Abstract
Genetic and biochemical studies have established a central role for α-synuclein accumulation in the pathogenesis of Parkinson disease. Here, two microRNAs, namely mir-7 and mir-153, have been identified to regulate α-synuclein levels post-transcriptionally. These microRNAs bind specifically to the 3′-untranslated region of α-synuclein and down-regulate its mRNA and protein levels, with their effect being additive. They are expressed predominantly in the brain with a pattern that mirrors synuclein expression in different tissues as well as during neuronal development, indicating that they play a tuning role in the amount of α-synuclein produced. Overexpression of mir-7 and mir-153 significantly reduces endogenous α-synuclein levels, whereas inhibition of mir-7 and mir-153 enhances translation of a luciferase construct bearing the α-synuclein 3′-untranslated region in primary neurons. These findings reveal a significant additional mechanism by which α-synuclein is regulated and point toward new therapeutic regimes for lowering endogenous α-synuclein levels in patients with familial or sporadic Parkinson disease.
Keywords: MicroRNA, Neurodegeneration, Neurological Diseases, Neuron, Neuroscience, Synuclein, Parkinson Disease
Introduction
The importance of α-synuclein (SNCA)2 in neurodegeneration is seminal. Point mutations and gene duplication and triplication events in the SNCA locus have been identified in a number of families with autosomal dominant early onset Parkinson disease (PD) (1). Moreover, SNCA is the major component of Lewy bodies in sporadic PD and dementia with Lewy bodies and also of inclusions found in both glial and neuronal cells in multiple system atrophy (2). These data suggest that modulation of SNCA levels, whether wild-type or mutant, is critical for the neurodegenerative processes in PD and other diseases, collectively termed synucleinopathies. The subject of SNCA regulation has been rather controversial. Some studies have shown that the proteasome is important, although others, which have failed to detect accumulation of SNCA upon proteasomal inhibition, propose a lysosomal autophagic pathway instead (3–6).
Although most genes encode proteins to carry out their biological functions, a recently discovered class of genes transcribing small noncoding RNAs, namely microRNAs, was found to be important in regulating normal development and homeostasis in diverse genomes, from worms to animals and plants. MicroRNAs (miRNAs) are small 19–23 RNA nucleotides that can regulate gene expression through RNA-induced silencing complex-mediated mRNA sequestration and translational repression by binding minimally to complementary “seed” sequences in the 3′-untranslated regions (UTR) of the target mRNAs. Each miRNA is estimated to regulate several closely related target genes, and the combinatorial action of miRNAs is expected to regulate the expression of hundreds of mRNAs (7). They have a wide variety of expression patterns, and many are expressed differentially during development or disease (8, 9). MiRNAs confer robustness to developmental genetic programs by two specific ways. In the first case, the miRNA and its target mRNA set are highly expressed in mutually exclusive tissues where the miRNA functions to block translation of the unwanted mRNAs expressed from the inherently leaky promoters. In the second case, both the miRNA and its target mRNA set are co-expressed in the same tissues where the miRNA acts as rheostat to dampen protein translation to optimal levels, enabling customized expression (10, 11).
Based on the fact that (i) proteasomal or lysosomal degradation of SNCA appears to be limited (3–6), (ii) SNCA possesses a long 3J-UTR longer than the coding region itself, (iii) several SNPs (single-nucleotide polymorphisms), located in the 3J-UTR of SNCA, have strong association with sporadic PD (12–14), and (iv) SNCA is a synaptic protein, with neuronal proteins likely to be under stronger selection for miRNA regulation (15), it was hypothesized that SNCA is a brain miRNA target. It is shown here that SNCA is directly targeted by brain-enriched mir-7 and mir-153, resulting in a significant decrease in mRNA and protein levels and that this effect is cumulative. Further, expression of miRNAs and SNCA mRNA and protein in different tissues and ages overlapped, indicating that the mir-7 and mir-153 role is to control fluctuations in endogenous SNCA protein levels.
EXPERIMENTAL PROCEDURES
Generation of DNA Constructs
The murine full-length SNCA DNA (GenBank accession no. NM_001042451 transcript variant 1, bases 300–1257, coding region plus 3′-UTR) was amplified by proofreading RT-PCR from postnatal day 1 mouse brain total RNA by using the primers 5′-GAATTCACCACCATGGATGTGTTCATGAAAGGACTTT-3′ and 5′-GCGGCCGCTTATTTTATTTTCCACC-3′. The PCR product was cloned into pcDNA3.1 vector (Invitrogen). The murine “short” SNCA DNA (coding region alone) was amplified by proofreading PCR from full SNCA by using the primers 5′-GAATTCACCACCATGGATGTGTTCATGAAAGGACTTT-3′ and 5′-GCGGCCGCAATGACATTCTTAGGCT-3′. This PCR product was also cloned into pcDNA3.1 vector. The murine SNCA 3′-UTR (NM_001042451, bases-1257) was amplified by proofreading RT-PCR from postnatal day 1 mouse brain total RNA by using the primers 5′-CTCGAGGAATGTCATTGCACCCA-3′ and 5′-GCGGCCGCTTATTTTATTTTCCACC-3′. The PCR product was cloned into psiCHECK2 vector (Promega, Madison, WI) downstream from the Renilla luciferase coding sequence. Expression vectors directing the synthesis of mmu-mir-7a-2 (miRBase accession no. MI0000729) and mmu-mir-153 (miRBase accession no. MI0000175) were prepared by introducing annealed oligonucleotides corresponding to the precursor microRNA sequences (pri-miRNA) into the pcDNA6.2-GW/EmGFP vector (Invitrogen). The oligonucleotide sequences were as follows: mmu-mir-7a-2 sense, 5′-TGCTGGGTCGGGCCAGCCCCGTTTGGAAGACTAGTGATTTTGTTGTTGTGTCTCTGTATCCAACAACAAGTCCCAGTCTGCCACATGGTGCTGGTCATTTCA-3′ and mu-mir-7a-2 antisense, 5′-CCTGTGAAATGACCAGCACCATGTGGCAGACTGGGACTTGTTGTTGGATACAGAGACACAACAACAAAATCACTAGTCTTCCAAACGGGGCTGGCCCGACCC-3′ mmu-mir-153 sense, 5′-TGCTGCGGTGTCATTTTTGTGACGTTGCAGCTAGTAATATGAGCCCAGTTGCATAGTCACAAAAGTGATCATTG-3′ and mmu-mir-153 antisense, 5′-CCTGCAATGATCACTTTTGTGACTATGCAACTGGGCTCATATTACTAGCTGCAACGTCACAAAAATGACACCGC-3′.
