Abstract
The minichromosome maintenance (Mcm) 2–7 complex is the replicative helicase in eukaryotic species, and it plays essential roles in the initiation and elongation phases of DNA replication. During late M and early G1, the Mcm2–7 complex is loaded onto chromatin to form prereplicative complex in a Cdt1-dependent manner. However, the detailed molecular mechanism of this loading process is still elusive. In this study, we demonstrate that the previously uncharacterized C-terminal domain of human Mcm6 is the Cdt1 binding domain (CBD) and present its high resolution NMR structure. The structure of CBD exhibits a typical “winged helix” fold that is generally involved in protein-nucleic acid interaction. Nevertheless, the CBD failed to interact with DNA in our studies, indicating that it is specific for protein-protein interaction. The CBD-Cdt1 interaction involves the helix-turn-helix motif of CBD. The results reported here provide insight into the molecular mechanism of Mcm2–7 chromatin loading and prereplicative complex assembly.
Keywords: DNA Replication, NMR, Protein Assembly, Protein Structure, Protein-Protein Interactions, Cdt1, Mcm6, Pre-RC
Introduction
For the maintenance of genetic integrity, initiation of eukaryotic DNA replication is tightly controlled to ensure that DNA replication occurs exactly once in each cell cycle. Replication begins by the formation of pre-RCs4 on replication origins during late M and G1 phases (1, 2). For pre-RC assembly, the six-subunit origin recognition complex first binds replication origin on newly synthesized chromatin. The origin recognition complex serves as an origin marker and recruits the initiation factors Noc3p, Cdc6, and Cdt1 to origins for the chromatin loading of the heterohexameric Mcm2–7 complex (3–5). Once the Mcm complex is loaded onto chromatin and pre-RC is formed, the cell is licensed for DNA replication, awaiting additional signals for the activation of the licensed origins (6).
The Mcm2–7 complex was first identified as a set of genes required for minichromosome maintenance in budding yeast (7). The Mcm proteins are members of the highly diversified AAA+ (ATPases associated with a variety of cellular activities) protein family (8). They are conserved in eukaryotes and play essential roles in DNA replication initiation and elongation (9–11). Previous studies suggested that the majority of Mcm2–7 proteins form stoichiometric heterohexamers in vivo (12). The Mcm2–7 complex was shown to possess helicase activity in vitro and in vivo (13–15). All eukaryotic Mcm proteins contain a conserved central nucleotide binding domain, an AAA+ ATPase domain (MCM domain), and the N- and C-terminal domains that are unique to each of the Mcm proteins (12).
Cdt1 is a critical member of pre-RC, and its main function is to load Mcm2–7 helicase onto chromatin to license the DNA for replication in the subsequent S phase (16). Overexpression of Cdt1 alone in many types of mammalian cells is sufficient to induce rereplication (17–19). Previous studies have broadly defined three functional domains of Cdt1: a domain in the middle of the molecule containing the major Geminin interaction site; an N-terminal domain, which is required for ubiquitin-mediated proteolysis and contains a second interaction site for Geminin; and a C-terminal domain, which is required for association with Mcm proteins (12). Interactions between Cdt1 and individual members of the Mcm2–7 complex have been examined, and Cdt1 was found to interact with Mcm2 and Mcm6 (16, 21–23). The existence of a stoichiometric complex between Cdt1 and Mcm2–7 was recently reported, which is consistent with earlier biochemical and genetic investigations (24). However, the detailed molecular mechanism underlying the chromatin loading of the Mcm2–7 complex through Cdt1 remains elusive.
In this report, we demonstrate that the conserved C-terminal domain of human Mcm6 directly interacts with the C-terminal region of Cdt1. We also present the solution structure of the Cdt1 binding domain (CBD) of human Mcm6 determined by high resolution NMR spectroscopy. The structure of CBD adopts a typical “winged helix” fold consisting of three α-helices and two β-strands. We also mapped the residues of CBD involved in Cdt1 binding by NMR titration and verified the results by mutagenesis studies. Biochemical characterization and structure determination of the Cdt1 binding domain of Mcm6 provide a basis for understanding the molecular mechanism of Mcm2–7 chromatin loading and pre-RC assembly.
