by multiple phosphorylation events rather than a single one as commonly found and that responsiveness was transferable. The C-terminal control region sequences of MafA and MafB spanned both conserved and nonconserved sequences, and we presently do not know which specific amino acids are essential to this response. We propose that interactions between the phospho-rich activation domain and b-Zip region modulate MafA structure and DNA-binding activity, presumably also creating unique interfaces for co-regulatory proteins.
A key issue is whether the C-terminal phosphoregulatory sequences of MafA defined here in vitro serve in control in vivo. Large Maf proteins play roles in development and tissue differentiation within the brain (MafB) (17), eye (NRL and MafA/L-Maf) (10, 16), cartilage (c-Maf) (18), kidney (MafB) (46), and pancreas (MafB) (19, 47). However, their significance in these situations may reflect differences in expression rather than fundamental activation properties. Evidence supporting distinctions in activation come from overexpression studies showing that MafA is capable of inducing the normally silent endogenous insulin gene in islet α (glucagon+) cells (48), liver cells (49, 50), and intestinal cells (51, 52). Under similar experimental conditions, MafB activates glucagon (and not insulin) in islet β cells. Most pertinent to the present studies, analysis of MafA/B chimeras in the chick in ovo electroporation assay demonstrated that insulin gene activation specifically depended upon MafA C-terminal region sequences (i.e. MafBAA, Fig. 4) (34). Notably, MafB+insulin+ cells generated during human embryonic stem cell differentiation were dysfunctional unless becoming MafA+insulin+ (53), supporting further efforts to understand how MafA and MafB regulate (at least) islet β cell function.
Supplementary Material
Acknowledgments
We thank Drs. Hayes McDonald and Amy-Joan L. Ham for support and guidance in the mass spectrometry analysis and Drs. Chad Hunter and Brian Wadzinski for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 DK42502 (to R. S.). This work was also supported by Postdoctoral Research Training Grant in Diabetes and Endocrinology 5T32 DK007061-34 (to N. L. V.), Juvenile Diabetes Foundation Postdoctoral Training Grant 3-2009-555 (to S. G.), and the Vanderbilt University Diabetes Research and Training Center Molecular Biology Core Laboratory (Public Health Service Grant P60 DK20593).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–6.
- b-Zip
- basic-leucine zipper
- aa
- amino acids
- CMV
- cytomegalovirus
- CIAP
- calf intestine alkaline phosphatase
- STAT
- signal transducers and activators of transcription.
REFERENCES
- 1.Hunter T. (2007) Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 2.Hunter T., Karin M. (1992) Cell 70, 375–387 [DOI] [PubMed] [Google Scholar]
- 3.Eychene A., Rocques N., Pouponnot C. (2008) Nat. Rev. 8, 683–693 [DOI] [PubMed] [Google Scholar]
- 4.Takagi Y., Kobayashi M., Li L., Suzuki T., Nishikawa K., Yamamoto M. (2004) Biochem. Biophys. Res. Commun. 320, 62–69 [DOI] [PubMed] [Google Scholar]
- 5.Vinson C., Myakishev M., Acharya A., Mir A. A., Moll J. R., Bonovich M. (2002) Mol. Cell. Biol. 22, 6321–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motohashi H., Shavit J. A., Igarashi K., Yamamoto M., Engel J. D. (1997) Nucleic Acids Res. 25, 2953–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujiwara K. T., Kataoka K., Nishizawa M. (1993) Oncogene 8, 2371–2380 [PubMed] [Google Scholar]
