Abstract
The process of adipocyte differentiation is driven by a highly coordinated cascade of transcriptional events that results in the development of the mature adipocyte and in lipid accumulation. One of the early events of differentiation is the up-regulation of CCAAT/enhancer-binding protein β (C/EBPβ) expression. C/EBPβ then acts to up-regulate the expression of adipogenic factors such as C/EBPα, which control the late stage of adipogenesis. Retinoic acid (RA) is a potent inhibitor of adipogenesis, and its action appears to block C/EBPβ transcriptional potential early during differentiation. Using preadipocytes and mesenchymal stem cell models, we show that RA specifically blocks the occupancy of C/EBPβ of the Cebpa promoter, thereby abrogating the differentiation process. RA does not act directly on C/EBPβ but rather stimulates the expression of the transforming growth factor β-effector protein Smad3, which can interact with C/EBPβ via its Mad homology 1 domain and can interfere with C/EBPβ DNA binding. The RA-induced increase in Smad3 expression results in increased cytoplasmic and nuclear Smad3, an important event as ectopic expression of Smad3 in preadipocytes in the absence of RA treatment only modestly inhibits adipogenesis and C/EBPβ DNA binding, suggesting that Smad3 alone is not sufficient to completely recapitulate the effects of retinoic acid treatment during differentiation. However, in the absence of Smad3, RA is not able to inhibit adipocyte differentiation or to elicit a decrease in C/EBPβ DNA occupancy suggesting that Smad3 is necessary to convey the inhibitory effects of retinoic acid during adipogenesis.
Keywords: Cell/Differentiation, DNA/Transcription, Gene/Promoters, Protein/Binding/DNA, Tissue/Organ Systems/Adipocyte, Transcription/C/EBP, Transcription/SMAD, Vitamins and Cofactors/Vitamin A
Introduction
In times of caloric restriction, an organism relies on stores of fat to provide the energy necessary for survival. Adipocytes are capable of storing large amounts of lipid, and when caloric intake far exceeds the energy requirement of the organism, adipocytes can increase in size and in number to accommodate the excess.
The process of adipocyte differentiation is driven by a highly coordinated cascade of transcriptional events that results in the development of the mature adipocyte and in lipid accumulation. When confluent preadipocytes are treated with a hormonal induction mixture containing insulin, a cyclic AMP phosphodiesterase inhibitor (methylisobutylxanthine (MIX)),2 and glucocorticoids, they differentiate efficiently into mature adipocytes within 7–10 days (1, 2). Treatment with this induction mixture also causes the rapid and transient induction of the early transcriptional regulators C/EBPβ and C/EBPδ (3). Transcription by these two factors leads to the induction of other factors involved in the development of the mature adipocyte phenotype, notably C/EBPα and PPARγ.
C/EBPβ and C/EBPδ are members of the CCAAT/enhancer-binding protein family of bZIP transcription factors (4). Although ablation of Cebpb in vivo results in reduced white adipose tissue in mice and increased insulin sensitivity, the loss of Cebpd is without effect (5, 6). Although C/EBPδ can compensate partially for the loss of C/EBPβ in vivo, the regulation of C/EBPβ transcriptional activity is an important control point during adipogenesis (7, 8). In this regard, fibroblastic cell lines such as NIH 3T3, which express only low levels of endogenous C/EBPβ, are unable to differentiate into adipocytes if ectopic C/EBPβ is not provided (7, 9).
In addition to a role in adipogenesis, a role for C/EBPβ activity has been identified in numerous other biological processes, including osteoblast differentiation, mammary gland development, female reproduction, and liver regeneration (3, 11–16). Because of its important role in differentiation processes, C/EBPβ activity is tightly regulated (17). In particular, the transcriptional activity of C/EBPβ is modulated by members of the nuclear hormone receptor superfamily so that steroid hormone receptors, such as the glucocorticoid and progesterone receptor, enhance its activity (8), and retinoic acid receptors α/γ attenuate C/EBPβ-mediated transcription (13, 18).
