Abstract
We characterized the properties of Drosophila melanogaster DAAM-FH2 and DAAM-FH1-FH2 fragments and their interactions with actin and profilin by using various biophysical methods and in vivo experiments. The results show that although the DAAM-FH2 fragment does not have any conspicuous effect on actin assembly in vivo, in cells expressing the DAAM-FH1-FH2 fragment, a profilin-dependent increase in the formation of actin structures is observed. The trachea-specific expression of DAAM-FH1-FH2 also induces phenotypic effects, leading to the collapse of the tracheal tube and lethality in the larval stages. In vitro, both DAAM fragments catalyze actin nucleation but severely decrease both the elongation and depolymerization rate of the filaments. Profilin acts as a molecular switch in DAAM function. DAAM-FH1-FH2, remaining bound to barbed ends, drives processive assembly of profilin-actin, whereas DAAM-FH2 forms an abortive complex with barbed ends that does not support profilin-actin assembly. Both DAAM fragments also bind to the sides of the actin filaments and induce actin bundling. These observations show that the D. melanogaster DAAM formin represents an extreme class of barbed end regulators gated by profilin.
Keywords: Actin, Cytoskeleton, Fluorescence, Microscopic Imaging, Protein Assembly, Actin Nucleation Factor, Microfilament, Molecular Mechanisms, Profilin Gating, Protein Interactions
Introduction
The actin cytoskeleton fulfills its various biological functions under the tight and well controlled balance of regulatory systems. The regulation in many cases is manifested by actin-binding proteins (for reviews, see Refs. 1–3). Among these proteins, the actin nucleation factors play critical roles in actin assembly by initiating the formation of new actin filaments in a spatially and temporally controlled fashion. Formins are actin nucleation factors known to assist the formation of unbranched actin structures by catalyzing processive assembly of actin filaments (for a review, see Ref. 4). Formins consist of several conserved domains (5), including the signature formin homology domains (FH1 and FH2) and domains thought to be responsible for the regulation (6). The FH2 domain is both necessary and sufficient to nucleate actin in vitro (7, 8), and it has been shown to remain associated with the barbed end of the growing filament (9, 10). The proline-rich FH1 domain can serve as a docking site for the G-actin-binding protein profilin (11–14) and also for other regulators, such as members of the Src family (15).
Phylogenetic analyses of a large set of FH2 domains from various species revealed that metazoan formins segregate into seven subfamilies (16). Three of these groups, Dia (diaphanous), DAAM (Dishevelled-associated activator of morphogenesis), and FRL (formin-related gene in leukocytes), also exhibit similarities outside of the FH2 domain and have been termed DRL (Diaphanous-related) formins. The activity of these proteins is thought to be regulated by an autoinhibitory mechanism involving the intramolecular association of the N-terminal DID (Diaphanous inhibitory domain) with the C-terminal DAD (diaphanous autoregulatory domain) (17–19). This interaction can be relieved upon Rho GTPase binding to the GBD (GTPase binding domain) adjacent to the DID, leading to the activation of the formin protein (17, 20).
The in vivo function of some formins, in particular DAAM family members, has been extensively studied (21–26). These studies suggested that human and Xenopus DAAM may have important roles in non-canonical Wnt signaling and affect convergent extension, an early embryonic morphogenetic process (26). The crystal structure of the FH2 domain of the human DAAM1 protein was recently solved (27). In Drosophila, dDAAM is required to organize apical actin filaments into parallel bundles in the tracheal system (25), whereas in the embryonic neurites, dDAAM plays a role in axon growth by regulating filopodia formation in the growth cone (28). However, the molecular mechanism supporting these biological functions of DAAM formins remains unclear. Former studies concluded that the overall structure of the FH2 domain is likely to be conserved (27, 29–31). However, the large variations in the rates of nucleation and of barbed end growth and in the effect of profilin (4, 32) might account for the functional diversity within the formin family (32). Therefore, to better understand how DAAM subfamily formins exert essential biological functions (e.g. in convergent extension or axonal growth), it is important to characterize the biochemical and biophysical properties of these proteins in detail.
Here we have undertaken the biochemical and biophysical analysis of the FH2 and FH1-FH2 domains of Drosophila DAAM. We show that in living cells, the ectopically expressed FH1-FH2 of dDAAM behaves like an activated formin, whereas the FH2 domain alone does not appear to affect cellular actin dynamics. In vitro, the DAAM-FH2 and DAAM-FH1-FH2 domains have similar effects on the kinetic and thermodynamic parameters of actin polymerization in the absence of profilin but different activities in the presence of profilin, indicating that the FH1 domain is essential for the interaction of DAAM with profilin-actin. In biomimetic assays, the bead-immobilized DAAM-FH1-FH2 but not the DAAM-FH2 nucleates actin and processively elongates actin filaments from profilin-actin. In addition, cosedimentation assays show that the DAAM fragments bind to the sides of the actin filaments. Together, these observations established that the dDAAM behaves as a bona fide formin possessing a number of properties previously reported for other members of the formin family.
EXPERIMENTAL PROCEDURES
Protein Purifications and Modifications
Actin was prepared from rabbit skeletal muscle (33). The concentration of G-actin was determined spectrophotometrically using a Shimadzu UV-2100 spectrophotometer, with the absorption coefficient of 0.63 mg−1 ml cm−1 at 290 nm (34). A relative molecular weight of 42,300 was used for G-actin (35). The actin was stored in 4 mm Tris-HCl, 0.1 mm CaCl2, 0.2 mm ATP, 0.5 mm DTT, pH 7.3 (buffer A). For the polymerization assays, actin was labeled with N-(1-pyrene)iodoacetamide (pyrene)5 (Sigma) on Cys374 as described previously (36). The concentration of the fluorescence dye in the protein solution was determined using the absorption coefficient of 2.2 × 104 m−1 cm−1 at 344 nm for pyrenyl-actin. For total internal reflection fluorescence microscopy (TIRFM) and biomimetic motility assays, actin was labeled with Alexa Fluor 488 or 568 carboxylic acid succinimidyl ester (Alexa 488 or Alexa 568) or 5-(and 6)-carboxytetramethylrhodamine succinimidyl ester (rhodamine) (Molecular Probes) as described (37).
The FH2 and the FH1-FH2 fragments of Drosophila melanogaster DAAM were prepared as by Shimada et al. (29). Briefly, the FH2 and the FH1-FH2 fragments were expressed as glutathione S-transferase fusion proteins in the Escherichia coli BL21(DE3)pLysS strain (Novagen). The protein expression was induced with isopropyl-β-d-thiogalactopyranoside (1 mm). The cell lysate was centrifuged at 100,000 × g at 4 °C for 1 h, and the supernatant was loaded onto a GSH column (Amersham Biosciences). The DAAM fragments were cleaved with thrombin and eluted. Sephacryl S-300 column was used for size exclusion as further purification. The extinction coefficient was calculated with ProtParam (available on the ExPASy Proteomics Server) and was determined to be ϵ280 = 22,920 m−1 cm−1 for DAAM-FH2 and 22,982.5 m−1 cm−1 for DAAM-FH1-FH2 at 280 nm. The molecular mass of the FH2 and FH1-FH2 fragments was taken to be 47.9 and 54.7 kDa, respectively. The purified protein was frozen in liquid nitrogen and stored at −80 °C.
Full-length yeast profilin subcloned in pHAT2-His-tagged expression vector (a gift from Pekka Lappalainen, Institute of Biotechnology, University of Helsinki (Helsinki, Finland)) was expressed in BL21(DE3)pLysS E. coli (Novagen). Profilin was purified under native conditions by Ni2+ affinity chromatography using Ni2+-nitrilotriacetic acid-agarose (Qiagen, Valencia, CA) according to the instructions of the manufacturer. Profilin from bovine spleen and recombinant human actin-depolymerizing factor (ADF) and gelsolin were purified as described (38). The purified profilin ADF and gelsolin were stored at −80 °C.
DNA Techniques, Transfection, and Immunohistochemistry
DNA constructs for transgenic flies and transfection experiments were created by the Gateway cloning system (ABI-Invitrogen). To create the entry clones, we PCR-amplified the appropriate portions of the dDAAM cDNA that were subsequently inserted into pENTR1. The FH1-FH2 fragment tested encodes amino acids 568–1053, whereas the FH2 contains amino acids 637–1053. As destination clones, we used pAVW for transfection experiments and pTVW or pTMW (Drosophila Gateway Collection) for transgenic flies. For bacterial protein expression, we used the same dDAAM subfragments as above, inserted into a pGEX-2T vector (Amersham Biosciences).
Drosophila S2 cells were transfected with the Effectene transfection kit (Qiagen) and incubated in Drosophila Schneider's medium (Lonza) for 24 h before fixation. S2 cells were fixed in 4% formaldehyde in phosphate-buffered saline for 10 min, and permeabilized in phosphate-buffered saline plus 0.1% Triton X-100 for 3 min before staining. Primary antibodies were applied for 1 h at room temperature, and after three 5-min washes in phosphate-buffered saline, cells were incubated with secondary antibodies for another 1 h. We used Rb-anti-green fluorescent protein (1:1000; Molecular Probes) to detect the Venus-tagged proteins. As secondary antibody, we used anti-Rb-Alexa 488, and actin was stained with rhodamine/phalloidin (1:100; Molecular Probes). Confocal images were collected with an Olympus FV1000 LSM microscope, and images were edited with Adobe Photoshop version 7.0CE and Olympus FW10-ASW version 1.7a.
dsRNA Treatment
Drosophila S2 cells (2 × 106) were plated into a Petri dish in 1 ml of serum-free medium (Sigma). 10 μg of dsRNA was added directly to the medium. The cells were incubated for 30 min at room temperature, followed by the addition of 2 ml of Schneider's medium (Sigma) containing fetal bovine serum (Sigma). The cells were incubated for an additional 3 days and then transfected with pAVW-FH1-FH2, and 15 μg of dsRNA was added also. These cells were incubated in Drosophila Schneider's medium (with fetal bovine serum) for 24 h before fixation. When following the same protocol, cells were treated with 15 μg of dsRNA each day; most cells died by the time of fixation.
