Abstract
Mycolic acids are key cell wall components for the survival, pathogenicity, and antibiotic resistance of the human tubercle bacillus. Although it was thought that Mycobacterium tuberculosis tightly regulates their production to adapt to prevailing environmental conditions, the molecular mechanisms governing mycolic acid biosynthesis remained extremely obscure. Meromycolic acids, the direct precursors of mycolic acids, are synthesized by a type II fatty acid synthase from acyl carrier protein-bound substrates that are extended iteratively, with a reductive cycle in each round of extension, the second step of which is catalyzed by the essential β-ketoacyl-acyl carrier protein reductase, MabA. In this study, we investigated whether post-translational modifications of MabA might represent a strategy employed by M. tuberculosis to regulate mycolic acid biosynthesis. Indeed, we show here that MabA was efficiently phosphorylated in vitro by several M. tuberculosis Ser/Thr protein kinases, including PknB, as well as in vivo in mycobacteria. Mass spectrometric analyses using LC-ESI/MS/MS and site-directed mutagenesis identified three phosphothreonines, with Thr191 being the primary phosphor-acceptor. A MabA_T191D mutant, designed to mimic constitutive phosphorylation, exhibited markedly decreased ketoacyl reductase activity compared with the wild-type protein, as well as impaired binding of the NADPH cofactor, as demonstrated by fluorescence spectroscopy. The hypothesis that phosphorylation of Thr191 alters the enzymatic activity of MabA, and subsequently mycolic acid biosynthesis, was further supported by the fact that constitutive overexpression of the mabA_T191D allele in Mycobacterium bovis BCG strongly impaired mycobacterial growth. Importantly, conditional expression of the phosphomimetic MabA_T191D led to a significant inhibition of de novo biosynthesis of mycolic acids. This study provides the first information on the molecular mechanism(s) involved in mycolic acid regulation through Ser/Thr protein kinase-dependent phosphorylation of a type II fatty acid synthase enzyme.
Keywords: Bacteria, Bacterial Protein Kinases, Cell Wall, Prokaryotic Protein Kinases, Prokaryotic Signal Transduction, Mycobacterium tuberculosis
Introduction
Tuberculosis is a leading cause of death from infectious disease worldwide (1). Mycobacterium tuberculosis, the etiologic agent of tuberculosis, is perfectly adapted to the human host, having evolved a vast array of mechanisms that promote immune evasion, permitting latent infection to occur (2). M. tuberculosis has a complex lifestyle comprising different developmental stages correlated with the various steps in infection. The success of this pathogen largely stems from its remarkable capacity to survive within the infected host, where it can persist for several decades. The presence of its unusual cell wall is a key factor in this survival (3). Despite extensive literature on the biosynthesis, structure, and biological function(s) of the major cell wall components of M. tuberculosis, very little is known regarding the mechanisms allowing the bacterium to modulate and adapt expression of its cell wall components in response to environmental changes. Therefore, uncovering cell wall regulatory processes represents a crucial step toward understanding the physiology and physiopathology of M. tuberculosis, as well as the interactions between mycobacteria and their environment in general (3).
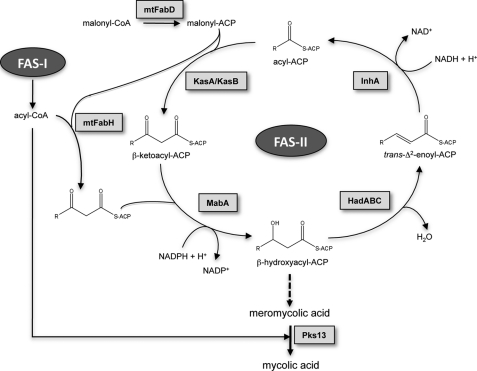
Mycolic acids are essential components of the lipid-rich cell envelope of M. tuberculosis and related mycobacteria (4, 5). They are found either covalently attached to the terminal arabinose residues of the mycolyl arabinogalactan-peptidoglycan complex or as extractable glycolipids, including trehalose monomycolate and trehalose dimycolate. Recent studies have also revealed free mycolic acids in M. tuberculosis biofilms (6). Mycolic acids are very long-chain α-alkyl β-hydroxylated fatty acids (4, 5) that play an important role in reduced cell wall permeability (3, 7), virulence (8–13), and acid fastness characteristic of M. tuberculosis (13). The biosynthesis of mycolic acids depends on two distinct systems (Fig. 1): the eukaryotic-like type I fatty acid synthase (FAS-I)3 and the prokaryotic-like type II fatty acid synthase (FAS-II). FAS-I is a polypeptide that performs de novo biosynthesis of medium length acyl-CoAs (C16 and C24–C26) (14, 15). These are used as primers by the FAS-II system and iteratively condensed with malonyl-ACP in a reaction catalyzed by mtFabH, the β-ketoacyl-ACP synthase III of M. tuberculosis (16–18). During the second step of the elongation cycle, the resulting β-ketoacyl-ACP product is reduced by MabA, the NADPH-dependent β-ketoacyl reductase of M. tuberculosis (19, 20). The resulting β-hydroxyacyl-ACP is then dehydrated by a set of dehydratases, HadABC (21, 22), and finally reduced by the enoyl-ACP reductase, InhA, the primary target of isoniazid (23). The succeeding steps of condensation of the elongating chain with malonyl-ACP units are performed by the β-ketoacyl-ACP synthases KasA and KasB (8, 24, 25), leading to very long-chain meromycolyl-ACPs (up to C56), which are the direct precursors of mycolic acids. How this complex metabolic pathway is regulated, allowing M. tuberculosis to tightly adjust mycolic acid biosynthesis to allow survival under variable environmental conditions, is currently unknown. Only recently, a first step has been taken toward identifying a mycolic acid regulatory system that involves post-translational modification via phosphorylation of several FAS-II enzymes (26).
FIGURE 1.
Mycolic acid biosynthetic pathway. The malonyl-CoA:ACP transacylase mtFabD converts malonyl-CoA into malonyl-ACP, providing the elongation building blocks for the FAS-II. Cycles of elongation are initiated by the condensation of the FAS-I acyl-CoA products with malonyl-ACP, a reaction catalyzed by the β-ketoacyl-ACP synthase mtFabH. The second step in the elongation cycle is performed by the NADPH-dependent β-ketoacyl-ACP reductase MabA, generating a β-hydroxyacyl-ACP intermediate, which is subsequently dehydrated by the β-hydroxyacyl-ACP dehydratase HadABC to form a trans-Δ2-enoyl-ACP. The final step in the elongation is carried out by the NADH-dependent enoyl-ACP reductase InhA. Subsequent rounds of elongation are initiated by the elongation condensing enzymes KasA and KasB, giving raise to meromycolic acids, which are condensed with C26 acyl-CoAs by the termination condensing enzyme Pks13 to form mycolic acids.
Signal sensing leading to cellular responses must be tightly regulated to allow survival under variable conditions. Prokaryotes generally control their signal transduction processes through two-component (His/Asp) systems, but this can also be achieved by eukaryotic-like Ser/Thr protein kinases (STPKs). Protein phosphorylation/dephosphorylation represents a central mechanism for transduction of specific signals to various parts of the cell, such as regulation of growth, differentiation, mobility, and survival (27). It is becoming clear that in M. tuberculosis many of these kinases are involved in the regulation of metabolic processes, transport of metabolites, cell division, or virulence. Therefore, signaling through Ser/Thr phosphorylation has emerged as a key regulatory mechanism in pathogenic mycobacteria (28). In response to its environment, M. tuberculosis activates or represses the expression of a number of genes to rapidly adjust to new conditions. The infection process of M. tuberculosis involves cross-talk of signals between the host and the bacterium, resulting in reprogramming of the host signaling network (29). Many of these stimuli are transduced in the bacteria via sensor kinases present in the mycobacterial membrane, allowing the pathogen to adapt its cellular response to survive in hostile environments. In addition to two-component systems, M. tuberculosis contains 11 STPKs (30, 31), and most are being investigated for their physiological roles and potential application for future drug development to combat tuberculosis (28).