The mmu-mir-7/153 vector was prepared by inserting mmu-mir-7a-2 sequence after the mmu-mir-153 sequence in the pcDNA6.2-GW/EmGFP vector by restriction cloning. Scramble 1, provided by Invitrogen, is presumably predicted not to target any vertebrate gene. Scramble 2 was prepared by scrambling the mature sequence of brain mmu-mir-128b (miRBase accession no. MI0000726) before inserting it in the pcDNA6.2-GW/EmGFP vector. The oligonucleotide sequences for scramble 2 were as follows: mmu-mir-scramble2 sense, 5′-TGCTGCAGTGGGATGACTCTTGAGTATTGACAATGCGAGTGAGTAGCAGGTCGCTGTCAATAGCGAAGAGTCACCCTACTG-3′ and mmu-mir-scramble2 antisense, 5′-CCTGCAGTAGGGTGACTCTTCGCTATTGACAGCGACCTGCTACTCACTCGCATTGTCAATACTCAAGAGTCATCCCACTGC-3′. All vectors were checked by sequencing before use.
Site-directed Mutagenesis
Two point mutations, one for the mir-7 and the other for the mir-153 binding sites, were inserted in the SNCA 3′-UTR as described previously (16). Briefly, a set of three proofreading PCRs with mutagenized primers was carried out using the psiCHECK2-SNCA 3′-UTR vector as a template. The reactions were as follows (mutations are underlined): PCR1, external wtSNCA UTR F (5′-CTCGAGGAATGTCATTGCACCCA-3′) + mut-mir-7R (5′-ACTGCTGATGGTTGACTTTGAAACACA-3′); PCR2, internal mutSNCA UTR F (5′-CAACCATCAGCAGTGATCGGCGTC-3′) + internal mutSNCA UTR R (5′-AATGTCTCATGCTCACATAATT-3′); PCR3, mut-mir-153 F (5′-GTGAGCATGAGACATTGCACCTATAAATAT-3′) + external wtSNCA UTR R (5′-GCGGCCGCTTATTTTATTTTCCACC-3′). The PCR products were purified, and equal amounts were PCR-assembled using the external primers. The cycling conditions were as follows: five cycles with twice as long annealing and extension times (40 s instead of 20 s), to allow assembly of full mutant 3′-UTR, followed by 15 cycles of proofreading PCR to amplify the mutated full fragment. The mutagenized PCR product was then cloned into the psiCHECK2 vector and sequenced.
Lentivirus Production
The plasmids PLL3.7/EmGFP-scramble-1 and PLL3.7/EmGFP-mir-153/7 were constructed by replacing the U6 promoter, and GFP sequences between WRE and FLAP of the PLL3.7 vector (kindly provided by Dr. D. Thanos, Biomedical Research Foundation of the Academy of Athens) with the EmGFP-mir cassettes from the pcDNA6.2-GW/EmGFP-scramble-1 and pcDNA6.2-GW/EmGFP-mir-153/7 vectors, respectively. Viral particles were prepared by co-transfection of HEK293T cells with PLL3.7/EmGFP-mirs and helper pCMV-dR8.91 and pMD2.G plasmids at 10:7:3.5 μg of DNA ratios/10-cm plate by using the calcium phosphate method. After 48 h, the supernatants were spun at 2,000 rpm for 6 min, filtered at 0.45-μm pore size, and spun at 30,000 rpm (Sorvall TH-641 rotor) for 1 h 30 min and the pellets were resuspended in 50 μl of 1% bovine serum albumin. Lentiviral titers were determined by infection of HEK293 cells with serial dilutions of the viral stock for 48 h and followed by fluorescent-activated cytometric sorting (BD FACSAria sorter). Titers ranged from 3 to 6 × 107 infectious units.
Measurement of Mir-7, Mir-153, SNCA, and U6 RNAs
A semiquantitative RT-PCR assay was used to assess the levels of mir-7, mir-153, SNCA, and U6 in the various tissues. Total RNA was isolated using the mirVana miRNA isolation kit (Ambion, Austin, TX) and recovered in diethylpyrocarbonate (DEPC)-treated H2O. 0.5 μg of RNA was reverse transcribed for 1 h at 37 °C with superscript enzyme (Invitrogen) in a reaction containing the manufacturer's buffer and dithiothreitol supplemented with 0.5 mm dNTPs (Promega), 10 μm random hexanucleotides (Amersham Biosciences/GE Healthcare), and 5pmol of gene-specific primers for mature mmu-mir-7–1,2,3,b (CATGATCAGCTGGGCCAAGACAACAAAA) and mmu-mir-153 (5′-CATGATCAGCTGGGCCAAGATCACTTTT-3′). These primers included an extension sequence (underlined) to facilitate subsequent amplification and monitoring by PCR (modified from Refs. 17, 18).