- 8.Benkhelifa S., Provot S., Lecoq O., Pouponnot C., Calothy G., Felder-Schmittbuhl M. P. (1998) Oncogene 17, 247–254 [DOI] [PubMed] [Google Scholar]
- 9.Kim J. I., Li T., Ho I. C., Grusby M. J., Glimcher L. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3781–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A. (2001) Nat. Genet. 29, 447–452 [DOI] [PubMed] [Google Scholar]
- 11.Blanchi B., Kelly L. M., Viemari J. C., Lafon I., Burnet H., Bévengut M., Tillmanns S., Daniel L., Graf T., Hilaire G., Sieweke M. H. (2003) Nat. Neurosci. 6, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 12.Matsushima-Hibiya Y., Nishi S., Sakai M. (1998) Biochem. Biophys. Res. Commun. 245, 412–418 [DOI] [PubMed] [Google Scholar]
- 13.Vinson C., Acharya A., Taparowsky E. J. (2006) Biochim. Biophys. Acta 1759, 4–12 [DOI] [PubMed] [Google Scholar]
- 14.Motohashi H., Katsuoka F., Shavit J. A., Engel J. D., Yamamoto M. (2000) Cell 103, 865–875 [DOI] [PubMed] [Google Scholar]
- 15.Nishizawa M., Kataoka K., Vogt P. K. (2003) Oncogene 22, 7882–7890 [DOI] [PubMed] [Google Scholar]
- 16.Ogino H., Yasuda K. (1998) Science 280, 115–118 [DOI] [PubMed] [Google Scholar]
- 17.Cordes S. P., Barsh G. S. (1994) Cell 79, 1025–1034 [DOI] [PubMed] [Google Scholar]
- 18.MacLean H. E., Kim J. I., Glimcher M. J., Wang J., Kronenberg H. M., Glimcher L. H. (2003) Dev. Biol. 262, 51–63 [DOI] [PubMed] [Google Scholar]
- 19.Nishimura W., Kondo T., Salameh T., El Khattabi I., Dodge R., Bonner-Weir S., Sharma A. (2006) Dev. Biol. 293, 526–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Artner I., Le Lay J., Hang Y., Elghazi L., Schisler J. C., Henderson E., Sosa-Pineda B., Stein R. (2006) Diabetes 55, 297–304 [DOI] [PubMed] [Google Scholar]
- 21.Zhang C., Moriguchi T., Kajihara M., Esaki R., Harada A., Shimohata H., Oishi H., Hamada M., Morito N., Hasegawa K., Kudo T., Engel J. D., Yamamoto M., Takahashi S. (2005) Mol. Cell. Biol. 25, 4969–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami Y. I., Yatabe Y., Sakaguchi T., Sasaki E., Yamashita Y., Morito N., Yoh K., Fujioka Y., Matsuno F., Hata H., Mitsuya H., Imagawa S., Suzuki A., Esumi H., Sakai M., Takahashi S., Mori N. (2007) Am. J. Surg. Pathol. 31, 1695–1702 [DOI] [PubMed] [Google Scholar]
- 23.Morito N., Yoh K., Fujioka Y., Nakano T., Shimohata H., Hashimoto Y., Yamada A., Maeda A., Matsuno F., Hata H., Suzuki A., Imagawa S., Mitsuya H., Esumi H., Koyama A., Yamamoto M., Mori N., Takahashi S. (2006) Cancer Res. 66, 812–819 [DOI] [PubMed] [Google Scholar]
- 24.Hurt E. M., Wiestner A., Rosenwald A., Shaffer A. L., Campo E., Grogan T., Bergsagel P. L., Kuehl W. M., Staudt L. M. (2004) Cancer Cell 5, 191–199 [DOI] [PubMed] [Google Scholar]
- 25.Shao C., Cobb M. H. (2009) J. Biol. Chem. 284, 3117–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo S., Burnette R., Zhao L., Vanderford N. L., Poitout V., Hagman D. K., Henderson E., Ozcan S., Wadzinski B. E., Stein R. (2009) J. Biol. Chem. 284, 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocques N., Abou Zeid N., Sii-Felice K., Lecoin L., Felder-Schmittbuhl M. P., Eychène A., Pouponnot C. (2007) Mol. Cell 28, 584–597 [DOI] [PubMed] [Google Scholar]