In our previous studies, the three condensing enzymes mtFabH, KasA, and KasB of the mycobacterial FAS-II system were identified as specific substrates of Ser/Thr protein kinases both in vitro and in vivo, and it was found that phosphorylation of these enzymes modulates their condensing activity (26, 32). However, whether phosphorylation directly controls mycolic acid biosynthesis has not been clearly established. The present study was undertaken to determine whether mycolic acid synthesis in general might be influenced by STPK-dependent regulatory mechanisms. Here, we have identified MabA as a new substrate of the M. tuberculosis SPTKs and characterized its phosphorylation sites. This allowed us to address the role/contribution of phosphorylation in regulating the β-ketoacyl-ACP reductase (KAR) activity of MabA. Importantly, through the use of a genetically inducible system in mycobacteria expressing a phosphorylated MabA mimic, we provide, for the first time evidence that phosphorylation negatively regulates de novo biosynthesis of mycolic acids.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strains used for cloning and expression of recombinant proteins were Escherichia coli TOP10 (Invitrogen) and E. coli BL21(DE3)pLysS (Novagen) or BL21(DE3)Star (Novagen). All strains were grown in LB medium at 37 °C. Media were supplemented with ampicillin (100 μg/ml), hygromycin (200 μg/ml), or kanamycin (25 μg/ml), when required. Mycobacterium bovis BCG Pasteur 1173P2 strain was grown on Middlebrook 7H11 agar plates with OADC enrichment (Difco) or in Sauton's medium containing 0.025% tyloxapol (Sigma), supplemented with hygromycin (50 μg/ml) or kanamycin (25 μg/ml), when required.
Cloning, Expression, and Purification of Recombinant MabA and Mutant Proteins
The mabA gene was amplified by PCR using M. tuberculosis H37Rv chromosomal DNA as a template and a set of primers containing an NdeI and an NheI site (supplemental Table SI). This amplified product was then digested with NdeI and NheI and ligated into the pETPhos vector (33), generating pETPhos_mabA. Site-directed mutagenesis was directly performed on pETPhos_mabA using inverse PCR amplification with self-complementary primers (supplemental Table S2). All constructs were verified by DNA sequencing. Recombinant strains harboring the MabA wild-type or mutant-expressing constructs were used to inoculate 200 ml of LB medium supplemented with ampicillin and the resulting cultures were incubated at 37 °C with shaking until the A600 reached 0.5. Isopropyl 1-thio-β-d-galactopyranoside was then added at a final concentration of 1 mm and growth was continued for 3 h at 37 °C. Purification of recombinant wild-type His-MabA and derived MabA mutants was performed as follows. Cells were harvested by centrifugation, washed in buffer A (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm DTT, 1 mm EDTA, 10 mm imidazole) containing DNase, RNase, and a mixture of protease inhibitors (Roche Applied Science). Bacteria were disrupted in a French pressure cell and the resulting extract was centrifuged for 45 min at 10,000 × g at 4 °C. The supernatant was collected and mixed together with Ni2+-nitrilotriacetic acid-agarose beads (Qiagen) previously equilibrated with buffer A. Beads were then extensively washed with buffer B (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm DTT, 1 mm EDTA, 20 mm imidazole) and eluted with buffer C (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm DTT, 1 mm EDTA, 300 mm imidazole). Fractions containing pure MabA proteins were pooled, dialyzed when required, and stored at −20 °C until further use.
In Vitro Kinase Assay
In vitro phosphorylation was performed as described earlier (34) with 4 μg of MabA in 20 μl of buffer P (25 mm Tris-HCl, pH 7.0, 1 mm DTT, 5 mm MgCl2, 1 mm EDTA) with 200 μCi/ml of [γ-33P]ATP corresponding to 65 nm (PerkinElmer Life Sciences, 3000 Ci/mmol), and 0.6–4.2 μg of kinase to obtain the optimal autophosphorylation activity for each kinase for 30 min at 37 °C. Cloning, expression, and purification of the eight recombinant GST-tagged STPKs from M. tuberculosis were described previously (26).
Mass Spectrometry Analysis
Purified MabA_WT and mutant derivatives were subjected to in vitro phosphorylation by GST-tagged PknB as described above, except that [γ-33P]ATP was replaced with 5 mm cold ATP. Subsequent mass spectrometric analyses were performed as previously described (35). Mass spectrometry was performed using a Q-Star XL nanoESI Quadrupole/time-of-flight tandem mass spectrometer, nanoESI-qQ-TOF-MS/MS (Applied Biosystems, Courtaboeuf, France), coupled to an online nano-liquid chromatography system (Famos, Switchos, and Ultimate from Dionex, The Netherlands). The samples were loaded onto a trap column (PepMap100 C18; 5 μm; 100 Å; 300 μm × 5 mm, Dionex), washed for 3 min at 25 μl/min with 0.05% trifluoroacetic acid, 2% CH3CN, then eluted onto a C18 reverse phase column (PepMap100 C18; 3 μm; 100 Å; 7 m × 150 mm, Dionex). Peptides were separated at a flow rate of 0.300 μl/min with a linear gradient of 5–80% CH3CN in 0.1 n formic acid for 60 min. MS data were acquired automatically using Analyst QS 1.1 software (Applied Biosystems). A 1-s TOF-MS survey scan was acquired over 400–1600 atomic mass units, followed by three 3 s product ion scans over the mass range of 65–2000 atomic mass units. The three most intense peptides with charge state of 2–4 above a 20-count threshold were selected for fragmentation and dynamically excluded for 60 s with ±50 atomic unit mass tolerance. The collision energy was set by the software according to the charge and mass of the precursor ion. The MS and MS/MS data were recalibrated using internal reference ions from a trypsin autolysis peptide at m/z 842.51 [M + H]+ and m/z 421.76 [M + 2H]2+.
Overexpression in M. bovis BCG Pasteur and Purification of the Recombinant Protein
The mabA gene was amplified by PCR using M. tuberculosis H37Rv chromosomal DNA as template and a set of primers containing NdeI and EcoRI restriction sites (supplemental Table S2). This amplified product was then digested by NdeI and EcoRI and ligated into the pMK1 expression vector (supplemental Table S1). The resulting construct was introduced by electroporation into M. bovis BCG using a Bio-Rad Gene Pulser. Transformants were grown and harvested at early stationary phase and the purification protocol was performed as previously described (32).
In Vitro Ketoacyl Reductase Assay
KAR activity was assayed as described previously (20), by adding 22.5 μg/ml of His-MabA (wild-type or mutants) to a standard reaction mixture, which was kept at 25 °C (final volume 1 ml), consisting of 100 mm HEPES, pH 7.0, 100 μm NADPH (Sigma), 100 μm acetoacetyl-CoA (Sigma). Oxidation of NADPH was monitored spectrophotometrically at 340 nm. The baseline was established for 3 min prior to the addition of acetoacetyl-CoA.
Monitoring of NADPH Binding by Fluorescence Spectroscopy
Following purification, MabA proteins were dialyzed at 4 °C against dialysis buffer (100 mm HEPES, pH 7.0, 10% glycerol). Measurements were performed at room temperature using 56 μg/ml of protein in dialysis buffer in the absence or presence of increasing concentrations of NADPH in a final volume of 40 μl. Steady-state fluorescence anisotropy binding titrations were carried out on a Tecan Saphire II microplate reader, using a 295-nm LED for excitation, and a monochromator set at 340 nm (bandwidth 8 nm) for emission.
Circular Dichroism Analysis
UV-CD spectra were recorded using a Chirascan spectrophotometer (Applied Photophysics) at a protein concentration of ∼1 mg/ml in buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm DTT, 1 mm EDTA). The data (average of three spectra) were analyzed with the program CDNN to estimate the secondary structure composition (36).
Mycolic Acid Biosynthesis in M. bovis BCG
M. bovis BCG cultures carrying the acetamide-inducible plasmids derived from pSD26 (37) were grown in Sauton medium supplemented with 50 μg/ml of hygromycin and 0.2% succinate at 37 °C until the A600 nm reached 0.7–0.8. At this point, acetamide was added at a final concentration of 0.2%. After 16 h of incubation at 37 °C, 1 μCi/ml of [1,2-14C]acetate (50–62 mCi/mmol, Amersham Biosciences) was added to the cultures followed by an additional 6-h incubation. The 14C-labeled cells were harvested, then subjected to alkaline hydrolysis using 15% aqueous tetrabutylammonium hydroxide overnight at 100 °C, followed by the addition of 4 ml of CH2Cl2, 300 ml of CH3I, and 2 ml of water. The entire reaction mixture was then mixed for 60 min. The upper aqueous phase was discarded and the lower organic phase washed twice with water and evaporated to dryness. Methyl esters were re-dissolved in diethyl ether and the solution was again evaporated to dryness. The final residue was then dissolved in 800 μl of CH2Cl2 and counted. Equal volumes (40 μl) of the resulting solution of fatty acid methyl esters and mycolic acid methyl esters were subjected to TLC, using silica gel plates (5735 Silica Gel 60 F254; Merck, Darmstadt, Germany) developed in petroleum ether/acetone (95:5, v/v). Bands on TLC plates corresponding to each type of mycolates were quantified using a PhosphorImager system (STORM-Applied Biosystem).
