Abstract
The ape2 gene encoding a hypothetical O-acetylpeptidoglycan esterase was amplified from genomic DNA of Neisseria gonorrhoeae FA1090 and cloned to encode either the full-length protein or a truncated version lacking its hypothetical signal sequence. Expression trials revealed that production of the full-length version possessing either an N-terminal or C-terminal His6 tag was toxic to Escherichia coli transformants and that the host rapidly degraded the small amount of protein that was produced. An N-terminally truncated protein could be produced in sufficient yields for purification only if it possessed an N-terminal His6 tag. This form of the protein was isolated and purified to apparent homogeneity, and its enzymatic properties were characterized. Whereas the protein could bind to insoluble peptidoglycan, it did not function as an esterase. Phenotypic characterization of E. coli transformants producing various forms of the protein revealed that it functions instead to O-acetylate peptidoglycan within the periplasm, and it was thus renamed peptidoglycan O-acetyltransferase B. This activity was found to be dependent upon a second protein, which functions to translocate acetate from the cytoplasm to the periplasm, demonstrating that the O-acetylation of peptidoglycan in N. gonorrhoeae, and other Gram-negative bacteria, requires a two component system.
Keywords: Bacteria, Bacterial Metabolism, Carbohydrate Biosynthesis, Cell Surface Enzymes, Cell Wall, Acetylesterase, Acetyltransferase, Peptidoglycan
Introduction
The bacterial cell wall heteropolymer peptidoglycan (PG)2 is comprised of alternating N-acetylglucosaminyl-β-1,4-N-acetylmuramoyl residues with attached stem peptides. The existence of the peptides permits interpeptide cross-linking between neighboring strands of PG to provide a continuous three-dimensional macromolecule, the PG sacculus. This sacculus completely surrounds the bacterial cell over its cytoplasmic membrane to withstand the internal turgor pressure of the cell and thereby providing structural rigidity and shape to the cells. Given the essential role in the vitality of bacterial cells, the PG sacculus is the target of lytic enzymes such as lysozyme (muramidases), which are produced and released by eukaryotic hosts as the first line of defense against invasion. As found with the xylans of plant cell walls (reviewed in Ref. 1), the decoration of PG with simple aglycon moieties such as acetate provides protection from this lytic action.
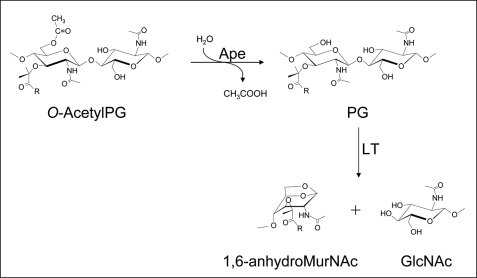
The O-acetylation of PG occurs specifically at the C6 hydroxyl group of muramoyl residues (Fig. 1) in many bacteria, both Gram-positive and Gram-negative, including a number of important human pathogens, such as Staphylococcus aureus, species of Campylobacter, Helicobacter, Neisseria, and Bacillus, including B. anthracis (2–4). The extent of this modification varies with species and strain, but ranges between 20 and 70% (relative to muramoyl residues). O-AcetylPG evades degradation by lysozymes in a concentration-dependent manner (5–9) through steric hindrance that precludes productive binding of PG in the enzyme binding cleft (5). In addition to protecting cells from lysis, the persistence of O-acetylated high molecular weight PG fragments within the mammalian host has serious pathobiological consequences, such as complement activation, pyrogenicity, somnogenesis, and arthrogenicity (reviewed in Ref. 2,3). The O-acetylation of PG also blocks the function of autolysins (10), endogenous enzymes involved in the biosynthesis and turnover of PG, insertion of appendages, and secretion systems (reviewed in Refs. 11, 12). This applies to the major class of autolysin involved with these processes, the lytic transglycosylases. These enzymes act on PG at the same site as lysozyme (viz. between MurNAc and GlcNAc residues) but instead of being hydrolases, the lytic transglycosylases cleave the β-1,4 linkage with the concomitant formation of 1,6-anhydormuramoyl residues (Fig. 1) (13). Thus, a free, unmodified C6 hydroxyl group on muramoyl residues is a strict requirement for lytic transglycosylase activity.
FIGURE 1.
Function of Ape and lytic transglycosylase (LT) on PG.
Despite its significance, very little is known about the process of PG O-acetylation, especially with Gram-negative bacteria (2, 3). Early studies with both Proteus mirabilis and Neisseria gonorrhoeae indicated O-acetylation to be a maturation event that closely follows after the incorporation and cross-linking of new strands into the PG sacculus (14 and references therein). This would require that a source of acetate from within the cytoplasm be transported across the cytoplasmic membrane prior to being attached to the newly ligated muramoyl residues in PG. An O-acetyltransferase (OatA) that serves these functions has been discovered in some Gram-positive bacteria (15, 16), but neither a homolog nor a paralog have been found in Gram-negative bacteria (2, 3).
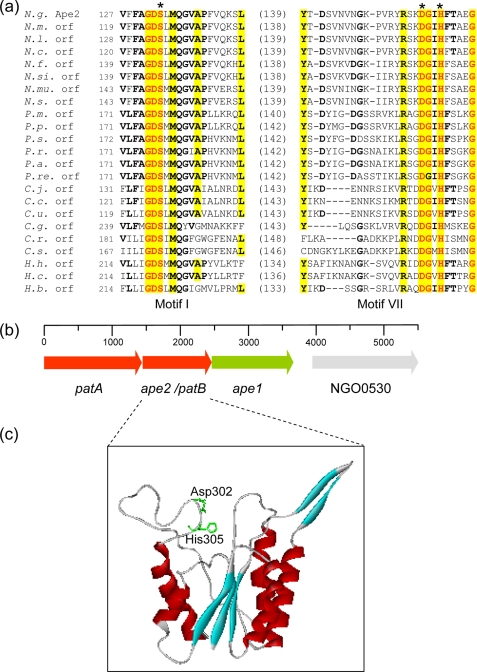
We recently reported the discovery of a gene cluster within the genomes of a number of bacteria that O-acetylate their PG, including N. gonorrhoeae, which encode hypothetical proteins that may participate in this process (Fig. 2) (4). We named this cluster poa for peptidoglycan O-acetylation. Sequence alignment searches suggested that one of the open reading frames associated with each poa encodes a hypothetical protein with similarity to Pseudomonas aeruginosa AlgI, a putative integral membrane protein thought to be responsible for the translocation of acetate for the O-acetylation of its extracellular polysaccharide alginate (17). These AlgI homologs were named Pat for peptidoglycan O-acetyltransferase. The other open reading frame(s) of these clusters were predicted to encode SGNH/GDSL-family esterases with similarity to the CAZy family CE-3 O-acetylxylan esterases (4). With the N. gonorrhoeae chromosome, two of these open reading frames are located immediately downstream from pat, and one (ape1a) was subsequently demonstrated to encode an O-acetylPG esterase (Ape), which serves to remove O-acetyl groups from PG (18). The second was named ape2 based on 20.6% amino acid identity (58.5% similarity) of its hypothetical amino acid sequence with Ape1a, but its true activity and function remained uncharacterized. A phenotypic study conducted by others involving the disruption of the genes comprising the N. gonorrhoeae poa cluster did demonstrate their involvement with PG O-acetylation (19), but the biochemical details of the process are still lacking. In this study, we demonstrate that N. gonorrhoeae Ape2 is, in fact, not an O-acetylPG esterase but rather functions as a PG O-acetyltransferase. Moreover, we present the first evidence in support of a two-component system for the O-acetylation of PG in Gram-negative bacteria.
