Abstract
The DNA mismatch repair protein MutS acts as a molecular switch. It toggles between ADP and ATP states and is regulated by mismatched DNA. This is analogous to G-protein switches and the regulation of their “on” and “off” states by guanine exchange factors. Although GDP release in monomeric GTPases is accelerated by guanine exchange factor-induced removal of magnesium from the catalytic site, we found that release of ADP from MutS is not influenced by the metal ion in this manner. Rather, ADP release is induced by the binding of mismatched DNA at the opposite end of the protein, a long-range allosteric response resembling the mechanism of activation of heterotrimeric GTPases. Magnesium influences switching in MutS by inducing faster and tighter ATP binding, allowing rapid downstream responses. MutS mutants with decreased affinity for the metal ion are impaired in fast switching and in vivo mismatch repair. Thus, the G-proteins and MutS conceptually employ the same efficient use of the high energy cofactor: slow hydrolysis in the absence of a signal and fast conversion to the active state when required.
Keywords: DNA/Protein Interaction, DNA/Repair, Enzymes/ATPases, Enzymes/Mechanisms, Enzymes/Structure, G-proteins, Protein/Metal Ion Interaction, Protein/Structure, DNA mismatch repair, Magnesium
Introduction
DNA mismatch repair (MMR)4 is responsible for the recognition and repair of mispaired bases and small insertion-deletion loops that are formed during DNA replication or recombination between non-identical DNA sequences. MMR activity lowers the mutation frequency in the genome by 2–3 orders of magnitude. MMR is important in maintaining genome integrity, as loss of function of one of the mismatch repair proteins results in a mutator phenotype and increased predisposition to hereditary colon cancer (HNPCC) (1–3).
The first step in mismatch repair, the recognition of the DNA pairing error, is carried out by homodimeric MutS in Escherichia coli or by heterodimeric MutSα (MSH2/MSH6) and MutSβ (MSH2/MSH3) in eukaryotes. Crystal structures of MutS and MutSα bound to different mismatches reveal that there is a common mode for mismatch recognition (4–7). The two subunits tightly embrace the DNA with the clamp and mismatch binding domains, sharply kinking and interrogating the DNA by inserting a phenylalanine next to the destabilized base pair and forming a hydrogen bond with a glutamate involved in allosteric signaling (4, 8, 9). Mismatch binding triggers the uptake of ATP in the nucleotide binding domains located at the opposite end of the protein. These ATP binding sites belong to the ABC superfamily of ATPases (10). Two ABC motifs form composite active sites, with the conserved signature loop from one subunit completing the active site of the opposite subunit in the dimer. The conserved Walker B motif (11) contains an aspartate (position 693 in E. coli MutS) that coordinates two of the water molecules in the hydration shell of the catalytic magnesium ion (4, 6). In E. coli MutS this aspartate is followed by a glutamate (position 694) that serves as the catalytic base during hydrolysis of ATP (12). Mutation of these carboxylates results in proteins with partially or completely impaired mismatch repair capabilities (13–15).
The ATPase sites in the two monomers of E. coli MutS are not equivalent (4). This asymmetry exists even in the absence of DNA. In homodimeric MutS, one high affinity nucleotide binding site and one low affinity nucleotide binding site are present (13, 16). Mismatch binding inhibits ATP hydrolysis in the high affinity nucleotide binding site (MSH6 in MutSα), which allows stable binding of ATP resulting in a mismatch-specific conformational change (9, 17–19). As a result, MutS releases from the DNA mismatch as a so-called sliding clamp that is able to diffuse along the DNA backbone (20). In MutS and MutSα, ATP binding is both necessary and sufficient to induce release of the DNA mismatch (21–25), and ATP hydrolysis is not required (20, 25, 26). This ATP-driven conformational change into a sliding clamp allows recruitment of repair protein MutL (MutLα in eukaryotes) and initiates the search for the strand discrimination signal. The mechanism of this search is under debate, and models vary from diffusional sliding along the DNA to active translocation, and DNA loop formation (12, 20, 22).
MutS and MutSα have been compared with the family of G-protein switches because, analogous to the G-proteins that are “off” and “on” in the GDP and GTP states, ATP binding and hydrolysis toggles the MutS protein between two different states, one in which it searches for a DNA mismatch (the ADP state in this model) and one in which it signals for repair (the ATP state). Just like guanine exchange factors (GEFs) do for G-proteins, mismatched DNA acts as an exchange factor for ADP release in MutS and MutSα, controlling the rate-limiting step in the ATPase cycle (23, 27, 28).
In the small G-proteins, the nucleotide-bound magnesium ion plays a crucial regulatory role in controlling the rate of nucleotide exchange. Nucleotide exchange occurs more efficiently in the absence of magnesium in RhoA, p21, and ARF1 (29–31). In the RhoA structure bound to GDP in the absence of magnesium, the switch I region opens up to allow fast nucleotide release (32). The GEFs exploit this effect by interfering sterically with the binding of the metal ion or by removing one of its ligands (33–35).
It is less clear how nucleotide exchange is regulated in the heterotrimeric GTPases, but it does not seem to happen through magnesium coordination (33, 36). Because of these mechanistic differences between the two G-protein families, we decided to explore which of them is relevant for the MMR switches. We therefore studied the mismatch binding, ATPase activity, and signaling properties of MutS in the presence and absence of magnesium, as well as that of variants D693N and D693V carrying mutations at the conserved Walker B aspartate. We found striking similarities as well as specific mechanistic differences between MutS and the G-proteins, and here we discuss the implications for mismatch repair and the role of fast signaling during the evolution of these distinct cellular pathways.
EXPERIMENTAL PROCEDURES
Proteins and DNA Substrates
MutS mutants D693N and D693V were constructed by converting the respective GAT codon of wild type MutS plasmid pMQ372 (full-length) or pM800 (deletion construct ΔC800, residues 1–800 of MutS) into AAT (Asn) or GTT (Val) using QuikChange (Stratagene). Wild type and mutant MutS were purified as described (5). MutL was purified as follows. Rosetta2(DE3) cells were transformed with the MutL expression plasmid pTX418 (37) and plated onto LB-agar with 50 μg/ml carbenicillin. Cells were grown in minimal medium supplemented with all amino acids (38) and carbenicillin at 37 °C up to OD 0.5, cooled to 20 °C, and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h. Cells were harvested and lysed in binding buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 5 mm imidazole, 10% glycerol, 1 mm β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride) with protease inhibitors (Roche Diagnostics) using an EmulsiFlex-C5 (Avestin, Ontario, Canada). The cleared supernatant was incubated with Talon resin (Clontech Laboratories) for 30 min on ice. Beads were washed using binding buffer with 1 m NaCl, and MutL was eluted with 300 mm imidazole in binding buffer. This fraction was loaded onto a heparin column (GE Healthcare) and eluted with a 0–1 m NaCl gradient in 20 mm Tris-HCl, pH 8.0, 10% glycerol, and 1 mm β-mercaptoethanol. MutL fractions were pooled, concentrated, and run on a Superdex 200 size exclusion column in 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10% glycerol, and 1 mm β-mercaptoethanol. Concentrated samples were flash-frozen in liquid nitrogen and stored at −80 °C. Protein concentrations were determined spectrophotometrically (ϵ280 nm = 73,605 m−1 cm−1 for MutS; 54,270 m−1 cm−1 for MutL). To deplete MutS from endogenous ADP, the protein was dialyzed against 25 mm Hepes, pH 7.5, 250 mm NaCl, 10 mm β-mercaptoethanol, 5 mm MgCl2, and 1.5 mg/ml apyrase (Sigma-Aldrich) for 30 min at room temperature and then for 2 h at 4 °C followed by overnight dialysis against MutS buffer. The dialyzed MutS was diluted 2-fold using buffer without salt, bound to a heparin column, and eluted using a 0.1–1 m NaCl gradient in 25 mm Hepes, pH 7.5, and 10 mm β-mercaptoethanol. The amount of endogenous ADP bound to MutS (0.24 μmol ADP/μmol MutS; 0.04 μmol ADP/μmol nucleotide-depleted MutS; and 0.62 μmol ADP/μmol D693N MutS) was determined as described (13). Oligonucleotides (Sigma-Aldrich) used in the assays are shown in supplemental Fig. S2.