EXPERIMENTAL PROCEDURES
Expression and Purification of Mcm6-CBD and Cdt1-MBD
The coding sequence of human CBD of Mcm6, corresponding to residues 708–821 (C721S), was inserted into the expression vector pET-28a(+)(Novagen). The protein was expressed in Escherichia coli BL21(DE3) at 37 °C for 5 h. Cell lysates were subject to affinity purification with the nickel-nitrilotriacetic acid resin (Qiagen) followed by the cleavage of the hexahistidine tag with thrombin. The resulting product (residues 708–821 of Homo sapiens Mcm6 with a 4-residue N-terminal extension remaining from the thrombin cleavage site) was further purified on a Mono Q HR 10/10 column (GE Healthcare). For the NMR studies, CBD samples uniformly labeled with 15N and/or 13C isotopes were expressed and purified as described above, except that the cells were grown in M9 medium containing [15N]ammonium chloride and d-glucose (or d-[13C6]glucose) (Cambridge Isotopes), respectively. Residual dipolar couplings were measured on a sample that was weakly aligned in the magnet by adding 12 mg/ml filamentous phage Pf1 (ASLA BIOTECH).
The coding sequence of human Mcm6 binding domain (MBD), corresponding to residues 410–445 of Cdt1, was inserted into the pET-32a(+) (Novagen)-derived expression vector, in which the thioredoxin tag was replaced by the 56-residue B1 immunoglobulin binding domain of Streptococcal protein G (GB1) to increase the solubility and stability of the fusion protein. The N-terminal GB1-His6-fused Cdt1-MBD was expressed in E. coli BL21(DE3) at 16 °C for 16 h. Cell lysates were first purified with nickel-nitrilotriacetic acid resin (Qiagen) followed by gel filtration purification with Superdex 75 10/300 GL (GE Healthcare).
GST Pulldown Assay
The prewashed glutathione-Sepharose 4B beads (GE Healthcare) were incubated with lysates of GST or GST-Mcm6-(708–821) at 4 °C for 5 h. The beads were washed three times, and the lysates of Cdt1 fragments were incubated with GST-Mcm6-(708–821) or GST-coupled beads at 4 °C for 4 h. Then, the beads were washed three times and eluted in 50 mm Tris (pH 7.9) containing 10 mm glutathione. One-fourth of the total elute was assayed on a 12% SDS-PAGE and visualized by Coomassie Blue staining.
NMR Structure Determination
NMR spectra were acquired at 37 °C on 750- and 500-MHz Varian NMR spectrometers. All spectra were processed using NMRPipe (25–26) and analyzed using Sparky 3 (47). The 1H, 15N, and 13C chemical shifts were assigned by standard methods (27–29). Distance restraints were obtained from 15N- or 13C-edited three-dimensional NOE spectroscopy spectra and two-dimensional NOE spectroscopy. Chemical shift perturbations (Δδ = [(ΔδH)2 + (0.16 ΔδN)2]½, in parts per million) were monitored from two-dimensional 1H-15N-HSQC spectra. NOE assignment and structure calculations were performed using the CANDID (30) module of the program CYANA2.1 (31). Unassigned resonances were represented by appropriate proxy residues (32). Dihedral restraints were generated from TALOS (33). Water refinement was performed by using CNS (34) and following the RECOORD protocol (35). Structures were validated using WHATCHECK (36) and PROCHECK (37) and analyzed by MOLMOL (38) (supplemental Table S1). The solvent accessibility was calculated by NACCESS (39). All of the figures representing the structures were generated by PyMOL. For further details, see the supplemental Experimental Procedures.