Production of Anti-MabA Antibodies and Immunoblotting Studies
Purified His-tagged MabA_WT was used to prepare an anti-MabA immune serum. Rats were injected four times with 25 μg of MabA mixed with an equal volume of adjuvant (monophosphoryl lipid A + trehalose dicorynemycolate, Sigma) at 1, 15, 30, and 60 days. The serum was collected at different time points after the last injection and the specificity of the antibodies tested by Western blotting. Rabbit anti-InhA antibodies were produced as described previously (38). M. bovis BCG bacteria were harvested, resuspended in phosphate-buffered saline, and disrupted using a Bead-beater (Retsch). Protein concentration was determined on total lysates using the BCA Protein Assay reagent kit (Pierce), according to the manufacturer's instructions. One hundred μg of total protein was subjected to SDS-PAGE with 12% acrylamide. Proteins were transferred to nitrocellulose membranes for Western blot analysis. For detection of MabA, membranes were incubated overnight with the rat antiserum at a dilution of 1:30,000, washed, and subsequently incubated with anti-rat antibodies conjugated to alkaline phosphatase (Promega) used at a 1:7,000 dilution. For the detection of InhA, the membrane was incubated overnight with rabbit anti-InhA antibodies at a 1:25,000 dilution, washed, and probed with anti-rabbit antibodies conjugated to alkaline phosphatase used at a 1:7,000 dilution.
RESULTS
MabA Is Phosphorylated in Vitro by Multiple Ser/Thr Kinases
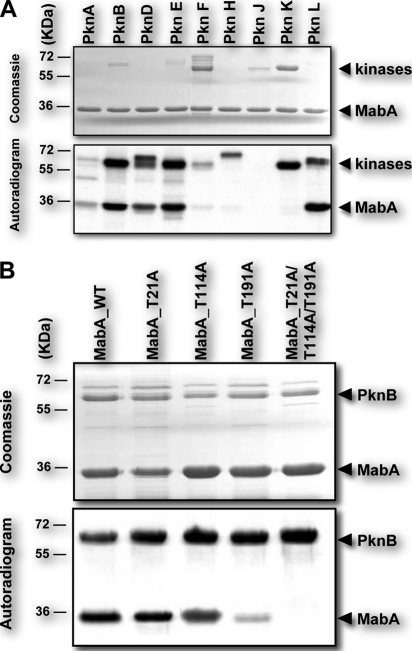
The NADPH-dependent β-ketoacyl-ACP reductase MabA is a key enzyme of the M. tuberculosis FAS-II system, catalyzing the conversion of β-ketoacyl-ACP to β-hydroxyacyl-ACP (20). As a first step to investigate whether this protein represents a regulatory checkpoint controlling mycolic acid production, we examined if MabA is post-translationally modified by phosphorylation in the presence of purified STPKs (PknA to PknL). All the kinases from M. tuberculosis were expressed as GST-tagged fusion proteins and purified from E. coli as reported earlier (26). The purified kinases were incubated with MabA and [γ-33P]ATP, resolved by SDS-PAGE, and their phosphorylation profiles analyzed by autoradiography. The presence of intense radioactive signals indicated that MabA was phosphorylated by multiple kinases, including PknA, PknB, PknD, PknE, and PknL (Fig. 2A). No signal was observed in the presence of PknF, PknH, PknJ, or PknK, which displayed various autokinase activities, as reported earlier (26). These results clearly indicated that MabA is a specific substrate and interacts with various STPKs in vitro, suggesting that this key protein of the mycolic acid biosynthetic pathway might be regulated in mycobacteria by multiple extracellular signals.
FIGURE 2.
In vitro phosphorylation of MabA. A, phosphorylation of MabA by multiple STPK. Nine recombinant STPKs (PknA to PknL) encoded by the M. tuberculosis genome were expressed and purified as GST fusions and incubated with purified His-tagged MabA and [γ-33]ATP. The quantity of the different STPKs varied from 0.6 to 4.2 μg to obtain the optimal autophosphorylation activity for each kinase. Samples were separated by SDS-PAGE and stained with Coomassie Blue (upper panel) and visualized by autoradiography after overnight exposure to a film (lower panel). The upper bands illustrate the autokinase activity of each STPK, and the lower bands represent phosphorylated MabA. B, in vitro phosphorylation of the single and triple MabA mutants by PknB. Four μg of purified MabA_WT, MabA_T21A, MabA_T114A, MabA_T191A, and MabA_T21A/T114A/T191A were incubated individually with PknB and [γ-33]ATP. Samples were separated by SDS-PAGE and stained with Coomassie Blue (upper panel) and visualized by autoradiography after overnight exposure to a film (lower panel). Upper bands reflect the intense autokinase activity of PknB, and the lower bands illustrate the phosphorylation state of each MabA variant following in vitro phosphorylation by PknB.
MabA Is Phosphorylated in Vitro on Three Threonine Residues
Mass spectrometry was used to identify the number and nature of the phosphorylation sites on MabA, a method that has been successfully used to elucidate the phosphoacceptors in a sequence-specific fashion for several other M. tuberculosis STPK substrates (32, 39). Briefly, MabA was incubated with cold ATP in the presence of PknB (one of the most active kinases for MabA, Fig. 2A) and subjected to mass spectrometric analysis after tryptic and chymotryptic digestion. Spectral identification and phosphorylation determination were achieved with the paragon algorithm from the ProteinPilot® 2.0 data base-searching software (Applied Biosystems) using the phosphorylation emphasis criterion against a homemade data base that included the sequences of MabA and derivatives. Afterward, the phosphopeptides identified by the software were validated by manual examination of the corresponding MS2 spectra. The sequence coverage of the protein was 86% and phosphorylation occurred only on peptide-(174–197). The MS/MS spectra unambiguously confirmed the presence of a phosphate group on Thr191 (Table 1 and supplemental Fig. S1). Then, to prevent in vitro phosphorylation on Thr191, this residue was changed to alanine by site-directed mutagenesis. The corresponding MabA_T191A mutant was expressed and purified as a His-tagged protein in E. coli BL21(DE3)Star harboring pETPhos_mabA_T191A. The resulting MabA_T191A mutant was purified and incubated with PknB in the presence of [γ-33P]ATP. Following separation by SDS-PAGE and analysis by autoradiography, a significant decrease in the phosphorylation signal was observed compared with MabA_WT (Fig. 2B). The fact that phosphorylation was not completely abrogated in MabA_T191A indicated the presence of additional phosphorylation sites. Therefore, the MabA_T191A protein was subjected to another round of mass spectrometric analysis after in vitro phosphorylation with PknB, which allowed us to identify Thr21 as an additional phosphorylation site (Table 1 and supplemental Fig. S2). The double mutant, MabA_T21A/T191A, was generated, purified, and used in the kinase assay (data not shown), and subjected to mass spectrometric analysis, revealing a third phosphorylation site, identified as Thr114 (Table 1 and supplemental Fig. S3). Importantly, phosphorylation of the triple mutant, MabA_T21A/T114A/T191A, was completely abrogated, compared with phosphorylation of MabA_WT (Fig. 2B), indicating that MabA is phosphorylated only on these three residues, at least in vitro, in the presence of PknB. This was further supported by analysis of an additional round of mass spectrometry of MabA_T21A/T114A/T191A pre-treated with ATP and PknB, which failed to identify any additional phosphate groups (data not shown). Taken together, these results indicate that Thr21, Thr114, and Thr191 are likely to play critical roles in the regulation of MabA activity. However, as shown in Fig. 2B, the Thr21 → Ala or Thr114 → Ala substitutions did not significantly reduce the phosphorylation signal of the mutants compared with MabA_WT, in contrast to the Thr191 → Ala substitution. This leads to the proposal that Thr191 represents the primary phosphorylation site, a conclusion strengthened by the fact that Thr191 was the first phosphoacceptor identified in our mass spectrometric analysis. Moreover, when assayed in the presence of PknD, PknE, or PknL, MabA_T191A generated a weak radioactive signal, similar to the one observed with PknB (data not shown). This confirms that Thr191 represents a critical phosphorylation site for several (if not all) STPKs, indicating that phosphorylation mechanisms are dependent on the phosphoacceptor, which is shared by all these kinases, thus highlighting the cross-talk previously observed with others substrates (26, 32).