Structure Determination
MutS ΔC800 in complex with 16-bp AA DNA was crystallized in 18–20% PEG 3350, 80–120 mm sodium citrate, 100 mm Bistris propane, pH 7.5, 2–5 mm MgCl2, and 100 μm ADP. Crystals were transferred to 22% PEG 3350, 20% glycerol, and 150 mm sodium citrate and flash-frozen in liquid nitrogen. ΔC800 MutS D693N in complex with 30-bp GT DNA was crystallized in 12–14% PEG 6000, 550–850 mm NaCl, 25 mm Hepes, pH 7.5, 10 mm MgCl2, and 100 μm ADP. Crystals were transferred to 16% PEG 6000, 30% glycerol, 600–800 mm NaCl, and 10 mm Hepes, pH 7.5, and flash-frozen in liquid nitrogen. Data were collected at beamlines ID14-EH3 (MutS, 16 AA) and ID14-EH2 (MutS D693N) at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) and processed using MOSFLM and SCALA (39) (Table 1). Both structures were solved using molecular replacement (AMoRe, CCP4 (40)). The structures were refined using REFMAC (41) and Phenix-refine (42) including TLS parameters determined with the TLSMD server (43). The model was built using Coot (44) and validated with MolProbity (45). Fig. 1 (c–f) was made using PyMOL (DeLano Scientific LLC). Coordinates have been deposited in the Protein Data Bank with accession numbers 2WTU (MutS, 16 AA) and 3K0S (MutS D693N, 30 GT).
TABLE 1.
X-ray data collection and refinement statistics
These statistics are for wild type ΔC800 MutS in complex with a 16-bp duplex DNA containing an AA mismatch and ΔC800 D693N MutS in complex with a 30-bp duplex containing a GT mismatch. Numbers within parentheses refer to the highest resolution shell. r.m.s.d., root mean square deviation.
| Statistics | Wild type MutS, 16 AA | D693N MutS, 30 GT |
|---|---|---|
| Crystallographic | ||
| Resolution range (Å) | 50.0–3.4 (3.5–3.4) | 50.0–2.2 (2.3–2.2) |
| Completeness (%) | 99.8 (99.8) | 96.4 (95. 6) |
| I/sig (I) | 13.6 (4.3) | 11.0 (2.9) |
| Rmerge (%) | 14.7 (46.7) | 8.6 (44.0) |
| Space group | P212121 | P212121 |
| Cell parameters (Å) | a = 91.10 | a = 89.60 |
| b = 137.90 | b = 90.79 | |
| c = 161.44 | c = 260.19 | |
| Total no. of observations | 205,712 (29,422) | 460,869 (66,417) |
| Total unique reflections | 28,641 (4,101) | 104,793 (15,010) |
| Multiplicity | 7.2 (7.2) | 4.4 (4.4) |
| Wilson B-factor | 72.072 | 36.638 |
| Refinement | ||
| No. of atoms (protein + DNA + ligands) | 12,753 | 13,671 |
| R (%) | 20.3 | 18.5 |
| Rfree (%) | 26.3 | 24.6 |
| r.m.s.d. bonds (Å) | 0.008 | 0.007 |
| r.m.s.d. angles (Å) | 1.111 | 1.089 |
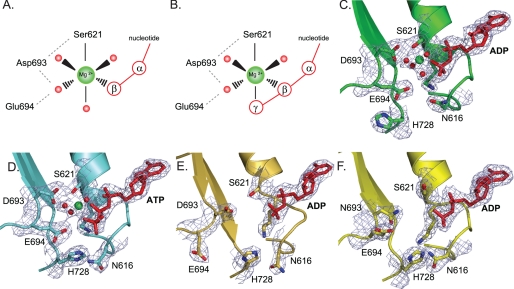
FIGURE 1.
Structural details of magnesium binding sites in wild type MutS and mutant D693N. A and B, schematic view of octahedral magnesium coordination by MutS bound to ADP (Protein Data Bank accession number 1E3M (4)) (A) and ATP (Protein Data Bank accession number 1W7A (13)) (B). The metal ion is coordinated by six ligands: the side chain hydroxyl group of Ser-621, the β-phosphate, and four ordered water molecules. In the case of ATP, the γ-phosphate has displaced one of the ordered water molecules. Asp-693 hydrogen bonds with Ser-621 and coordinates one of the ordered waters; the catalytic base, Glu-694, is in close proximity to this same water molecule. An extensive hydrogen bonding network is formed among the oxygens from the Walker B carboxylates, Ser-621, the nucleoside phosphate groups, and the hydration shell waters. C, magnesium coordination in the A subunit of wild type MutS bound to ADP (1E3M). D, magnesium coordination in the A subunit of wild type MutS bound to ATP (1W7A) and rotations of the His-728 and Asn-616 side chains. E, absence of magnesium and reordering of the Walker B motif in the 16 AA MutS crystal structure. F, absence of magnesium in the crystal structure of D693N MutS.
Nucleotide Binding
Equilibrium constants for nucleotide binding were obtained using the difference in fluorescence anisotropy of free and MutS bound TAMRA-derivatized nucleotide cofactor. Increasing concentrations of MutS were incubated with 1 nm TAMRA-derivatized ADP and ATP (Jena Biosciences) in 25 mm Hepes, pH 7.5, 150 mm NaCl, and 10 mm MgCl2 or 1 mm EDTA for 1 h at room temperature. Fluorescence anisotropy was measured on a Wallac EnVision 2101 multilabel reader. The unbound and bound nucleotide fractions as well as equilibrium binding constants were derived by fitting a single-site binding model to the data using GraphPad Prism 4 (46, 47). Filter retention assays using [α-32P]ATP and [γ-32P]ATP binding to increasing concentrations of MutS were performed as described (9).
Nucleotide Exchange
Nucleotide exchange was quantified using the difference in fluorescence of free versus MutS-bound mantADP (Invitrogen). Reactions contained 2.5 μm MutS, 10 μm ADP, and 0.05 μm mantADP in 25 mm Hepes, pH 7.5, 150 mm NaCl, and 10 mm MgCl2 or 1 mm EDTA and optionally 2.5 μm 30-bp mismatched DNA. Nucleotide exchange was initiated by the addition of 1 mm ATP. A function describing a single exponential decay was fitted to the data using GraphPad Prism 4.
Surface Plasmon Resonance (SPR)
SPR spectroscopy was performed on a Biacore T100 (GE Healthcare) with assay conditions as described (9). Wild type MutS, D693N, and D693V affinities for 41-bp mismatched DNA were compared in 25 mm Hepes, pH 7.5, 150 mm NaCl, and 0.05% Tween 20 and either 10 mm MgCl2 (SPR buffer) or 1 mm EDTA. To study ATP-induced mismatch release, MutS (200 nm) was bound to 41-bp mismatched DNA, and after the wash phase, ATP (0–10 mm) was added to induce dissociation. Dissociation traces are composed of at least three components, and therefore the estimated apparent half-lives of dissociation were derived manually. These values were converted to rate constants and plotted against ATP concentration. A model describing a 1:1 binding (GraphPad Prism 4) was fitted to these data, yielding maximum rate constants for ATP-induced release from the DNA mismatch and an apparent affinity constant for ATP (K½ATP). Sliding clamp formation was analyzed by comparing ATP-induced release from DNA with one free end versus DNA with the end blocked using a fluorescein-antifluorescein complex (48). Ternary complex formation was studied by binding 200 nm wild type MutS or D693N, 400 nm MutL, and 1 mm ATP to a 100-bp homoduplex and mismatched DNA in SPR buffer.