FIGURE 2.
Sequence, genetic organization, and structure of Ape2. a, partial sequence alignment of hypothetical family 2 Ape involving consensus motifs I and VII. The residues in bold face and yellow highlight denote greater than 50 and 80% identity, respectively, while the residues in red are invariant within the family. The values in parentheses denote the number of residues between the respective partial sequences, and the asterisks denote the putative catalytic residues. Abbreviations (accession numbers): N.g., N. gonorrhoeae Ape2 (YP_207683); N.m., N. menningitidis (NP_284201); N.l., N. lactamica (ZP_03722920); N.c., N. cinerea (ZP_03744458); N.f., N. flavescens (EEG32498); N.s., N. subflava (ZP_03750678); N.mu., N. mucosa (ACDX02000005.1); N.si., N. sicca (EET44325); P.m., P. mirabilis (YP_002152370); P.p., P. penneri (EEG87531); P.s., Providencia stuartii (ZP_02961974); P.r., Prov. rustigianii (ZP_03315886); P.a., Prov. alcalifaciens (EEB46552); P.re., Prov. rettgeri (EEF72588); C.j., Campylobacter jejuni (NP281793); C.c., C. coli (ZP_00366779); C.u., C. upsaliensis (ZP_00369961); C.g., C. gracilis (EEV17613); C.r., C. rectus (EEF14732); C.s., C. showae (EET79176); H.h., Helicobacter hepaticus (NP860614); H.c., H. cinaedi (ABQT01000013); H.b., H. bilis (ACDN01000074). b, genetic organization of the poa cluster on the N. gonorrhoeae chromosome including the gene encoding NGO0530, a hypothetical acyl-CoA synthetase. c, three-dimensional structure of the N. gonorrhoeae Ape2 as predicted using PHYRE involving residues Asp-125—Pro-326. The Ape2 residues were threaded onto the SGNH hydrolase thioesterase I (GenBankTM Acc. No. D1jrla) with 17% identity and an E-value of 1.6−19. The position of the putative catalytic Asp-302 and His-305 residues are illustrated within a groove through the middle of the enzyme; the N-terminal segment containing Ser-133 was not modeled by the algorithm.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Acrylamide, glycerol, pyridine, and Luria-Bertani (LB) growth medium were obtained from Fisher Scientific. DNase I, RNase A, Pronase, isopropyl β-d-1-thiogalactopyranoside (IPTG), and EDTA-free protease inhibitor tablets were purchased from Roche Molecular Biochemicals (Laval, PQ). All other growth media were acquired from Difco Laboratories (Detroit, MI). MonoS 5/5 was purchased from Amersham Biosciences (Uppsala, Sweden) while Qiagen (Valencia, CA) supplied Ni2+-NTA-agarose. Recombinant N-acetylmuramoyl-l-alanine amidase B (AmiB) from P. aeruginosa and Ape1a from N. gonorrhoeae were isolated and purified from E. coli transformants as previously described (18, 20). T4 DNA ligase and all restriction enzymes were from New England Biolabs (Mississauga, ON) and, unless stated otherwise, all other reagents and chemicals were purchased from Sigma.
Bacterial Strains and Growth
Sources of plasmids and bacterial strains used or constructed in this study, together with their genotypic description, are listed in Table 1. pBAD-based constructs were screened, maintained, and overexpressed in E. coli Top10 while the Top10 and DH5α strains were used for pGEMT-Easy based plasmids. For optimal production of Ape2/PatB, transformed E. coli Top10 was cultured in Super Broth (32 g/liter tryptone peptone; 20 g/liter yeast extract; 5 g/liter NaCl; pH 7.0) at 37 °C, and gene expression was induced with 0.1% arabinose.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic | Ref. or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21[λDE3] codonPlus | F−ompT hsdSB (rB− mB−) dcm met gal(λDE3)endA Hte[argU, ileY,leuW] TetR | Novagen |
| BL21[λDE3] pLysS | F−ompT hsdSB (rB− mB−) gal dcm met (λDE3) pLysS (CmR) | Novagen |
| DH5α | K-12 ϕ80d lacZΔM15 endA1 hsdR17 (rK− mK−) supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF) U169 | Invitrogen |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu) 7697 galU galrpsL (StrR) endA1 nupG | Invitrogen |
| JW3533-1 | Δ(araD-araB)567 Δ(rhaD-rhaB)568 lacZ487::rmB-3 LAM- rph-1 hsdR514 wecH744::kan lacZ4787 (KmR) | Yale CGSC |
| N. gonorrhoeae FA1090 | Serum-resistant, proline-requiring strain isolated from a patient with probable disseminated gonococcal infection | 41 |
| B. cereus 10987 | NRS 248; genome sequenced strain | ATCC |
| Plasmids | ||
| pET28▵+) | IPTG-inducible T7 expression vector; N- and C- terminal His6 tag; KmR | Novagen |
| pET30▵+) | IPTG-inducible T7 expression vector; N- and C- terminal His6 tag; KmR | Novagen |
| pET32 Ek/LIC | IPTG-inducible T7 expression vector, N- and C- terminal His6 tag; LIC direction cloning strategy; KmR | Novagen |
| pGEM-T Easy | T-overhang assisted cloning, lacZ gene for blue/white screening; AmR | Novagen |
| pBAD 18-Cm | Arabinose-inducible araBAD expression vector, no RBS or purification tag in vector MCS; CmR | 42 |
| pBAD His-A | Arabinose-inducible araBAD expression vector, N-terminal His6 tag; AmR | Invitrogen |
| pACPM16g | pET30▵+) derivative containing ape2 from FA1090 with a G inserted at the 4th base pair (frame shift mutation) on a NdeI/HindIII fragment; KmR | This study |
| pACPM17 | pBAD His-A derivative containing ape2 from FA1090 with an N-terminal His6-tag on a XhoI/HindIII fragment; AmR | This study |
| pACPM19 | pBAD 18-CM derivative containing ape2 from FA1090 with a C-terminal His6 tag on a NheI/HindIII fragment; CmR | This study |
| pACPM20 | A pACPM19 derivative encoding Ape2 truncated by its N-terminal 26 amino acid residues; CmR | This study |
| pACPM22 | A pACPM21 derivative encoding Ape2 truncated by its N-terminal 36 amino acid residues; AmR | This study |
N. gonorrhoeae FA1090 was grown on GCB agar or in GCBL broth, both supplemented with Kellogg's defined supplement (21, 22), at 37 °C in a humid, 5% CO2 incubator as previously described (18). Kanamycin (Km) at 18 μg/ml final concentration was added when appropriate.
Isolation and Purification of PG
Samples of insoluble PG were extracted from strains of B. cereus, E. coli, and N. gonorrhoeae using the boiling 4% SDS procedure as described by Holye and Beveridge (23) taking care to preserve any natural levels of O-acetylation (24). The PG preparations were purified by enzyme treatment (amylase, DNase, RNase, Pronase) as described by Clarke (24).
Determination of Extent of PG O-Acetylation
Isolated insoluble PG was resuspended in 25 mm sodium phosphate buffer, pH 6.0 and fragmented by continuous sonication for 4 min using an XL2020 Heat Systems sonicator prior to treatment with 500 mm NaOH (final concentration) for 30 min at ambient temperature to release any ester-linked acetate. Quantification of released acetate in supernatants following ultracentrifugation (100,000 × g, room temperature) using a Beckman Airfuge (Beckman Coulter, Mississauga, ON) was performed by either HPLC-based organic acid analysis using an Aminex HPX-87H Bio-Rad column as previously described (8) or using the Megazyme Acetic Acid Assay kit (Megazyme International Ireland Ltd., Wicklow, Ireland). The extent of O-acetylation is presented as a percentage of muramic acid content, which was determined by quantitative aminosugar analysis (25) of associated insoluble PG pellets after its hydrolysis in 4 m HCl at 96 °C in vacuo for 18 h.