TABLE 1.
Sequence of the phosphorylated peptides identified in MabA as determined by mass spectrometry
Phosphorylated residues are indicated in bold and underlined.
| Phosphorylated tryptic and chymotryptic peptide sequence | Number of detected phosphate groups | Phosphorylated residue(s) |
|---|---|---|
| LC-ESI/MS/MS | ||
| 174ANVTANVVAPGYIDTDMTRALDER197 | 1 | Thr191 |
| 174ANVTANVVAPGYIDTDMTRALDERIQQGALQFIPAKR210 | 1 | Thr191 |
| 17SVLVTGGNR25 | 1 | Thr21 |
| 108VINANLTGAFR118 | 1 | Thr114 |
In Vivo Phosphorylation of MabA
To corroborate the in vitro results, it was necessary to confirm the phosphorylation state of the MabA protein in vivo. Therefore, the mabA gene was cloned into the pMK1 mycobacterial expression vector under control of the strong promoter hsp60. The resulting construct was used to transform M. bovis BCG Pasteur to allow overproduction of recombinant His-tagged MabA, which was purified to homogeneity by affinity chromatography on nickel-containing beads and subjected to mass spectrometric analysis. LC-MS/MS identified an 80-Da mass increase corresponding to the peptide-(174–197), ANVTANVVAPGYIDTDMTRALDER, supporting the conclusion that MabA is phosphorylated in vivo in M. bovis BCG. Although we could not directly localize the phosphorylation site in this peptide, it is very likely that Thr191 corresponds to the phosphoacceptor in vivo, despite the presence of two other threonine residues in this peptide (Thr177 and Thr188). Once again, this result supports the conclusion that Thr191 corresponds to the primary phosphorylation site both in vitro and in vivo.
Localization of the Thr191 Residue on the MabA Structure
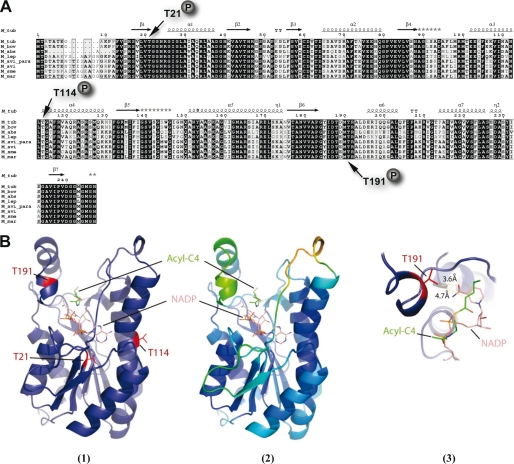
Multiple sequence alignments showed that MabA is a ubiquitous and highly conserved protein present in all mycobacterial species, as would be expected for an essential protein. Except for Thr114, which is substituted by an Ile in Mycobacterium abcessus, the three phosphorylation sites Thr21, Thr114, and Thr191 were found to be conserved in all the species shown in Fig. 3A, including pathogenic and non-pathogenic mycobacteria. This suggests that, if phosphorylation modulates the activity of MabA, this may represent a general and conserved regulatory mechanism.
FIGURE 3.
Conservation of phosphoacceptors in MabA orthologues. A, multiple sequence alignment of MabA sequences from various mycobacterial species. The alignment was performed using ClustalW and Espript (M_tub, M. tuberculosis; M_bov, M. bovis; M_abs, M. abcessus; M_lep, Mycobacterium leprae; M. avi_para; Mycobacterium paratuberculosis; M_avi, Mycobacterium avium; M_sme, Mycobacterium smegmatis; M_mar, Mycobacterium marinum). Residues conserved in all species are shown in black boxes. The three phosphorylation sites of MabA_Mtb are indicated. Protein secondary element assignments are represented above the sequences. Numbering of amino acids corresponds to the MabA protein from M. tuberculosis. B, localization of the identified phosphorylation sites in the three-dimensional structure of MabA. (1) MabA in complex with NADPH and the acyl-C4 substrate. The model of the ternary complex MabA-cofactor-substrate was obtain after superposition of the crystal structure of MabA with different homologous ternary complexes. The three phosphorylation sites identified by mass spectrometry are represented in red. (2) The same complex has been redrawn with the color representing crystal structure B factor of the MabA holo-form (PDB code 1UZM). The residues with the lower B factor and mobility are represented in blue, whereas those with the higher B factor and mobility are in green and yellow. Only Thr191 lies in a high mobility zone and probably accessible to the kinase. (3) Close-up of the substrate and cofactor binding pocket centered on residue Thr191. The lateral chain of Thr191 is 4.7 Å from the substrate and 3.6 Å from the cofactor. Addition of the phospho group on the oxygen of the lateral chain of Thr191 of MabA is incompatible with substrate binding.
MabA belongs to the family of short-chain dehydrogenases/reductases (SDR) (40), and its three-dimensional structure revealed a conserved Rossman-fold and the Ser-Tyr-Lys catalytic triad conserved among short-chain dehydrogenases/reductase members (41). To gain insight into the possible effect(s) of Thr21, Thr114, and Thr191 phosphorylation on the enzymatic behavior of MabA, we determined the positions of all three residues with respect to the substrate-binding site and the catalytic residues, taking advantage of the available three-dimensional structure of the protein (41). Interestingly, Thr191 lies close to the substrate- and cofactor-binding site, implying that it may potentially be involved in β-ketoacyl-ACP reductase activity (41). In fact, in the MabA structure complexed with NADPH, the lateral chain of Thr191 points toward the substrate binding pocket in a hydrophobic environment. Given this strategic position, one may predict that the introduction of the negative charge of a phosphate group by phosphorylation could destabilize this region, ultimately affecting binding of both the substrate and NADPH (Fig. 3B1). Moreover, Thr191 is part of a mobile α′ α-helix (residues 189–192) (Fig. 3B2). In the structure of the apo-form of MabA, the α′ α-helix is very mobile and residues 189–201 are not visible in one of the two MabA monomers, thus making Thr191 accessible to Ser/Thr kinases (Fig. 3B3).
Thr21 is part of the conserved glycine-rich loop (TGXXXGXG) involved in cofactor phosphate binding (42), so a mutation at this position is very likely to alter cofactor binding activity. Also, Thr21 lies in the N-terminal part of the buried β sheet β1, making it difficult to explain how a kinase interacts and phosphorylates this particular site. However, it is reasonable to hypothesize that partial or total unfolding of the MabA structure may occur upon phosphorylation of a first site (for instance, Thr191), leading to the exposure of Thr21 to the kinase.
Thr114 lies in the MabA α4 helix (residues 102–130). The lateral chain of Thr114 is part of a H-bond network with the residues located on the same face of the helix, Asn110 and Arg116. These interactions produce a kink, present in all the short-chain dehydrogenases/reductase α4 helices. The role of such a kink is to position the lateral chain of the highly conserved Asn112 on the opposite face of the α-helix. Indeed, the lateral chain of Asn112 plays a major role in stabilizing the α4/β5 loop after the conformational change induced by cofactor binding (43).
In conclusion, mapping the phosphorylation sites on the three-dimensional MabA structure emphasized the strategic roles of all three threonine residues for MabA activity. However, it is noteworthy that localization of the phosphorylated sites was performed using the available crystal structure corresponding to the unphosphorylated form of MabA. Determining the definitive and more precise role of these sites awaits resolution of the structure of the phosphorylated isoform(s) of the protein.