Fly Strains and Genetics
For trachea-specific overexpression of the appropriately tagged FH2 and FH1-FH2 domains, we created w; pTMW-FH2 and w; pTVW-FH1-FH2 transgenic flies. Expression in the trachea was driven by btl-Gal4. As wild type controls, we used Oregon-R and w1118. The tracheal system was visualized in second instar larvae, and bright field images were collected on a Zeiss Axioskop MOT2 microscope with Axiocam HR.
Kinetic and Steady-state Measurements of Actin Assembly
Polymerization Assay
Monomeric calcium-actin was kept in buffer A after the purification. Before the experiments, the actin monomer solution was clarified by ultracentrifugation (328,000 × g, 4 °C, 30 min) in a Beckman ultracentrifuge. Then the bound calcium was replaced with magnesium by adding 200 μm EGTA and 50 μm MgCl2 and incubating the samples for 5–10 min. The polymerization of magnesium-actin was initiated by the addition of 1 mm MgCl2 and 50 mm KCl either in the presence or absence of formin fragments. The actin concentration was 3.5 μm in the measurements. The time course of actin polymerization (5% pyrenyl-labeled) was measured by monitoring the change in pyrenyl fluorescence (λex = 365 nm/λem = 407 nm) in the presence of various concentrations of DAAM-FH2 or DAAM-FH1-FH2. The elongation rate was determined from the slope of the linear fit to the pyrene fluorescence curves at half-maximum polymerization.
Depolymerization Assay
The depolymerization of actin filaments (5 μm, 70% pyrenyl-labeled) in the presence of various concentrations of DAAM-FH2 or DAAM-FH1-FH2 was followed after dilution to 0.1 μm in polymerization buffer (buffer A supplemented with 50 mm KCl and 1 mm MgCl2). The depolymerization rate was determined from the linear fit to the initial part of the time dependence of the pyrene fluorescence curves and normalized using the rate of actin alone as a standard.
Barbed End Growth Assay
The effect of DAAM-FH2 and FH1-FH2 on filament barbed end growth in the absence or presence of 2.6 μm profilin was monitored using 1.1 nm spectrin-actin seeds, 1 μm G-actin (2% pyrenyl-labeled), and variable amounts of DAAM fragments. The polymerization was initiated by adding 100 mm KCl, 1 mm MgCl2, and 0.2 mm EGTA to the solution of Mg-ATP-G-actin. The kinetics of actin assembly was measured by monitoring the change in either pyrene fluorescence (in the absence of profilin) or in light scattering (in the presence of profilin) (λex = 310 nm/λem = 310 nm). Initial barbed end elongation rates were derived from the linear fit of the polymerization curves and normalized with the rate of actin alone or with the rate of profilin-actin if profilin was present. To derive the value of the equilibrium dissociation constant (KD) of DAAM formins for the barbed ends, the initial rate of fluorescence/light scattering increase was analyzed as a function of DAAM concentration using Equation 1,
where V, V0, and Vmin are the rates measured in the presence of DAAM fragments at a concentration of [D], in the absence of DAAM fragments and in the presence of a saturating amount of DAAM fragments, respectively. It was assumed that the concentration of barbed end-bound DAAM was negligible as compared with the concentration of free DAAM.
Pointed End Growth Assay
The effect of DAAM-FH2 and FH1-FH2 on actin filament pointed end growth was monitored using 20 nm gelsolin-actin seeds (GA2), 1.25 μm G-actin (2% pyrenyl-labeled), and variable amounts of DAAM fragments. GA2 was prepared by mixing G-actin (1.3 μm) with gelsolin (0.5 μm) in buffer G (5 mm Tris-HCl, pH 7.8, 1 mm DTT, 0.1 mm CaCl2, 0.2 mm ATP, 0.01% (w/v) NaN3) supplemented with 2.5 mm CaCl2. The G-actin-bound calcium was replaced with magnesium by adding 200 μm EGTA and 20 μm MgCl2 and incubating the samples for 5–10 min. The polymerization was initiated by adding 100 mm KCl and 2 mm MgCl2 to the solution of Mg-ATP-G-actin. Initial barbed end elongation rates were derived from the linear fit of the polymerization curves and normalized with the rate of actin alone.
Determination of the Critical Concentration
Actin (5% pyrenyl-labeled) was incubated at various concentrations in polymerization buffer overnight. The pyrene fluorescence intensities were measured and plotted as a function of the total actin concentration. The value of the critical concentration was determined by fitting the Equation 2 to the plots,
 |
where I is the pyrene fluorescence intensity at various actin concentrations, [A] is the actin concentration, cc is the critical concentration for actin assembly, I0 is the ordinate value at [A] = cc, and SL and SR are the slopes of the intensity versus actin concentration curves before and after the breaking point, respectively.
The effect of profilin on the steady-state amount of F-actin was measured similarly following overnight incubation of samples of F-actin (1.97 μm, 2% pyrenyl-labeled) in the absence or presence of either 6.67 nm gelsolin or 0.7, 0.9, and 1.4 μm DAAM-FH2 or 0.7 μm DAAM-FH1-FH2 and increasing amounts of profilin. The sequestering activity of profilin and the value of the equilibrium dissociation constant for profilin binding to G-actin (Kd) were derived using Equation 3 (39),
where [PA] and [P]0 are the profilin-actin complex and the total profilin concentration, respectively, and cc is the critical concentration of actin assembly. The slope of the decrease in F-actin versus [P]0 is cc/(cc + Kd).
Stopped-flow Experiments
The polymerization of actin was followed with a stopped-flow instrument (SX.18MV-R Stopped Flow Reaction Analyzer, Applied Photophysics). Actin (7 μm, 5% pyrenyl-labeled) was mixed with buffer containing 200 μm EGTA and 50 μm MgCl2 (concentrations established after the mixing) to exchange the bound calcium to magnesium. Then the sample was mixed with polymerization buffer to establish the actin concentration of 3.5 μm and salt concentrations of 1 mm MgCl2 and 50 mm KCl. The experiments were done in the absence or presence of various concentrations of DAAM-FH1-FH2.
Cosedimentation Assays
To determine the affinity of DAAM-FH2 and FH1-FH2 for the actin filaments, we polymerized 1.5 μm actin at room temperature in the presence of different concentrations of DAAM-FH2 or FH1-FH2. After 2 h, the samples were centrifuged with a Beckman Optima MAX bench top ultracentrifuge (TLA-100 rotor, 20 °C, 30 min at 400,000 × g). The supernatants were separated from the pellets, and both were analyzed by 12% SDS-PAGE. After staining with Coomassie Blue, the band intensities were determined with a Syngene bioimaging system. The band intensities were corrected for the molecular weights of the proteins, and the ratios of the formin and actin band intensities measured in the pellets (D) were plotted as a function of the formin concentration and analyzed by using Equation 4 (40),
where [D]0 and [A]0 are the total formin and actin concentrations, respectively, KD is the dissociation equilibrium constant for formin binding to actin, and D is the fraction of bound formin.
In Vitro Microscopy of Actin Assembly
Epifluorescence Microscopy
Actin (1 μm) was polymerized in 4 mm Tris-HCl, pH 7.0, 0.1 mm CaCl2, 0.2 mm ATP, 0.5 mm DTT, 1 mm MgCl2, 50 mm KCl, and 1 mm EGTA for 2 h. Actin filaments were labeled with rhodamine-phalloidin in a 1:1 molar ratio in the absence or presence of 500 nm DAAM-FH2 or FH1-FH2 for 1 h and then diluted to 5 nm in a microscope buffer (4 mm Tris-HCl, pH 7.0, 1 mm EGTA, 50 mm KCl, 1 mm MgCl2, 0.2 m DTT, 15 mm glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 0.5% (w/v) methylcellulose). The samples were applied between a slide and coverslip and visualized with an Olympus IX81 inverted fluorescence microscope using a ×100 objective (numerical aperture 1.4) and a CCD camera (Orca ERG Hamamatsu). The images were analyzed using ImageJ (available on the National Institutes of Health Web site).
TIRFM
Glass flow cells (length ∼20 mm, width ∼8 mm, height ∼0.3 mm, volume ∼50 μl) were incubated with 1 volume of N-ethylmaleimide myosin (4.31 mg/ml) for 2 min, washed extensively with 4 volumes of 10% (w/v) BSA, and equilibrated with 4 volumes of TIRFM buffer (0.5% (w/v) methylcellulose (M-0512, Sigma), 10% (w/v) BSA, 1 mm 1,4-diazabicyclo-[2,2,2]octane (D2522, Sigma), 10 mm DTT in buffer F (5 mm Tris-HCl, pH 7.8, 1 mm DTT, 0.2 mm ATP, 0.1 mm CaCl2, 50 mm KCl, 1 mm MgCl2, 0.2 mm EGTA)).
A mixture of unlabeled and Alexa 488-labeled (10%) G-actin was mixed with DAAM formins and/or profilin in TIRFM buffer (for exact concentrations, see legends for Figs. 3B and 4B) and transferred to a flow cell sealed with Vaseline, lanolin, and paraffin at a 1:1:1 ratio for imaging. Images were captured every 20–60 s with an Olympus IX70 microscope equipped with a two-color TIRFM system using a ×60 oil objective (numerical aperture 1.45) and a CCD camera (Cascade II 512, Photonics). Time lapse images were analyzed with ImageJ. Filament growth was quantified by either measuring the length of each filament over several frames or by kymograph analysis. Filament length was converted to subunits using 330 subunits/μm.