Steady State ATP Hydrolysis
Steady state ATP hydrolysis at 100 mm MgCl2 was monitored as a function of ATP concentration using spectrophotometric analysis (13). Because of the magnesium dependence of the pyruvate kinase in this coupled assay, magnesium dependence was analyzed using hydrolysis of radiolabeled [α-32P]ATP. At increasing concentrations of magnesium, the ionic strength in the samples was maintained by appropriately adjusting the NaCl concentration to the increase in MgCl2. 1 μm MutS was incubated with 0–200 mm MgCl2, 0–165 mm NaCl, 25 mm Hepes, pH 7.5, 1 mm ATP, and optionally 5 μm 30-bp mismatched DNA. After 30 min at room temperature, reactions were quenched with 0.5% SDS and 10 mm EDTA, spotted in triplo onto TLC-PEI cellulose (Merck), developed in 1 m orthophosphoric acid, pH 3.8, and analyzed by phosphorimaging.
In Vivo Mismatch Repair
Mismatch repair complementation assays were performed as described (9, 49). Mutation frequencies and 95% confidence intervals were calculated according to Rosche and Foster (50) and verified according to Stewart (51).
RESULTS
Structural Details of Magnesium Coordination in E. coli MutS
Nucleotide-binding proteins coordinate magnesium via six ligands in an octahedral arrangement (Fig. 1A) (52). In the high affinity nucleotide binding site (subunit A) of E. coli MutS, the magnesium is contacted directly by the side chain hydroxyl of serine 621 and the β-phosphate of ADP (Fig. 1C) (4). In addition it is coordinated by four ordered water molecules. These waters interact with the α- and β-phosphates of the nucleotide and the carboxylate side chains of Walker B aspartate 693 and glutamate 694. In the Mg2+-ATP-containing structure of MutS, one of the ordered water molecules is replaced with the γ-phosphate of ATP (Fig. 1, B and D) (53).
We crystallized MutS on a 16-bp DNA duplex containing an AA mismatch in a new crystal form in which the MutS DNA complex forms different crystal contacts with its neighboring molecules (Table 1). Nevertheless, the overall protein fold and the details of DNA binding and mismatch recognition are unchanged, indicating that the observed mode of mismatch recognition and DNA kinking are not imposed by the crystal lattice. This validates our previous conclusion that mismatch recognition by MutS relies on a single binding mode and that different mismatches adapt their conformation to achieve optimal interaction with the MutS protein (5).
As in the 30-bp GT DNA structure, a single ADP is bound to subunit A. However, the catalytic metal ion is absent (Fig. 1E) even though high concentrations of magnesium were present during crystallization (2–5 mm). The absence of Mg2+ may be due to new crystal contacts in this region that induce a rearrangement of the Walker B motif. In this new conformation, the aspartate 693 side chain is stabilized by a main chain hydrogen bond with glycine 658, and the glutamate 694 side chain approaches the amine nitrogens on the side chain of arginine 697 of the same monomer (not shown). Interestingly, we observed an ordering of the signature loop comprising residues 659–670 from the opposite subunit B (not shown). This disorder-to-order transition had been observed previously in response to ATP binding (53). Concomitantly, the side chains of Asn-616 and His-728 rotate and form a hydrogen bond (Fig. 1, D and E). Clearly, occupation of the catalytic metal ion binding site is correlated to the structural plasticity of both the Walker B and signature motifs involved in nucleotide binding.
The Walker B Aspartate Is Involved in Magnesium Coordination
To study the importance of correct magnesium coordination by the Walker B aspartate, we constructed two MutS variants in which this residue was mutated to asparagine or valine (MutS D693N and D693V). Both mutations removed the negative charge, with D693N retaining hydrogen bonding capacity. Both mutants were purified after overexpression in E. coli with yields similar to the wild type protein. Both bound mismatched DNA in the presence and absence of magnesium (Table 2). Although MutS D693V failed to crystallize, D693N produced crystals in complex with 30-bp GT DNA that diffracted to 2.2 Å (Table 1). Crystal packing, overall structure, and the details of mismatch binding were similar to that of the previously reported wild type structure on 30-bp DNA (4), except for a small rotation of the B subunit with respect to the mismatch binding subunit A. ADP is bound in the A subunit, but the metal ion binding pocket is empty and there are structural rearrangements in the immediate surroundings (Fig. 1F). The hydrogen bond between Walker B residue 693 and Walker A serine 621 is broken, the serine side chain has rotated, and both Walker B residues move away from the nucleotide. As in the ATP-bound structure we found side chain rotation and hydrogen bond formation between His-728 and Asn-616 as well as signature loop ordering, albeit to a lesser extent than induced by ATP binding (53). Signature loop Ser-668, the last residue on an α-helix in the opposing subunit, delivers the positive charge of the helix dipole to the nucleoside triphosphate (53, 54). In our new structures the absence of magnesium resulted in a local net excess of negative charge. This may induce signature loop ordering via helix dipole stabilization, just like the extra negative charge of the γ-phosphate in the case of ATP binding. Rearrangements in this area led to a slight tilting of the B subunit, which centers on the local area of the missing magnesium ion. To summarize, our two new structures indicate that conformational changes in the nucleotide binding sites, and therefore the allosteric response upon ATP binding, are tightly regulated by a delicate balance of electrostatic forces.
TABLE 2.
Equilibrium dissociation constants
These are the equilibrium dissociation constants for the binding of mismatched DNA (mmDNA) and nucleotide cofactors in the absence and presence of mismatched DNA for wild type, D693N, and D693V MutS in the presence and absence of magnesium. ND, not determined.
| MutS | KDmmDNA |
KDATP |
KDADP |
||
|---|---|---|---|---|---|
| No DNA | mmDNA | No DNA | mmDNA | ||
| nm | μm | μm | |||
| Wild type | |||||
| Mg2+ | 48.8 ± 0.19 | 0.034 ± 0.002 | 0.023 ± 0.004 | 0.274 ± 0.067 | 0.570 ± 0.14 |
| EDTA | 29.6 ± 1.7 | 0.320 ± 0.032 | 0.469 ± 0.042 | 1.10 ± 0.15 | 2.35 ± 0.58 |
| D693N | |||||
| Mg2+ | 94.1 ± 7.0 | 0.505 ± 0.025 | 0.452 ± 0.028 | 0.646 ± 0.14 | 0.897 ± 0.17 |
| EDTA | 141 ± 6.6 | 0.400 ± 0.033 | 0.463 ± 0.022 | 1.33 ± 0.22 | 2.11 ± 0.45 |
| D693V | |||||
| Mg2+ | 104 ± 10 | 20.7 ± 1.6 | ND | 2.62 ± 0.80 | ND |
| EDTA | 176 ± 23 | 16.2 ± 0.84 | ND | 10.3 ± 3.8 | ND |
Magnesium Is Required for High Affinity Nucleotide Binding
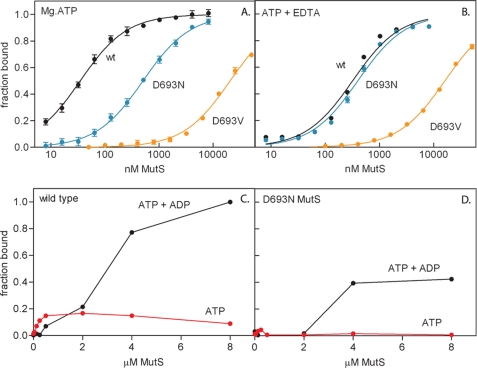
To quantify the effects of magnesium on nucleotide binding, we followed the change in fluorescence anisotropy of TAMRA-labeled nucleotide cofactors upon binding to MutS (Table 2 and Fig. 2). We first analyzed the binding of ADP in the presence and absence of magnesium and DNA. In agreement with previous observations (55), we found that the high affinity nucleotide binding site of wild type MutS binds Mg2+-ADP with low micromolar affinity (KD = 0.27 μm). The affinity is not influenced by the presence of endogenous ADP in the high affinity site of the purified protein (13); we found identical binding of TAMRA-ADP to nucleotide-depleted MutS (supplemental Fig. S1). DNA consistently weakens the affinity for ADP but at most 2-fold. Magnesium contributes to this affinity but not to a large extent (a 4-fold change at most). In agreement with this, we found that mutant D693N, which has a lower affinity for magnesium (the metal ion is absent from the crystal structure), has an affinity for ADP similar to that of wild type MutS. In contrast, D693V MutS binds ADP with significantly reduced affinity, especially in the presence of EDTA. It is likely that the loss of hydrogen bonds upon introduction of the hydrophobic valine results in an additional conformational change. This has also been reported for the same mutation in human MSH6 (14).