RESULTS
The Conserved C-terminal Domain of Human Mcm6 Binds with Cdt1
We conducted an extensive search to identify the Cdt1 binding domains in human Mcm2–7 complex by the yeast two-hybrid method and found that the interaction between human Mcm6 and Cdt1 is much stronger than that between Mcm2 and Cdt1, which is consistent with mouse study (21). To identify the binding domain of human Cdt1 and that of human Mcm6, different fragments of Mcm6 and Cdt1 were used in the yeast two-hybrid analysis. Our results showed that the C terminus of Mcm6 spanning residues 708–821 (the CBD) could bind with residues 392–471 of Cdt1. The direct physical interaction between these two domains was confirmed by pulldown assays in which GST-fused Mcm6-CBD and thioredoxin-fused Cdt1-(392–471) were used (Fig. 1A). The physical and functional interactions between binding domains of Cdt1 and Mcm6 were also verified by co-immunoprecipitation and dominant negative assays in human cells.5
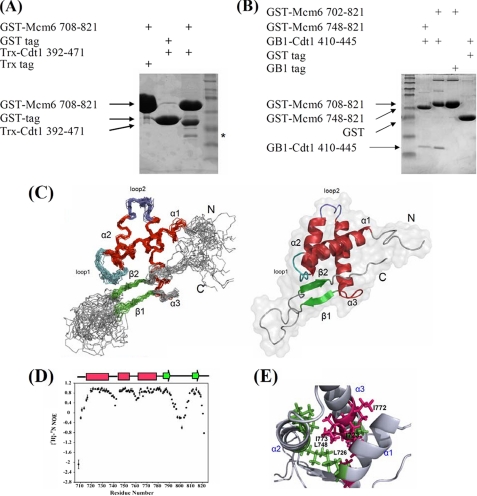
FIGURE 1.
In vitro interaction of Mcm6-CBD with Cdt1-MBD and solution structure of CBD of human Mcm6. A, the C terminus of Mcm6 interacts with the C-terminal region of Cdt1. Thioredoxin (Trx)-Cdt1-(392–471) was pulled down by GST-Mcm6-(708–821) and visualized by Coomassie Blue staining. The asterisk indicates the Cdt1 degradation product. B, the minimal region of Cdt1 required for Mcm6 interaction. GB1-Cdt1-(410–445) was pulled down by GST-Mcm6-(748–821) or GST-Mcm6-(708–821) and visualized by Coomassie Blue staining. C, the left panel shows the backbone superposition of the 20 lowest energy NMR structures of Mcm6-CBD. Secondary structural elements are indicated by color coding: α-helices (red), β-strands (green), and loops (cyan, gray). N-terminal and C-terminal ends are indicated as N and C. The right panel shows a ribbon representation of the same structure of CBD using the coordinates of the lowest energy structure. D, the {1H}-15N heteronuclear NOEs of each amino acid are depicted. Secondary structural elements are indicated with color coding as in B on the top of the figure. E, the structure of the hydrophobic cage of CBD is shown with the side chains of important residues such as Leu-726 and Leu-748 presented in green and residues Ile-723, Ile-772, and Ile-773 shown in red.
As we observed that the Cdt1-(392–471) was unstable and unsuitable for structural study, several truncation mutations of this Cdt1 fragment fused with GB1 tag (40, 41) were constructed in an attempt to make them suitable for NMR analysis. We found that the region of Cdt1 spanning residues 410–445 is the core MBD, which is sufficient to interact with CBD (Fig. 1B). The interaction was also confirmed through a yeast two-hybrid assay. A recent investigation reported that mouse Cdt1 with alanine substitution of two lysines, Lys-441 and Lys-445 (corresponding to human Cdt1 Lys-429 and Lys-433), failed to form a complex with Mcm2–7 (23) and also confirms the importance of the 410–445 region of human Cdt1 in the Mcm6-Cdt1 interaction. Taken together, our data demonstrate that the conserved C-terminal domain of human Mcm6 (residues 707–821) physically interacts with Cdt1 (residues 410–445), the MBD.