FIGURE 3.
The effect of DAAM-FH2 and DAAM-FH1-FH2 on the polymerization and depolymerization of actin. A, the polymerization rate of actin (3.5 μm, 5% pyrenyl-labeled) at half-maximum polymerization as a function of DAAM concentration derived from pyrenyl or stopped-flow measurements as described under “Experimental Procedures.” Both DAAM-FH2 and DAAM-FH1-FH2 reach their maximal effect at 1 μm. Inset, the time course of actin polymerization (3.5 μm, 5% pyrenyl-labeled) monitored by the change in pyrene fluorescence in the absence or presence of various concentrations of DAAM-FH2, as indicated. The linear fit to each polymerization curve at 50% completion of the polymerization is shown by dashed lines in the corresponding color. B, time lapse evanescent wave fluorescence microscopy of the effect of DAAM formins on the polymerization of actin. Left panels, time lapse micrographs of actin assembly (1.2 μm, 10% Alexa 488-labeled) in the absence or presence of DAAM-FH1-FH2 and DAAM-FH2. Elapsed time (s) is shown. Scale bar, 10 μm. In the presence of DAAM fragments, a 2.67 × 2.67-μm area of the field (marked by a small green square in the first frame) with a single growing filament is enlarged 3-fold and shown at the bottom left corner of the subsequent images. Right panels, changes in filament length as a function of time. The elongation rate of individual filaments (v) was derived from the linear fit to the data. C, barbed end elongation from spectrin-actin seeds (SA; 1.1 nm) at 1 μm G-actin (2% pyrenyl-labeled) in the presence of DAAM-FH2 or DAAM-FH1-FH2, as indicated. Initial barbed end elongation rates were derived from the polymerization curves (shown in the inset) as described under “Experimental Procedures.” The dashed lines are calculated best fit binding curves (see “Experimental Procedures”), leading to half-saturation formin concentrations of 50 ± 6 and 11 ± 4 nm for the FH2 and FH1-FH2 fragments, respectively. Inset, kinetics of barbed end elongation of G-actin from spectrin-actin seeds in the absence or presence of DAAM-FH1-FH2. The yellow and orange curves show controls: time course of actin assembly (1 μm, 2% pyrenyl-labeled) in the absence of spectrin-actin seeds and in the absence or presence of DAAM-FH1-FH2, as indicated. Note that there is no detectable nucleation by DAAM-FH1-FH2 under these experimental conditions. D, pointed end elongation (20 nm gelsolin-actin seeds (GA2)) at 1.25 μm G-actin (2% pyrenyl-labeled) in the presence of DAAM-FH2 or DAAM-FH1-FH2. Initial elongation rates were derived from the polymerization curves (shown in the inset) as described under “Experimental Procedures.” Inset, kinetics of pointed end elongation of G-actin from GA2 in the absence and presence of DAAM-FH2 or DAAM-FH1-FH2. The gray line shows the time course of actin assembly (1.25 μm, 2% pyrenyl-labeled) in the absence of both GA2 and formins. E, dilution-induced depolymerization of F-actin (70% pyrenyl-labeled) in the presence of various concentrations of DAAM-FH2 or DAAM-FH1-FH2. Hyperbola fits to the plots (dashed lines) gave half-saturation formin concentrations of 13 ± 5 and 1 ± 2 nm for the FH2 and FH1-FH2 fragments, respectively.
FIGURE 4.
The interaction of actin, formin, and profilin. A, the polymerization rate of actin (3.5 μm, 5% pyrenyl-labeled) as a function of the formin concentration in the absence or presence of 5 μm profilin. Shown are the data for DAAM-FH2 and FH1-FH2 in the absence (as in Fig. 3A) or presence of profilin, as indicated. B, time lapse evanescent wave fluorescence microscopy of the effect of DAAM formins on the barbed end growth from profilin-actin. Left panels, time lapse micrographs of actin assembly (0.3 μm actin, 10% Alexa 488-labeled; green) from F-actin seeds (10% Alexa 568-labeled; red) in the presence of profilin (0.72 μm) and in the absence or presence of DAAM-FH1-FH2 and DAAM-FH2. The barbed end of typical filaments growing from a red seed or nucleated in solution are marked by red and green arrows in the subsequent images, respectively. Elapsed time (s) is shown. Scale bar, 10 μm. Right panels, kymographs of the length (y axis) of the marked filaments versus time (x axis). The elongation rate of individual filaments (v) was derived from kymograph analysis. C, barbed end elongation from spectrin-actin seeds (SA; 1.1 nm) at 1 μm G-actin in the presence of 2.6 μm profilin and DAAM-FH2 or DAAM-FH1-FH2 derived from the pyrenyl polymerization curves as described under “Experimental Procedures.” The dashed lines are calculated best fit binding curves (see “Experimental Procedures”). For comparison, the values obtained in the absence of profilin are shown in open symbols for DAAM-FH2 and DAAM-FH1-FH2 (see also Fig. 3C). Hyperbola fit to the data obtained in the presence of DAAM-FH2 gave a half-saturation formin concentration of 31 ± 5 nm. D, dependence of barbed end (BE) elongation rate from spectrin-actin seeds (1.1 nm) on profilin-actin (PA) concentration, with either free barbed ends, DAAM-FH2-bound barbed ends (0.53 μm), or DAAM-FH1-FH2-bound barbed ends (0.52 μm), as indicated Profilin concentration was 18.7 μm. The reference plot obtained with free G-actin and free barbed ends is shown in open circles. Values of the association rate constants (k+) derived from the slopes (dashed lines) are shown in Table 1. E, the effect of profilin on the depolymerization rate of actin as a function of the formin concentration. The data are presented for DAAM-FH2 and for DAAM-FH1-FH2 in the absence (as in Fig. 3E) or presence of profilin (5 μm), as indicated.
Two-color TIRFM experiments were carried out as follows. G-actin (0.3 μm, 10% Alexa 568-labeled) was polymerized in the flow cell for 10–15 min to form “red” actin seeds, and then unpolymerized actin was washed out by 4 volumes of TIRFM buffer. A mixture of G-actin (0.3 μm, 10% Alexa 488-labeled) and/or DAAM formins and profilin in TIRFM buffer was injected into the flow cell sealed with Vaseline, lanolin, and paraffin at a 1:1:1 ratio. Images were acquired using a dual view system (MAG Biosystems).
Motility Assay with Formin-coated Beads
Motility assays using formin-coated beads were performed as described (10). Carboxylated polystyrene microspheres (2-μm diameter, Polysciences Inc., 2.5% solid) were functionalized by incubating with 2.1 μm mDia1-FH1-FH2, 76.6 μm DAAM-FH2, or 141.5 μm DAAM-FH1-FH2 in Xb buffer (10 mm HEPES, pH 7.8, 100 mm KCl, 1 mm MgCl2, 0.1 mm CaCl2, 1 mm ATP) for 1 h on ice. The reaction was stopped by the addition of BSA (10% (w/v) for 15 min), and then the beads were washed by centrifugation (Beckman Microfuge 22R, 21,920 × g, 4 °C, 5 min) and stored on ice in Xb buffer supplemented with 0.1% (w/v) BSA. To determine the amount of protein bound to the beads after functionalization, formin-coated beads were washed five times in Xb buffer and processed for 12.5% SDS-PAGE. The amount of protein bound to the beads was visualized by Coomassie staining.
The motility medium was prepared by mixing 7 μm F-actin (5% rhodamine-labeled), ADF, profilin (for exact concentration, see legend for Fig. 6), 0.5% (w/v) BSA, 0.18% (w/v) methylcellulose (M-0512, Sigma), 6.6 mm DTT, 0.15 mm 1,4-diazabicyclo-[2,2,2]octane (D2522, Sigma), 2 mm ATP, and 4 mm MgCl2 in buffer F. The samples were incubated for ∼5 min before the addition of the beads (final concentration 0.01%). Samples of 4 μl were placed between a slide and coverslip sealed with Vaseline, lanolin, and paraffin at a 1:1:1 ratio. The beads were observed in a fluorescence microscope (AX70, Olympus) using a ×20 objective (numerical aperture 0.5) and a CCD camera (Orca II ERG Hamamatsu). The average rate of movement was determined by recording time lapse series of freely moving beads. The template recognition-based tracking option of Methamorph version 6.0 (Universal Imaging Corp.) was used to measure mean velocities, calculated for 15–23 beads selected from 3–4 different fields. Kymographs were generated using ImageJ.
FIGURE 6.
DAAM-FH1-FH2-functionalized beads move in the reconstituted biomimetic motility assay. A, left panels, time lapse recording of the propulsive movement of typical beads coated with the FH1-FH2 domain of mDia1 or of DAAM. Right panels, the trajectory of beads. The green, white, and red arrows indicate the initial, intermediate, and final positions of beads, respectively. Conditions were as follows: 7 μm F-actin (5% rhodamine-labeled), 16 μm profilin, 15 μm ADF. Scale bar, 20 μm. Elapsed time (s) is shown. B, kymographs generated using the trajectory of beads (y axis, length; x axis, time). Conditions were as in A. C, time courses of actin polymerization (2 μm, 2% pyrenyl-labeled) monitored by the change in pyrenyl fluorescence in the absence (black line) and in the presence of different formin fragments, as indicated. D, in the absence of profilin, DAAM-FH1-FH2-coated beads do not initiate actin comets in the motility assay. Conditions were as follows: 7 μm F-actin (5% rhodamine-labeled), 15 μm ADF. Scale bar, 10 μm. E, the amount of formin FH1-FH2 bound to the beads after functionalization visualized by Coomassie staining of SDS-polyacrylamide gels.