Conditions for PCR
Chromosomal DNA template for PCR was isolated from N. gonorrhoeae FA1090. All oligonucleotide primers used in this study (listed in Table 1 of supplemental materials) were acquired from the Guelph Molecular Supercenter (University of Guelph, Guelph, ON). PCR amplifications were achieved in 50-μl volumes using a Perkin Elmer GeneAmp PCR system 2400. Conditions were optimized for each primer pair using the Expand Long Template PCR system (Roche Molecular Biochemicals). Purification of PCR products was performed using the MinElute PCR Purification kit (Qiagen) or the High Pure PCR Product Purification kit (Roche Molecular Biochemicals).
Cloning of N. gonorrhoeae ape2
For cloning into pET30a(+), pET28a(+), pBADHis-A, and pBAD18-Cm vectors, PCR products, and vectors were digested with the appropriate restriction enzymes according to the manufacturer's instructions and verified by agarose gel electrophoresis. The MinElute Reaction Cleanup kit was used to purify the DNA from these reactions. Ligation reactions were performed in a 1× T4 DNA ligase buffer with insert to vector ratios of ∼3:1 or 5:1. When cloning into the pET32 Ek/LIC system, ligation reactions were performed according to the manufacturer's specifications.
With the pGEMT-Easy (Promega, Madison, WI) vector system, thymine overhangs on the vector enabled rapid ligation of PCR products with adenine overhangs generated by the activity of the polymerase(s) present in the Expand Long Template PCR system. Direct ligations of PCR products to the linear pGEMT-Easy vector provided were performed according to the manufacturer's instructions. Individual constructs were isolated from transformants, screened for the correct size insert, and were completely sequenced to confirm nucleotide identity.
Site-directed Mutagenesis of ape2
Site-directed mutagenesis was performed on pACPM16g to repair a deliberately introduced mutation into the full-length ape2 gene. The QuickChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA), including pfu Turbo DNA polymerase (Stratagene), was used according to the manufacturer's instructions. Constructs were transformed into E. coli DH5α and clones were screened by DNA sequencing.
Expression of ape2
Various plasmid constructs and E. coli hosts were tested for optimal production of Ape2 under different culture and induction conditions. E. coli strains freshly transformed with plasmid DNA were inoculated into LB supplemented with 50 μg/ml kanamycin and incubated at 37 °C until early exponential phase (A600 ∼ 0.5). With the pET-based plasmid pACPM16g, expression was induced with freshly prepared IPTG at a final concentration of 0.1–1 mm, and growth was continued for a further 3 h at either 15, 25, or 37 °C. ape2 was also overexpressed from pACPM17 or pACPM19 transformed into cells of E. coli Top10 or E. coli JW3533–1. These pBAD-based plasmids were repressed with 0–0.4% glucose and induced with 0.0004–0.4% arabinose at 15, 25, 30, and 37 °C for 3, 6, and 24 h. In each case, 1-ml samples of cells were collected for analysis of protein production by SDS-PAGE with Western immunoblot detection.
SDS-PAGE and Western Immunoblotting
SDS-PAGE analysis of protein samples was performed following the method of Laemmli (26) using 12% polyacrylamide gels and staining with Coomassie Brilliant Blue. Western immunoblotting for the detection of His6-tagged proteins was performed as previously described (24) using a 1:1000 dilution of a commercially available mouse anti-His6 antibody (Santa Cruz Biotechnology) and a 1:2000 dilution of the secondary antibody, goat anti-mouse IgG + IgM alkaline phosphatase-conjugated antibody (Bio-Rad).
Purification of Ape2
Cells collected by centrifugation (5,000 × g, 15 min, 4 °C) were washed and resuspended in IMAC buffer (50 mm sodium phosphate buffer, pH 8.0, 500 mm NaCl, and 0.5% Triton X-100) containing Complete EDTA-free protease inhibitor mixture tablets, 10 μg/ml RNase A, 5 μg/ml DNase I, and 1 mg/ml hen egg-white lysozyme. These cell suspensions were incubated on ice for 30 min prior to disruption by either sonication using an Ultrasonic Liquid Processor (Mandel Scientific Instruments, Guelph, ON) (intervals of 5-s bursts with 10-s cooling periods for a total of 2 min) or at least three passages through a French Pressure Cell Press (American Instruments Co., Silver Spring, MD) (18,000 psi). The resulting cell lysates were clarified by centrifugation (5,000 × g, 15 min, 4 °C) and the collected soluble cell fractions were mixed with Ni2+-NTA-agarose (0.5 ml/liters starting culture) for 1–5 h at 4 °C on a Clay-Adams nutator. The resin slurry was packed into a disposable column and the flow-through fractions were collected. Contaminating proteins were removed from the resin by washing with 10 column volumes each of cold IMAC buffer, IMAC buffer at pH 7, and IMAC buffer at pH 6. Ape2 was eluted in 10 ml of IMAC buffer at pH 4.5 and immediately subjected to exhaustive dialysis at 4 °C against 2 × 4 liters of 50 mm sodium phosphate buffer, pH 8.0 containing 0.1% Triton X-100.
His6-tagged Ape2 was further purified by cation-exchange chromatography on MonoS, previously equilibrated in 25 mm sodium phosphate buffer, pH 7.0. Protein samples were loaded onto the column at a flow rate of 0.70 ml/min and eluted by applying a linear gradient of 0–1 m NaCl over 30 min. Eluted proteins were collected and dialyzed against 4 liters of 50 mm sodium phosphate buffer, pH 8.0.
Cellular Localization of Ape2
The cellular localization of Ape2-His6 was investigated by Western immunoblot analysis of isolated fractions of cells from 1 liter of culture. Cell fractionation was conducted as described previously (18).
PG Binding Assay
The ability of Ape2 to bind different PGs was assessed as previously described (20) using 0.15 mg samples of B. cereus or E. coli PG suspended in 300 μl of 50 mm sodium phosphate buffer, pH 6.0. Binding was assessed by SDS-PAGE with Western immunoblotting detection after samples of purified protein (15 μg) were incubated with PG for 1 h at 4 °C. In some experiments, the O-acetyl groups were removed from the B. cereus PG by mild-base hydrolysis prior to assay.
Acetylesterase Activity Assays
For the routine assay of acetylesterase activity, 2 mm p-nitrophenylacetate (pNP-acetate) in 50 mm sodium phosphate buffer, pH 6.5 was used as a substrate in a 96-well microtiter plate assay as previously described (18).
Insoluble N. gonorrhoeae PG was used as substrate for the specific detection of O-acetylpeptidoglycan esterase activity. Prior to assay, a 5 mg/ml suspension of the O-acetylated PG in 50 mm sodium phosphate buffer, pH 7 containing 0.01% Triton X-100 was subjected to sonication to evenly disperse the insoluble material. Samples of purified Ape2 were incubated with the substrate at 37 °C for appropriate periods of time and then the insoluble PG was removed by ultracentrifugation (100,000 × g, 20 min, room temperature) using a Beckman Airfuge. The resulting supernatants and PG pellets were analyzed for released and bound O-linked acetate, respectively, as described above. Control samples consisted of the above reaction without the addition of enzyme and were analyzed for spontaneous release of O-linked acetate.