Three-dimensional Structure of the Cdt1 Binding Domain of Mcm6
The three-dimensional structure of the CBD of human Mcm6 (residues 708–821) was determined by the standard multidimensional heteronuclear NMR spectroscopy and well defined (27–29) (supplemental Table S1; see also “Experimental Procedures”). A superimposed ensemble of the 20 water-refined NMR structures and the ribbon plot of the lowest energy structure are shown in Fig. 1C. The Ramachandran plot of the superimposed ensemble showed that 81.7% of the meaningful residues fell into the most favorable regions of torsion angle space, and 16.5% of the residues were in other allowed and generously allowed regions. The remaining 1.8% of the residues, all of which were in the disordered terminal and loop regions of the protein, fell in the so-called disallowed region of the Ramachandran plot. The structure of CBD adopts a winged helix-turn-helix fold, which is a typical DNA binding motif and may also participate in protein-protein interaction (42). Residues 718–736 (α1), 745–756 (α2), and 763–781 (α3) constitute the canonical three-helix bundle, which is packed against two short antiparallel β-strands denoted as β1 (residues 787–790) and β2 (residues 810–813). The N terminus (residues 708–715), the region between β1 and β2 (residues 791–809), and the C terminus (residues 814–821) are flexible based on heteronuclear NOE data and are less well defined in the NMR ensemble due to high internal mobility (Fig. 1D). By contrast, loop 1 (residues 737–744) and loop 2 (residues 757–762) appear to be appreciably rigid because the three-helix bundle is tied with the β-sheet through loop 1. The rigidity is confirmed by the {1H}-15N heteronuclear NOE results (Fig. 1D) and is consistent with the calculated Random Coil Index (43, 44). The helix α1 and the helix-turn-helix (HTH) motif (α2-loop 2-α3) establish the stable helix bundle by creating a hydrophobic core through the interactions between conserved residues Ile-726, Leu-748, and Ile-773 (Fig. 1E). A network of NOEs among the core residues in the three helices was observed (residues Glu-740–Val-812, Ser-741–Ile-788, and Ala-742–Val-812), revealing the formation of a compact architecture. The C terminus of CBD, including the antiparallel stranded β-sheet and the conserved Tyr-817, contributes significantly to the fold of CBD as deletion or substitution of these residues led to precipitation in solution or no expression of CBD in E. coli.
To validate the three-dimensional structural fold of CBD in solution, we measured 1HN-15N residual dipolar couplings (RDCs) (46) with CBD protein weakly aligned in Pf1. The 52 experimental RDCs (not used in the structure calculations) of the well resolved resonances of the amino acids in the secondary structural elements agreed nicely with the values back-calculated from the structure that was determined based on the NOEs and the dihedral angle constraints, with the quality factor equal to 0.32. The surface residues and hydrogen-bond network exhibited in the NMR structures were confirmed from a paramagnetic mapping experiment5 and hydrogen exchange data obtained from solvent-exposed amides-HSQC experiment (45). Taken together, data from the backbone dynamics study, RDC, and amide hydrogen exchange measurements demonstrate that the first few turns of the helix α1 are not as tightly packed as the remaining secondary structural elements in CBD.
Mapping Cdt1 Binding Sites of the CBD Domain
To identify the binding sites, we conducted an NMR titration experiment on CBD with GB1-tagged MBD. Remarkably, the addition of MBD induced significant chemical shift perturbations. As shown in Fig. 2A, signals that display large chemical shift changes (Δδ > 0.12 ppm) correspond to amino acids in the helix-turn-helix region formed by helix α2 (residues Trp-751, Lys-754, Glu-755, and Ile-756), loop 2 (residues Glu-757 and Ser-758), and helix α3 (residues Glu-764, Glu-765, and Arg-771) (Fig. 2, B–D). This observation is consistent with the pulldown results shown in Fig. 1B. Residues Leu-753, Glu-763, and Leu-766 completely disappeared due to line broadening after the CBD was titrated by MBD at a ratio of ∼1:2. The solvent accessibility calculated from NACCESS 2.1 and the amide exchange measurements from the SEA-HSQC experiment (45) indicate that either the main chain or the side chain of residues Lys-754, Glu-755, Glu-757, Ser-758, Glu-763, Glu-764, Glu-765, and Leu-766 is found to be on the surface of CBD (Fig. 2, C and D). These solvent-exposed residues form an MBD binding site. All available chemical shift perturbations were quantitatively analyzed to derive the dissociation constant, Kd, which is about 10−4 m.
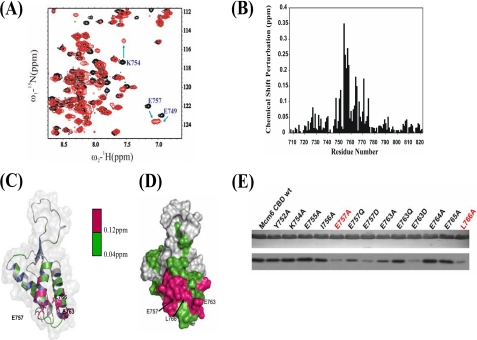
FIGURE 2.