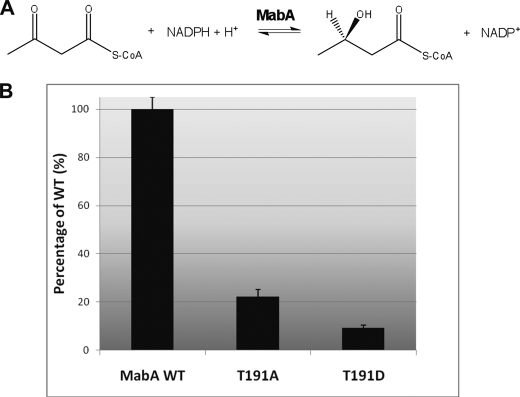
KAR Activity Is Reduced in MabA_T191A and MabA_T191D Mutants
The above compelling data suggest that Thr191 is the primary phosphoacceptor in MabA: (i) this site was the first site identified in our mass spectrometric analyses on several independent occasions; (ii) the Thr191 → Ala replacement, but neither the Thr21 → Ala nor the Thr114 → Ala substitutions, was accompanied by a strong decrease in the phosphorylation signal; (iii) Thr191 belongs to the peptide identified as being phosphorylated in vivo; and (iv) residues around Thr191 appear to be accessible to the kinase based on the crystal structure. This prompted us to evaluate and compare the KAR activity of MabA_WT and MabA_T191A. In vitro MabA activity was determined using acetoacetyl-CoA (C4) and NADPH as reported earlier (20). Under these conditions, MabA catalyzes the reduction of acetoacetyl-CoA to β-hydroxybutyryl-CoA, concomitantly with reduction of NADPH to NADP+ (Fig. 4A). The disappearance of NADPH to form NADP+ can be monitored spectrophotometrically at 340 nm. The activity of each MabA protein was assessed in the presence of acetoacetyl-CoA and NADPH, both at 100 μm. The activity of MabA_WT was around 0.75 ± 0.04 μmol/min/mg, in agreement with previously published results (44). When the activity of MabA_WT was arbitrarily fixed at 100%, the activity of the MabA_T191A mutant was around 22% (Fig. 4B). These results clearly indicate that Thr191 plays a critical role in the catalysis, as supported by its strategic location on the MabA three-dimensional structure. Replacement of Thr191 by a neutral Ala residue may possibly prevent formation of a hydrogen bond between residue 191 and the MabA substrate and/or NADPH, resulting in reduced KAR activity. It can also be inferred that post-translational modification of the hydroxyl group of Thr191 via phosphorylation may strongly impact on the enzymatic activity.
FIGURE 4.
KAR activity of MabA_WT and mutant derivatives. A, reaction scheme for the reduction of acetoacetyl-CoA by MabA. B, MabA_WT, as well as MabA_T191A and MabA_T191D mutants, were purified from recombinant E. coli, dialyzed, and assayed for KAR activity in the presence of acetoacetyl-CoA and NADPH in 100 mm HEPES, pH 7.0, at 25 °C. Conversion of NADPH to NADP was monitored spectrophotometrically at 340 nm. The activity of MabA_WT was arbitrarily fixed at 100%. The values are means of triplicates and are representative of two sets of experiments done with independent protein preparations.
Previous studies have shown that acidic residues such as Asp or Glu qualitatively mimic the effect of phosphorylation with regard to functional activity (32). Following a strategy that has been successfully used previously to demonstrate that regulation of Wag31 via phosphorylation is important during active growth in mycobacteria (45), or in elucidating the role of phosphorylation on the condensing activity of mtFabH (32), we expressed, purified, and determined the KAR activity of MabA_T191D, the phosphorylation mimic. This inhibitory effect on KAR activity was even more pronounced for MabA_T191D than for MabA_T191A, with an activity lower than 10% of MabA_WT (Fig. 4B), presumably due to the negative charge carried by the Asp residue. Indeed, introducing a negative charge, for example, via a phosphate group, may destabilize the α helix and prevent correct binding of both substrate and NADPH.
To exclude the possibility that the T191A or T191D mutations affected the correct folding of the protein, which might explain the reduced activity of the proteins compared with the MabA_WT, circular dichroism was performed to check the protein folding and estimate secondary structures (supplemental Fig. S4). α-Helices represented 27–30% of the residues, whereas 20–24% of the residues were present in β-sheets in the MabA_WT, MabA_T191A, and MabA_T191D structures. This indicated that replacement of MabA_T191A by either Ala or Asp did not significantly affect the folding of the protein. However, circular dichroism is not a technique indicative of fine structural/conformational changes expected to be induced following a phosphorylation event. Therefore, we reasoned that replacement of Thr191 by either Ala or Asp is very likely to affect catalysis of MabA by preventing binding to the cofactor and/or the substrate, rather than altering the overall structure of the protein.
MabA_T191A and MabA_T191D Mutants Are Affected in Binding the Cofactor NADPH
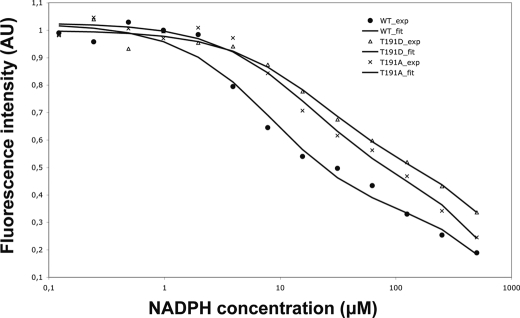
MabA participates in the reductive cycle associated with the elongation of fatty acyl chains coupled to the acyl carrier protein (AcpM) up to C56 (4). However, because such substrates are not commercially available, the assay relies on the use of acetoacetyl-CoA, although it has a poor affinity for the enzyme (19, 20). Because KAR activity can be assessed by monitoring conversion of NADPH to NADP+ spectro photometrically, we examined if replacement of the Thr191 primary phosphoacceptor by either Ala or Asp might affect binding to the NADPH cofactor. The presence of a unique Trp145 residue in the MabA substrate binding pocket offers the possibility to measure binding of NADPH for each mutant by fluorescence spectroscopy, as reported previously (20). It was demonstrated that upon excitation at 300 nm, the emission spectrum displayed a maximum at about 340 nm, indicating that the tryptophan is partially buried (20). In the case of MabA_WT and upon the addition of increasing concentrations of NADPH, a decreased fluorescence intensity was observed (Fig. 5), suggesting that the substrate binding pocket closes upon NADPH addition, as reported earlier (20), leading to determination of an affinity constant for NADPH of 7.4 ± 3 μm, consistent with the value reported elsewhere (20). Importantly, the fluorescence intensity curve of MabA_T191A was shifted to the right, reflecting a lower affinity value, which was estimated at 17 ± 5 μm, perhaps explaining the decreased KAR activity (Fig. 5). An even more shifted fluorescence intensity curve was obtained with MabA_T191D, with an affinity for NADPH of 25 ± 6 μm (Fig. 5). This result is consistent with the very low KAR activity of this mutant and with the hypothesis that mutation of Thr191 impairs the binding to the cofactor. Nevertheless, considering the dramatic drop in KAR activity and the relatively weak decrease in NADPH affinity of the MabA_T191D mutant with respect to the wild-type protein, it is very likely that phosphorylation of Thr191 also alters binding of the substrate. This is supported by the structure of MabA in complex with NADPH and the acetoacetyl-CoA substrate (Fig. 3). In this model, the addition of a phosphate group to the Thr191, in our case its substitution by an Asp residue, generates a steric clash and charge repulsion, thus hindering substrate binding.
FIGURE 5.
Fluorescence emission of MabA_WT and Thr191 derivatives in the presence of increasing concentrations of NADPH. The fluorescence emission of MabA_WT was compared with those of the MabA_T191A and MabA_T191D mutants. AU, arbitrary unit. Similar data were obtained using two independent protein preparations.
A Growth Defect of M. bovis BCG Overexpressing the Phosphomimetic MabA_T191D
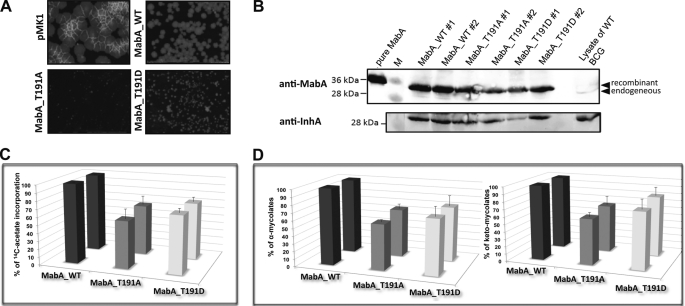
M. bovis BCG cells were transformed with the empty pMK1 vector, pMK1_mabA_WT, pMK1_mabA_T191A, or pMK1_mabA_T191D to allow constitutive expression of the mabA alleles under control of the strong hsp60 promoter. Transformed mycobacteria were selected on Middlebrook 7H10 agar plates. Fig. 6A illustrates the morphology and size of M. bovis BCG colonies after incubation at 37 °C. Overexpression of MabA_WT is accompanied by a decrease in the growth rate compared with the parental strains carrying the empty construct. More importantly, overexpression of either MabA_T191A or MabA_T191D severely altered bacterial growth compared with the strain overproducing MabA_WT (Fig. 6A), a phenotype that was observed in two independent experiments. These results suggest that the impaired KAR activity of MabA_T191A or MabA_T191D is likely to be responsible for this growth defect, presumably by inhibiting FAS-II activity. Because mycolic acids are essential cell wall-associated lipids (4, 5), we reasoned that the growth inhibitory effect in strains overexpressing MabA mutants may be attributed to an alteration of mycolic acid biosynthesis.