Following reverse transcription, duplicate measurements of 2 μl of 2.5 × diluted cDNA were made in 10-μl PCRs containing 1 × buffer (Hytest, Turku, Finland), 0.5–1.5 mm MgCl2, 0.2 mm dNTPs, 4 pmol of primers, and 1.25 units of Taq (Hytest). The forward primers were as follows: mir-7–1,2,3 F, 5′-GCTGGGTTGGAAGACTAGTGAT-3′; mir-153 F, 5′-GCCGGGCTTGCATAGTCACAA-3′; mmu-SNCA F, 5′-GGAAGGAGTGGTTCATGG-3′; hs/mmu-GAPDH F, 5′-GCACCACCAACTGCTTAG-3′; and hs/mmu-U6 F, 5′-CGCTTCGGCAGCACATATAC-3′. The reverse primers were as follows: universal reverse (UR) for both mir-7 and mir-153, 5′-CATGATCAGCTGGGCCAAGA-3′; mmu-SNCA R, 5′-TCCTCCAACATTTGTCACTT-3′; hs/mmu-GAPDH R, 5′-GCCATCCACAGTCTTCTG-3′ and hs/mmu-U6 R, 5′-TTCACGAATTTGCGTGTCAT-3′. cDNAs were amplified by cycling at 95 °C for 35 s, followed by 35 s at 52 °C, followed by 35 s at 72 °C. The reaction was then completed with a 10-min extension at 72 °C. Cycles were falling within the linear range of amplification for each primer pair. These were 20–21 cycles for U6 RNA and 25–28 cycles for mir-7, mir-153, and SNCA mRNA. The PCR products were next separated on 8% nondenaturing polyacrylamide gels. The gels were subsequently stained with SyberGold (Invitrogen), and images were captured with the Dolphin gel documentation system (Wealtec, Sparks, Nevada).
Cell Line Culture and Transfection
HEK293 cells were maintained in low glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Biowest). For both reporter assay and Western blotting, HEK293 cells were transfected 1 day after plating by using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). To ensure that transfection efficiencies were uniform across conditions, both Lipofectamine 2000 reagent and SNCA constructs (CHECK-2 and pcDNA3) were prepared as master mix before aliquoting into tubes containing the different microRNAs. Transfection efficiencies were at 80–90% as indicated by GFP.
Neuron Culture and Transfection
Dissociated, embryonic day 16–17 rat or murine cortical neurons (>95% pure), were grown in Neurobasal medium with B-27 supplement (Invitrogen) and Glutamax (Invitrogen) in poly-l-lysine (Sigma)-coated tissue culture dishes (Greiner, Kremsmünster, Austria) in the absence of trophic factors (19). Neurons were either (i) lipofected straight after dissociation with the antisense 2′-O-methyl oligonucleotides plus luciferase vector according to the manufacturer's instructions (Invitrogen), (ii) infected at 4 days after plating with miRNA lentiviruses at approximately multiplicity of infection 10 or (iii) lipofected at 5 days after plating with miRNA expression vectors according to the manufacturer's instructions (Invitrogen). The antisense 2′-O-methyl RNA sequences were: for scramble, uagugaguaacaucucaaguugaauauguugu; for mir-7, uaaaaacaacaaaaucacuagucuuccacacag; and for mir-153, ucaaugaucacuuuugugacuaugcaacuggg.
Astrocyte Culture
Embryonic day 17 murine cerebral tissues, free of meninges, were treated with 0.05% trypsin solution (Worthington) for 5 min at 37 °C, then dissociated using a fire-polished Pasteur glass pipette and plated in tissue culture dishes (Greiner) containing RPMI 1640 medium (Biosera, East Sussex, UK) and 10% fetal bovine serum (Biowest). After ∼1 week of culture, astrocytes were trypsinized, dispersed in RPMI 1640 medium plus 10% fetal bovine serum, and replated at one-third the original density. They were harvested for RNA isolation after becoming confluent again.
Luciferase Reporter Assay
Luciferase assays were performed 48 h after transfection with the Dual-Luciferase reporter kit (Promega) and measured with the Lumat LB9507 reader (Berthold Technologies, Bad Wildbad, Germany). Changes in expression of Renilla luciferase (target) were calculated relative to firefly luciferase (internal control).
Western Blotting
Western blotting was used to assay SNCA protein levels in freshly dissected murine tissues and in cells treated with the different miRNAs. With respect to treatments, embryonic day 16 cortical neurons were infected with the appropriate lentiviral miRNA at 4 days after plating (SNCA levels pick up after several days in culture) (4), whereas HEK293 cells were co-transfected with the appropriate miRNA expression vector and SNCA vector a day after plating by using Lipofectamine 2000. Four (for cortical neurons) and 2 (for HEK293 cells) days later, whole cell protein extracts were harvested with urea lysis buffer (8 m urea, 0.5% SDS, 1 mm Na3VO4) and boiled for 5 min.
For Western blotting, 10–30 μg of each protein extract was loaded onto denatured 15% polyacrylamide gel and transferred to Protran® nitrocellulose membrane (Whatman). The membrane was incubated in 5% skimmed milk for 1 h followed by overnight incubation with SNCA (BD Biosciences) and GAPDH (Fitzgerald Industries, Concord, MA) antibodies. The next day, the membrane was incubated for 1 h with anti-mouse secondary antibody conjugated with either alkaline phosphatase or horseradish peroxidase antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) depending on the expected SNCA levels. Alkaline phosphatase signals were detected by combining the 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium substrates (AppliChem, Darmstadt, Germany) whereas horseradish peroxidase signals were detected by the Western blotting luminol reagent (Santa Cruz Biotechnology).