Unlike glucocorticoids, cotreatment of preadipocytes with induction mixture and retinoic acid (RA) leads to the inhibition of differentiation (18, 19). In fact, RA is a potent repressor of adipocyte differentiation, both in vivo and in vitro (18–22). Pharmacological use of oral retinoids for the treatment of skin conditions (acne and psoriasis) or cancer causes weight loss in humans (23, 24). Obese rats fed diets supplemented with vitamin A exhibit a decrease in adiposity without change in food intake (20), whereas a vitamin A-deficient diet elicits an increase in both adiposity and body weight (22). Dietary supplementation with all-trans-RA reduces adipose marker expression, including C/EBPα (22), and in mice, RA treatment triggers a remodeling of white adipose tissue depots such that they contain less lipid and express markers of brown adipose tissue (21).
Treatment of preadipocytes with RA during the early stages of differentiation triggers a profound inhibition of both adipocyte gene expression and lipid accumulation (19). Interestingly, the period of sensitivity to RA appears to be confined to early differentiation, which overlaps with the period of C/EBPβ activity, suggesting that retinoid signaling directly interferes with early transcriptional events (19). Indeed, although both C/EBPβ and C/EBPδ are normally induced in RA-treated cells, expression of C/EBPα (a C/EBPβ target gene) as well as other adipocyte markers is abrogated (18, 19).
In mesenchymal stem cell (MSC) lines, such as C3H10T1/2, ectopic expression of C/EBPβ stimulates adipogenesis and potently represses osteoblast differentiation (13) during which C/EBPβ prevents the expression of the master osteoblast regulator runx2 via direct promoter interaction (13). Osteoblastic differentiation of these cells can be evoked by treating confluent cultures continuously with RA for a period of 3–4 weeks. Within 48 h of RA treatment, there is a loss of C/EBPβ occupancy at its negative response element in the Runx2 promoter (13) and results in RunX2 expression. Interestingly, the effect of RA, although mediated by retinoic acid receptor α/γ, occurs in the absence of interaction with the RunX2 promoter or C/EBPβ itself and requires 48 h of treatment to be induced. These results suggest that the effect of RA on C/EBPβ DNA occupancy is dependent on an intermediary.
Here, we demonstrate that retinoic acid can interfere with C/EBPβ occupancy of the Cebpa promoter during adipogenesis of MSCs and 3T3-L1 preadipocytes. RA acts to specifically stimulate the expression of Smad3, its nuclear accumulation, and its transcriptional activity. We provide evidence that Smad3 is necessary for the inhibition of adipogenesis by RA and does so by specifically interfering with C/EBPβ DNA occupancy of the Cebpa promoter.
EXPERIMENTAL PROCEDURES
Plasmids
The retroviral vector pLPCX-Smad3 (Addgene plasmid 12638), pGEX-Smad3 (Addgene plasmid 12630), and pEXL-FLAG-Smad3 (Addgene plasmid 10920) were purchased through Addgene and were kindly deposited by Dr. Rik Derynck and Dr. Bob Weinberg, respectively (25–27). pLXSN, pLXSN-C/EBPβ, RSV-βgal, and the wild type and mutant Cebpa-reporter construct have been described previously (8). Smad transcriptional activity was monitored using the CignalTM Smad Reporter (Luc) kit (SABiosciences) according to the manufacturer's instructions. A specific small hairpin RNA (shRNA) directed against the Smad3 sequence (5′-TGGAAAGGACGAAACACCGGTGCGAGAAGGCGGTCAAGAGActcgagTCTCTTGACCCCTTCTCGCACCTTTTTGAATTCGCCAGCACAGTGGT-3′) was cloned into pMKO.1 (Addgene plasmid 8452), which was kindly deposited by Dr. Bob Weinberg (28). This Smad3 target sequence has been used previously (29) and is known to not affect the expression of other Smad factors.
Antibodies
Smad expression was evaluated with the following antibodies: Smad2/3 FL-425, Smad1/5/8 N-18, and Smad4 H-552 all from Santa Cruz Biotechnology. To evaluate adipocyte differentiation, the following antibodies were used: C/EBPβ C-19, C/EBPα 14AA, adipsin M-120, and PPARγ H-100 (all from Santa Cruz Biotechnology), and anti-actin and anti-tubulin (Sigma).
Cell Culture and Differentiation
3T3-L1 preadipocytes and NIH 3T3 fibroblasts (ATCC) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% calf serum in a humidified incubator at 10% CO2. C3H10T1/2 mesenchymal stem cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in a humidified incubator at 5% CO2. Replication-incompetent retroviruses were generated in Phoenix Ampho packaging cells (ATCC). The 10-cm dishes of 50% confluent target cells were infected using 1 ml of viral supernatant in the presence of 4 μg/ml Polybrene (Sigma). Cells were selected in media containing 400 μg/ml G418 (Sigma) or 1 μg/ml puromycin (Sigma) for 10 days before differentiation to ensure retroviral expression in all cells.