Cell-free Assay for PG O-Acetylation
The toluene-based permeabilization method for preparing cell-free fractions of wall components of P. mirabilis and assay for PG O-acetylation described previously (27, 28) was adapted for use with E. coli Top10 transformants harboring pBAD18-Cm, pBADHis-A, pACPM17, or pACPM19. Immediately following toluene treatment, 10 ml of E. coli cells expressing ape2, as well as control cells containing an empty pBAD18-Cm or pBADHis-A vector, in 100 mm Tris-HCl buffer, pH 6.8 containing 30 mm MgCl2 were incubated with 15 mm acetyl-CoA at 30 °C for 30 min. The insoluble fraction was collected by centrifugation (5,000 × g, 15 min, room temperature) and the associated PG was isolated for determination of O-acetyl content and muramic acid concentration as described above. In some experiments, the isolated PG was resuspended in 25 mm sodium phosphate buffer, pH 6.0 containing 0.02% sodium azide and incubated overnight at 37 °C with 80 units of mutanolysin with and without 50 μg of freshly purified AmiB. Any remaining insoluble PG was removed by ultracentrifugation using a Beckman Airfuge (100,000 × g, 20 min, room temperature). The supernatants were evaporated in vacuo, resuspended in a minimal volume of 0.1% trifluoroacetic acid and desalted using C18-ZipTips (Millipore Co., Billerica, MA) according to the manufacturer's specifications prior to MALDI-TOF mass spectrometry analysis.
MALDI-TOF Mass Spectrometry
PG digests and purified protein samples were dried in vacuo for 2 h at 50 °C and then desalted using C18-ZipTips. Eluted samples in 1–2 μl of 0.05% trifluoroacetic acid, 25% acetonitrile were submitted for MALDI-TOF mass spectrometric analysis. Mass spectrometry was carried out at the Biological Mass Spectrometry Facility (University of Guelph, Guelph, ON) on a Bruker Reflex III MALDI-TOF mass spectrometer in linear mode using a 337 nm nitrogen laser (set to 109 to 121 μJ output).
Other Analytical Techniques
Identification of open reading frames, protein translations, and isoelectric points (pI) were performed using Clone Manager 5. Signal sequence predictions were performed using SignalP 3.0 and TMpred. Homology searches for Ape2-like proteins were carried out using the online BLASTP program to search the NCBI data base and analysis of sequence data were performed using ClustalW2 software (29). Secondary structure was predicted using JPred (30) and PSIpred (31) while PHYRE was used to predict tertiary structure (32, 33). PCR products as well as plasmids were sequenced on an ABI 377 DNA sequencing system (Applied Biosystems) at the Guelph Molecular Supercenter (University of Guelph, Guelph, ON). Protein concentration was determined by the bicinchoninic acid (BCA) assay (Pierce Biotechnology, Inc.) essentially according to the manufacturer's instructions.
RESULTS
Identification and Characterization of the ape2 Gene
The nucleotide sequence of ape2 (GenBankTM Acc. No.YP_207683) was obtained from the N. gonorrhoeae FA1090 genome data base at NCBI. This sequence was used by Weadge et al. (4) to identify eight homologs from other Gram-negative bacteria and organize them into family 2 of their classification system. We conducted a more recent analysis of the genome databases, which revealed at least 33 other sequences that fit into this family, all from Gram-negative bacteria including pathogenic species of Campylobacter, Helicobater, Proteus, Providencia, and Treponema (Fig. 2 and supplemental Fig. S1). This list thus includes the causative agents of diarrheal diseases, stomach ulcers, urinary tract infections, and syphilis, in addition to meningitis and gonorrhoeae.
The FA1090 ape2 gene encodes a moderately sized hypothetical protein of 335 amino acids with a calculated molecular mass of 37,146 Da and a relatively high estimated pI value of 8.75. The region of similarity with other Ape2 proteins involves the C-terminal two-thirds of the FA1090 sequence, which includes motifs I and VII that assign it as a member of the SGNH/GDSL hydrolases (4). Thus, the Ser-133 of motif I and Asp-302 and His-305 of motif VII of the hypothetical protein (Fig. 2) are predicted to form a catalytic triad.
The SignalP 3.0 server predicted that Ape2 carries a periplasmic-localizing, cleavable signal peptide at its N terminus. The Neural Network (NN) model predicted that this peptide would be cleaved between positions Ala-36 and Tyr-37. In contrast, the Hidden Markov Model (HMM) suggested the most likely cleavage point as being between Ala-26 and Val-27, but it also indicated that the Ala-36/Tyr-37 position was a possibility. Cleavage between the latter two residues would result in a protein with a theoretical molecular mass of 33,206.62 Da and a pI value of 7.96.
The two secondary structure prediction programs Signalp3.0 and PSI Pred were generally in agreement that the protein is mainly α-helical (34–36%) with some (9–10%) β-strands. The sequence was analyzed with the Phyre algorithm, which provided a prediction of its tertiary structure modeled on thioesterase I from E. coli (PDB: 1JRL) with an E-value score of 1.6 × 10−19 (Fig. 2). This protein has 17% identity with the aligned portion of Ape2; the structure predicted by Phyre does not include the N-terminal 124 amino acids nor the C-terminal nine amino acids. Nonetheless, the portion that is modeled generally reflects the secondary structures predicted by SignalP 3.0 and PSI Pred and moreover, presents the structure of an α/β-fold characteristic of SGNH hydrolases.
Cloning of ape2 from N. gonorrhoeae
The entire 1005-bp sequence of ape2 was targeted for cloning from N. gonorrhoeae FA1090. Initially, efforts were made to clone ape2 into various pET-type vectors. Unlike the situation with ape1a (18), however, this was not met with any success with respect to the generation of a final construct despite ensuring the precision of the DNA sequences. Thus, whereas specific primers used in PCR reactions successfully amplified the correct gene sequence from genomic DNA, repeated attempts to clone into pET28a (+) or pET30a (+) resulted in no successful transformations of ligation products. To assess this problem, a pET30a(+) construct was engineered that contained a guanine insertion at the fourth nucleotide of the coding sequence that shifted the entire ape2 gene out of frame. This cloning attempt was immediately successful and the construct was named pACPM16g. However, repeated attempts to remove this introduced mutation by site-directed mutagenesis failed. These data thus suggested that the un-induced, low-level expression of ape2 from the pET vectors led to toxicity issues for the E. coli host.
To better control background expression of cloned genes, the pBAD vector system was selected for further studies. Using this system, amplification was initially successful but subsequent ligation and transformation experiments required the introduction of glucose to the growth medium which serves to inhibit transcription in the pBAD system. The initial construct generated, pACPM17, involved the pBAD His-A vector which provides an N-terminal His6 tag on protein products. Because of complications with purification (described below), another construct was prepared utilizing the pBAD18-Cm vector. This vector lacks a ribosomal binding site (RBS) and a purification tag, and thus both were engineered into the ape2 PCR fragment. The RBS was adapted from pBAD His-A, and a C-terminal His6 tag was also included. PCR amplification was performed as described above and the fragment was successfully cloned into the pBAD18-Cm vector, generating pACPM19. A truncated version of ape2 lacking coding for its N-terminal 26 amino acids was also prepared and cloned into this vector generating pACPM20. A second truncated version of ape2 lacking coding for its N-terminal 36 amino acids was also prepared, but this was cloned into pBAD His-A generating pACPM22 and thus encoding the truncated Ape2 with an N-terminal His6 tag. Nucleotide sequencing was used to verify the accuracy of each of the cloned constructs.