Immunocytochemistry and Densitometry
Embryonic day 16 cortical neurons were plated on poly-l-lysine (Sigma) borosilicate glass coverslips (VWR) in Neurobasal medium for 5 days before being lipofected with scramble 1 or mir-153/7 vectors according to the manufacturer's instructions (Invitrogen). Neurons were fixed 40 h later with freshly made 4% (w/v) paraformaldehyde solution (Sigma) and briefly permeabilized with 0.4% Triton X-100 (Sigma). Cells were subsequently blocked with 5% of secondary serum (normal goat serum) and incubated overnight at 4 °C with primary SNCA antibody (1:100, BD Biosciences). The next day, cells were incubated for 1 h with goat anti-mouse rhodamine secondary antibody (1:200, Santa Cruz Biotechnology) in the presence of the DNA stain 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and mounted with Vectashield (Invitrogen) medium. Images were taken with a Leica DMRA2 upright microscope using a 60 × objective. SNCA levels in scramble- or mir-153/7-transfected cells were estimated by comparing the average intensity of the five brightest for rhodamine neurons in each field with the miRNA transfected neuron in the same field (80 fields in total per miRNA condition per experiment). Duplicate transfections were set up for the each condition, and the data were compiled from four separate experiments. Statistical significance was assessed using Student's paired t test (two-tailed distribution).
Bioinformatics Analysis
Conserved miRNA targets were predicted using TargetScan software (20). Protein targets with a total context score below −0.15 were then selected for the analysis. There were 209 and 326 predicted targets for mir-7 and mir-153, respectively. The sets were then analyzed for enriched Bio-categories and signaling pathways using the Ingenuity software (IPA version 7.0; Ingenuity Systems, Redwood City, CA). Fisher's exact test was used to calculate p values.
RESULTS
Mir-7 and Mir-153 Post-transcriptionally Control SNCA Expression via Its 3′-UTR
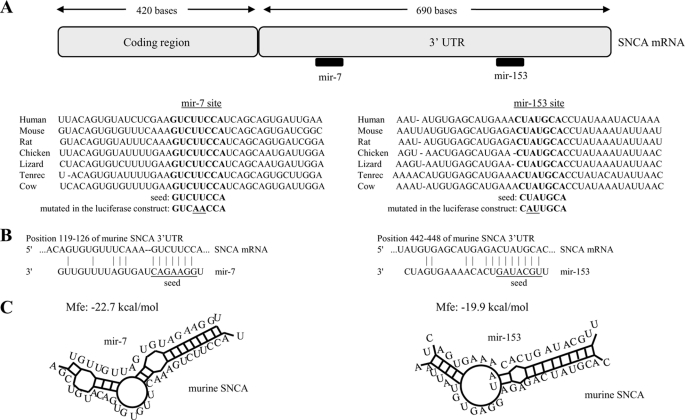
By using the sequence analysis software TargetScan (20), miRanda (21), and Pictar (22), two putative binding sites for mir-7 and mir-153 on SNCA gene 3′-UTR were identified; importantly, these sites are conserved across vertebrate species (Fig. 1A). The seed sequences of mir-7 and mir-153 are complementary to nucleotides 119–126 and 442–448 of the murine SNCA 3′-UTR, respectively (Fig. 1B), with a predicted duplex formation as shown in Fig. 1C. The minimum free energies of the interaction between mir-7 and mir-153 with their corresponding SNCA 3′-UTR binding sites are −22.7 kcal/mol and −19.9 kcal/mol, respectively. This interaction is likely to be further facilitated by the abundance of AU residues in the flanking regions of the mir-7 and mir-153 binding sites (20).
FIGURE 1.
Diagram of the miRNA binding sites within the SNCA 3′-UTR. A, two conserved sites, one for mir-7 and another for mir-153, are found in the SNCA 3′-UTR. B, bioinformatics prediction of miRNA and murine SNCA transcript interactions. C, predicted hybridization of miRNAs (gray) and SNCA (black) transcript using the RNAhybrid algorithm (36). The minimum free energy required for the hybridization is indicated.
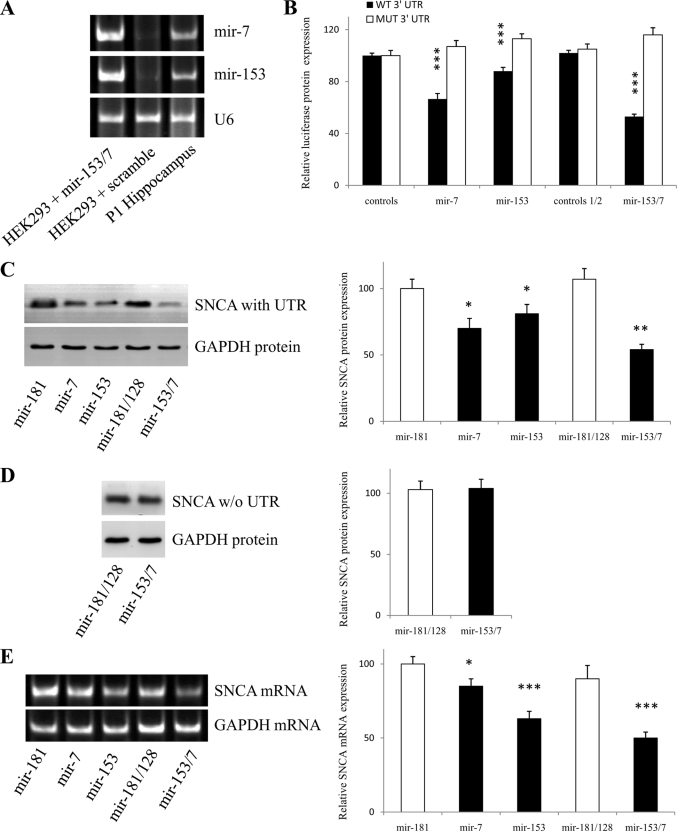
To investigate the potential interaction experimentally, the mouse SNCA 3′-UTR was cloned after the Renilla luciferase coding sequence and co-transfected together with expression plasmids for pri-mir-7 and pri-mir-153 into HEK293 cells. HEK293 cells were chosen because they have comparatively little endogenous expression of mir-7 and mir-153, unlike neural tissues, and could therefore provide a quantitative cell system for the assay (Fig. 2A). Mir-7, mir-153, and mir-153/7 (a vector that expresses both miRNAs in tandem) produced a 33% (p < 0.001), a 12% (p < 0.001), and a 47% (p < 0.001) decrease in luciferase activity compared with control vectors, respectively (Fig. 2B). In contrast, point mutations in the corresponding seed regions abolished completely this down-regulation (Fig. 2B). These findings indicate that (i) mir-7 and mir-153 interact directly with the specified regions of SNCA 3′-UTR to inhibit translation from the chimeric transcript and (ii) their effect is cumulative.