NMR and mutation studies of the interaction between Mcm6-CBD and Cdt1-MBD. A, a part of the overlaid 1H-15N-HSQC spectra of 15N-labeled CBD in free form (black) with that titrated with non-labeled GB1-Cdt1 410–445 (red) at a molar ratio of 1:4. Residues that undergo significant changes in chemical shifts upon formation of the complex with Cdt1-MBD are highlighted by arrows and labeled with peak assignments. B, chemical shift differences between the free-form and MBD saturated CBDs. C, chemical shift perturbations in the presence of MBD are colored onto the structure of CBD in ribbon representation. Residues with chemical shift perturbations ranging from 0.04 to 0.12 ppm are colored in green, whereas residues with chemical shift perturbations larger than 0.12 ppm are shown in red. Key residues, Glu-757, Glu-763, and Leu-766, are shown in the stick model. D, surface presentation of CBD with residues showing chemical shift perturbations as described in B. E, the effect of a single-point mutation in Mcm6-CBD on in vitro Cdt1-MBD interaction. The upper panel indicates a relatively equal amount of GST-Mcm6-CBD mutant proteins used in the GST pulldown assay. The immunoblot with anti-His antibodies shows the binding of GB1-His-MBD to the wild-type (wt) or mutant CBD (lower panel). Mutants of CBD highlighted in red exhibited a significant reduction in binding to Cdt1-MBD.
Mutagenesis Studies of the Interaction between MBD and CBD
The sequence alignment reveals that the CBD contains a series of acidic residues that form a negatively charged region encompassing α2-loop 2-α3, where the residues experienced major chemical shift changes in the NMR titration experiments (Fig. 2, A–D). A series of point mutants in CBD were constructed to evaluate the contributions of these residues to the CBD-MBD interaction. These point mutants, Y752E, K754A, E755A, I756A, E757A, E757Q, E757D, E763A, E763Q, E763D, E764A, E765A, and L766A, were expressed and purified to homogeneity. Pulldown assay results (Fig. 2E) indicated that the highly conserved residues, Glu-757, Glu-763, and Leu-766, make major contributions to the interaction between MBD and CBD. The electrostatic surface potential of CBD shows that these residues may bind MBD through charge-charge interactions and hydrophobic interactions. To assess whether point mutations affect the structure of CBD, we recorded two-dimensional 1H-15N-HSQC spectra of mutants E757A, E763A, and L766A. The spectra showed similar patterns and line widths, indicating that the structural folds were not disrupted by mutations. These observations are consistent with the fact that these residues are on the surface of the protein.
To further confirm the importance of residues Glu-757 and Leu-766 in the Mcm6-Cdt1 interaction, we co-transfected HA-tagged wild-type or alanine substitution mutants of full-length Mcm6 and full-length Cdt1 into 293T cells. As shown in supplemental Fig. S1, HA-wtMcm6 and Myc-Cdt1 co-localized in the nucleus of 293T cells, whereas HA-Mcm6 mutants, such as E757A and L766A, failed to co-localize with Myc-Cdt1, although they still remained in the nucleus. As mutations on Glu-757 and Leu-766 disrupted Mcm6-Cdt1 co-localization without influencing the nuclear import of the Mcm6 mutants, the results indicate that these 2 residues were specific and pivotal for the Mcm6-Cdt1 interaction.
DISCUSSION
In all eukaryotic cells, a highly conserved regulatory mechanism ensures that genomic DNA replicates exactly once in each cell cycle. Cdt1-mediated loading of the Mcm2–7 onto chromatin is a critical and accurately regulated event during the initiation of replication. The Mcm2–7 complex chromatin loading might be achieved through direct interaction between Cdt1 and subunits of Mcm2–7 complex. The newly reported results showed that single heptamers of Cdt1-Mcm2–7 are loaded onto DNA cooperatively (24). However, the detailed domains of Cdt1 and Mcm2–7 involved in the cooperative binding are missing. To fill this missing link, we have applied the yeast two-hybrid method to map the interacting domains between human Cdt1 and human Mcm2–7. We have characterized the C terminus of Mcm6 as the CBD and determined the high resolution NMR structure of CBD. We also identified and verified the binding surface and key residues of Mcm6-CBD involved in the Cdt1 interaction.
Structural studies showed that the CBD contains a winged helix fold, which consists of an HTH motif and a wing (β-loop-β) motif. The core structure of the CBD spans residues Ser-718–His-782. The NMR titration experiment showed that the Cdt1 binding sites are located in the HTH region (helix α2-loop 2-helix α3). The GST pulldown assay with CBD mutants confirmed that Glu-757 in conjugation with Glu-763 and Glu-766 form a small cluster of residues in α2, loop 2, and α3 of CBD whose single-point mutation disrupts the interaction with Cdt1-MBD.