- 28.Han S. I., Aramata S., Yasuda K., Kataoka K. (2007) Mol. Cell. Biol. 27, 6593–6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benkhelifa S., Provot S., Nabais E., Eychène A., Calothy G., Felder-Schmittbuhl M. P. (2001) Mol. Cell. Biol. 21, 4441–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao L., Cissell M. A., Henderson E., Colbran R., Stein R. (2000) J. Biol. Chem. 275, 10532–10537 [DOI] [PubMed] [Google Scholar]
- 31.Soufi A., Noy P., Buckle M., Sawasdichai A., Gaston K., Jayaraman P. S. (2009) Nucleic Acids Res. 37, 3288–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sehgal P. B. (2000) Cell. Signal. 12, 525–535 [DOI] [PubMed] [Google Scholar]
- 33.Shuai K., Horvath C. M., Huang L. H., Qureshi S. A., Cowburn D., Darnell J. E., Jr. (1994) Cell 76, 821–828 [DOI] [PubMed] [Google Scholar]
- 34.Artner I., Hang Y., Guo M., Gu G., Stein R. (2008) J. Endocrinol. 198, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuoka T., Zhao L., Stein R. (2001) J. Biol. Chem. 276, 22071–22076 [DOI] [PubMed] [Google Scholar]
- 36.Kurokawa H., Motohashi H., Sueno S., Kimura M., Takagawa H., Kanno Y., Yamamoto M., Tanaka T. (2009) Mol. Cell. Biol. 29, 6232–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover J. N., Harrison S. C. (1995) Nature 373, 257–261 [DOI] [PubMed] [Google Scholar]
- 38.Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. (1992) Cell 71, 1223–1237 [DOI] [PubMed] [Google Scholar]
- 39.Kerppola T. K., Curran T. (1994) Oncogene 9, 3149–3158 [PubMed] [Google Scholar]
- 40.Sassone-Corsi P., Ransone L. J., Lamph W. W., Verma I. M. (1988) Nature 336, 692–695 [DOI] [PubMed] [Google Scholar]
- 41.Landschulz W. H., Johnson P. F., McKnight S. L. (1988) Science 240, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 42.Kouzarides T., Ziff E. (1988) Nature 336, 646–651 [DOI] [PubMed] [Google Scholar]
- 43.Ferdous A., O'Neal M., Nalley K., Sikder D., Kodadek T., Johnston S. A. (2008) Mol. Biosyst. 4, 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braunstein J., Brutsaert S., Olson R., Schindler C. (2003) J. Biol. Chem. 278, 34133–34140 [DOI] [PubMed] [Google Scholar]
- 45.Stancato L. F., David M., Carter-Su C., Larner A. C., Pratt W. B. (1996) J. Biol. Chem. 271, 4134–4137 [DOI] [PubMed] [Google Scholar]
- 46.Sadl V., Jin F., Yu J., Cui S., Holmyard D., Quaggin S., Barsh G., Cordes S. (2002) Dev. Biol. 249, 16–29 [DOI] [PubMed] [Google Scholar]
- 47.Artner I., Blanchi B., Raum J. C., Guo M., Kaneko T., Cordes S., Sieweke M., Stein R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuoka T. A., Artner I., Henderson E., Means A., Sander M., Stein R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2930–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song Y. D., Lee E. J., Yashar P., Pfaff L. E., Kim S. Y., Jameson J. L. (2007) Biochem. Biophys. Res. Commun. 354, 334–339 [DOI] [PubMed] [Google Scholar]
- 50.Muniappan L., Ozcan S. (2007) BMC Physiol. 7, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuoka T. A., Kaneto H., Stein R., Miyatsuka T., Kawamori D., Henderson E., Kojima I., Matsuhisa M., Hori M., Yamasaki Y. (2007) Mol. Endocrinol. 21, 2764–2774 [DOI] [PubMed] [Google Scholar]
- 52.Nomura S., Nakamura T., Hashimoto T., Nishio Y., Maegawa H., Kudo M., Kashiwagi A. (2006) Biochem. Biophys. Res. Commun. 349, 136–143 [DOI] [PubMed] [Google Scholar]
- 53.Kroon E., Martinson L. A., Kadoya K., Bang A. G., Kelly O. G., Eliazer S., Young H., Richardson M., Smart N. G., Cunningham J., Agulnick A. D., D'Amour K. A., Carpenter M. K., Baetge E. E. (2008) Nat. Biotechnol. 26, 443–452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.