FIGURE 2.
Mir-7 and mir-153 directly reduce SNCA protein expression. A, nonneuronal cell line HEK293 expresses low levels of mir-7 and mir-153. Representative gel shows the products of RT-PCRs amplified with primers specific for mir-7 and mir-153 in postnatal day (P1) murine hippocampus and HEK293 cells transfected with scramble or mir-153/7 vectors. The amount of starting template for each condition was equilibrated relative to U6 RNA. B, luciferase activity assays demonstrated that mir-7 and mir-153 directly suppress SNCA expression by targeting the identified seed regions in the SNCA 3′-UTR. HEK293 cells were co-transfected with both the reporter gene (either wild-type or mutated) and miRNA expression vectors, and luciferase activity was measured 48 h later. For control 1 and control 1/2, the average value of three (scramble1, mir-181, mir-218) and two (mir-181/128, mir-218/377) miRNA constructs expressing miRNA predicted not to bind SNCA 3′-UTR were used, respectively. Data show the mean ± S.E. (error bars) from six independent transfections (***, p < 0.001). C and D, representative Western blot analysis of SNCA protein demonstrating that mir-7 and mir-153 reduce SNCA protein levels through interaction with the 3′-UTR. HEK293 cells were co-transfected with either a full-length (coding region plus 3′-UTR) or a shorter version (coding region alone) SNCA construct and miRNA expression vectors. Cells were harvested 48 h later, and 15 μg of whole cell lysate was loaded in each lane. GAPDH is used as an internal control for loading. Data show the mean ± S.E. (error bars) from four independent experiments (*, p < 0.05; **, p < 0.01). E, representative RT-PCR analysis of SNCA mRNA demonstrating that both mir-7 and mir-153 regulate SNCA mRNA levels. HEK293 cells were co-transfected with a full-length SNCA construct and miRNA expression vectors. Cells were harvested 48 h later, RNA was purified, and RT-PCR for SNCA mRNA was carried out. GAPDH and U6 were used as an internal control for loading. Data show the mean ± S.E. (error bars) from four independent experiments (*, p < 0.05; ***, p < 0.001).
To dissect the interaction further, the effect of mir-7 and mir-153 on SNCA protein levels was examined. For this, HEK293 cells were co-transfected with a plasmid expressing either full-length SNCA (coding region + 3′-UTR, “full SNCA”) or a shorter version of SNCA (coding region alone, “short SNCA”) and the miRNA expression plasmids. Western blots from protein extracts using the full SNCA-expressing construct revealed a significant reduction in SNCA levels with mir-7 (30%, p < 0.05) or mir-153 (19%, p < 0.05) alone and a more pronounced (46%, p < 0.01) reduction after treatment with both miRNAs (Fig. 2C). In contrast, mir-153/7 did not regulate SNCA protein levels produced from the short SNCA construct (Fig. 2D). These findings indicate that mir-7 and mir-153 (i) are synergistic in their effect, (ii) require the 3′-UTR of SNCA mRNA to regulate SNCA protein expression, (iii) do not interact with the coding region of SNCA to regulate SNCA protein expression, and (iv) act at the pretranslational level.
Next, the effect of mir-7 and mir-153 on SNCA mRNA levels was determined. For this, HEK293 cells were co-transfected with the plasmid expressing full-length SNCA and the miRNA expression plasmids. RT-PCR from RNA extracts using SNCA-specific primers revealed that SNCA mRNA expression was significantly reduced by mir-7 (15%, p < 0.05), mir-153 (37%, p < 0.001), and mir-153/7 overexpression (50%, p < 0.001) (Fig. 2E), indicating that mir-7 and mir-153 (i) regulate SNCA protein levels in part by inducing degradation of SNCA mRNA, (ii) use different kinetics to regulate SNCA protein levels, with mir-7 showing a preference for sustained translation inhibition while mir-153 for transient mRNA degradation, and (iii) act synergistically to lower SNCA mRNA levels. Because neither of the miRNAs is fully complementary to SNCA mRNA, to allow endonucleolytic cleavage by Argonaute proteins, the degradation effect is likely to stem from the accelerated level of deadenylation and decapping of SNCA mRNA induced by the mir-7 and mir-153 binding to 3′-UTR (23–26).