FIGURE 6.
In vivo effects of M. bovis BCG strains overexpressing MabA_T191A and MabA_T191D. A, alteration of mycobacterial growth. Electrocompetent M. bovis BCG 1173P2 cells were transformed with the empty pMK1 construct, the pMK1_mabA_WT, pMK1_mabA_T191A, or pMK1_mabA_T191D to allow constitutive expression of the mabA alleles under control of the strong hsp60 promoter. Transformed mycobacteria were plated and incubated at 37 °C for 3 (for pMK1) or 4 weeks (for pMK1_mabA_WT, pMK1_mabA_T191A, or pMK1_mabA_T191D). B, MabA and InhA expression levels in the M. bovis BCG strains. Western blot analysis of M. bovis BCG cells were transformed with pSD26_mabA_WT, pSD26_mabA_T191A, or pSD26_mabA_T191D and grown in the presence of 50 μg/ml of hygromycin. At an A600 of 0.8, expression was induced by adding 0.2% acetamide for 16 h. Bacteria were harvested at various time points, resuspended in PBS, and disrupted. Equal amounts of crude lysates (100 μg) were then loaded onto a 12% acrylamide gel, subjected to electrophoresis, and transferred onto a membrane for immunoblot analysis using either rat anti-MabA antibodies or rabbit anti-InhA antibodies. Endogenous and recombinant MabA proteins are indicated by arrowheads. The results for two individual clones with each construct are presented. C, the inhibitory effect on the incorporation of [1,2-14C]acetate was assayed using cultures induced for 16 h after addition of acetamide. After a 6-h period of labeling, cells were recovered and hydrolyzed by adding 15% tetrabutylammonium hydroxide at 100 °C overnight. 14C-Labeled lipids were extracted, resuspended in CH2Cl2, and counted in the presence of scintillation liquid. Counts for the strain overexpressing wild-type MabA were arbitrarily fixed at 100%. Bars represent [14C]acetate incorporation of two individual clones overexpressing either MabA_T191A or MabA_T191D and are expressed as the mean of three independent experiments ± S.D. D, radiolabeled lipids were subjected to TLC using petroleum ether/acetone (95:5, v/v). Spots corresponding to α- and keto-mycolic acids were quantified using a PhosphorImager system for each strain. Bars represent the percentage of α- and keto-mycolic acids of each strain, with respect to the WT, arbitrarily fixed at 100%. The results for two individual clones overexpressing either MabA_T191A or MabA_T191D are presented and expressed as the mean of three independent experiments ± S.D.
Inhibition of Mycolic Acid Biosynthesis in M. bovis BCG Strain Overproducing the Phosphomimetic MabA_T191D
Comparison of de novo biosynthesis of mycolic acids in different mycobacterial strains can be made only if these strains exhibit a comparable growth rate, because the assay used to quantify mycolic acid production is based on the incorporation of [14C]acetate as a precursor of fatty acids. As mentioned above, strains producing MabA mutants under the constitutive expression system displayed impaired growth, thus precluding the use of these strains for a thorough comparison of mycolic acid biosynthesis. Therefore, a conditional expression system was generated to control expression of the various MabA proteins, where the mabA genes were placed under control of the acetamidase-inducible promoter in the pSD26 vector (37). Addition of 0.2% acetamide to the culture medium for 16 h led to high expression levels of MabA_WT, MabA_T191A, or MabA_T191D compared with the endogenous MabA, as determined by immunoblotting using anti-MabA antibodies (Fig. 6B). Importantly, similar expression levels were also found in all cell lysates overexpressing a MabA protein, enabling direct comparison of the effect of the MabA mutation with respect to mycolic acid biosynthesis in vivo. We also investigated the possibility that overproduction of MabA might influence expression of other FAS-II enzymes, particularly InhA, whose gene is adjacent to mabA in M. tuberculosis and M. bovis BCG. Immunoblotting was performed by probing the membrane with anti-InhA antibodies (Fig. 6B). Bands of comparable intensity were observed in each lysate, indicating that the high expression levels of MabA did not affect InhA expression.
Bacterial growth was monitored spectrophotometrically before and after addition of acetamide to the culture medium, and no differences in growth were perceptible within the first hours following MabA expression (data not shown). We therefore pulsed the cultures with [14C]acetate for 6 h only, and measured incorporation of this radioactive precursor into the mycobacterial lipids. Incorporation of [14C]acetate was significantly reduced in the MabA_T191A and -T191D overexpressing strains, compared with the MabA_WT overexpressing strain (Fig. 6C). Because labeling was performed for a short period of time (before manifestation of a growth defect), it appears reasonable to hypothesize that reduced [14C]acetate incorporation in the MabA mutant strains was due to altered lipid biosynthetic activity rather than a diminished metabolic activity caused by a slow growth rate between the mycobacterial strains.
Because mycolic acids are essential components of the mycobacterial cell wall and are produced by FAS-II, these experimental conditions were also used to investigate if de novo synthesis of mycolic acids was inhibited in the M. bovis BCG strain overproducing MabA_T191D. Mycolic acid methyl esters were extracted, subjected to thin-layer chromatographic (TLC) analysis, and quantified using a PhosphorImager. As shown in Fig. 6D, induction of either MabA_T191A or MabA_T191D affected synthesis of both α-mycolates (left panel) and keto-mycolates (right panel) in M. bovis BCG, compared with the control strain expressing MabA_WT. Typically, a 25–40% reduction in de novo synthesis of mycolic acids was observed following induction with acetamide and [14C]acetate labeling. These results strongly suggest that overexpression of the phosphomimetic T191D impaired de novo biosynthesis of mycolic acids in mycobacteria.
DISCUSSION
M. tuberculosis has a complex lifestyle comprising different developmental stages matched to different environments. Its success results from its remarkable capacity to survive within the infected host, where it can persist in a non-replicating state for several decades, which is linked to the presence of its unique cell envelope. Paradoxically, whereas the structures, biological functions, and biosynthetic pathways of the major cell wall components have been extensively studied over recent decades, very little is known regarding the capacity of the bacterium to modulate and adapt expression of cell wall components in response to environmental changes. Nevertheless, among the few mycobacterial kinase substrates identified and investigated for their physiological roles, several were found to be connected to cell shape/division and cell envelope biosynthesis, supporting the notion that Ser/Thr phosphorylation plays an important role in the regulation of major cell envelope components (26, 32, 34, 46–49), as well as in cell shape/division (45, 50–55).
In our previous studies we demonstrated that all of the condensing enzymes of the M. tuberculosis FAS-II system (mtFabH, KasA, KasB, and Pks13) are STPK substrates (8, 26, 32). In particular, we showed that mtFabH is phosphorylated, in vitro and in vivo, on a single Thr residue (Thr45), which alters the accessibility of the substrate and thus negatively affects mtFabH enzymatic activity (32). We have also shown that mtFabD, catalyzing the malonyl-CoA:AcpM transacylase activity that provides the malonyl-AcpM substrates necessary for mycolic acid building, was also phosphorylated by several STPKs, although it is not known whether phosphorylation regulates malonyl-CoA:AcpM transacylase activity positively or negatively (26). Here, we provide a clear demonstration that the β-ketoacyl-ACP reductase of the FAS-II system is also regulated by mycobacterial STPKs. To our knowledge, this study represents the first demonstration of phosphorylation of a ketoacyl reductase in prokaryotes. MabA has been reported to be essential in mycobacteria (56), and this study demonstrates that overexpression of MabA_WT under a strong promoter was also associated to a reduced M. bovis BCG colony size compared with the control strain. It was previously reported that each monomer of MabA can interact with each condensing enzyme and that its high propensity to multimerization (MabA is a tetramer in solution) might maintain the macromolecular organization of FAS-II complexes (57). Thus, a large excess of MabA may alter FAS-II activity, resulting in a slower growth rate.