RESULTS
To characterize the D. melanogaster DAAM formin, we analyzed the interactions of its FH2 and FH1-FH2 fragments with actin and profilin by using various biophysical methods. First we carried out fluorescence microscopy experiments to characterize the role of DAAM in living cells.
In Vivo Effects of DAAM Formin Fragments
To examine the in vivo activity of the DAAM fragments, we used cultured Drosophila S2 cells and the Drosophila tracheal system, where DAAM is known to be required for cuticle patterning (25). Untransfected Drosophila S2 cells do not express the endogenous DAAM protein at a detectable level.6 When YFP-tagged DAAM-FH2 or DAAM-FH1-FH2 was expressed in S2 cells, both isoforms could be detected in the cytoplasm of the cells (Fig. 1). Although DAAM-FH2 had no significant effect on the organization of the actin filaments in S2 cells (Fig. 1, A–C), the DAAM-FH1-FH2-expressing cells often exhibited an increased filament level (Fig. 1, D–F). In addition, cells expressing DAAM-FH2 did not exhibit morphological changes, whereas about 25% of the cells expressing DAAM-FH1-FH2 were flattened and displayed lamellipodial and/or filopodial protrusions (Fig. 1G). These observations indicate that the presence of the FH1 domain dramatically modifies the in vivo activity of the actin-binding DAAM-FH2 domain. The FH2 domain alone does not appear to affect actin dynamics in S2 cells, but the FH1 and FH2 domains together behave as a typical activated formin with a profound effect on actin-based motile processes and cell shape.
FIGURE 1.
The expression of dDAAM FH2 or FH1-FH2 in Drosophila S2 cells. A–C, S2 cells transfected with YFP::FH2. Note that cells expressing YFP::FH2 (white in A, green in C) are round in shape, such as the non-expressing cells (arrows in B and C). Moreover, both cell types exhibit a similar pattern and level of actin staining (white in B, red in C). D–F, S2 cells transfected with YFP-tagged FH1-FH2 (YFP::FH1-FH2). Note that cells expressing YFP-tagged FH1-FH2 (white in D, green in F) are larger than non-expressing cells (arrows in E and F) and often exhibit a large number of filopodia-like protrusions. Additionally, the F-actin level is highly increased in YFP-tagged FH1-FH2-transfected cells (white in E, red in F). The nucleus is stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue in C and F). G, statistical analyses of the cell shape changes exhibited by S2 cells expressing YFP-tagged FH2 (YFP::FH2), YFP-tagged FH1-FH2, or YFP alone. H, statistical analyses of the cell shape changes exhibited by S2 cells expressing YFP-tagged FH1-FH2 in the absence or presence of profilin (chic) specific dsRNA. Scale bars, 5 μm.
Previously, we have shown that C-DAAM, another activated version of DAAM including the FH1 and FH2 domains and the entire C-terminal half of the protein, impairs actin organization and cuticle structure when expressed in the Drosophila larval tracheal system (25). The comparison of the effect of DAAM-FH2 and DAAM-FH1-FH2 in this system has led to the conclusion that whereas DAAM-FH2 does not affect tracheal development (Fig. 2B), the overexpression of DAAM-FH1-FH2 resulted in phenotypic effects very similar to those of C-DAAM with severe impairment of cuticular fold formation (Fig. 2C), often leading to the collapse of the tracheal tube. Moreover, the trachea-specific expression of DAAM-FH2 does not affect viability; however, the presence of DAAM-FH1-FH2 induces lethality in the larval stages. Thus, the in vivo overexpression tests together strongly suggest that both the FH1 and FH2 domain of DAAM are essential for its biological activity. Although several molecular mechanisms can account for the different behaviors of FH2 and FH1-FH2, one of the most likely possibilities is that the actin monomers are in complex with profilin in these cells, and the effective utilization of the profilin-actin pool requires the presence and active contribution of the FH1 domain of DAAM. To test this hypothesis, we examined if the DAAM-FH1-FH2-induced cell shape changes in S2 cells are profilin-dependent. The almost complete depletion of profilin levels by dsRNA resulted in cell lethality; however, an incomplete removal of profilin did not significantly affect cell viability (supplemental Fig. S1). The overexpression of DAAM-FH1-FH2 in cells with decreased profilin level was still able to induce cell shape changes; however, the efficiency dropped by 27% as compared with control cells (Fig. 1H). These results support the view that DAAM-FH1-FH2 acts in a profilin-dependent manner, which is consistent with our previous findings that dDAAM and chic (profilin) mutants exhibit a dominant genetic interaction in the central nervous system and that the proteins can be co-immunoprecipitated from S2 cells expressing activated dDAAM (28).
FIGURE 2.
The expression of dDAAM FH2 or FH1-FH2 in the Drosophila tracheal system. A, the cuticle structure of a wild type (wt) Drosophila tracheal tube from a second instar larvae. The cuticle of the main airway is characterized by taenidial folds, running perpendicular to the tube axis. B, the tracheal cuticle of an FH2-expressing larvae is essentially identical to that of the wild type shown in A. C, tracheal tubes in which FH1-FH2 is expressed exhibit a strongly impaired cuticle pattern, often leading to the flattening and collapse of the tubes. Scale bar, 50 μm.
DAAM Nucleates Actin Assembly but Markedly Slows Barbed End Elongation in Vitro
To understand the molecular bases of our in vivo observations, Drosophila DAAM-FH2 and DAAM-FH1-FH2 fragments were assessed for their ability to assemble actin filaments in vitro by polymerization assays using pyrenyl-labeled actin (see “Experimental Procedures”). Both DAAM-FH2 and DAAM-FH1-FH2 accelerated the polymerization of actin in bulk solutions (Fig. 3A, inset), indicating that, as in vivo for the whole protein, the purified DAAM fragments interact with actin (25). We did not observe differences between the effect of FH2 and FH1-FH2 on the polymerization rate (Fig. 3A), indicating that in the absence of profilin, the FH1 does not play a major role in the interaction between actin and formin. The formin concentration dependence of the polymerization rate showed a saturating behavior, reaching its maximum at ∼1 μm formin. To test whether the limitations of the steady-state fluorescence spectroscopic method are responsible for the apparent saturation of the polymerization rate, we carried out rapid kinetic experiments using a stopped-flow apparatus. In these experiments, the dead time of the measurements is substantially shorter (∼1 ms) than in the case of manual mixing (∼20–40 s), and the time resolution of the instrument is higher. The formin concentration dependence of the polymerization rate monitored using the stopped-flow apparatus was similar to the data obtained using the steady-state fluorescence method (Fig. 3A). This control experiment corroborated our conclusions that the rate of actin polymerization is limited at greater formin concentrations.
Formins are generally known to affect both the nucleation and elongation of actin filaments (4). From bulk solution spontaneous assembly assays (Fig. 3A), it is not possible to determine how each of these polymerization steps was affected by the DAAM fragments. To overcome this limitation, we studied the effect of DAAM-FH2 and FH1-FH2 on actin polymerization using TIRFM, which allows real-time visualization of the assembly of individual actin filaments. The TIRFM assay revealed numerous short filaments in the presence of DAAM-FH2 and FH1-FH2 compared with control samples, which were initiated immediately after the addition of formin to the actin solution and therefore nucleated by the DAAM fragments (supplemental Movie 1 and Fig. 3B, left panels). The DAAM-FH2 and FH1-FH2 nucleated actin filaments elongated at a much slower rate (0.59 ± 0.22 subunit/s (n = 10) and 0.99 ± 0.32 subunit/s (n = 10), respectively) than spontaneously nucleated actin filaments (12.68 ± 0.96 subunits/s (n = 10)) (Fig. 3B, right panels). These observations show that the acceleration of the polymerization observed in our bulk solution pyrene assays was due to the nucleation activity of the DAAM fragments.
To investigate whether the slow growth of DAAM-nucleated actin filaments is solely due to pointed end elongation or to a residual assembly at barbed ends, the effect of DAAM-FH2 and FH1-FH2 on barbed or pointed end elongation selectively was monitored using spectrin-actin or gelsolin-actin seeds, respectively (Fig. 3, C and D). To keep the number of nuclei constant and minimize the contribution of the nucleating activity of DAAM formins, we used a low concentration of actin monomers (1 or 1.25 μm, respectively) and relatively high (compared with the conditions of spontaneous actin polymerization, where the concentration of filament ends typically falls below 1 nm) concentration of spectrin-actin or gelsolin-actin seeds (1.1 or 20 nm, respectively). Control experiments showed that under these experimental conditions, DAAM formins do not have detectable nucleating activity (Fig. 3C, inset). Both DAAM-FH2 and FH1-FH2 strongly inhibited but did not block completely filament barbed end elongation from actin monomers (Fig. 3C). The same level of inhibition (90%) was reached at saturation by either FH2 or FH1-FH2. Half-inhibition of barbed end growth was recorded at 50 ± 6 nm DAAM-FH2 or 11 ± 4 nm DAAM-FH1-FH2. Neither DAAM-FH2 nor FH1-FH2 affected pointed end growth from gelsolin-actin seeds (Fig. 3D). These results show that DAAM-nucleated actin filaments grow at both their barbed and pointed ends.