FIGURE 2.
Nucleotide binding by wild type and mutant MutS. A and B, equilibrium binding curves of wild type, D693N, and D693V MutS binding to TAMRA-derivatized ATP determined by fluorescence anisotropy spectroscopy in the presence of magnesium (A) and the absence of magnesium (B). Lines represent the fit of a single-site binding model to the data. C and D, filter retention of [α-32P]ATP and [γ-32P]ATP by wild type MutS (C) and D693N MutS (D). Using [α-32P]ATP, simultaneous binding of ADP and ATP is detected; using [γ-32P]ATP only ATP is detected. Lines serve to connect the data points.
We next analyzed the equilibrium binding curves upon the addition of Mg2+-ATP to wild type MutS (Fig. 2A). The observed nucleotide affinity (0.034 μm) is an order of magnitude tighter than for Mg2+-ADP. This was also found for nucleotide-depleted MutS (supplemental Fig. S1). In contrast, D693N MutS does not display such a high affinity, because the binding of ATP is similar to the binding of ADP. Furthermore the affinity of D693V MutS for ATP is so low that cofactor specificity is reversed in this mutant (Table 2). Interestingly, the mutation of the equivalent residue in p21H-ras to alanine has the same effect (30), indicating the importance of the Walker B aspartate in enforcing nucleotide di- or triphosphate specificity.
Because the TAMRA modification was on the ribose moiety of the nucleotide, we could not distinguish between the binding of ATP and its hydrolysis product. To make this distinction, we used radioactively labeled nucleotides in a filter retention assay, which monitored the binding of ATP only (using [γ-32P]ATP) or the simultaneous binding of ADP and ATP (using [α-32P]ATP). Wild type MutS binds [γ-32P]ATP with nanomolar affinity, indicating binding of ATP without hydrolysis (Fig. 2C). Binding of wild type MutS to [α-32P]ATP is biphasic. The nucleotide that was bound in the nanomolar range was probably ATP, because the [γ-32P]ATP also was retained in this range. The nucleotide that was bound in the low micromolar range was probably ADP, because there was no corresponding signal with [γ-32P]ATP. For D693N MutS, we did not observe binding of [γ-32P]ATP, but after the addition of [α-32P]ATP, ADP was retained as in wild type MutS (Fig. 2D). So in this mutant ATP was bound but was hydrolyzed immediately to ADP, indicating that high affinity binding of ATP and inhibition of its hydrolysis require correct magnesium coordination. To summarize, although magnesium does not play a major role in the equilibrium binding of ADP, correct magnesium coordination is necessary for high affinity binding of ATP and the regulation of its hydrolysis.
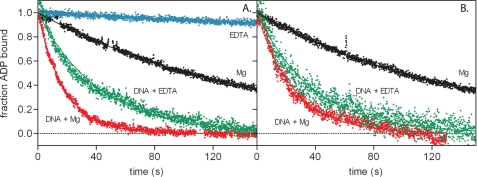
Magnesium Is Not Required for Mismatch-induced Fast Nucleotide Exchange
We then studied the role of magnesium in nucleotide exchange. Analogous to the GEFs, mismatched DNA switches MutS and MutSα from an ADP- to an ATP-bound state by increasing the rate of ADP release (23, 48). Consequently, we found slow rates for ADP release from E. coli MutS in the absence of DNA and a 10-fold acceleration of this reaction in the presence of mismatched DNA (Table 3 and Fig. 3A). In the absence of DNA, magnesium stimulates nucleotide exchange. This is in contrast to the large and inhibitory effect of magnesium on the nucleotide exchange rates in many small G-proteins (29–31). In the presence of DNA, magnesium has only a marginal effect; DNA alone is responsible for most of the observed acceleration. This finding was confirmed with mutant D693N (Fig. 3B), which has a lower affinity for magnesium but displays exchange rates similar to those of wild type MutS in the presence of the metal ion (Table 3). The nucleotide exchange signal was too weak for significant data analysis for D693V or for D693N without DNA in the presence of EDTA.
TABLE 3.
Rate constants
These are rate constants for ADP release in the absence and presence of mismatched DNA (mmDNA) and the rate and apparent affinity constants for release from mismatched DNA for wild type and D693N MutS in the absence and presence of magnesium. ND, not determined.
| MutS | ADP exchange, koffADP |
Release from mismatch |
||
|---|---|---|---|---|
| No DNA | mmDNA | Rate | K½ATP | |
| s−1 | s−1 | s−1 | mm | |
| Wild type | ||||
| Mg2+ | 0.0072 ± 0.0006 | 0.076 ± 0.029 | 0.24 ± 0.01 | 0.019 ± 0.003 |
| EDTA | 0.00068 ± 0.0 | 0.049 ± 0.04 | 0.12 ± 0.01 | 1.2 ± 0.2 |
| D693N | ||||
| Mg2+ | 0.0066 ± 0.0004 | 0.091 ± 0.07 | 0.16 ± 0.006 | 0.42 ± 0.05 |
| EDTA | ND | 0.033 ± 0.01 | 0.032 ± 0.003 | 0.18 ± 0.03 |
FIGURE 3.
DNA- and magnesium-dependent nucleotide exchange in wild type MutS (A) and mutant D693N (B). The fraction of ADP-bound MutS is plotted against time, and the lines represent the fit of a first order exponential decay to the data. For D693N with EDTA in the absence of DNA, ADP release could not be determined because of the lack of change in fluorescent properties of the nucleotide upon incubation with the protein.
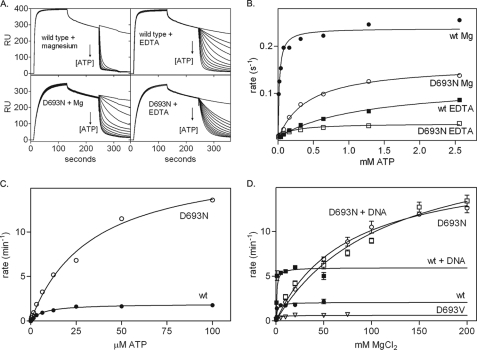
Magnesium Is Required for Fast ATP-induced Release from a DNA Mismatch
Next we analyzed whether magnesium is required for the ATP-induced conformational change that allows MutS to release the DNA mismatch (9, 22, 24, 25). We studied this release using SPR by binding MutS to immobilized mismatched DNA and inducing dissociation by the addition of ATP (Fig. 4A) (9, 48, 56). The dissociation rate is dependent on the ATP concentration, and we found an apparent affinity constant for ATP of 19 μm for wild type MutS (Fig. 4B and Table 3). This is similar to the affinity of the low affinity nucleotide binding site (13, 57), suggesting that MutS does not release the mismatch until both sites have bound ATP. This is in agreement with observations from yeast MutSα (19, 58). The affinity for ATP is greatly reduced in the absence of magnesium (K½ATP >1 mm in EDTA) and in mutant D693N (K½ATP 0.42 mm). When the electrostatic interactions are balanced again by the removal of both the aspartate carboxylate and the magnesium ion, ATP binding is partially restored (K½ATP is 0.18 mm for D693N in the presence of EDTA (Fig. 4A and Table 3)). Despite its ability to bind ATP at 1 mm concentrations, at least in its high affinity site (Fig. 2A), D693V is not released from mismatched DNA upon the addition of ATP (Fig. 5D).