This study provides a fundamental insight into the molecular mechanism(s) controlling mycolic acid biosynthesis through STPKs-dependent phosphorylation of a FAS-II enzyme. First, KAR activity of the phosphomimetic MabA_T191D was markedly reduced compared with MabA_WT, which may be (at least partially) explained by its impaired binding to the NADPH cofactor necessary for catalysis to occur. Second, that this decreased KAR activity is relevant in vivo in mycobacteria was subsequently demonstrated by: (i) impaired mycobacterial growth of an M. bovis BCG strain transformed with a multicopy vector carrying the mabA_T191D allele and (ii) inhibition of de novo biosynthesis of mycolic acids following conditional expression of the phosphomimetic MabA_T191D in this strain. The fact that only a limited inhibition of de novo mycolic acid biosynthesis was observed in the MabA_T191D-overproducing strain can be inferred to the presence of endogenous MabA functionally operating, thus, preventing higher inhibition levels to be achieved. One can therefore hypothesize that complete inhibition, leading to mycolic acid cessation and cell death, is likely to occur in the context where endogenous MabA protein would be missing. Such a strain is, however, not available due to the essentiality of the mabA gene (56).
The fact that the MabA_T191A and MabA_T191D mutants behaved similarly in in vitro assays as well as in vivo points to Thr191 as a critical residue in catalysis, an observation that had not been made previously. The importance of this residue is also strengthened by its conservation in all mycobacterial MabA sequences analyzed and its surface-exposed location that enables direct contact with the kinase(s). Therefore, by adding a phosphate group to Thr191, M. tuberculosis has developed an original and powerful strategy by dampening the activity of MabA, subsequently leading to partial inhibition of mycolic acid production.
The fact that most (if not all) FAS-II enzymes are regulated by phosphorylation is a rather unexpected observation. Why would M. tuberculosis phosphorylate multiple enzymes that are known to interact together (57) and to act repetitively to ensure mycolic acid elongation? One explanation of the biological significance of this specific regulation could be that altering the activity of most of these enzymes may lead to a tightly regulated system to allow survival and/or adaptation under variable growth conditions. If phosphorylation of each individual FAS-II enzyme is accompanied by only a partial reduction of activity, one could assume that the cumulative effects could result in a complete cessation of mycolic acid production. However, it remains to be addressed whether the remaining FAS-II components, notably the β-hydroxyacyl-ACP dehydratase complex HadABC, as well as the enoyl-ACP reductase InhA, which represent potential regulatory checkpoints, are also possible STPK substrates. In M. tuberculosis, the inhA operon, consists of three genes, mabA, inhA, and hemZ, which are cotranscribed as a polycistronic message (19). Therefore, and by analogy with the gene products encoded by the cell division cluster carrying pbpA, Rv0019c, and Rv0020c, which are all phosphorylated by multiple kinases (47, 50, 58), one can hypothesize that InhA, like MabA, is regulated by phosphorylation. Similarly, mtFabD, KasA, and KasB, whose genes belong to the same operon, were all three found to be phosphorylated (26). Work is currently in progress to address whether the activity of these enzymes might be influenced by STPK-dependent regulatory mechanisms and to test the hypothesis that several or all members of a given biosynthetic gene cluster are specifically regulated by Ser/Thr phosphorylation.
An interesting further observation in the present study relies on the identification of three phosphoacceptors following in vitro phosphorylation, although Thr191 appeared to be the primary site of phosphorylation. Thr191 was the first site identified by mass spectrometric analysis and the two other sites (Thr21 and Thr114) were identified only after substitution of Thr191 by an Ala residue. In addition, Thr191 is part of the peptide identified in MabA phosphorylated in vivo. This could be, at least partly, explained by the fact that Thr191, but neither Thr21 nor Thr114, is accessible to the kinase, although one cannot rule out the possibility that phosphorylation of a primary site induces conformational changes, ultimately exposing and uncovering additional sites of phosphorylation. More importantly, the Thr191 → Ala, but neither the Thr21 → Ala nor the Thr114 → Ala replacements, were accompanied by a severe decrease of phosphorylation. A similar situation has been observed recently in M. tuberculosis GroEL1, for which two phosphoacceptors, Thr25 and Thr54, were originally identified by mass spectrometry (39). Whereas a significant decrease of the phosphorylation signal was observed with the GroEL_T25A mutant, phosphorylation of the GroEL_T54A mutant was not affected, suggesting that Thr25 and Thr54 were not equivalent with respect to phosphorylation by PknF, as Thr25 appears to be the primary phosphorylation site (39).
To our knowledge this is the first description of a molecular mechanism employed by M. tuberculosis to adjust/regulate mycolic acid biosynthesis in response to environmental cues. This study provides a framework for further investigations on a seemingly important functional linkage between STPKs and the FAS-II system. Future work is required to identify the extracellular signals leading to MabA phosphorylation so as to comprehend how M. tuberculosis senses its environment and mediates its response in a coordinated manner to regulate cell wall mycolic acid biosynthesis. This will be a very challenging task because recent findings demonstrate cross-talk between mycobacterial STPKs and their substrates (26, 32, 58, 59). It is anticipated that similar strategies involving STPK-dependent mechanisms are used by mycobacteria to regulate expression of other cell wall lipids/glycolipids to face the various signals encountered during the infection or latency processes.
Finally, the identification and validation of mycobacterial STPKs as novel drug targets for tuberculosis therapeutics is currently being explored, underpinning programs for screening and designing specific inhibitors against M. tuberculosis STPKs (28, 29, 59). The mycobacterial STPK family members, which are highly homologous between themselves, are sufficiently different from their human counterparts to represent attractive drug targets. Because mycolic acids are essential lipids of M. tuberculosis participating in multiple physiological and physiopathological processes, such as survival within the infected host, interfering with phosphorylation-dependent mechanisms regulating the activity of FAS-II enzymes represents an attractive therapeutic option.
Supplementary Material
Acknowledgments
We thank M. Becchi and A. Cornut (Institut de Biologie et Chimie des Protéines, Lyon) for excellent technical assistance in mass spectrometric analysis and Sir D. Hopwood for critical reading the manuscript.
This work was supported by National Research Agency Grants ANR-06-MIME-027-01 (to V. M. and L. K.) and ANR-JC07_203251 (to M. C.-G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1 and S2.
- FAS
- fatty acid synthase
- ACP
- acyl carrier protein
- STPK
- Ser/Thr protein kinase
- KAR
- ketoacyl-ACP reductase
- DTT
- dithiothreitol
- GST
- glutathione S-transferase
- WT
- wild-type
- TOF-MS
- time of flight-mass spectrometry.