Mir-7 and Mir-153 Have Similar Tissue Distribution with SNCA mRNA and Protein
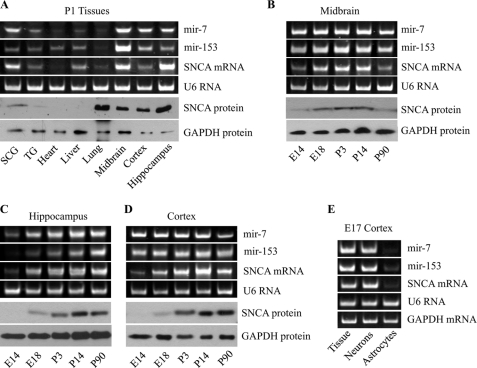
To begin to characterize the physiological interaction among mir-7, mir-153, and SNCA, the levels of mature mir-7 and mir-153 as well as SNCA mRNA and protein were quantified in different tissues of postnatal day 1 mice. To compare the relative levels of these RNAs and SNCA protein in the different tissues, the RNA and protein levels for the ubiquitously expressed U6 RNA and GAPDH protein were also quantified, respectively. Fig. 3A shows that the expression of mir-7 and mir-153 has distribution similar to both SNCA mRNA and protein in the different tissues of postnatal day 1 mice. Specifically, mir-7, mir-153, and SNCA mRNA and protein show highest expression in neural tissues, such as midbrain, hippocampus, and cortex and least in nonneural tissues such as lung and heart. Interestingly, all three RNA species show highest expression in midbrain, indicating that deregulation of their expression levels may be important in the pathogenesis of PD. SNCA protein is also detected in the lung as a result of the high amount of peripheral blood present in this tissue.
FIGURE 3.
Mir-7 and mir-153 show similar distribution to SNCA mRNA and protein. A, representative gels of the PCR amplification products of mir-7, mir-153, and SNCA mRNA and Western blot analysis of SNCA levels. Different murine tissues of postnatal day 1 (P1) animals are shown. SCG, superior cervical ganglion; TG, trigeminal ganglion. B, midbrain of different ages. C, hippocampus of different ages. D, cortex of different ages. E, representative gels of the PCR amplification products of mir-7, mir-153, and SNCA mRNA in E17 cortical tissue, 3-day neuronal, and 15-day astrocytic cultures. The amount of starting template for each condition was normalized to U6 RNA and GAPDH mRNA or protein.
Further, to determine whether mir-7 and mir-153 expression correlates with SNCA expression throughout development, the levels of mature mir-7, mir-153, and SNCA mRNA and protein were also quantified in midbrain, cortex, and hippocampus at different ages. Fig. 3, B–D, shows that the levels of mir-7 and mir-153 correspond overall to the expression of SNCA mRNA, with high levels of SNCA mRNA associated with increased levels of mir-7 and mir-153 expression. Moreover, the expression profiles of both SNCA mRNA and protein are similar, indicating that during development a constant level of the miRNA-regulated SNCA mRNA is translated into protein.
Because central neural tissues are a mixed population of predominantly neurons and astrocytes, the levels of mature mir-7, mir-153, and SNCA mRNA were also quantified in these cells. Fig. 3E shows that the levels of mir-7, mir-153, and SNCA mRNA are significantly higher in neurons than astrocytes (protein levels were particularly low and could not be assayed). These results indicate that mir-7 and mir-153 are co-expressed with SNCA in neurons to regulate its levels through a transcription feed-forward loop (11) that fine-tunes rather than blocks SNCA mRNA expression and translation.
Mir-7 and Mir-153 Regulate Endogenous SNCA Protein Levels in Neurons
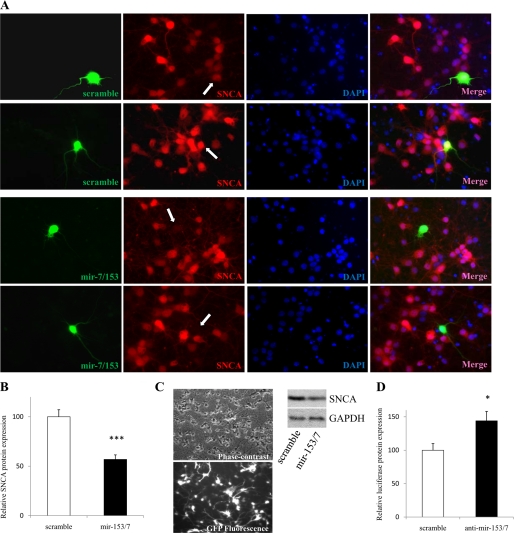
Based on aforementioned experimental and expression data, it was hypothesized that changes in miRNA levels in neurons would impact endogenous SNCA protein levels. To test this, cortical neurons, a population that is directly implicated in Lewy body dementia (27), were cultured for 5 days, to boost endogenous SNCA levels (4), and then lipofected with mir-153/7 vector for 40 h before fixed with paraformaldehyde solution. SNCA protein levels were then assayed by immunofluorescence and estimated by ImageJ densitometry. An average decrease of 43% in endogenous SNCA protein levels with mir-153/7 overexpression was measured in cortical neurons (p < 0.001, Fig. 4, A and B). To analyze a different neuronal population that expresses similarly high levels of endogenous SNCA protein (Fig. 3A) and provides a large number of cells required for the assay, hippocampal neurons were infected with a lentiviral vector expressing mir-153/7, and SNCA protein levels were assayed 4 days later by Western blotting. An ∼30–40% reduction of endogenous SNCA protein levels was again observed (Fig. 4C). These findings indicate that (i) endogenous mir-7 and mir-153 expression levels are not saturated in neurons, thus allowing a controlled amount of SNCA mRNA to be translated in accordance with the tuning mechanism and (ii) the endogenous miRNA maturation machinery in neurons can significantly accommodate the processing of additional pri-miRNAs.
FIGURE 4.