To induce the differentiation of 3T3-L1 preadipocytes, 2-day post-confluent cultures were treated with regular culture media supplemented with 100 nm insulin, 1 × 10−6 m dexamethasone, and 500 μm MIX (induction mixture) with the addition of vehicle or 1 × 10−6 m all-trans-RA as indicated in figure legends. Cells were refed every 2 days with media containing 100 nm insulin, and vehicle or RA as required for a total of 8–10 days.
Adipogenic induction of C3H10T1/2 cells was achieved by induction with insulin, dexamethasone, and MIX as described above or by the treatment of subconfluent cultures with 3 μm 5-azacytidine (Sigma) for 48 h, after which cells were refed with growth media containing vehicle or 10−6 m RA as indicated in figure legends for a total of 14 days.
Transient Transcription Assays
NIH 3T3 or C3H10T1/2 cells were transiently transfected with the indicated plasmids using Lipofectamine 2000TM (Invitrogen) according to the manufacturer's instructions. Twenty four hours after transfection, media were changed to include vehicle, 1 × 10−6 m RA, or 1 ng/ml TGFβ (Sigma) as indicated for 24 h, after which cells were harvested and processed for luciferase activity and β-galactosidase activity according to standard procedures. Relative light units were corrected for transfection efficiency using the cotransfected RSV-βgal construct. Results are representative of three independent experiments. Error bars are the means ± S.E.
GST Pulldown
GST and GST-C/EBPβ were expressed in Escherichia coli strain BL21. Bacteria were lysed, and GST constructs were captured on glutathione-Sepharose beads (Sigma) and washed extensively in lysis buffer before use. Full-length Smad3 protein and the N-terminally truncated Smad3 lacking the MH1 domain (ΔMH1) were produced by in vitro translation following the manufacturer's protocol (Promega). Each binding assay was achieved by mixing 0.5 μg of GST protein with 10 μl of Smad3 translation product followed by incubation with rotation at 4 °C for 90 min. After extensive washing in binding buffer, precipitates were separated by SDS-PAGE, and detection of bound Smad3 protein was achieved by Western analysis (anti-Smad2/3 antibody, FL-425, Santa Cruz Biotechnology). Results are representative of three independent experiments.
Nuclear Localization Studies
Differentiating 3T3-L1 preadipocytes were harvested in phosphate-buffered saline and immediately fractionated into cytoplasmic and nuclear extracts according to standard procedures. Equal amounts of protein were analyzed by Western blotting as determined by the Bradford assay. Integrity of the nucleus was verified using tubulin expression as a cytoplasmic marker and C/EBPβ as a nuclear marker. Results are representative of three independent experiments.
Avidin Biotin-conjugated DNA Assays
Five hundred nanograms of whole cell lysates from C3H10T1/2 cells was incubated with 2 μg of 5′ biotin-tagged double-stranded oligonucleotide in binding buffer (20 mm HEPES (pH 7.7), 50 mm KCl, 20% glycerol, 0.1% Nonidet P-40, 2 μg of sheared salmon sperm DNA) in the presence of recombinant GST or GST-Smad3. Oligonucleotides were then immunoprecipitated with streptavidin-conjugated magnetic beads (Dynal, Invitrogen), and precipitates were washed extensively in binding buffer and resolved by SDS-PAGE. Binding was evaluated by Western blot analysis using anti-C/EBPβ antibody (C-19; Santa Cruz Biotechnology). Results are representative of three independent experiments.
Semi-quantitative RT-PCR
For RT-PCR, RNA was extracted using the RNeasyTM kit (Qiagen) and was reverse-transcribed using oligodeoxythymidine and Superscript IIITM (Invitrogen) according to the manufacturer's instructions. PCRs were optimized to determine the linear phase of amplification, and results were compared with glyceraldehyde 3-phosphate message. Primer sequences used for amplification are available upon request. Results are representative of three independent experiments.