Overexpression of ape2
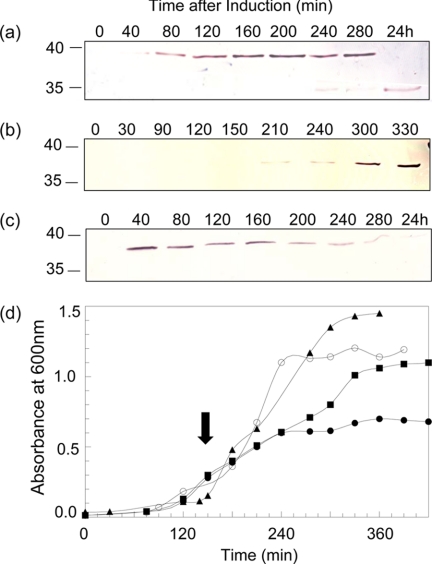
Optimum conditions for glucose repression and arabinose induction of ape2 expression in E. coli Top10 transformed with either pACPM17, pACPM19, pACPM20, or pACPM22 were determined. These experiments took advantage of the presence of the His6 tags on produced Ape2 which could be readily detected by Western immunoblot analysis using a commercially available anti-His6 tag antibody. As seen in Fig. 3a for pACPM19, 0.4% (w/v) glucose was required for complete repression of ape2 expression whereas 0.04% (w/v) arabinose was sufficient for its induction. A single band with an apparent molecular mass of 39.7 kDa was detected, which corresponds closely with the theoretical molecular mass of 37,969 Da for the His6-tagged Ape2. Similar results were obtained with the other three transformants harboring pACPM17, pACPM20, and pACPM22 (data not shown).
FIGURE 3.
Expression of ape2 in E. coli transformants. The expression of ape2 from pACPM19 transformed into (a) E. coli Top10 and (b) E. coli JW3533–1, and (c) from pACPM22 transformed into E. coli Top10 was monitored with time by SDS-PAGE and Western immunoblot analysis of cell culture samples using a polyclonal anti-His6 antibody. d, growth of E. coli Top10 harboring (■) pBAD-HisA, (●) pACPM19, and (▲) pACPM22, and (○) E. coli JW3533–1 harboring pACPM19 in LB at 37 °C. The solid arrow denotes induction of ape2 expression with the addition of 0.4% arabinose.
A typical growth curve for E. coli Top10 over-expressing ape2 from pACPM19 under these conditions is presented in Fig. 3c. From this experiment, it was evident that growth arrested immediately after induction of ape2 expression. Moreover, SDS-PAGE and Western immunoblot analysis of the proteins produced during the course of induction indicated that accumulation of Ape2 led to its intracellular degradation (Fig. 3a). This resulted typically in the production of one to six different Ape2 degradation products ranging in apparent mass from 39.7 to 14.3 kDa. Attempts to prevent this degradation by reducing incubation temperature, in addition to inducer concentrations, were unsuccessful. Similar results were obtained with transformants harboring pACPM17 and pACPM20. In contrast, expression of ape2 from pACPM22 did not affect the growth of the E. coli host and the N-terminally His6-tagged, truncated protein produced was not degraded over time to the same extent (Fig. 3). These data thus suggested that the only form of the protein that was not deleterious to the growth of E. coli was one that would not be transported to the periplasm but instead remained within the cytoplasm. Optimal production of the N-terminally truncated and tagged protein was obtained with transformed E. coli Top10 grown in Super Broth and induced with 0.1% arabinose.
Localization of Ape2 in E. coli
As noted above, Ape2 is predicted to be a soluble protein that possesses a signal sequence for transport to the periplasm. The localization of Ape2 by E. coli was investigated by analyzing for its presence in cytoplasmic and periplasmic fractions by SDS-PAGE coupled with Western immunoblotting. The results indicated that in addition to be retained in the cytoplasm, a large fraction of the overproduced protein was exported to the periplasm (data not shown).
Isolation and Purification of Ape2 from E. coli Top10[pACPM22]
Attempts to isolate Ape2 with an N-terminal His6-tag produced in E. coli Top10 transformed with the pACPM17 construct by immobilized metal affinity chromatography on Ni2+-NTA agarose were met with limited success because of the apparent processing of the protein and export to the periplasm followed by its rapid degradation. The addition of detergent, such as 0.1% Triton X-100, varying the amount of NaCl, or adjusting the pH of the binding buffer, did not provide any improvement. In contrast, N-terminally His-tagged, truncated Ape2 produced from the pACPM22 vector in E. coli Top10 could be isolated with modest yields. The relatively low stringency of the buffer required to retain Ape2 on the Ni2+-NTA-agarose affinity matrix sometimes resulted in the retention of a few contaminating proteins, which subsequently co-eluted with the His-tagged protein.
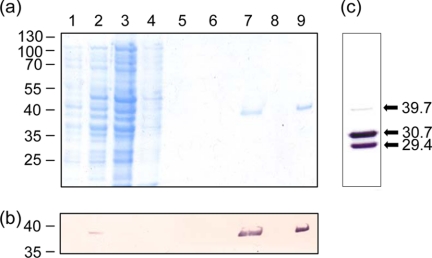
With its relatively high pI value, further purification of Ape2 was achieved, when necessary, by cation exchange on MonoS. The protein eluted from the column in approx. 0.56 m NaCl, which was subsequently removed by dialysis. Analysis by SDS-PAGE indicated that the Ape2 preparations with the expected mass of 38 kDa for full-length enzyme were greater than 95% homogeneous (Fig. 4) and overall yields of ∼0.6 mg of protein were obtained from a liter of culture.
FIGURE 4.
Purification of Ape2. a, SDS-PAGE analysis of fractions obtained during purification of Ape2 by affinity chromatography on Ni-NTA-agarose and cation-exchange chromatography on Mono S. Lane 1, whole cell lysate from uninduced E. coli Top10[pACPM22]; lane 2, whole cell lysate from E. coli Top10[pACPM22] induced with 0.1% arabinose; lane 3, flow-through fraction from Ni-NTA-agarose; lane 4, elution with 20 mm imidazole in Elution Buffer (50 mm sodium phosphate containing 500 mm NaCl and 0.1% Triton X-100), pH 8.0; lane 5, 10 mm imidazole in elution buffer, pH 7.0; lane 6, elution buffer, pH 6.0; lane 7, elution buffer, pH 4.5; lane 8, flow-through fraction from Mono S; lane 9, fraction obtained following application of linear gradient of NaCl. b, Western immunoblot analysis of SDS-PAGE fractions using an anti-His6 antibody. c, Western immunoblot analysis of full-length Ape2 possessing a C-terminal His6 tag purified from E. coli Top10[pACPM19]. The positions of molecular mass markers and the calculated molecular masses of identified fractions in kDa are indicated on the left and right, respectively.
Despite repeated attempts, purification of C-terminally His-tagged Ape2 was not possible without losing most of it to proteolysis even when various cocktails of protease inhibitors were included in the isolation buffer. The most abundant fragments detected by the SDS-PAGE were analyzed by MALDI-TOF mass spectrometry and found to have m/z ratios of 30,744 and 29,377. These represented ∼77 and 81% of the expected size of the protein, respectively. As these Ape2 derivatives were detected by Western immunoblotting analysis using an anti-His tag antibody (Fig. 4c), they must represent the C-terminal region of Ape2 and that cleavage occurred beyond the expected N-terminal processing site.