Mir-153/7 reduces endogenous SNCA protein levels in neurons. A, representative images of transfected cortical neurons stained with SNCA antibody and DAPI. Merged images are shown in the fourth panel. Embryonic day 16 rat cortical neurons were lipofected with scramble1 or mir-7/153 at 5 days after plating by using Lipofectamine 2000. Immunocytochemistry was carried out 40 h after transfection. B, average decrease in SNCA protein levels. 80 neurons were analyzed per experiment. Data show the mean ± S.E. (error bars) from four independent experiments (***, p < 0.001). C, hippocampal neurons were infected with a lentivirus expressing GFP-mir-7/153 (multiplicity of infection, 10) at 4 days after plating. Neurons were harvested 4 days later, and 30 μg of whole cell lysate was loaded in each lane. GAPDH is used as an internal control for loading. D, freshly dissociated cortical neurons were transfected with SNCA luciferase vector and antisense 2′-O-methyl oligonucleotides of mir-7 and mir-153 and assayed 48 h later. Data show the mean ± S.E. (error bars) from four independent experiments (*, p < 0.05).
Next, the ability of endogenously expressed mir-7 and mir-153 to regulate the levels of the luciferase construct bearing the SNCA 3′-UTR was evaluated in primary cultures of neurons. Antisense 2′-O-methyl mir-7 and mir-153 oligonucleotides were lipofected together with the luciferase construct in freshly dissociated (to increase transfection efficiency) cortical neurons, and luciferase activity was assayed 48 h later. A 44% (p < 0.05) increase in luciferase levels was measured with the antisense mir-7 and mir-153 oligonucleotides (Fig. 4D), indicating that endogenous mir-7 and mir-153 bind to mRNAs bearing SNCA 3′-UTR and block translation. A similar experiment in which antisense 2′-O-methyl mir-7 and mir-153 oligonucleotides were lipofected in either freshly dissociated cortical neurons, SH-SY5Y cells, or Neuro-2a cells was also performed, but endogenous SNCA protein levels were too low for detection by Western blotting.
DISCUSSION
This study has revealed a previously unknown mechanism by which SNCA levels are regulated in the nervous system. Specifically, two brain-enriched miRNAs, mir-7 and mir-153, have been shown to bind directly to the 3′-UTR of SNCA mRNA and significantly reduce its levels. This finding has been based on (i) conservation analysis of the miRNA seed binding sites on SNCA 3′-UTR which show that they are conserved throughout vertebrate species; (ii) in vitro experiments in which the seed regions have been mutagenized, resulting in the abrogation of protein degradation of a chimeric luciferase transcript bearing the SNCA 3′-UTR; (iii) in vitro experiments in which the 3′-UTR has been removed from the SNCA mRNA transcript, resulting in the loss of miRNA regulation but also proving that they do not bind the SNCA coding region; (iv) RNA and protein expression analysis that show developmental and tissue co-expression among mir-7, mir-153, and SNCA; (v) in culture neuron experiments in which overexpression of mir-153/7 resulted in down-regulation of endogenous SNCA protein levels; and (vi) in culture neuron experiments in which inhibition of endogenous mir-7 and mir-153 by antisense oligonucleotides promoted the expression of chimeric luciferase mRNA bearing SNCA 3′-UTR.
By considering the two different means of miRNA function, these findings strongly suggest that mir-7 and mir-153 play a role in modulating/buffering SNCA protein levels in the nervous system. This is further reiterated by recent findings showing that the physiological role of mir-7 is to impart robustness in protein expression output against environmental fluctuations during neural development (28). Although little is known about mir-7 and mir-153 function, two other reports have associated mir-7 expression with neurosecretory cell development and evolution (29) and mir-153 levels with neuroepithelial response to ethanol toxicity (30). These findings are particularly interesting from a SNCA perspective because they link mir-7 with the role of SNCA in synapses and they link the lower levels of mir-153 with increased levels of SNCA in alcoholic subjects (31).
Based on the inherent property of miRNAs to regulate several closely related target genes, bioinformatics was used to reveal protein networks that mir-7 and mir-153 may regulate and in which presynaptic SNCA is likely to participate. This was done by categorizing the mir-7 and mir-153 predicted conserved targets using the Ingenuity Pathways Analysis software (supplemental Table S1). Accordingly, the top two enriched Biocategories for mir-7 identified were “Behavior” and “Nervous System Development and Function,” whereas for mir-153 they were “Nervous System Development and Function” and “Organismal Survival.” Within these categories several important neural processes, with among the lowest p values (p < 0.001), were distinguished that include “Learning” and “Paired-pulse Facilitation of Synapse” for mir-7 and “Branching,” “LTP,” and “Neurotransmission” for mir-153. These preliminary findings are supported by the miRNA expression data and indicate that mir-7 and mir-153 are likely to play an important role in synaptic plasticity in which SNCA has already been implicated.
Given the centrality of SNCA in PD, delivery of both mir-7 and mir-153 may represent an appealing approach for therapy. Evidence for systemic use and successful medical intervention based on miRNAs has already been provided in cornerstone studies to lower plasma cholesterol levels in rodents and primates (32–34). In this direction, overexpression of both miRNAs in primary cultures of hippocampal and cortical neurons significantly lowered endogenous SNCA protein levels, indicating that the endogenous neuronal miRNA machinery can accommodate increased miRNA processing and targeting. Furthermore, promising unpublished data indicate that mir-153/7 overexpression significantly promotes neurite outgrowth and is neuroprotective against classical PD insults in in culture models.
Overall, the findings suggest that SNCA expression is regulated by brain miRNAs at the post-transcriptional level. This sets the stage for further evaluations of the role of mir-7 and mir-153 in the PD pathophysiological process; in particular, it would be important to determine, noting the high mir-7 and mir-153 expression in midbrain and cortex, whether human aging and/or environmental factors alter miRNA expression levels and therefore contribute to increased SNCA protein levels and PD risk (35). In view of the seminal role of SNCA in both inherited and sporadic PD, it may be important to also sequence PD patients' DNA for polymorphisms or mutations in both SNCA 3′-UTR and mir-7/153 loci that may affect SNCA miRNA regulation.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- SNCA
- α-synuclein
- PD
- Parkinson disease
- miRNA
- microRNA
- UTR
- untranslated region
- RT
- reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- EmGFP
- emerald green fluorescent protein
- SNP
- single-nucleotide polymorphism
- DEPC
- diethylpyrocarbonate.