A DALI search using CBD as input rendered more than 500 structurally related proteins (z-score > 2.0). Most closely related protein structures are those having nucleic acid binding activity, including the transcription factor E2F-4 (1CF7; z-score = 6.0), the penicillinase repressor (1XSD; z-score = 4.2), and the putative AphA-like transcription factor (2RKH; z-score = 4.8). The analysis of structural homologs suggests that CBD could potentially bind with double-stranded-DNA through the short sequence (KRIIEK) in the third helix. We therefore examined whether or not CBD binds DNA with the use of electrophoretic mobility shift assays and an NMR titration experiment using 16-bp double-stranded-DNA and single-stranded-DNA with low and high salt concentrations. However, we failed to observe any shift change in either experiment (data not shown). This may be explained based on the analysis of surface charge distributions of CBD, E2F-4, and penicillinase repressor. The surface charge distribution of CBD does not have the characteristic positively charged patches required for DNA binding (42). The third helix (α3) of CBD is less positively charged than the DNA binding regions of E2F-4 and penicillinase repressor (supplemental Fig. S2), probably making its binding with DNA impossible.
To summarize, we have identified and characterized the Cdt1-interacting domain in the subunits of the human Mcm2–7 complex and the Mcm6 binding domain of Cdt1. We also determined the high resolution solution structure of the Mcm6-CBD. The results here represent an important progress toward the understanding of the molecular mechanism of chromatin loading of the Mcm2–7 complex by Cdt1, which is a critical step during eukaryotic DNA replication initiation. Structural and functional studies on the CBD-MBD complex currently in progress will provide more details of the molecular mechanism for Cdt1-mediated Mcm2–7 complex chromatin loading.
Supplementary Material
Acknowledgment
We thank Feng Rui for helping to set up the NMR experiments.
This work was supported by a grant from the Hong Kong Research Grants Council (Grant HKUST6436/06M to G. Z. and C. L.).
The atomic coordinates and structure factors (code 2KLQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Table S1, and Figs. S1 and S2.
Z. Wei, C. Liu, X. Wu, N. Xu, B. Zhou, C. Liang, and G. Zhu, unpublished data.
- pre-RC
- prereplicative complex
- CBD
- Cdt1 binding domain of MCM6
- Mcm
- minichromosome maintenance
- MBD
- Mcm6 binding domain of Cdt1
- HTH
- helix-turn-helix
- GST
- glutathione S-transferase
- RDC
- residual dipolar coupling
- HA
- hemagglutinin
- NOE
- nuclear Overhauser effect
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1.Bell S. P., Dutta A. (2002) Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 2.Diffley J. F. (2004) Curr. Biol. 14, R778–R786 [DOI] [PubMed] [Google Scholar]
- 3.Maiorano D., Moreau J., Méchali M. (2000) Nature 404, 622–625 [DOI] [PubMed] [Google Scholar]
- 4.Liang C., Weinreich M., Stillman B. (1995) Cell 81, 667–676 [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Yu Z., Fu X., Liang C. (2002) Cell 109, 849–860 [DOI] [PubMed] [Google Scholar]
- 6.Masai H., Sato N., Takeda T., Arai K. (1999) Front. Biosci. 4, D834–D840 [DOI] [PubMed] [Google Scholar]
- 7.Maine G. T., Sinha P., Tye B. K. (1984) Genetics 106, 365–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999) Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 9.Chong J. P., Mahbubani H. M., Khoo C. Y., Blow J. J. (1995) Nature 375, 418–421 [DOI] [PubMed] [Google Scholar]
- 10.Aparicio O. M., Weinstein D. M., Bell S. P. (1997) Cell 91, 59–69 [DOI] [PubMed] [Google Scholar]
- 11.Labib K., Tercero J. A., Diffley J. F. (2000) Science 288, 1643–1647 [DOI] [PubMed] [Google Scholar]