Characterization of Ape2
The binding specificity of Ape2 for different PGs was investigated using a previously developed pull-down assay. Purified samples of Ape2 were incubated for 1 h at 4 °C with PG from different sources, both O-acetylated and non-O-acetylated. The insoluble material was then collected by centrifugation and all associated protein was extracted from the PG in 4% SDS and analyzed by SDS-PAGE with Western immunoblot detection. Ape2 exhibited strong binding to the non-O-acetylated PG from E. coli or base-treated PG from B. cereus (Table 2). In contrast, the protein only weakly bound to native N. gonorrhoeae PG and did not bind at all to O-acetylated B. cereus PG. These data thus indicated that Ape2 preferentially, and surprisingly, binds PG of the A1γ chemotype that is non-O-acetylated.
TABLE 2.
Binding of Ape2 to PG
| PG source | Chemotype | Acetylation | Ape2 Recovered ina |
||
|---|---|---|---|---|---|
| Supernatant | Wash | SDS | |||
| % | |||||
| E. coli Top10 | A1γ | 0.02 | − | − | ++++ |
| N. gonorrhoeae 1291 | A1γ | 40 | +++ | − | + |
| B. cereus 10987 | A1α | 54b | ++++ | − | − |
| A1α | 0 | − | +++ | + | |
a Ape2 was incubated with insoluble PG in 50 mm sodium phosphate buffer, pH 6.0 for 1 h at 4 °C and then subjected to ultracentrifugation. The amount of PatB recovered in the supernatant (unbound), buffer wash (weakly retained), and 4% SDS extract (bound) was estimated by SDS-PAGE with Western immunoblot analysis.
b J. Pfeffer, personal communication.
The observation that Ape2 binds preferentially to non-O-acetylated PG suggested that it may not function as an O-acetylPG esterase. To test for this, freshly prepared Ape2 was assayed using two protocols involving p-NP-acetate and O-acetylated PG as substrates, respectively. Using 2 mm p-NP-acetate in 50 mm sodium phosphate buffer, pH 7.0 at 25 °C, the specific activity of PatB as an esterase was calculated to be 0.099 ± 0.021 μmol·min−1·mg−1 protein. Under these conditions, purified Ape1a from N. gonorrhoeae was found to have a specific activity of 5.26 ± 0.28 μmol·min−1·mg−1 protein. Thus, the specific activity of Ape2 was only 1.9% of that of Ape1a from the same bacterium. Furthermore, no acetate release was detected in supernatants following extensive incubation of Ape2 with insoluble O-acetylPG under conditions optimized for Ape1a activity (18). These data thus, suggested that the true function of Ape2 is not as an O-acetylPG esterase.
Phenotypic Analysis of E. coli Top10[pACPM19]
Given its amino acid sequence similarity with Ape1a, the finding that Ape2 does not function as an O-acetylPG esterase was surprising. However, unlike the situation with Ape1a (18), overexpression of ape2 in E. coli led to both cessation of cell growth and rapid degradation of the protein indicating a significant difference in their respective activities. One explanation, albeit unlikely, could be that if functioning as an esterase, Ape2 catalyzes the hydrolysis of one or more vital cellular metabolites. Alternatively, Ape2 may not be an esterase but instead functions as an O-acetyltransferase to add acetyl groups to PG. This activity indeed would be detrimental to E. coli given it does not naturally O-acetylate its PG and thus does not possess the appropriate housekeeping enzymes to manage this modification. To test for this latter possibility, the PG was isolated from E. coli Top10 transformed with pACPM19 and assayed for O-acetyl content. Mild-base hydrolysis of these PG preparations and quantification of released acetate did indicate the presence of O-acetylation but at the low level of 1.19 ± 0.11% relative to muramic acid content (n = 3). Nonetheless, this level was considered significant because PG isolated from E. coli Top10 harboring an empty pBADHis-A vector was found, as expected, to be virtually devoid of the modification (0.025 ± 0.03%). To further investigate this activity, cell-free preparations of the two E. coli strains were obtained by toluene treatment (28) and they were incubated with exogenously added acetyl-CoA. Following incubation for 30 min at 30 °C, the PG was isolated from these reaction mixtures and assayed for O-acetylation. With the PG from E. coli Top10[pACPM19], the level of O-acetylation increased to 2.52 ± 0.06% while that from the control E. coli Top10[pBADHis-A] cells remained unaltered at 0.027 ± 0.03%.
MALDI-TOF mass spectrometry was used to confirm the presence of N,O-diacetylmuramoyl residues in the PG of E. coli Top10 producing Ape2. The PG obtained from the cell-free assay systems was digested with mutanolysin or with both mutanlysin and N-acetylmuramoyl-l-alanine amidase B (the latter to remove stem peptides from muroglycans). Following their desalting, these digests were subjected to MALDI-TOF mass spectrometric analysis using two different matrices, 2,5-dihydroxybenzoic acid (DHB), which is typically used for peptide analysis, and α-cyanohydroxycinnamic acid (CCA) for carbohydrate analysis. The data, presented in Table 3, showed unequivocally the existence of O-acetylated muropeptides uniquely associated with E. coli Top10 producing Ape2. The chemical structures of these muropeptides are presented in Fig. 2 of supplemental materials. These data confirmed that Ape2 contributes to the O-acetylation of PG and it was thus renamed PatB for PG O-acetyltransferase B.
TABLE 3.
Unique muropeptide/glycans detected in PG isolated from E. coli Top10[pACPM19]
| Muropeptide/glycana |
m/z |
|||
|---|---|---|---|---|
| Calculated | Observed | Δ | ||
| PG + mutanolysin | ||||
| G-OAcM(Tri)b | [M+Na]+ | 934.87 | 934.57 | 0.30 |
| G-OAcM(Tetra) | [M+H]+ | 982.44 | 982.96 | 0.52 |
| [M+Na]+ | 1005.95 | 1005.55 | 0.40 | |
| G-OAcM(Penta) | [M+H]+ | 1054.04 | 1053.62 | 0.42 |
| PG + mutanolysin + amidase | ||||
| G-OAcM | [M+H]+ | 539.21 | 539.20 | 0.01 |
| G-OAcM-G | [M+H]+ | 741.36 | 742.28 | 0.93 |
| [M+Na]+ | 764.41 | 765.27 | 0.87 | |
| G-OAcM-G-Mc | [M+H]+ | 1040.38 | 1041.55 | 1.17 |
| G-OAcM-G-anhM | [M+H]+ | 994.37 | 994.46 | 0.08 |
| G-OAcM-G-OAcM | [M+H]+ | 1060.00 | 1061.55 | 1.17 |
a PG isolated from E. coli strains Top10[pACPM19] and Top10[pBAD18-Cm] was treated initially with mutanolysin (generating muropeptides) and then N-acetylmuramoyl-l-alanine amidase B from P. aeruginosa (generating muroglycans) and subjected to MALDI-TOF MS. Listed are the detected muropeptide/glycans that were unique to strain Top10[pACPM19].
b G, GlcNAc; M, MurNAc; OacM, O-acetylMurNAc; anhM, 1,6-anhydro-MurNAc; Tri, tripeptide; Tetra, tetrapeptide; Penta, pentapeptide.
c The position of the O-acetyl group could be on either the first or second MurNAc residue.