REFERENCES
- 1.Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) Science 302, 841 ( abstr.) [DOI] [PubMed] [Google Scholar]
- 2.Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6469–6473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett M. C., Bishop J. F., Leng Y., Chock P. B., Chase T. N., Mouradian M. M. (1999) J. Biol. Chem. 274, 33855–33858 [DOI] [PubMed] [Google Scholar]
- 4.Vogiatzi T., Xilouri M., Vekrellis K., Stefanis L. (2008) J. Biol. Chem. 283, 23542–23556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 6.Paxinou E., Chen Q., Weisse M., Giasson B. I., Norris E. H., Rueter S. M., Trojanowski J. Q., Lee V. M., Ischiropoulos H. (2001) J. Neurosci. 21, 8053–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D. P., Chen C. Z. (2004) Nat. Rev. Genet. 5, 396–400 [DOI] [PubMed] [Google Scholar]
- 8.Lai E. C. (2005) Curr. Biol. 15, R458–R460 [DOI] [PubMed] [Google Scholar]
- 9.Wienholds E., Plasterk R. H. (2005) FEBS Lett. 579, 5911–5922 [DOI] [PubMed] [Google Scholar]
- 10.Hornstein E., Shomron N. (2006) Nat. Genet. 38, (suppl) S20–S24 [DOI] [PubMed] [Google Scholar]
- 11.Peterson K. J., Dietrich M. R., McPeek M. A. (2009) Bioessays 31, 736–747 [DOI] [PubMed] [Google Scholar]
- 12.Winkler S., Hagenah J., Lincoln S., Heckman M., Haugarvoll K., Lohmann-Hedrich K., Kostic V., Farrer M., Klein C. (2007) Neurology 69, 1745–1750 [DOI] [PubMed] [Google Scholar]
- 13.Mizuta I., Satake W., Nakabayashi Y., Ito C., Suzuki S., Momose Y., Nagai Y., Oka A., Inoko H., Fukae J., Saito Y., Sawabe M., Murayama S., Yamamoto M., Hattori N., Murata M., Toda T. (2006) Hum. Mol. Genet. 15, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 14.Simon-Sanchez J., Schulte C., Bras J. M., Sharma M., Gibbs J. R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S. W., Hernandez D. G., Kruger R., Federoff M., Klein C., Goate A., Perlmutter J., Bonin M., Nalls M. A., Illig T., Gieger C., Houlden H., Steffens M., Okun M. S., Racette B. A., Cookson M. R., Foote K. D., Fernandez H. H., Traynor B. J., Schreiber S., Arepalli S., Zonozi R., Gwinn K., van der Brug M., Lopez G., Chanock S. J., Schatzkin A., Park Y., Hollenbeck A., Gao J., Huang X., Wood N. W., Lorenz D., Deuschl G., Chen H., Riess O., Hardy J. A., Singleton A. B., Gasser T. (2009) Nat. Genet. 41, 1308–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2746–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge L., Rudolph P. (1997) BioTechniques 22, 28–30 [DOI] [PubMed] [Google Scholar]
- 17.Raymond C. K., Roberts B. S., Garrett-Engele P., Lim L. P., Johnson J. M. (2005) RNA 11, 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lao K., Xu N. L., Yeung V., Chen C., Livak K. J., Straus N. A. (2006) Biochem. Biophys. Res. Commun. 343, 85–89 [DOI] [PubMed] [Google Scholar]
- 19.Doxakis E., Huang E. J., Davies A. M. (2004) Curr. Biol. 14, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 20.Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright A. J., John B., Gaul U., Tuschl T., Sander C., Marks D. S. (2003) Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 23.Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. (2009) RNA 15, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishima Y., Giraldez A. J., Takeda Y., Fujiwara T., Sakamoto H., Schier A. F., Inoue K. (2006) Curr. Biol. 16, 2135–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. (2006) Genes Dev. 20, 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behm-Ansmant I., Rehwinkel J., Izaurralde E. (2006) Cold Spring Harb. Symp. Quant. Biol. 71, 523–530 [DOI] [PubMed] [Google Scholar]
- 27.Trojanowski J. Q., Goedert M., Iwatsubo T., Lee V. M. (1998) Cell Death Differ. 5, 832–837 [DOI] [PubMed] [Google Scholar]
- 28.Li X., Cassidy J. J., Reinke C. A., Fischboeck S., Carthew R. W. (2009) Cell 137, 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tessmar-Raible K., Raible F., Christodoulou F., Guy K., Rembold M., Hausen H., Arendt D. (2007) Cell 129, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 30.Sathyan P., Golden H. B., Miranda R. C. (2007) J. Neurosci. 27, 8546–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker S. J., Grant K. A. (2006) Alcohol 38, 1–4 [DOI] [PubMed] [Google Scholar]
- 32.Krützfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005) Nature 438, 685–689 [DOI] [PubMed] [Google Scholar]
- 33.Elmén J., Lindow M., Schütz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjärn M., Hansen H. F., Berger U., Gullans S., Kearney P., Sarnow P., Straarup E. M., Kauppinen S. (2008) Nature 452, 896–899 [DOI] [PubMed] [Google Scholar]
- 34.Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjärn M., Hansen J. B., Hansen H. F., Straarup E. M., McCullagh K., Kearney P., Kauppinen S. (2008) Nucleic Acids Res. 36, 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu Y., Kordower J. H. (2007) Neurobiol. Dis. 25, 134–149 [DOI] [PubMed] [Google Scholar]
- 36.Krüger J., Rehmsmeier M. (2006) Nucleic Acids Res. 34, W451–W454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.