FIGURE 4.
Rates of MutS release from mismatched DNA and ATP hydrolysis. A, ATP-induced release of wild type and D693N MutS from 41-mer duplex DNA containing a GT mismatch determined using SPR in the absence and presence of magnesium. B, apparent half-lives for release of the mismatch were plotted against ATP concentration. Lines represent the fit of a single-site binding model to the data. C, steady state ATP hydrolysis rates for wild type and D693N MutS in the absence of DNA as a function of ATP concentration at 100 mm MgCl2. D, steady state ATP hydrolysis rates for wild type and mutant MutS in the absence and presence of DNA as a function of magnesium concentration at 1 mm ATP. Lines represent fits of a Michaelis-Menten model to the data. Kinetic constants for ATP hydrolysis are summarized in supplemental Table 1.
FIGURE 5.
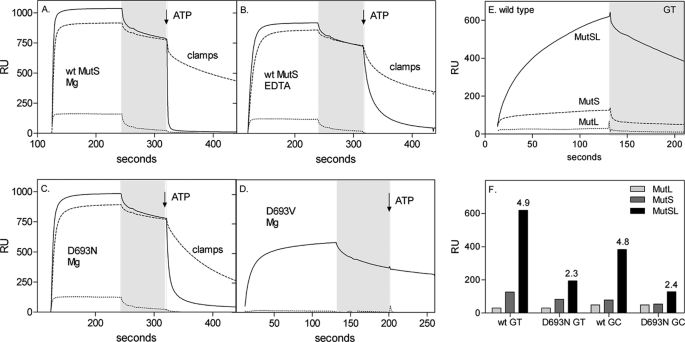
ATP-induced sliding clamp formation and MutL recruitment by wild type MutS and mutants determined by SPR. A–D, the formation of sliding clamps was analyzed by flowing 1 μm MutS wild type (wt) or mutants over a chip derivatized with a 41-bp mismatched DNA containing one open end (—) or blocked with anti-fluorescein (–––). After a buffer wash (gray area), buffer containing 1 mm ATP was added (dissociation phase). The control lane contains 41-mer correctly paired DNA with its end blocked by anti-fluorescein (···). The difference in response in the dissociation phase reflects the fraction of MutS trapped as sliding clamps on the blocked DNA after the addition of 1 mm ATP. A, wild type MutS with magnesium in the assay buffer. B, wild type MutS with EDTA in the assay buffer. C, D693N MutS with magnesium in the assay buffer. D, D693V MutS with magnesium in the assay buffer. E, MutL recruitment was monitored using SPR analysis of 200 nm wild type MutS, 400 nm MutL, and 1 mm ATP on a 100-bp heteroduplex (GT) or homoduplex (GC) DNA. F, efficiency of MutL recruitment by wild type and D693N MutS on heteroduplex and homoduplex DNA. The response at 120 s was plotted, and the -fold amplification of the signal due to the presence of MutL is indicated above the MutSL signal bar.
At saturating ATP concentrations, wild type MutS releases the mismatch more rapidly with magnesium than with EDTA and faster than the D693N mutant (Fig. 4B and Table 3). Either ATP binding is slower in D693N, or a downstream reaction step could become rate-limiting for the release of the mismatched DNA (for example the propagation of the conformational change induced by ATP binding). Release of D693N MutS from the mismatch in EDTA supports this latter possibility; although the affinity for ATP is partially restored, the maximum release rate from the mismatch is nevertheless reduced even further. This indicates that magnesium coordination by Asp-693 is required for correct allosteric coupling between ATPase and DNA binding sites within MutS.
Magnesium Coordination Controls the Rate-limiting Step of Basal ATPase Activity
MutS has a slow steady state rate for ATP hydrolysis of 1.9 ± 0.04 min−1 (Fig. 4, C and D, and supplemental Table 1). The Km value for ATP (6.8 ± 0.6 μm) reflects the affinity of the low affinity nucleotide binding site (13). The apparent Km for magnesium (1.2 mm) reflects the chelation of the metal ion by 1 mm ATP in the reaction. The steady state ATPase activity is stimulated by mismatched DNA (Fig. 4D and supplemental Table 1), which increases the rate of ADP release from the high affinity site.
The reduced affinity for ATP of D693N MutS is evident from the increase in the Km for ATP to 34 μm (Fig. 4C and supplemental Table 1). At 10 mm magnesium, the steady state ATP hydrolysis rate of D693N MutS is the same as for wild type (1.9 min−1; Fig. 4D); however, mismatched DNA is unable to further increase this rate. Because mismatched DNA increases the rate of ADP release from D693N just as in wild type MutS (see above), this indicates that in the mutant, in the presence of DNA, an alternative rate-limiting step controls ATP hydrolysis. Interestingly, this step is very dependent on magnesium, because at higher metal ion concentrations D693N is a much more active ATPase than wild type MutS (kcat 18.3 ± 1.0) (Fig. 4, C and D). The steady state ATPase activity of D693V MutS is greatly reduced (kcat 0.62 ± 0.05). In conclusion, magnesium coordination plays a profound role in regulating the ATPase activity of MutS.
Magnesium Is Not Essential for the Formation of the MutS Sliding Clamp
Next we determined whether magnesium is required for the formation of the sliding clamp that travels along the DNA. This was visualized using the SPR sensor with DNA blocked by a fluorescein-antibody complex at the free end of the DNA (48). The fraction of MutS that is retained on the blocked DNA has formed a stable closed conformation, encircling the DNA, and cannot slide off the DNA because of the end block. Wild type MutS forms these clamps efficiently (Fig. 5A) (9). A similar fraction of MutS forms sliding clamps in the absence of magnesium (Fig. 5B), indicating that despite the reduced signaling capacity, the correct conformational change still takes place and does not require ATP hydrolysis. Clamps are also formed by D693N MutS, although their rate of dissociation from the blocked DNA is slightly faster (Fig. 5C). This is in contrast to human MutSα, which, upon the addition of ATP in the absence of the metal ion, releases the mismatch by direct dissociation (14, 20). This could reflect the intrinsic differences between prokaryotic and eukaryotic MutS or the inherent differences in the assays, highlighting the lower stability of the magnesium-independent clamps more prominently in gel shifts than in the SPR analysis.
D693V does not release from DNA upon the addition of ATP (Fig. 5D). Again, this contrasts with human MutSα; the mutation analogous to our D693V variant in human MSH6 (14) causes direct dissociation of MutSα from the mismatch. Here the different subunit composition of these two enzymes should be considered. Whereas in the MutSα mutant the MSH6 subunit is defective, intact MSH2 can still drive (an incorrect) conformational change. In E. coli D693V both subunits are simultaneously mutated, which in the case of the valine substitution is fatal.