Expression of patB in E. coli JW3533-1
As PatB would appear to function within the periplasm as a peripheral membrane protein, it would have to be dependent upon the activity of a second component for the translocation of an activated acetate source from the cytoplasm for presentation within the periplasm. We have proposed that PatA (Fig. 2), the AlgI homolog, would serve this function (2, 4). E. coli does not naturally O-acetylate its PG, and thus it does not encode PatA, but it is evident that a source of acetate was nonetheless available to recombinant PatB in E. coli transformants for PG O-acetylation. However, E. coli does O-acetylate its enterobacterial common antigen (ECA), a cell surface glycolipid. This O-acetylation involves a putative integral cytoplasmic membrane protein, WecH, which is a member of an acyltransferase family of enzymes (34). Hence, we postulated that WecH could act as a paralog of PatA/AlgI and provide functional equivalency for acetate translocation in E. coli transformants. To test this, competent cells of E. coli JW3533-1, which lacks a functional copy of WecH, were prepared and transformed with pACPM19 and their growth was monitored over time. As seen in Fig. 3, growth continued at the same rate following arabinose induction as cells harboring the empty vector, pBAD His A. Moreover, SDS-PAGE analysis of isolated cells indicated that the PatB produced did not undergo any degradation but rather appeared to remain intact with an apparent molecular mass of 37.4 kDa. Finally, despite an apparent high level of PatB production, these cells were found to be devoid of PG O-acetylation.
DISCUSSION
The hypothetical N. gonorrhoeae protein Ape2 had been identified previously as the archetype for a second family of O-acetylPG esterases based on sequence alignments and the presence of consensus motifs that define the GDSL/SGNH family of esterases (4). In a separate study involving the phenotypic characterization of knock-out mutants, the ape2 (pacB) gene had been shown to be associated with PG O-acetylation (19), but its function and role remained unknown. In this study, we demonstrated that Ape2, in fact, does not possess esterase activity but rather functions as a periplasmic protein for the O-acetylation of PG, the first to be discovered in any Gram-negative bacterium. The protein was renamed PG O-acetyltransferase B and, for consistency with other acetyltransferases, we recommend the acronym PatB. In addition, we confirmed that the PG O-acetylating activity of PatB was dependent upon the presence of an integral membrane protein. These data thus provide the first biochemical evidence in support of a model, first postulated over a decade ago (2), for a two-component system responsible for PG O-acetylation in N. gonorrhoeae.
The two-component model for the O-acetylation of PG was originally proposed (2) based on the process for the O-acetylation of alginate in P. aeruginosa (15). According to this model, acetate is translocated from cytoplasmic pools by an integral membrane protein through the cytoplasmic membrane and presented on its periplasmic face (Fig. 5). A specific periplasmic protein then would transfer the available acetate to the C6 hydroxyl groups of muramoyl residues on the PG sacculus. This process would thus be different to that observed in some Gram-positive bacteria for the O-acetylation of their PG. As first discovered in Staphylococcus aureus (15) and later found in Streptococcus pneumoniae (16), a single integral membrane enzyme, OatA, would appear to both translocate acetate from the cytoplasm and then transfer it to PG, but this has not been demonstrated biochemically. Indeed, despite considerable effort by ourselves and others, including those investigating the Gram-positive bacteria (15, 16), an in vitro kinetic assay has yet to be developed for any PG O-acetyltransferase and thus no quantitative data have been reported. This is understandable given the complexities that are associated with accommodating in vitro an integral membrane protein acting in concert with a peripheral membrane protein for the modification of a totally insoluble substrate. Nonetheless, in this study we were able to combine the phenotypic characterization of E. coli transformants with a crude cell-free membrane assay system developed previously to provide the first quantitative data on a PG O-acetyltransferase. These results confirmed the identity of PatB as the periplasmic PG acetyltransferase. Although crude, these assays were successfully used also to demonstrate that PG O-acetylation would only occur with E. coli transformants expressing patB in the presence of a functional acetate translocating protein WecH. (It should be noted that the E. coli genome databases does not encode homologs of AlgI or PatA.) Thus, it would appear that this integral membrane protein associated with the O-acetylation of the extracellular ECA of E. coli provides functional equivalency for the translocation of acetate to periplasm for use by recombinant PatB. In N. gonorrhoeae, this activity has been ascribed to PatA (2, 4) which is encoded immediately upstream from patB (Fig. 2) and apparently transcribed from the same promoter. Like WecH (and AlgI), hypothetical PatA is a member of the membrane-bound O-acyltransferase (MBOAT) (35) family of proteins. Unfortunately, however, the cloning of patA has yet to be reported and the biochemical properties of the protein product remain unknown. We have initiated these experiments, but we anticipate significant technical challenges for the reasons described above. Finally, the source of acetate for PatA/PatB activity would likely be cytoplasmic acetyl-CoA and interestingly, a hypothetical acyl-CoA synthetase is encoded immediately downstream from the poa cluster on the N. gonorrhoeae chromosome (Fig. 2).
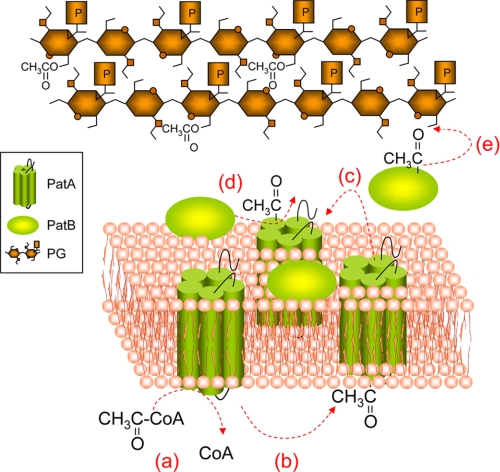
FIGURE 5.
Pathway proposed for the O-acetylation of PG in N. gonorrhoeae. Acetyl-CoA present in the cytoplasm (a) would provide the source of acetate for its translocation by integral membrane protein PatA (b) to the periplasm (c) and thereby made available to PatB (d) for transfer to PG (e).
The finding that PatB functions as an acetyltransferase was somewhat surprising given its prediction as both a member of the GDSL hydrolases and the α/β hydrolase structural superfamily. Members of the α/β hydrolase structural superfamily catalyze a variety of divergent reactions that include thioesterase, haloperoxidase, dehalogenase activities, in addition to that of acetylesterases (36). Recently, however, this family was also found to include homoserine transacetylase (37, 38), an enzyme that commits homoserine to the biosynthesis of methionine.
PatB (PacB) was previously identified as being similar to type II membrane proteins based on amino acid sequence alignments (19). Type II membrane proteins are localized to the periplasm but remain anchored to the outer leaflet of the cytoplasmic membrane by an uncleaved N-terminal signal sequence (39). Our proteomic analysis predicted that PatB possesses a cleavable signal sequence and indeed, we demonstrated experimentally that the majority of the recombinant enzyme remains free in the periplasm. This discrepancy is explained by an apparent misidentification of the start codon for the originally reported pacB gene (GenBankTM accession number AAY67447). This error resulted in the truncation of the hypothetically translated protein by eight amino acids at its N terminus. Instead, the full-length protein (accession numbers YP_002002024 and ACF30073) has a 36-amino acid leader peptide and cleavage between Ala-36 and Tyr-37 results in a His-tagged protein of 34,029 Da.
Amino acid sequence searches suggested that, in addition to N. gonorrhoeae, homologs of PatB are encoded in the genomes of a number of other important human pathogens, including N. menningitidis, P. mirabilis, and species of Helicobacter, Campylobacter, and Treponema. These bacteria are the causative agents of meningitis, urinary tract, and digestive tract infections, and syphilis. With the exception of Treponema, each of these bacteria is known to O-acetylate their respective PGs (2–4); the PG of Treponema species has yet to be characterized for O-acetylation. Without exception, homologs of PatA appear to be encoded on the chromosomes of each of these bacteria immediately upstream of PatB. This would suggest that, as with N. gonorrhoeae, a two-component system is used also by each of them for PG O-acetylation.