We have also studied the effects of DAAM-FH2 and FH1-FH2 fragments on the depolymerization rate of actin filaments. Pyrenyl-labeled actin (5 μm, 70% labeled) was polymerized overnight, and then filaments were diluted to 100 nm (below the barbed end critical concentration) in the absence or presence of formins. Both DAAM-FH2 and DAAM-FH1-FH2 fragments decreased the rate of depolymerization at submicromolar concentrations, consistent with their binding to barbed ends (Fig. 3E). At saturating formin concentrations, the rate of depolymerization decreased to less than 5% of the value observed in the absence of formins. The half-effect was detected at 13 ± 5 nm DAAM-FH2 or 1 ± 2 nm DAAM-FH1-FH2 (Fig. 3E). In conclusion, both DAAM-FH2 and FH1-FH2 enhance the nucleation of actin filaments but substantially slow barbed end dynamics from G-actin with high affinities (Kd of 10–50 nm).
Profilin Acts as a Molecular Switch in DAAM Function by Stimulating Actin Assembly Driven by FH1-FH2 but Not by FH2
It is believed that one of the key functions of formin is to make use of the intracellular profilin-actin complexes in the processive polymerization of actin filaments. To fulfill this function, the FH1 domain of formins participates in a mechanism in which the profilin-actin complex is binding to the actin filament-bound formin (4). This consensus was supported by our in vivo experiments (Fig. 1) showing the importance of both the FH1 domain and profilin for the biological function of DAAM. The role of profilin in DAAM function was further addressed by in vitro experiments. In our assay, profilin (5 μm) inhibited actin assembly (3.5 μm) in the presence of DAAM-FH2 (Fig. 4A). In contrast, in the presence of profilin, the DAAM-FH1-FH2-stimulated actin polymerization was faster than in its absence (Fig. 4A).
To characterize the effect of profilin on the DAAM-FH2- and FH1-FH2-mediated actin assembly in more detail, we used TIRFM. To selectively monitor barbed end growth, the elongation of preassembled F-actin seeds (10% Alexa 568-labeled) was monitored in the presence of a low amount of G-actin (0.3 μm, below the pointed end critical concentration, 10% Alexa 488-labeled) in a dual wavelength fluorescence assay (see “Experimental Procedures”). In the presence of profilin (0.72 μm), barbed end growth of Mg-ATP-G-actin was slowed down by ∼27%, from 3.98 ± 0.61 subunits/s (n = 32) (data are not shown) to 2.91 ± 0.50 subunits/s (n = 69) (Fig. 4B), in good agreement with previous observations (39, 41). Under these experimental conditions, barbed end growth was severely slowed down by DAAM-FH2 (0.2 ± 0.05 subunit/s (n = 31)). In striking contrast, profilin has a dramatic influence on DAAM-FH1-FH2, allowing barbed end assembly at a rate similar to the value measured in the absence of formins (2.35 ± 0.28 subunits/s (n = 62)) (supplemental Movie 2 and Fig. 4B).
To support these observations, we also measured the effect of DAAM-FH2 and FH1-FH2 on barbed end growth from profilin-actin using spectrin-actin seeds in a bulk solution assay. The affinity of profilin for unmodified actin is higher than for actin monomers modified with pyrene at Cys374 (42); thus, the fluorescence of pyrenyl-actin does not accurately reflect the total amount of actin polymer formed from profilin-actin (10, 43). Therefore, the kinetics of polymerization was followed by measuring the change in light scattering. Control experiments showed that under these experimental conditions, DAAM formins do not have detectable nucleating activity (data not shown). In agreement with the results from TIRFM, DAAM-FH2 inhibited barbed end elongation from profilin-actin with a half-effect measured at 31 ± 5 nm DAAM-FH2 (Fig. 4C). In contrast, profilin relieved the inhibiting activity of DAAM-FH1-FH2 and allowed full-speed barbed end growth (Fig. 4C). The compared kinetic parameters for barbed end growth from profilin-actin at free, FH2-bound, and FH1-FH2-bound barbed ends were derived from the J(c) plot shown in Fig. 4D. Data (summarized in Table 1) show that profilin-actin associates with free and with FH1-FH2-bound barbed ends at a comparable rate. These observations further indicate that the FH1 domain is essential for the interaction of DAAM formin with profilin, and the FH1-FH2 fragment is effective in assisting the polymerization of actin filaments from profilin-actin monomers, in agreement with previous results (10, 32, 44).
TABLE 1.
Actin and profilin-actin assembly rates in the presence of DAAM formins
The data were derived from TIRFM (Fig. 3B) and bulk solution (Fig. 4D) measurements. For comparison, the values of the corresponding parameters obtained with mDia1 are shown in parentheses (10).
In the absence of formin fragments, dilution-induced depolymerization of actin filaments appeared faster in the presence of 5 μm profilin than in its absence (data not shown), in agreement with previous results (45, 46). In the presence of both profilin and either DAAM-FH2 or DAAM-FH1-FH2, the depolymerization rate was faster than in the absence of profilin and decreased with increasing formin concentrations (previously similar observations were made for Cdc12-FH1-FH2 (10, 47)) (Fig. 4E). These observations suggest that profilin-actin dissociates from the barbed end of the filaments faster than actin alone, and the formin fragments, by binding to barbed ends, slow the dissociation of the profilin-actin complexes.
DAAM-FH2 Inhibits Whereas DAAM-FH1-FH2 Allows Steady-state Monomer-Polymer Exchange at Barbed Ends from Profilin-Actin
Based on the strong effects of the DAAM fragments on the elongation and depolymerization rates, one would expect that by blocking the dynamics of the monomer exchange at the barbed end, the DAAM formin fragments can shift the critical concentration to values closer to the critical concentration of pointed ends. Previously, the FH2 fragment from mDia3 or the FH1-FH2 fragment of Cdc12p was reported to increase the critical concentration to values of the pointed end critical concentration (29, 48), whereas neither the FH2 nor the FH1-FH2 fragment of mDia1 altered the value of this parameter (10).
The critical concentration of actin assembly was determined using the pyrenyl-actin fluorescence assay (see “Experimental Procedures”). In the absence of formin fragments, the critical concentration was 0.17 ± 0.04 μm (data are not shown), similar to the value obtained previously for the critical concentration of magnesium-bound actin (e.g. see Ref. 49). In the presence of 100 nm DAAM-FH2 or DAAM-FH1-FH2, critical concentrations of 0.21 ± 0.09 μm and 0.25 ± 0.07 μm were obtained, respectively (Fig. 5A). These values indicate that the DAAM fragments have little effect on the critical concentration of actin assembly. To obtain further details regarding the formin effect, we carried out experiments at fixed pyrenyl-actin concentration (1 μm) by changing the concentration of the formin fragments. In these experiments, the pyrene intensity measured in the absence of formin corresponded to the difference between the total actin concentration (1 μm) and the critical concentration (∼0.2 μm) (i.e. to the mass amount of F-actin of ∼0.8 μm). These results also showed that there is an approximately 0.1 μm decrease in the concentration of assembled F-actin at greater formin concentrations (Fig. 5A, inset), which indicates a slight increase in the critical concentration. The decrease in F-actin concentration occurred between 20 and 40 nm formin concentration in accordance with our data (Fig. 3), showing that the affinity of DAAM-FH2 and DAAM-FH1-FH2 for the barbed ends is high.
FIGURE 5.
The effect of DAAM-FH2 and DAAM-FH1-FH2 on the steady-state actin assembly dynamics. A, critical concentration plots of F-actin (5% pyrenyl-labeled) polymerized in the presence of 100 nm DAAM-FH2 or DAAM-FH1-FH2, as indicated. Fitting Equation 2 to the curves gave values of critical concentration of 0.21 ± 0.09 and 0.25 ± 0.073 μm in the presence of DAAM-FH2 or DAAM-FH1-FH2, respectively. Inset, actin (1 μm, 5% pyrenyl-labeled) was polymerized in the presence of various DAAM-FH2 or FH1-FH2 concentrations, as indicated. The concentrations of polymerized actin were derived from pyrenyl fluorescence measurements (see “Experimental Procedures”) and plotted as a function of DAAM concentration. B, the pyrene fluorescence intensity of samples containing different concentrations of actin (5% pyrenyl-labeled) polymerized in the presence of 100 nm DAAM-FH2 or DAAM-FH1-FH2 and profilin (5 μm), as indicated. The critical concentration was found to be 0.42 ± 0.08 μm and 0.22 ± 0.03 μm in the presence of DAAM-FH2 or DAAM-FH1-FH2, respectively. C, the amount of F-actin assembled at steady state (1.97 μm total actin, 2% pyrenyl-labeled) was measured in the absence and presence of either gelsolin, DAAM-FH1-FH2, or DAAM-FH2 and increasing amounts of profilin, as indicated. The data show that whereas the binding of DAAM-FH1-FH2 to barbed ends allows profilin-actin to maintain barbed end dynamics, DAAM-FH2 causes depolymerization of actin by profilin. The capping of barbed ends by gelsolin results in sequestration of actin by profilin. The linear decrease in F-actin upon the addition of profilin in the presence of gelsolin is consistent with the value of 0.56 μm for the critical concentration at pointed ends and a value of 0.36 μm for the equilibrium dissociation constant of profilin-actin complex. Dashed lines in the corresponding colors show the linear fit to the data. a.u., arbitrary units.
The above thermodynamic measurements of the effect of DAAM formin on the assembly of free G-actin confirm the conclusions from the kinetic assays, indicating that although the monomer-polymer exchanges are greatly slowed down by the binding of DAAM-FH2 or FH1-FH2 to barbed ends, the filaments are not strongly capped by formin, and the measured steady-state concentration of G-actin is lower than the critical concentration at pointed ends (0.6 μm).