REFERENCES
- 1.Frieden T. R., Sterling T. R., Munsiff S. S., Watt C. J., Dye C. (2003) Lancet 362, 887–899 [DOI] [PubMed] [Google Scholar]
- 2.Flynn J. L., Chan J. (2003) Curr. Opin. Immunol. 15, 450–455 [DOI] [PubMed] [Google Scholar]
- 3.Daffé M., Draper P. (1998) Adv. Microb. Physiol. 39, 131–203 [DOI] [PubMed] [Google Scholar]
- 4.Kremer L., Baulard A. R., Besra G. S. (2000) in Molecular Genetics of Mycobacteria (Hatfull G. F., Jacobs W. R., Jr. eds) pp. 173–190, ASM Press, Washington, DC [Google Scholar]
- 5.Takayama K., Wang C., Besra G. S. (2005) Clin. Microbiol. Rev. 18, 81–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ojha A. K., Baughn A. D., Sambandan D., Hsu T., Trivelli X., Guerardel Y., Alahari A., Kremer L., Jacobs W. R., Jr., Hatfull G. F. (2008) Mol. Microbiol. 69, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan P. J., Nikaido H. (1995) Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 8.Bhatt A., Molle V., Besra G. S., Jacobs W. R., Jr., Kremer L. (2007) Mol. Microbiol. 64, 1442–1454 [DOI] [PubMed] [Google Scholar]
- 9.Dubnau E., Chan J., Raynaud C., Mohan V. P., Lanéelle M. A., Yu K., Quémard A., Smith I., Daffé M. (2000) Mol. Microbiol. 36, 630–637 [DOI] [PubMed] [Google Scholar]
- 10.Glickman M. S., Cox J. S., Jacobs W. R., Jr. (2000) Mol. Cell 5, 717–727 [DOI] [PubMed] [Google Scholar]
- 11.Rao V., Gao F., Chen B., Jacobs W. R., Jr., Glickman M. S. (2006) J. Clin. Invest. 116, 1660–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dao D. N., Sweeney K., Hsu T., Gurcha S. S., Nascimento I. P., Roshevsky D., Besra G. S., Chan J., Porcelli S. A., Jacobs W. R. (2008) PLoS Pathog. 4, e1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatt A., Fujiwara N., Bhatt K., Gurcha S. S., Kremer L., Chen B., Chan J., Porcelli S. A., Kobayashi K., Besra G. S., Jacobs W. R., Jr. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5157–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimhony O., Vilchèze C., Jacobs W. R., Jr. (2004) J. Bacteriol. 186, 4051–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloch K. (1975) Methods Enzymol. 35, 84–90 [DOI] [PubMed] [Google Scholar]
- 16.Choi K. H., Kremer L., Besra G. S., Rock C. O. (2000) J. Biol. Chem. 275, 28201–28207 [DOI] [PubMed] [Google Scholar]
- 17.Scarsdale J. N., Kazanina G., He X., Reynolds K. A., Wright H. T. (2001) J. Biol. Chem. 276, 20516–20522 [DOI] [PubMed] [Google Scholar]
- 18.Brown A. K., Sridharan S., Kremer L., Lindenberg S., Dover L. G., Sacchettini J. C., Besra G. S. (2005) J. Biol. Chem. 280, 32539–32547 [DOI] [PubMed] [Google Scholar]
- 19.Banerjee A., Sugantino M., Sacchettini J. C., Jacobs W. R., Jr. (1998) Microbiology 144, 2697–2704 [DOI] [PubMed] [Google Scholar]
- 20.Marrakchi H., Ducasse S., Labesse G., Montrozier H., Margeat E., Emorine L., Charpentier X., Daffé M., Quémard A. (2002) Microbiology 148, 951–960 [DOI] [PubMed] [Google Scholar]
- 21.Brown A. K., Bhatt A., Singh A., Saparia E., Evans A. F., Besra G. S. (2007) Microbiology 153, 4166–4173 [DOI] [PubMed] [Google Scholar]
- 22.Sacco E., Covarrubias A. S., O'Hare H. M., Carroll P., Eynard N., Jones T. A., Parish T., Daffé M., Bäckbro K., Quémard A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilchèze C., Wang F., Arai M., Hazbón M. H., Colangeli R., Kremer L., Weisbrod T. R., Alland D., Sacchettini J. C., Jacobs W. R., Jr. (2006) Nat. Med. 12, 1027–1029 [DOI] [PubMed] [Google Scholar]
- 24.Schaeffer M. L., Agnihotri G., Volker C., Kallender H., Brennan P. J., Lonsdale J. T. (2001) J. Biol. Chem. 276, 47029–47037 [DOI] [PubMed] [Google Scholar]
- 25.Kremer L., Dover L. G., Carrère S., Nampoothiri K. M., Lesjean S., Brown A. K., Brennan P. J., Minnikin D. E., Locht C., Besra G. S. (2002) Biochem. J. 364, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molle V., Brown A. K., Besra G. S., Cozzone A. J., Kremer L. (2006) J. Biol. Chem. 281, 30094–30103 [DOI] [PubMed] [Google Scholar]
- 27.Stock J. B., Ninfa A. J., Stock A. M. (1989) Microbiol. Rev. 53, 450–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wehenkel A., Bellinzoni M., Graña M., Duran R., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cerveñansky C., Cole S. T., Alzari P. M. (2008) Biochim. Biophys. Acta 1784, 193–202 [DOI] [PubMed] [Google Scholar]
- 29.Walburger A., Koul A., Ferrari G., Nguyen L., Prescianotto-Baschong C., Huygen K., Klebl B., Thompson C., Bacher G., Pieters J. (2004) Science 304, 1800–1804 [DOI] [PubMed] [Google Scholar]
- 30.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 31.Av-Gay Y., Everett M. (2000) Trends Microbiol. 8, 238–244 [DOI] [PubMed] [Google Scholar]
- 32.Veyron-Churlet R., Molle V., Taylor R. C., Brown A. K., Besra G. S., Zanella-Cléon I., Fütterer K., Kremer L. (2009) J. Biol. Chem. 284, 6414–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canova M. J., Kremer L., Molle V. (2008) Plasmid 60, 149–153 [DOI] [PubMed] [Google Scholar]
- 34.Molle V., Kremer L., Girard-Blanc C., Besra G. S., Cozzone A. J., Prost J. F. (2003) Biochemistry 42, 15300–15309 [DOI] [PubMed] [Google Scholar]
- 35.Fiuza M., Canova M. J., Zanella-Cléon I., Becchi M., Cozzone A. J., Mateos L. M., Kremer L., Gil J. A., Molle V. (2008) J. Biol. Chem. 283, 18099–18112 [DOI] [PubMed] [Google Scholar]
- 36.Böhm G., Muhr R., Jaenicke R. (1992) Protein Eng. 5, 191–195 [DOI] [PubMed] [Google Scholar]
- 37.Daugelat S., Kowall J., Mattow J., Bumann D., Winter R., Hurwitz R., Kaufmann S. H. (2003) Microbes Infect. 5, 1082–1095 [DOI] [PubMed] [Google Scholar]
- 38.Kremer L., Dover L. G., Morbidoni H. R., Vilchèze C., Maughan W. N., Baulard A., Tu S. C., Honoré N., Deretic V., Sacchettini J. C., Locht C., Jacobs W. R., Jr., Besra G. S. (2003) J. Biol. Chem. 278, 20547–20554 [DOI] [PubMed] [Google Scholar]
- 39.Canova M. J., Kremer L., Molle V. (2009) J. Bacteriol. 191, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., Shafqat J., Nordling E., Kallberg Y., Persson B., Jörnvall H. (2003) Chem. Biol. Interact. 143–144, 247–253 [DOI] [PubMed] [Google Scholar]
- 41.Cohen-Gonsaud M., Ducasse S., Hoh F., Zerbib D., Labesse G., Quemard A. (2002) J. Mol. Biol. 320, 249–261 [DOI] [PubMed] [Google Scholar]
- 42.Oppermann U. C., Filling C., Jörnvall H. (2001) Chem. Biol. Interact. 130–132, 699–705 [DOI] [PubMed] [Google Scholar]
- 43.Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., Oppermann U. (2002) J. Biol. Chem. 277, 25677–25684 [DOI] [PubMed] [Google Scholar]
- 44.Cohen-Gonsaud M., Ducasse-Cabanot S., Quemard A., Labesse G. (2005) Proteins 60, 392–400 [DOI] [PubMed] [Google Scholar]
- 45.Kang C. M., Nyayapathy S., Lee J. Y., Suh J. W., Husson R. N. (2008) Microbiology 154, 725–735 [DOI] [PubMed] [Google Scholar]
- 46.Singh A., Gupta R., Vishwakarma R. A., Narayanan P. R., Paramasivan C. N., Ramanathan V. D., Tyagi A. K. (2005) J. Bacteriol. 187, 4173–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta M., Sajid A., Arora G., Tandon V., Singh Y. (2009) J. Biol. Chem. 284, 34723–34734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar P., Kumar D., Parikh A., Rananaware D., Gupta M., Singh Y., Nandicoori V. K. (2009) J. Biol. Chem. 284, 11090–11099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma K., Gupta M., Pathak M., Gupta N., Koul A., Sarangi S., Baweja R., Singh Y. (2006) J. Bacteriol. 188, 2936–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dasgupta A., Datta P., Kundu M., Basu J. (2006) Microbiology 152, 493–504 [DOI] [PubMed] [Google Scholar]
- 51.Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., Husson R. N. (2005) Genes Dev. 19, 1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parikh A., Verma S. K., Khan S., Prakash B., Nandicoori V. K. (2009) J. Mol. Biol. 386, 451–464 [DOI] [PubMed] [Google Scholar]
- 53.Thakur M., Chakraborti P. K. (2006) J. Biol. Chem. 281, 40107–40113 [DOI] [PubMed] [Google Scholar]
- 54.Thakur M., Chakraborti P. K. (2008) Biochem. J. 415, 27–33 [DOI] [PubMed] [Google Scholar]
- 55.Molle V., Kremer L. (2010) Mol. Microbiol. 75, 1064–1077 [DOI] [PubMed] [Google Scholar]
- 56.Parish T., Roberts G., Laval F., Schaeffer M., Daffé M., Duncan K. (2007) J. Bacteriol. 189, 3721–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veyron-Churlet R., Guerrini O., Mourey L., Daffé M., Zerbib D. (2004) Mol. Microbiol. 54, 1161–1172 [DOI] [PubMed] [Google Scholar]
- 58.Grundner C., Gay L. M., Alber T. (2005) Protein Sci. 14, 1918–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molle V., Reynolds R. C., Alderwick L. J., Besra G. S., Cozzone A. J., Fütterer K., Kremer L. (2008) Biochem. J. 410, 309–317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.