E. coli represents one of the few exceptions of a human commensal or pathogenic bacterium investigated to date that does not O-acetylate its PG; the other notable exception is P. aeruginosa (2). Whereas E. coli appears to provide functional equivalency for PatA activity involving WecH, this bacterium does not produce an esterase with specificity for O-acetylPG (4). Such esterases are essential to cells to control the level of O-acetylation and make cites available for both the biosynthesis of new PG and cell division (18). These enzymes are encoded in the genomes of Gram-negative bacteria that O-acetylate their PG within poa clusters immediately downstream of associated pat genes (4) (Fig. 2). Hence, E. coli transformants harboring patB would be unable to clear any O-acetylation from their PG and this deficiency would thus account for the lethal affects observed with the E. coli hosts in this study when attempts were made to clone and express patB. Even with the relatively tight control of expression provided by the pBAD system, the immediate expression of patB resulted in both cessation of cell growth and degradation of produced protein. This consequence underscores the level of control that O-acetylation has on the metabolism of PG.
The O-acetylation of PG has been demonstrated to inhibit the activity of lysozyme in a concentration-dependent manner (5–9), including that of the innate immune system. As a result, large fragments of undegraded PG persist to cause a number of pathobiological effects, including rheumatoid arthritis (reviewed in Refs. 2, 40). Given the strict requirement for a free hydroxyl group at C6 of muramoyl residues, PG O-acetylation also completely precludes the activity of the lytic transglycosylases, the major bacterial enzymes responsible for lysis of PG concomitant with its biosynthesis as well as cell replication and division. With the significance of this modification to PG metabolism, it follows that PatB and its homologous PG O-acetyltransferases may present a new target for the development of a novel class of antibacterials.
Supplementary Material
This work was supported by Canadian Institutes for Health Research Grant MOP81223 (to A. J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
- PG
- peptidoglycan
- Am
- ampicillin
- Ape
- O-acetylpeptidoglycan esterase
- Cm
- chloramphenicol
- GlcNAc
- N-acetylglucosamine
- IPTG
- isopropyl β-d-thiogalactopyranoside
- MurNAc
- N-acetylmuramic acid
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- Pat
- peptidoglycan acetyltransferase
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight.
REFERENCES
- 1.Clarke A. J. (1996) Biodegradation of Cellulose: Enzymology and Biotechnology, CRC Press, Boca Raton, FL [Google Scholar]
- 2.Clarke A. J., Strating H., Blackburn N. T. (2000) in Glycomicrobiology (Doyle R. J. ed) pp. 187–223, Plenum Publishing Co. Ltd., New York [Google Scholar]
- 3.Vollmer W. (2008) FEMS Microbiol. Rev. 32, 287–306 [DOI] [PubMed] [Google Scholar]
- 4.Weadge J. T., Pfeffer J. M., Clarke A. J. (2005) BMC Microbiology 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupont C., Clarke A. J. (1991) Eur. J. Biochem. 195, 763–769 [DOI] [PubMed] [Google Scholar]
- 6.Swim S. G., Gfell M. A., Wilde C. E., 3rd, Rosenthal R. S. (1983) Infect. Immun. 42, 446–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenthal R. S., Folkening W. J., Miller D. R., Swim S. C. (1983) Infect. Immun. 40, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenthal R. S., Blundell J. K., Perkins H. R. (1982) Infect. Immun. 37, 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blundell J. K., Smith G. J., Perkins H. R. (1980) FEMS Microbiol. Lett. 9, 259–261 [Google Scholar]
- 10.Payie K. G., Strating H., Clarke A. J. (1996) Microbiol. Drug Resistance 2, 135–140 [DOI] [PubMed] [Google Scholar]
- 11.Koraimann G. (2003) Cell Mol. Life Sci. 60, 2371–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheurwater E., Reid C., Clarke A. J. (2008) Intl. J. Biochem. Cell Biol. 40, 586–591 [DOI] [PubMed] [Google Scholar]
- 13.Höltje J. V., Mirelmen D., Sharon N., Schwarz U. (1975) J. Bacteriol. 124, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snowden M. A., Perkins H. R., Wyke A. W., Hayes M. V., Ward J. B. (1989) J. Gen. Microbiol. 135, 3015–3022 [DOI] [PubMed] [Google Scholar]
- 15.Bera A., Herbert S., Jakob A., Vollmer W., Götz F. (2005) Mol. Microbiol. 55, 778–787 [DOI] [PubMed] [Google Scholar]
- 16.Crisóstomo M. I., Vollmer W., Kharat A. S., Inhülsen S., Gehre F., Buckenmaier S., Tomasz A. (2006) Mol. Microbiol. 61, 1497–1509 [DOI] [PubMed] [Google Scholar]
- 17.Franklin M. J., Ohman D. E. (1996) J. Bacteriol. 178, 2186–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weadge J. T., Clarke A. J. (2006) Biochemistry 45, 839–851 [DOI] [PubMed] [Google Scholar]
- 19.Dillard J. P., Hackett K. T. (2005) Infect. Immun. 73, 5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheurwater E. M., Clarke A. J. (2008) J. Biol. Chem. 283, 8363–8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellogg D. S., Jr., Peacock W. L., Jr., Deacon W. E., Brown L., Pirkle D. I. (1963) J. Bacteriol. 85, 1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagotto F. J., Salimnia H., Totten P. A, Dillon J. R. (2000) Gene 244, 13–19 [DOI] [PubMed] [Google Scholar]
- 23.Hoyle B. D., Beveridge T. J. (1984) Can. J. Microbiol. 30, 204–211 [DOI] [PubMed] [Google Scholar]
- 24.Clarke A. J. (1993) Anal. Biochem. 212, 344–350 [DOI] [PubMed] [Google Scholar]
- 25.Blackburn N. T., Clarke A. J. (2002) Biochemistry 41, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 26.Laemmeli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 27.Deutscher M. P. (1974) J. Bacteriol. 118, 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupont C., Clarke A. J. (1991) J. Bacteriol. 173, 4618–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuff J. A., Clamp M. E., Siddiqui A. S., Finlay M., Barton G. J. (1998) Bioinformatics 14, 892–893 [DOI] [PubMed] [Google Scholar]
- 31.Bryson K. L., McGuffin J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. (2005) Nucleic Acids Res. 33, W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett-Lovsey R. M., Herbert A. D., Sternberg M. J., Kelley L. A. (2008) Proteins 70, 611–625 [DOI] [PubMed] [Google Scholar]
- 33.Kelley L. A., MacCallum R. M., Sternberg M. J. (2000) J. Mol. Biol. 299, 499–520 [DOI] [PubMed] [Google Scholar]
- 34.Kajimura J., Rahman A., Hsu J., Evans M. R., Gardner K. H., Rick P. D. (2006) J. Bacteriol. 188, 7542–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann K. (2000) Trends Biochem. Sci. 25, 111–112 [DOI] [PubMed] [Google Scholar]
- 36.Holmquist M. (2000) Curr. Opin. Struct. Biol. 9, 209–235 [Google Scholar]
- 37.Mirza I. A., Nazi I., Korczynska M., Wright G. D., Berghuis A. M. (2005) Biochemistry 44, 15768–17773 [DOI] [PubMed] [Google Scholar]
- 38.Wang M., Liu L., Wang Y., Wei Z., Zhang P., Li Y., Jiang X., Xu H., Gong W. (2007) Biochem. Biophys. Res. Commun. 363, 1050–1056 [DOI] [PubMed] [Google Scholar]
- 39.Pugsley A. P. (1993) Microbiol. Rev. 57, 50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke A. J., Dupont C. (1992) Can. J. Microbiol. 38, 85–91 [DOI] [PubMed] [Google Scholar]
- 41.Black W. J., Schwalbe R. S., Nachamkin I., Cannon J. G. (1984) Infect. Immun. 45, 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.