To investigate the effect of profilin on the steady-state actin assembly dynamics mediated by DAAM formins, the effect of profilin on the steady-state amount of F-actin was measured in the presence of DAAM-FH2 or FH1-FH2. In the presence of 5 μm profilin, the critical concentration for polymerization with DAAM-FH1-FH2 (100 nm) is close to the value measured in the absence of profilin (Fig. 5, B and C), indicating that FH1-FH2-bound barbed ends support assembly from profilin-actin, as free barbed ends do. In contrast, with DAAM-FH2 (100 nm), the addition of 5 μm profilin caused partial depolymerization of F-actin, indicating that FH2-bound barbed ends do not support assembly from profilin-actin (Fig. 5B). To address this point in deeper detail, the amount of F-actin at steady state was measured at a fixed concentration of actin, as a function of profilin concentration in the absence and presence of DAAM fragments or gelsolin (Fig. 5C). The following observations were made. 1) Control assays show that profilin maintains monomer-polymer level as well as actin when barbed ends are free, in agreement with previous reports (50). At high profilin concentration, the routinely observed decrease in F-actin is attributed to the weak barbed end capping activity of profilin (41). 2) In contrast, profilin sequesters G-actin, thus causing total depolymerization, when barbed ends are strongly capped by gelsolin (50). The slope of the linear decrease in F-actin was consistent with the value of the critical concentration of pointed ends (0.56 μm) and a value of 0.3 μm for the equilibrium dissociation constant of the profilin-actin complex (see Equation 3). 3) When barbed ends were saturated by FH1-FH2 (0.7 μm), a new steady state was established, corresponding to an increase in unassembled actin from 0.29 μm in the absence of profilin to 0.51 μm in the presence of a low amount of profilin. With increasing profilin concentration, the level of F-actin assembly in the presence of DAAM-FH1-FH2 formin was identical to the level obtained in the absence of formin. Notably, in the range of high concentrations of profilin (>10 μm), DAAM-FH1-FH2 established a higher F-actin level than that resulting from the weak capping activity of profilin on free barbed ends. We tentatively attribute this change to the fact that FH1-FH2 prevents the capping of barbed ends by profilin. 4) In contrast, when barbed ends were saturated by DAAM-FH2 (0.7–1.4 μm), profilin no longer stabilized filaments but caused depolymerization. The slope of the linear decrease in F-actin was consistent with a critical concentration of 0.012 ± 0.001 μm actin for FH2-bound barbed ends, which is appreciably lower than the value of 0.4 μm found in the absence of profilin.
In conclusion, both thermodynamic data and kinetic data support the view that neither DAAM-FH2 nor FH1-FH2 strongly caps barbed ends, but both allow free G-actin to maintain monomer-polymer exchange at barbed ends. In contrast, profilin-actin maintains barbed end dynamics when DAAM-FH1-FH2 but not FH2 is bound to filament ends.
DAAM-FH1-FH2-functionalized Beads Propel in the Biomimetic Motility Assay
One of the important characteristics of most formins is to associate persistently with the barbed ends of the actin filaments and simultaneously permit subunit addition, processively moving with the growing filament end (9, 10, 51). To find out whether this function can be attributed to the DAAM formin, we tested the FH2 and FH1-FH2 fragments of DAAM in the reconstituted biomimetic motility assay (10). In this assay, beads functionalized with formins are placed in a medium consisting of actin filaments at steady state with profilin and ADF. This chemostat maintains a stationary amount of profilin-actin via the regulated treadmilling of actin filaments to supply barbed end growth (52). In this assay, beads coated with the FH1-FH2 domain of mDia1 initiate processive filament assembly from profilin-actin at the bead surface and propel in the motility medium (10) (Fig. 6A, top).
DAAM-FH1-FH2-functionalized beads nucleated actin filaments and moved steadily at 5.54 ± 0.97 μm/min (n = 23) in the motility assay, in the presence of 7 μm F-actin, 16 μm profilin, and 15 μm ADF (see supplemental Movie 3 and Fig. 6A, bottom). The propulsion of DAAM-FH1-FH2-coated beads was ∼4-fold slower, and the actin tails formed by the beads were less dense than those of mDia1-FH1-FH2-coated beads (see supplemental Movie 3 and Fig. 6, A and B). In accordance with these observations, control pyrene polymerization assays also showed a difference between the activities of these formins, in agreement with previous studies (53) (Fig. 6C).
In the absence of profilin, filaments nucleated by bead-bound DAAM-FH1-FH2 were released in solution, and the beads remained bare. In this case, no bead movement was observed (Fig. 6D). DAAM-FH2-coated beads also remained bare and did not initiate the formation of actin tails, in the absence as well as in the presence of profilin (data not shown). SDS-PAGE analysis confirmed that the FH2 and the FH1-FH2 fragments of DAAM were bound to the beads (Fig. 6E). These observations demonstrate that immobilized DAAM-FH1-FH2, but not the DAAM-FH2, nucleates and processively elongates actin filaments from profilin-actin. Both the FH1 domain and profilin are required for sustained bead propulsion mediated by DAAM, as previously observed with mDia1 (10). These findings are in good agreement with our results from bulk solution measurements in showing that profilin acts as a switch in DAAM-FH1-FH2 function that allows DAAM-FH1-FH2 to effectively assemble filaments.
DAAM-FH2 and DAAM-FH1-FH2 Bind to the Sides of the Actin Filaments
To further investigate the interactions of DAAM fragments with actin, we applied co-sedimentation assays to study the binding of these formins to actin filaments (Fig. 7). Actin filaments (1.5 μm) were mixed with DAAM-FH2 or DAAM-FH1-FH2 fragments at different concentrations, and the samples were centrifuged at 400,000 × g for 30 min. The pellets and supernatants were subjected to SDS-PAGE analyses. The results showed that formin fragments sedimented with the actin filaments. In control samples, neither DAAM-FH2 nor DAAM-FH1-FH2 appeared in the pellets in the absence of actin (Fig. 7, left). Because the concentration of the DAAM fragments in the pellets was much higher (100–900 nm) than would be expected from their sole binding to the filament ends (8–15 nm), these observations showed that formin fragments bound to the sides of the actin filaments. The ratio of the formin and actin band intensities measured in the pellets were determined and plotted as a function of the formin concentration, and the plots were analyzed by hyperbola fits using Equation 4 (Fig. 7, right). The analyses gave equilibrium dissociation constants of 7.0 ± 2.5 and 2.1 ± 0.7 μm for DAAM-FH2 and DAAM-FH1-FH2, respectively, indicating that the DAAM-FH1-FH2 fragment bound slightly more tightly than the FH2 fragment to the sides of the actin filaments. Affinities of 2–7 μm are similar to those previously obtained for other formin fragments of mDia1, mDia3, and Bni1 formins (6, 29, 54).
FIGURE 7.
DAAM-FH2 and DAAM-FH1-FH2 bind to the sides of actin filaments. Left, gels of pellets (P) and supernatants (SN) obtained with either 3 μm DAAM-FH2 or FH2 in the absence of actin. Right, the fraction of DAAM formins bound to F-actin as a function of formin concentration, as indicated. Equation 1 was fitted to the data and gave equilibrium dissociation constants for binding of DAAM fragments to the sides of actin filaments of 7.0 ± 2.5 and 2.1 ± 0.7 μm for the DAAM-FH2 and DAAM-FH1-FH2 fragments, respectively.
DAAM-FH2 and DAAM-FH1-FH2 Cross-link Actin Filaments
Several formins were reported to potentially serve as cross-linkers, with variable efficiencies among the formin families (55–59). We have shown here that DAAM-FH2 and DAAM-FH1-FH2 bind to the sides of actin filaments (Fig. 7), providing one of the criteria for the cross-linking function. To test if these DAAM fragments can cross-link the actin filaments, we carried out fluorescence microscopy experiments. Rhodamine-phalloidin-labeled actin filaments (5 nm) were visualized in the absence and presence of DAAM fragments (Fig. 8A). In the presence of DAAM fragments, the filaments were shorter than in control samples (Fig. 8A). Similar observations were made with fragments from other formins (47, 60).
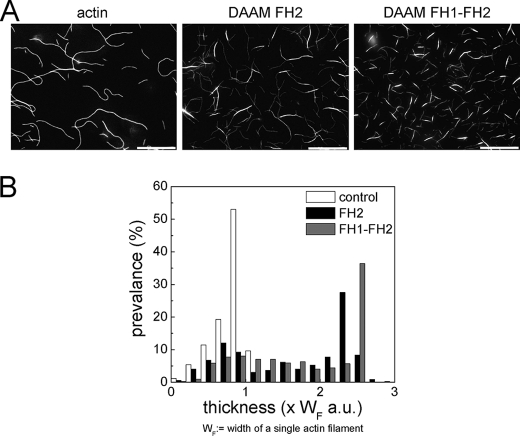
FIGURE 8.
DAAM-FH2 and DAAM-FH1-FH2 cross-link actin filaments. A, gallery of typical images of actin filaments (1 μm) polymerized in the absence or presence of either 500 nm DAAM-FH2 or FH1-FH2 (from left to right) visualized by rhodamine phalloidin fluorescence. B, diagram showing the distribution of the thickness of the filament structures formed in the absence or presence of DAAM-FH2 and FH1-FH2, as indicated. a.u., arbitrary units.
In the presence of DAAM fragments, the filaments also appeared thicker, and supramolecular actin structures were formed. The formin-induced changes were quantified by measuring the thickness of the actin filaments and/or actin structures (Fig. 8B). The width of the single actin filaments (WF) was used as a reference. In the absence of formins, a single class of filament width was observed (at ∼1 × WF), which corresponded to the size of the single actin filaments. In the presence of DAAM-FH2 or DAAM-FH1-FH2, two classes appeared in the width distribution. The first, lower peak distributed similarly to the one observed in the absence of formins and corresponded to the width of single filaments (at ∼0.8 × WF). The second peak shifted toward greater thickness, indicating actin structures that were ∼2–3 times thicker than the single actin filaments. These observations show that both DAAM-FH2 and FH1-FH2 induce actin filament bundling by cross-linking the filaments.