- 12.Forsburg S. L. (2004) Microbiol. Mol. Biol. Rev. 68, 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey M. J., Indiani C., O'Donnell M. (2003) J. Biol. Chem. 278, 4491–4499 [DOI] [PubMed] [Google Scholar]
- 14.Schwacha A., Bell S. P. (2001) Mol. Cell 8, 1093–1104 [DOI] [PubMed] [Google Scholar]
- 15.Bochman M. L., Schwacha A. (2008) Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]
- 16.Ferenbach A., Li A., Brito-Martins M., Blow J. J. (2005) Nucleic Acids Res. 33, 316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arentson E., Faloon P., Seo J., Moon E., Studts J. M., Fremont D. H., Choi K. (2002) Oncogene 21, 1150–1158 [DOI] [PubMed] [Google Scholar]
- 18.Tatsumi Y., Sugimoto N., Yugawa T., Narisawa-Saito M., Kiyono T., Fujita M. (2006) J. Cell Sci. 119, 3128–3140 [DOI] [PubMed] [Google Scholar]
- 19.Seo J., Chung Y. S., Sharma G. G., Moon E., Burack W. R., Pandita T. K., Choi K. (2005) Oncogene 24, 8176–8186 [DOI] [PubMed] [Google Scholar]
- 20.Deleted in proof
- 21.Yanagi K., Mizuno T., You Z., Hanaoka F. (2002) J. Biol. Chem. 277, 40871–40880 [DOI] [PubMed] [Google Scholar]
- 22.Teer J. K., Dutta A. (2008) J. Biol. Chem. 283, 6817–6825 [DOI] [PubMed] [Google Scholar]
- 23.You Z., Masai H. (2008) J. Biol. Chem. 283, 24469–24477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., Diffley J. F. (2009) Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 26.Zhu G., Bax A. (1992) J. Magn. Reson. 100, 202–207 [Google Scholar]
- 27.Bax A., Grzesiek S. (1993) Acc. Chem. Res. 26, 131–138 [Google Scholar]
- 28.Sattler M., Schleucher J., Griesinger C. (1999) Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 29.Clore G. M., Gronenborn A. M. (1998) Trends Biotechnol. 16, 22–34 [DOI] [PubMed] [Google Scholar]
- 30.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 31.Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 32.AB E., Pugh D. J., Kaptein R., Boelens R., Bonvin A. M. (2006) J. Am. Chem. Soc. 128, 7566–7571 [DOI] [PubMed] [Google Scholar]
- 33.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 34.Brunger A. T., Adams P. D., Clore G. M., Delano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (2001) Crystallography and NMR System (CNS), Version 1.1, Yale University, New Haven, CT: [DOI] [PubMed] [Google Scholar]
- 35.Nederveen A. J., Doreleijers J. F., Vranken W., Miller Z., Spronk C. A., Nabuurs S. B., Güntert P., Livny M., Markley J. L., Nilges M., Ulrich E. L., Kaptein R., Bonvin A. M. (2005) Proteins 59, 662–672 [DOI] [PubMed] [Google Scholar]
- 36.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 38.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graphics 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 39.Hubbard S. J., Thornton J. M. (1993) NACCESS computer program, Department of Biochemistry and Molecular Biology, University College London, UK [Google Scholar]
- 40.Gronenborn A. M., Filpula D. R., Essig N. Z., Achari A., Whitlow M., Wingfield P. T., Clore G. M. (1991) Science 253, 657–661 [DOI] [PubMed] [Google Scholar]
- 41.Zhou P., Lugovskoy A. A., Wagner G. (2001) J. Biomol. NMR 20, 11–14 [DOI] [PubMed] [Google Scholar]
- 42.Gajiwala K. S., Burley S. K. (2000) Curr. Opin. Struct. Biol. 10, 110–116 [DOI] [PubMed] [Google Scholar]
- 43.Berjanskii M. V., Wishart D. S. (2005) J. Am. Chem. Soc. 127, 14970–14971 [DOI] [PubMed] [Google Scholar]
- 44.Wishart D. S., Sykes B. D. (1994) J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 45.Lin D., Sze K. H., Cui Y., Zhu G. (2002) J. Biomol. NMR 23, 317–322 [DOI] [PubMed] [Google Scholar]
- 46.Tjandra N., Bax A. (1997) Science 278, 1111–1114 [DOI] [PubMed] [Google Scholar]
- 47.Goddard T. D., Kneller D. G. (2007) Sparky 3, University of California, San Francisco, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.