Correct Magnesium Coordination Is Required for Fast MutL Recruitment
Subsequently we considered whether MutL recruitment is influenced by magnesium. Recruitment of MutL by ATP-bound MutS on a 100-bp duplex DNA was monitored using SPR (Fig. 5E). The simultaneous addition of MutS, MutL, and ATP resulted in a response that far exceeded the response of MutS alone in the presence of Mg2+-ATP. This MutS-MutL complex formation is completely dependent on MutS (Fig. 5E), is more pronounced on DNA containing a GT mismatch than on homoduplex DNA (Fig. 5F), and is similar to those reported for the prokaryotic and eukaryotic complexes monitored in other SPR and total internal reflection fluorescence (TIRF) studies (25, 59).
The magnesium dependence of MutS-MutL complex formation cannot be analyzed directly because, in the presence of EDTA, MutL binds to DNA with high affinity, like eukaryotic MutLα (59). As expected from its inability to respond to nucleotide binding, D693V MutS was unable to bind MutL (results not shown). MutS D693N was able to recruit MutL only marginally; the amplification of the signal in the presence of MutL was significantly less for the mutant than for wild type MutS (Fig. 5F). This indicates that magnesium coordination is indeed important for MutL recruitment by MutS either via a direct effect on MutL binding or because the ATP-induced conformational change, and therefore MutL recruitment, is slower in the mutant.
Incorrect Magnesium Coordination Results in Reduced in Vivo Mismatch Repair
We determined the effect of the Walker B mutations on the ability of these MutS variants to complement a MutS-deficient strain in vivo. In this assay the absence of efficient mismatch repair results in increased mutation rates that lead to increased acquired rifampicin resistance (95% confidence interval of mutation frequency, 11.2–14.6 * 10−9). Heterologous expression of wild type MutS reduces this frequency to ∼1 * 10−9 (0.80–1.69 * 10−9). MutS D693V is completely inactive (16.3–22.4 * 10−9). MutS D693N has an intermediate phenotype (6.25–8.83 * 10−9), indicating that it is partially able to correct DNA mismatches. These findings are in good agreement with in vivo repair efficiencies reported earlier for D693N and a related D693A MutS variant (15). The severity of the phenotype of these two mutants correlates with their ability to respond to ATP and initiate downstream signaling in the in vitro assays.
DISCUSSION
Nucleotide cofactor binding and hydrolysis by proteins from the ABC ATPase family invariably result in local conformational rearrangements in the nucleotide binding regions of the protein, which are subsequently relayed to other parts such as the DNA binding domains (60, 61). In this study we have defined the role of the catalytic metal ion in controlling these conformational changes and will relate this to the role of magnesium in the G-protein switches.
We found that in the absence of magnesium, MutS still binds and exchanges ADP in the presence of mismatched DNA. However, high affinity ATP binding is lost and sliding clamp formation is slow. This magnesium effect is confirmed in mutant D693N, which has a lower affinity for the metal ion and shows similar characteristics: intact ADP binding and exchange, no high affinity ATP binding, and a slow conformational response. As a result, D693N is defective in MutL recruitment and mismatch repair. Further destabilization of the metal binding site in D693V results in a loss of specificity for ATP over ADP. D693V is unable to undergo any conformational response upon ATP binding and is completely inactive in mismatch repair. In summary, in addition to catalyzing the chemistry of ATP hydrolysis, magnesium plays an important role in relaying the rapid allosteric response upon ATP binding to MutS, which signals the presence of a DNA mismatch.
Our findings emphasize the importance of two aspects of the molecular switch model (20, 23): the necessity for fast switching to the active ATP state upon mismatch recognition and a correlation between sliding clamp formation and MutL recruitment. The observation that sliding clamps are formed in the absence of magnesium argues against active translocation by MutS (22). We and others have shown previously that ATP is used to verify mismatch binding (9, 12). We are currently investigating how these aspects from different models are combined in the search for the strand discrimination signal.
Role of Magnesium in Nucleotide Binding and Hydrolysis
Upon addition of substoichiometric amounts of TAMRA-labeled Mg2+-ATP, we observed a nanomolar affinity for the nucleotide, an order of magnitude tighter than observed upon the addition of Mg2+-ADP. We had observed a nanomolar affinity previously using filter binding experiments and radiolabeled Mg2+-ATP (13). This high affinity requires the negative carboxylate group of aspartate 693, as well as neighboring arginine 697 that is involved in positioning the Walker B motif of the opposite subunit in the MutS dimer in such a way that nucleotide binding is possible (13). Therefore the correct coordination of magnesium by the Walker B motif of MutS is required for high affinity nucleotide binding during the ATPase cycle.
At least part of the ATP that is bound with high affinity is not hydrolyzed immediately. Possibly MutS requires a low affinity nucleotide binding step or a hydrolysis event in the second nucleotide binding site of the dimer before the first site is able to hydrolyze its bound ATP. This second step may not have happened in our experimental setup because the assay required substoichiometric concentrations of TAMRA-ATP for the proper determination of nucleotide affinities. The requirement for such a succession of steps fits with the observation that in the absence of DNA, the steady state ATPase rate is governed by the rate of ADP release from the high affinity site, whereas the Km for ATP hydrolysis reflects the nucleotide affinity of the low affinity site (13, 57). Tight coupling between the composite ATPase sites has also been described for Taq MutS and the heterodimeric MutS homolog from yeast (57, 58, 62).
In the absence of DNA, ADP release is rate-limiting for steady state MutS ATPase activity (17). Mismatched DNA increases the rate of ADP release 10-fold; however, ATPase activity is increased only 3-fold. Therefore, ADP release is no longer rate-limiting in the presence of DNA. Nucleotide exchange rates for wild type and D693N MutS are similar, but the mutant is a more active ATPase. This indicates that in D693N, ADP release is not rate-limiting for steady state hydrolysis. Indeed, nucleotide exchange in the mutant is still stimulated by mismatched DNA, whereas steady state ATP hydrolysis is not. This can be explained by changes in the coupling between the composite ATPase sites in the MutS dimer (4, 58). Because the rate of ADP release from the high affinity site governs the overall steady state ATPase activity, the Walker B aspartate and the tight binding of magnesium may be important for correct coupling between the sites. In the mutant D693N these allosteric interactions may be partially destroyed, leading to an increased ATPase activity in one of the ATPase subunits that has become independent from the other. This conclusion is supported by increased steady state ATPase activity for D693N, loss of high affinity ATP binding, and inhibition of its hydrolysis, as well as by the crystal structure of D693N showing a small rearrangement of one subunit relative to the other.
The effect of mutating the Walker B aspartate is not the same in different ATPases. Mutation of this residue to asparagine in the E. coli UvrD helicase rescues the ATPase deficiency of the Walker B glutamate mutant (63). In F1 ATPase, the same mutation abolishes the inhibition of steady state ATPase activity by high magnesium concentrations (64). We have shown here that this residue is important for regulating the rate-limiting step and inhibition of ATP hydrolysis in MutS. The apparent uniform “activating” capability of this asparagine substitution exemplifies the regulatory role of this residue in controlling both motor and switch ATPase proteins.
Nucleotide Exchange and the Molecular Switch Concept in Mismatch Repair
Magnesium plays an important role in regulating nucleotide binding in the small G-proteins (33, 65). In numerous monomeric GTPases, magnesium is a GDP dissociation inhibitor (65). The removal of magnesium results in increased nucleotide exchange rates. It is, at least in part, this property that is used by GEF proteins to accelerate nucleotide exchange on GTPases; binding of the GEF to the GTPase interferes with magnesium ion binding (33–35). The absence of magnesium results in a lower affinity for nucleoside diphosphate and increased dissociation rates. In fact, this regulatory role of magnesium in controlling nucleotide binding is not restricted to the small G-proteins; in adenosine phosphate-binding proteins such as actin, myosin, and Rad51, magnesium is also described as influencing nucleotide dissociation (66–68).