DISCUSSION
In this work, we investigated the properties of D. melanogaster DAAM-FH2 and DAAM-FH1-FH2 formin fragments and their functional interactions with actin and profilin. We found that overexpression of the DAAM-FH2 fragment in Drosophila S2 cells did not induce specific actin-based cellular activities (Figs. 1 and 2). In contrast, the overexpression of DAAM-FH1-FH2 resulted in the flattening of the cells and the formation of actin-rich structures, such as filopodial protrusions, that are one of the functional signatures of constitutively active formin (Fig. 1) (61–63). Excess DAAM-FH1-FH2 also induced structural defects in the tracheal cuticle that led to collapse of the tracheal tube (Fig. 2). This strongly impaired phenotype of the respiratory system probably accounts for the observed lethality in the larval stages subsequent to trachea-specific overexpression of DAAM-FH1-FH2. The cell shape changes resulting from massive actin assembly induced by overexpression of DAAM-FH1-FH2 were less pronounced in cells that contained a lowered amount of profilin than in cells with wild type profilin level (Fig. 1H). These results suggest that, like in the case of other formins, profilin binding to the FH1 domain of DAAM formin appears to be required for the in vivo function of DAAM. The in vitro characterization of FH2 and FH1-FH2 of DAAM brings quantitative support to their respective interactions with actin and profilin and their role in processive actin assembly.
In the absence of profilin, the results from bulk solution spontaneous assembly and single filament TIRFM assays consistently show that both FH2 and FH1-FH2 of DAAM catalyze actin nucleation. However, both fragments also severely slow barbed end growth and depolymerization of filaments at concentrations in the range of 0–20 nm, indicating tight equilibrium binding to barbed ends (Fig. 3, C and E).
The decreased rate of depolymerization is consistent with the pseudofilament structure of FH2-actin, in which each FH2 arm of one formin dimer links two adjacent actin subunits, preventing their dissociation (17). The almost complete inhibition of actin depolymerization can be explained by the following alternative molecular mechanism. It is possible that the fragments change the conformation of the actin filaments in a way that does not favor the dissociation of the actin monomers. Formin-induced conformational changes were reported previously (64–67). This explanation does not require that the fragments spend most of the time associated with the filament ends, but the lifetime of the formin-induced actin conformations must be long in the time scale of depolymerization of the filaments (i.e. the filaments must remember the binding of formins). Further experiments are required to determine the importance of this mechanism.
DAAM-FH2 and FH1-FH2 nucleate actin assembly with identical efficiencies and dose dependences. At high DAAM concentrations (above ∼1 μm), the overall polymerization rate reached a plateau (Fig. 3A). This may indicate that there is a step in the polymerization that reaches its maximal rate at ∼1 μm formin or that at higher formin concentrations, the formation of prenucleus actin dimers becomes rate-limiting in formin-induced nucleation. Similar observations were made with other actin nucleation factors (7, 68).
Biomimetic motility assays showed (Fig. 6) that the DAAM fragments do not remain permanently bound to the filament ends in the absence of profilin. To explain the large but not total inhibition of both the elongation and depolymerization of actin filaments at saturation by formins, one can assume that monomer-polymer exchange reactions can occur slowly on a formin-bound end. This view is corroborated by the fact that the critical concentration for actin assembly was only slightly influenced by the presence of the DAAM fragments (Fig. 5A). The complete and permanent blocking of the barbed end, in contrast, would shift the apparent critical concentration of actin toward the critical concentration of the pointed ends, as promoted by classic barbed end capping proteins, such as gelsolin or CapG. The fact that the DAAM fragments only slightly increase the critical concentration for actin assembly at barbed ends further indicates that these fragments are not strong cappers of barbed ends, and the rates of G-actin association to and dissociation from the barbed end remained comparable with those recorded at the minus end.
Other formin fragments were shown to decrease the rate of elongation and depolymerization to only ∼50% of the value measured in the absence of formin and were called leaky cappers (7, 29, 60, 69). The DAAM formin thus represents an extreme case of the leaky cappers. Similarly, Cdc12 was proposed to behave as a strong capper in the absence of profilin; however, the complete blockage of barbed end dynamics by Cdc12 was questioned recently (9). Previously, mDia3-FH2 was also reported to completely inhibit depolymerization and shift the value of critical concentration to that of the pointed ends; however, the half-effect was observed at higher formin concentrations (∼2 μm) (29).
Although DAAM-FH2 and FH1-FH2 possess similar properties in the absence of profilin, the presence of profilin clearly generates functional differences between FH2 and FH1-FH2. Although DAAM-FH1-FH2 interacts with profilin-actin and provides effective acceleration of the polymerization, the DAAM-FH2 domain lacking the FH1 domain fails to utilize the profilin-actin complexes to assemble actin filaments (Figs. 4 and 5). In the TIRFM and spectrin-seeded actin assays, the elongation rate was greatly decreased by FH1-FH2 in the absence of profilin but was not significantly altered when profilin was present (Fig. 4, B and C). In contrast, binding of DAAM-FH2 to barbed ends inhibited barbed end growth from either actin or profilin-actin. In our interpretation, the DAAM-FH1-FH2 fragment can bind profilin in its complex with actin through the FH1 domain. In correlation with these observations, in the biomimetic assay, the DAAM-FH1-FH2-functionalized beads initiated actin tails and moved continuously in the presence of profilin, which was not the case for the DAAM-FH2-functionalized beads or for the DAAM-FH1-FH2 in the absence of profilin (Fig. 6). The 4-fold slower propulsion of DAAM-FH1-FH2-functionalized beads as compared with mDia1-coated beads correlates with the lower value of k+ for profilin-actin association to barbed ends (Fig. 4D and Table 1). Both in vitro and in vivo experiments support the view that the FH1 domain allows formins to use profilin-actin complexes for processive filament assembly, and the processivity requires both profilin and the FH1 domain (10, 45).
Both DAAM fragments bound to the sides of actin filaments with micromolar affinity (Fig. 7). We further showed that they could cross-link the actin filaments to form bundles (Fig. 8). Similar actin filament bundling activity was previously reported for other formins (55–59, 70, 71).
The filaments appeared shorter in the presence of the DAAM fragments (Fig. 8). The length of the filaments at steady state is influenced by the nucleation rate, the monomer association and dissociation rates, and also by the effects of annealing and filament fragmentation. Although we do not know all of the corresponding kinetic parameters, it is likely that in the presence of DAAM fragments, the filaments appeared shorter due to the enhanced nucleation, the slower kinetics of the monomer binding and dissociation to and from the barbed ends, and the inhibition of reannealing by binding of formin to barbed ends, which allowed the effect of the fragmentation to become more dominant.
In conclusion, the present biophysical experiments established that the DAAM-FH2 and the DAAM-FH1-FH2 fragments behave as bona fide formin fragments possessing many of the properties previously reported for other members of the formin family, in particular with Cdc12, which plays a role in assembly of the cytokinetic ring in Schizosaccharomyces pombe (11). Whereas Drosophila DAAM is the first insect formin to be examined in such detail, further investigations will be required to clarify how the molecular mechanisms maintained and assisted by the DAAM formins are related to their specific biological functions in flies and in other organisms.
Supplementary Material
Acknowledgments
We thank Miklós Erdelyi for providing fly stocks and reagents, and we are grateful to Anikó Berente and Margit Pál for technical assistance.
This work was supported by grants from the Hungarian Science Foundation (OTKA Grants K60968 and K77840 (to M. N.)) and from the Hungarian National Office for Research and Technology (Grants GVOP-3.2.1-2004-04-0190/3.0 and GVOP-3.2.1-2004-04-0228/3.0). This work was also supported by “Science, Please! Research Team on Innovation” Grant SROP-4.2.2/08/1/2008-0011.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Movies 1–3.