MutS and its homologs have been compared with the G-protein switches because of increased nucleotide exchange rates upon effector binding and switching between an active and inactive state based on nucleotide occupancy (23, 48). From our analysis it became clear that the actual role of magnesium in MutS is significantly different from its role in the small G-proteins. In contrast to GDP release in the G-proteins, ADP release in MutS is increased by magnesium in the absence of DNA and is insensitive to the presence of the metal ion in the presence of DNA. Magnesium increases the affinity for ATP and accelerates the propagation of the subsequent conformational change. In fact, the MMR switches more closely resemble the heterotrimeric G-proteins. GDP release from trimeric GTPases seems to be unaffected by magnesium (36). Trimeric GTPases release their nucleoside diphosphate after interaction with an activated membrane receptor, an interaction that occurs on the protein interface opposite from the nucleotide binding site. This resembles the long-range effect on ADP release of mismatched DNA binding at the opposite end of MutS from the ATPase sites and is unlike the direct interaction between the nucleotide binding site of small G-proteins and their respective GEFs. In the trimeric G-protein, subsequent GTP binding then induces release of the activated Gα subunit from the receptor and Gβγ subunits (33). Again, similarly, ATP binding induces a conformational change in the MMR switch that results in release from the DNA mismatch and activation of the repair reaction. So conceptually, MMR and heterotrimeric G-proteins are more similar than their large structural and functional differences would suggest.
However, a marked difference between the G-protein and MMR switches is the magnitude of the GEF-induced acceleration of nucleotide release. The increase in E. coli MutS is only around 10-fold (Ref. 48 and this study) and less than an order of magnitude higher for human MSH proteins (23, 69). This is smaller than the 2–3 orders of magnitude reported for heterotrimeric GTPases (70, 71) and far from the 5 orders of magnitude reported for Ras, Ran, and Rho (72–74).
Nevertheless, although the mechanistic details of magnesium dependence may differ considerably among these protein families, the overall result is the same in that magnesium ensures efficient use of the energy stored in the nucleoside triphosphate molecules: slow nucleotide hydrolysis in the absence of a signal and fast conversion to the active state when required (Fig. 6). The less pronounced acceleration of nucleotide exchange in the mismatch repair proteins may be based on the evolutionary pressure exerted on the pathways in which the individual proteins function. Whereas the G-proteins have to relay signals quickly, switching on MutS after mismatch recognition is certainly crucial but definitely not rate-limiting for the overall repair event.
FIGURE 6.
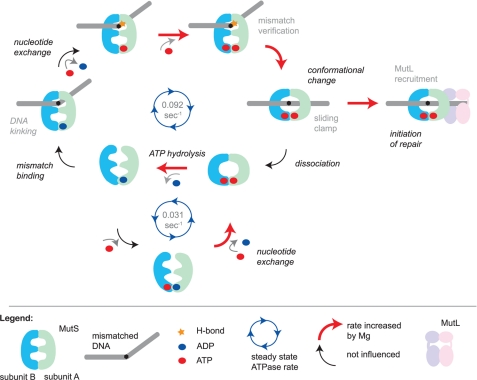
Model for initiation of DNA mismatch repair and the role of magnesium in specific reaction steps. MutS slowly hydrolyzes ATP when it is not bound to DNA. The rate-limiting step in this reaction is the release of ADP from the high affinity nucleotide binding site. The asymmetric ATPase sites are tightly coupled in a manner that depends on correct magnesium coordination by the Walker B motifs. When MutS recognizes a mismatch, it stably binds by kinking the DNA and forming a hydrogen bond between a conserved glutamate and one of the mismatched bases. This results in an increase in the release of ADP from the high affinity site, which is independent of magnesium. However, in the next step, the affinity for ATP strongly depends on magnesium. MutS then uses the hydrogen bond formed between Glu-38 and one of the mismatched bases to verify mismatch binding and undergo an ATP-dependent conformational change into a sliding clamp. If magnesium is not coordinated correctly, the sliding clamp can be formed but at a much reduced rate. Sliding clamps either recruit MutL and thereby initiate the repair reaction or, when idle, will ultimately dissociate from the DNA upon or followed by ATP hydrolysis. MutS is then reset into a conformation that is again able to search for a DNA mismatch.
Supplementary Material
Acknowledgments
We are very grateful to Rick Hibbert for critical reading of the manuscript, Meindert Lamers for contributions to a MutL purification protocol, Flora Groothuizen for experimental assistance, Krista Joosten for mutation frequency calculations, and M. Winkler for the gift of the pTX418 plasmid.
This work was supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under Grant 223545 ”mismatch2model,“the Nederlandse Organisatie voor Wetenschappelijk Onderzoek-Chemische Wetenschappen (VENI 700.53.407 to J. H. G. L.), the Koningin Wilhelmina Fonds (Project 04-3084), and the Center for Biomedical Genetics.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table 1.
The atomic coordinates and structure factors (codes 2WTU and 3K0S) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MMR
- mismatch repair
- ABC
- ATP binding cassette
- GEF
- guanine exchange factor
- SPR
- surface plasmon resonance
- PEG
- polyethylene glycol
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- TAMRA
- 6-carboxytetramethylrhodamine.
REFERENCES
- 1.Kunkel T. A., Erie D. A. (2005) Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. (2006) Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 3.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 4.Lamers M. H., Perrakis A., Enzlin J. H., Winterwerp H. H., de Wind N., Sixma T. K. (2000) Nature 407, 711–717 [DOI] [PubMed] [Google Scholar]
- 5.Natrajan G., Lamers M. H., Enzlin J. H., Winterwerp H. H., Perrakis A., Sixma T. K. (2003) Nucleic Acids Res. 31, 4814–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obmolova G., Ban C., Hsieh P., Yang W. (2000) Nature 407, 703–710 [DOI] [PubMed] [Google Scholar]
- 7.Warren J. J., Pohlhaus T. J., Changela A., Iyer R. R., Modrich P. L., Beese L. S. (2007) Mol. Cell 26, 579–592 [DOI] [PubMed] [Google Scholar]
- 8.Schofield M. J., Brownewell F. E., Nayak S., Du C., Kool E. T., Hsieh P. (2001) J. Biol. Chem. 276, 45505–45508 [DOI] [PubMed] [Google Scholar]
- 9.Lebbink J. H., Georgijevic D., Natrajan G., Fish A., Winterwerp H. H., Sixma T. K., de Wind N. (2006) EMBO J. 25, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins C. F., Linton K. J. (2004) Nat. Struct. Mol. Biol. 11, 918–926 [DOI] [PubMed] [Google Scholar]
- 11.Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junop M. S., Obmolova G., Rausch K., Hsieh P., Yang W. (2001) Mol. Cell 7, 1–12 [DOI] [PubMed] [Google Scholar]
- 13.Lamers M. H., Winterwerp H. H., Sixma T. K. (2003) EMBO J. 22, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iaccarino I., Marra G., Dufner P., Jiricny J. (2000) J. Biol. Chem. 275, 2080–2086 [DOI] [PubMed] [Google Scholar]
- 15.Junop M. S., Yang W., Funchain P., Clendenin W., Miller J. H. (2003) DNA Repair 2, 387–405 [DOI] [PubMed] [Google Scholar]
- 16.Bjornson K. P., Blackwell L. J., Sage H., Baitinger C., Allen D., Modrich P. (2003) J. Biol. Chem. 278, 34667–34673 [DOI] [PubMed] [Google Scholar]
- 17.Bjornson K. P., Allen D. J., Modrich P. (2000) Biochemistry 39, 3176–3183 [DOI] [PubMed] [Google Scholar]
- 18.Antony E., Hingorani M. M. (2003) Biochemistry 42, 7682–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazur D. J., Mendillo M. L., Kolodner R. D. (2006) Mol. Cell 22, 39–49 [DOI] [PubMed] [Google Scholar]
- 20.Gradia S., Subramanian D., Wilson T., Acharya S., Makhov A., Griffith J., Fishel R. (1999) Mol. Cell 3, 255–261 [DOI] [PubMed] [Google Scholar]
- 21.Iaccarino I., Marra G., Palombo F., Jiricny J. (1998) EMBO J. 17, 2677–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen D. J., Makhov A., Grilley M., Taylor J., Thresher R., Modrich P., Griffith J. D. (1997) EMBO J. 16, 4467–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gradia S., Acharya S., Fishel R. (1997) Cell 91, 995–1005 [DOI] [PubMed] [Google Scholar]
- 24.Grilley M., Welsh K. M., Su S. S., Modrich P. (1989) J. Biol. Chem. 264, 1000–1004 [PubMed] [Google Scholar]
- 25.Selmane T., Schofield M. J., Nayak S., Du C., Hsieh P. (2003) J. Mol. Biol. 334, 949–965 [DOI] [PubMed] [Google Scholar]
- 26.Jacobs-Palmer E., Hingorani M. M. (2007) J. Mol. Biol. 366, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fishel R. (1998) Genes Dev. 12, 2096–2101 [DOI] [PubMed] [Google Scholar]
- 28.Sprang S. R. (1997) Annu. Rev. Biochem. 66, 639–678 [DOI] [PubMed] [Google Scholar]
- 29.Zhang B., Zhang Y., Wang Z., Zheng Y. (2000) J. Biol. Chem. 275, 25299–25307 [DOI] [PubMed] [Google Scholar]
- 30.John J., Rensland H., Schlichting I., Vetter I., Borasio G. D., Goody R. S., Wittinghofer A. (1993) J. Biol. Chem. 268, 923–929 [PubMed] [Google Scholar]
- 31.Béraud-Dufour S., Robineau S., Chardin P., Paris S., Chabre M., Cherfils J., Antonny B. (1998) EMBO J. 17, 3651–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu T., Ihara K., Maesaki R., Kuroda S., Kaibuchi K., Hakoshima T. (2000) J. Biol. Chem. 275, 18311–18317 [DOI] [PubMed] [Google Scholar]
- 33.Cherfils J., Chardin P. (1999) Trends Biochem. Sci. 24, 306–311 [DOI] [PubMed] [Google Scholar]
- 34.Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J. (1998) Nature 394, 337–343 [DOI] [PubMed] [Google Scholar]
- 35.Kawashima T., Berthet-Colominas C., Wulff M., Cusack S., Leberman R. (1996) Nature 379, 511–518 [DOI] [PubMed] [Google Scholar]
- 36.Pan J. Y., Sanford J. C., Wessling-Resnick M. (1996) J. Biol. Chem. 271, 1322–1328 [DOI] [PubMed] [Google Scholar]
- 37.Feng G., Winkler M. E. (1995) BioTechniques 19, 956–965 [PubMed] [Google Scholar]
- 38.Budisa N., Steipe B., Demange P., Eckerskorn C., Kellermann J., Huber R. (1995) Eur. J. Biochem. 230, 788–796 [DOI] [PubMed] [Google Scholar]
- 39.Powell H. R. (1999) Acta Crystallogr. 55, 1690–1695 [DOI] [PubMed] [Google Scholar]
- 40.(1994) Acta Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 41.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 43.Painter J., Merritt E. A. (2006) Acta Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K. (2004) Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 45.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jameson D. M., Seifried S. E. (1999) Methods 19, 222–233 [DOI] [PubMed] [Google Scholar]
- 47.Stricher F., Martin L., Barthe P., Pogenberg V., Mechulam A., Menez A., Roumestand C., Veas F., Royer C., Vita C. (2005) Biochem. J. 390, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Acharya S., Foster P. L., Brooks P., Fishel R. (2003) Mol. Cell 12, 233–246 [DOI] [PubMed] [Google Scholar]
- 49.Wu T. H., Marinus M. G. (1999) J. Biol. Chem. 274, 5948–5952 [DOI] [PubMed] [Google Scholar]
- 50.Rosche W. A., Foster P. L. (2000) Methods 20, 4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart F. M. (1994) Genetics 137, 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A. (1989) Nature 341, 209–214 [DOI] [PubMed] [Google Scholar]
- 53.Lamers M. H., Georgijevic D., Lebbink J. H., Winterwerp H. H., Agianian B., de Wind N., Sixma T. K. (2004) J. Biol. Chem. 279, 43879–43885 [DOI] [PubMed] [Google Scholar]
- 54.Hol W. G. (1985) Prog. Biophys. Mol. Biol. 45, 149–195 [DOI] [PubMed] [Google Scholar]
- 55.Bjornson K. P., Modrich P. (2003) J. Biol. Chem. 278, 18557–18562 [DOI] [PubMed] [Google Scholar]
- 56.Blackwell L. J., Bjornson K. P., Allen D. J., Modrich P. (2001) J. Biol. Chem. 276, 34339–34347 [DOI] [PubMed] [Google Scholar]
- 57.Antony E., Hingorani M. M. (2004) Biochemistry 43, 13115–13128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antony E., Khubchandani S., Chen S., Hingorani M. M. (2006) DNA Repair 5, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendillo M. L., Mazur D. J., Kolodner R. D. (2005) J. Biol. Chem. 280, 22245–22257 [DOI] [PubMed] [Google Scholar]
- 60.Hopfner K. P., Tainer J. A. (2003) Curr. Opin. Struct. Biol. 13, 249–255 [DOI] [PubMed] [Google Scholar]
- 61.Lebbink J. H., Sixma T. K. (2005) Structure 13, 498–500 [DOI] [PubMed] [Google Scholar]
- 62.Hargreaves V. V., Shell S. S., Mazur D. J., Hess M. T., Kolodner R. D. (2010) J. Biol. Chem. Vol. 285, 9301–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brosh R. M., Jr., Matson S. W. (1995) J. Bacteriol. 177, 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Löbau S., Weber J., Wilke-Mounts S., Senior A. E. (1997) J. Biol. Chem. 272, 3648–3656 [DOI] [PubMed] [Google Scholar]
- 65.Pan J. Y., Wessling-Resnick M. (1998) Bioessays 20, 516–521 [DOI] [PubMed] [Google Scholar]
- 66.Kinosian H. J., Selden L. A., Estes J. E., Gershman L. C. (1993) J. Biol. Chem. 268, 8683–8691 [PubMed] [Google Scholar]
- 67.Rosenfeld S. S., Houdusse A., Sweeney H. L. (2005) J. Biol. Chem. 280, 6072–6079 [DOI] [PubMed] [Google Scholar]
- 68.Shim K. S., Tombline G., Heinen C. D., Charbonneau N., Schmutte C., Fishel R. (2006) DNA Repair 5, 704–717 [DOI] [PubMed] [Google Scholar]
- 69.Wilson T., Guerrette S., Fishel R. (1999) J. Biol. Chem. 274, 21659–21664 [DOI] [PubMed] [Google Scholar]
- 70.Muradov K. G., Artemyev N. O. (2000) Biochemistry 39, 3937–3942 [DOI] [PubMed] [Google Scholar]
- 71.Marin E. P., Krishna A. G., Sakmar T. P. (2001) J. Biol. Chem. 276, 27400–27405 [DOI] [PubMed] [Google Scholar]
- 72.Lenzen C., Cool R. H., Prinz H., Kuhlmann J., Wittinghofer A. (1998) Biochemistry 37, 7420–7430 [DOI] [PubMed] [Google Scholar]
- 73.Klebe C., Prinz H., Wittinghofer A., Goody R. S. (1995) Biochemistry 34, 12543–12552 [DOI] [PubMed] [Google Scholar]
- 74.Hutchinson J. P., Eccleston J. F. (2000) Biochemistry 39, 11348–11359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.