T. Matusek and J. Mihály, unpublished results.
- pyrene
- N-(1-pyrene)iodoacetamide
- TIRFM
- total internal reflection fluorescence microscopy
- rhodamine
- 5-(and 6)-carboxytetramethylrhodamine succinimidyl ester
- ADF
- actin-depolymerizing factor
- dsRNA
- double-stranded RNA
- DTT
- dithiothreitol
- BSA
- bovine serum albumin
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Le Clainche C., Carlier M. F. (2008) Physiol. Rev. 88, 489–513 [DOI] [PubMed] [Google Scholar]
- 2.Pollard T. D. (2007) Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 [DOI] [PubMed] [Google Scholar]
- 3.Chhabra E. S., Higgs H. N. (2007) Nat. Cell Biol. 9, 1110–1121 [DOI] [PubMed] [Google Scholar]
- 4.Goode B. L., Eck M. J. (2007) Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 5.Higgs H. N. (2005) Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 6.Li F., Higgs H. N. (2003) Curr. Biol. 13, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 7.Pring M., Evangelista M., Boone C., Yang C., Zigmond S. H. (2003) Biochemistry 42, 486–496 [DOI] [PubMed] [Google Scholar]
- 8.Sagot I., Klee S. K., Pellman D. (2002) Nat. Cell Biol. 4, 42–50 [DOI] [PubMed] [Google Scholar]
- 9.Kovar D. R., Pollard T. D. (2004) Nat. Cell Biol. 6, 1158–1159 [DOI] [PubMed] [Google Scholar]
- 10.Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M. F. (2004) Cell 119, 419–429 [DOI] [PubMed] [Google Scholar]
- 11.Chang F., Drubin D., Nurse P. (1997) J. Cell Biol. 137, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., Saito Y., Nakao K., Jockusch B. M., Narumiya S. (1997) EMBO J. 16, 3044–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evangelista M., Blundell K., Longtine M. S., Chow C. J., Adames N., Pringle J. R., Peter M., Boone C. (1997) Science 276, 118–122 [DOI] [PubMed] [Google Scholar]
- 14.Imamura H., Tanaka K., Hihara T., Umikawa M., Kamei T., Takahashi K., Sasaki T., Takai Y. (1997) EMBO J. 16, 2745–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tominaga T., Sahai E., Chardin P., McCormick F., Courtneidge S. A., Alberts A. S. (2000) Mol. Cell 5, 13–25 [DOI] [PubMed] [Google Scholar]
- 16.Higgs H. N., Peterson K. J. (2005) Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otomo T., Tomchick D. R., Otomo C., Panchal S. C., Machius M., Rosen M. K. (2005) Nature 433, 488–494 [DOI] [PubMed] [Google Scholar]
- 18.Lammers M., Rose R., Scrima A., Wittinghofer A. (2005) EMBO J. 24, 4176–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberts A. S. (2001) J. Biol. Chem. 276, 2824–2830 [DOI] [PubMed] [Google Scholar]
- 20.Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M. R., Wittinghofer A. (2005) Nature 435, 513–518 [DOI] [PubMed] [Google Scholar]
- 21.Franch-Marro X., Martín N., Averof M., Casanova J. (2006) Development 133, 785–790 [DOI] [PubMed] [Google Scholar]
- 22.Kida Y. S., Sato T., Miyasaka K. Y., Suto A., Ogura T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6708–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakaya M. A., Habas R., Biris K., Dunty W. C., Jr., Kato Y., He X., Yamaguchi T. P. (2004) Gene Expr. Patterns 5, 97–105 [DOI] [PubMed] [Google Scholar]
- 24.Kida Y., Shiraishi T., Ogura T. (2004) Brain Res. Dev. Brain Res. 153, 143–150 [DOI] [PubMed] [Google Scholar]
- 25.Matusek T., Djiane A., Jankovics F., Brunner D., Mlodzik M., Mihály J. (2006) Development 133, 957–966 [DOI] [PubMed] [Google Scholar]
- 26.Habas R., Kato Y., He X. (2001) Cell 107, 843–854 [DOI] [PubMed] [Google Scholar]
- 27.Yamashita M., Higashi T., Suetsugu S., Sato Y., Ikeda T., Shirakawa R., Kita T., Takenawa T., Horiuchi H., Fukai S., Nureki O. (2007) Genes Cells 12, 1255–1265 [DOI] [PubMed] [Google Scholar]
- 28.Matusek T., Gombos R., Szécsényi A., Sánchez-Soriano N., Czibula A., Pataki C., Gedai A., Prokop A., Raskó I., Mihály J. (2008) J. Neurosci. 28, 13310–13319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada A., Nyitrai M., Vetter I. R., Kühlmann D., Bugyi B., Narumiya S., Geeves M. A., Wittinghofer A. (2004) Mol. Cell 13, 511–522 [DOI] [PubMed] [Google Scholar]
- 30.Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., Eck M. J. (2004) Cell 116, 711–723 [DOI] [PubMed] [Google Scholar]
- 31.Lu J., Meng W., Poy F., Maiti S., Goode B. L., Eck M. J. (2007) J. Mol. Biol. 369, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovar D. R. (2006) Curr. Opin. Cell Biol. 18, 11–17 [DOI] [PubMed] [Google Scholar]
- 33.Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 34.Houk T. W., Jr., Ue K. (1974) Anal. Biochem. 62, 66–74 [DOI] [PubMed] [Google Scholar]
- 35.Elzinga M., Collins J. H., Kuehl W. M., Adelstein R. S. (1973) Proc. Natl. Acad. Sci. U.S.A. 70, 2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Criddle A. H., Geeves M. A., Jeffries T. (1985) Biochem. J. 232, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isambert H., Venier P., Maggs A. C., Fattoum A., Kassab R., Pantaloni D., Carlier M. F. (1995) J. Biol. Chem. 270, 11437–11444 [DOI] [PubMed] [Google Scholar]
- 38.Le Clainche C., Carlier M. F. (2004) Curr. Protoc. Cell Biol. Chapter 12, Unit 12.7 [DOI] [PubMed] [Google Scholar]
- 39.Gutsche-Perelroizen I., Lepault J., Ott A., Carlier M. F. (1999) J. Biol. Chem. 274, 6234–6243 [DOI] [PubMed] [Google Scholar]
- 40.Kurzawa S. E., Geeves M. A. (1996) J. Muscle Res. Cell Motil. 17, 669–676 [DOI] [PubMed] [Google Scholar]
- 41.Kinosian H. J., Selden L. A., Gershman L. C., Estes J. E. (2002) Biochemistry 41, 6734–6743 [DOI] [PubMed] [Google Scholar]
- 42.Vinson V. K., De La Cruz E. M., Higgs H. N., Pollard T. D. (1998) Biochemistry 37, 10871–10880 [DOI] [PubMed] [Google Scholar]
- 43.Paul A. S., Pollard T. D. (2008) Curr. Biol. 18, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vavylonis D., Kovar D. R., O'Shaughnessy B., Pollard T. D. (2006) Mol. Cell 21, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero S., Didry D., Larquet E., Boisset N., Pantaloni D., Carlier M. F. (2007) J. Biol. Chem. 282, 8435–8445 [DOI] [PubMed] [Google Scholar]
- 46.Bubb M. R., Yarmola E. G., Gibson B. G., Southwick F. S. (2003) J. Biol. Chem. 278, 24629–24635 [DOI] [PubMed] [Google Scholar]
- 47.Kovar D. R., Kuhn J. R., Tichy A. L., Pollard T. D. (2003) J. Cell Biol. 161, 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovar D. R., Wu J. Q., Pollard T. D. (2005) Mol. Biol. Cell 16, 2313–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wegner A., Isenberg G. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 4922–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perelroizen I., Didry D., Christensen H., Chua N. H., Carlier M. F. (1996) J. Biol. Chem. 271, 12302–12309 [DOI] [PubMed] [Google Scholar]
- 51.Higashida C., Miyoshi T., Fujita A., Oceguera-Yanez F., Monypenny J., Andou Y., Narumiya S., Watanabe N. (2004) Science 303, 2007–2010 [DOI] [PubMed] [Google Scholar]
- 52.Didry D., Carlier M. F., Pantaloni D. (1998) J. Biol. Chem. 273, 25602–25611 [DOI] [PubMed] [Google Scholar]
- 53.Higashi T., Ikeda T., Shirakawa R., Kondo H., Kawato M., Horiguchi M., Okuda T., Okawa K., Fukai S., Nureki O., Kita T., Horiuchi H. (2008) J. Biol. Chem. 283, 8746–8755 [DOI] [PubMed] [Google Scholar]
- 54.Harris E. S., Li F., Higgs H. N. (2004) J. Biol. Chem. 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- 55.Chhabra E. S., Higgs H. N. (2006) J. Biol. Chem. 281, 26754–26767 [DOI] [PubMed] [Google Scholar]
- 56.Harris E. S., Rouiller I., Hanein D., Higgs H. N. (2006) J. Biol. Chem. 281, 14383–14392 [DOI] [PubMed] [Google Scholar]
- 57.Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C. J., Blanchoin L. (2005) Plant Cell 17, 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moseley J. B., Goode B. L. (2005) J. Biol. Chem. 280, 28023–28033 [DOI] [PubMed] [Google Scholar]
- 59.Esue O., Harris E. S., Higgs H. N., Wirtz D. (2008) J. Mol. Biol. 384, 324–334 [DOI] [PubMed] [Google Scholar]
- 60.Zigmond S. H., Evangelista M., Boone C., Yang C., Dar A. C., Sicheri F., Forkey J., Pring M. (2003) Curr. Biol. 13, 1820–1823 [DOI] [PubMed] [Google Scholar]
- 61.Schirenbeck A., Arasada R., Bretschneider T., Schleicher M., Faix J. (2005) Biochem. Soc. Trans. 33, 1256–1259 [DOI] [PubMed] [Google Scholar]
- 62.Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S. (2003) Curr. Biol. 13, 534–545 [DOI] [PubMed] [Google Scholar]
- 63.Pellegrin S., Mellor H. (2005) Curr. Biol. 15, 129–133 [DOI] [PubMed] [Google Scholar]
- 64.Bugyi B., Papp G., Hild G., Lõrinczy D., Nevalainen E. M., Lappalainen P., Somogyi B., Nyitrai M. (2006) J. Biol. Chem. 281, 10727–10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papp G., Bugyi B., Ujfalusi Z., Barkó S., Hild G., Somogyi B., Nyitrai M. (2006) Biophys. J. 91, 2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kupi T., Gróf P., Nyitrai M., Belágyi J. (2009) Biophys. J. 96, 2901–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen K. K., Rubenstein P. A. (2009) J. Biol. Chem. 284, 16776–16783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Higgs H. N., Pollard T. D. (2001) Annu. Rev. Biochem. 70, 649–676 [DOI] [PubMed] [Google Scholar]
- 69.Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. (2002) Science 297, 612–615 [DOI] [PubMed] [Google Scholar]
- 70.Ishizaki T., Morishima Y., Okamoto M., Furuyashiki T., Kato T., Narumiya S. (2001) Nat. Cell Biol. 3, 8–14 [DOI] [PubMed] [Google Scholar]
- 71.Harris E. S., Higgs H. N. (2006) Methods Enzymol. 406, 190–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.