Chromatin Immunoprecipitation
C3H10T1/2 and 3T3-L1 cells were treated as indicated in the figure legends for 48 h, and cells were washed twice in serum-free media and treated with 1% formaldehyde at room temperature for 10 min. ChIP was performed essentially as described (12) using C/EBPβ C-19 (Santa Cruz Biotechnology) for precipitation at 4 °C overnight. A type-matched nonspecific antibody was used as a negative control. DNA fragments were purified using the QIAquickTM PCR purification kit (Qiagen) and amplified by PCR. Results shown are representative of a minimum of three independent experiments.
RESULTS
Retinoic Acid Is a Potent Inhibitor of Adipogenesis
Treatment of preadipocytes with RA, especially during the first 48 h of differentiation, results in a profound inhibition of adipogenesis (18). This blockade in differentiation has been attributed to abrogated C/EBPβ-mediated transcription that is normally responsible for initiating the differentiation process (6, 8, 30–32). However, the mechanism of inhibition has remained elusive. Given that retinoic acid has been shown to enhance osteoblastic differentiation of MSC cultures and that this activity has been attributed to a decrease in C/EBPβ occupancy of the Runx2 promoter, we asked whether RA treatment could inhibit the differentiation of these cells into adipocytes (13). To address this question, confluent cultures of C3H10T1/2 mesenchymal stem cells, which are known to differentiate into adipocytes, osteoblasts, chondrocytes, and skeletal muscle (33), were induced to differentiate into adipocytes with a mixture containing glucocorticoids, insulin, and a cAMP phosphodiesterase inhibitor (MIX) in the presence or absence of RA. Two days following induction, the cells were refed every 48 h with medium containing insulin and vehicle or RA and allowed to differentiate for a total of 8 days. It was noted that although C3H10T1/2 cells spontaneously differentiated into adipocytes when kept in culture, treatment with adipogenic mixture had little additional stimulatory effect, despite identical treatment evoking robust differentiation of preadipocytes such as 3T3-L1. Indeed, both mesenchymal stem cells and preadipocytes share many similarities in their adipogenic programs, including the transcriptional cascade, which drives adipogenesis and an early phase of clonal expansion. However, in preadipocytes, clonal expansion is a necessary step that is regulated by C/EBPβ and is required for the normal expression of the adipocyte master regulator PPARγ. Although MSCs undergo clonal expansion, this process can be blocked without impacting PPARγ expression (34).
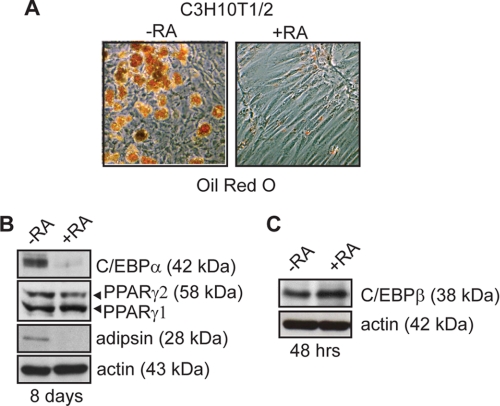
Despite the low levels of observed differentiation in C3H10T1/2 cells, pockets of lipid-laden cells could be observed in differentiating cultures in the absence of RA treatment (Fig. 1A), and adipocyte markers C/EBPα, PPARγ, and adipsin could be detected by Western blotting (Fig. 1B). The addition of RA to the induction mixture profoundly inhibited adipogenesis, resulting in a marked reduction in Oil Red O staining and adipocyte marker expression following 8 days of differentiation (Fig. 1, A and B). It should be noted that although RA treatment reduced the expression of PPARγ2, the PPARγ isoform associated with adipogenesis, it did not impact on PPARγ1 expression (Fig. 1B). As demonstrated previously, RA treatment did not impact C/EBPβ expression in these cells (Fig. 1C) (13).
FIGURE 1.
Retinoic acid treatment inhibits adipocyte differentiation of mesenchymal stem cells. A, Oil red O micrographs of C3H10T1/2 cells retrovirally transduced to express C/EBPβ or with empty virus and induced to differentiate with insulin, MIX, and dexamethasone for 2 days in the presence or absence of RA. Following induction, cells were refed every 2 days with growth media containing insulin and vehicle or RA as indicated for a total of 8 days. B, Western analysis of adipocyte marker expression 8 days after induction to differentiate in the presence or absence of RA. Actin is shown as a loading control. C, Western analysis of C/EBPβ expression following a 48-h retinoic acid treatment. Actin serves as a loading control.
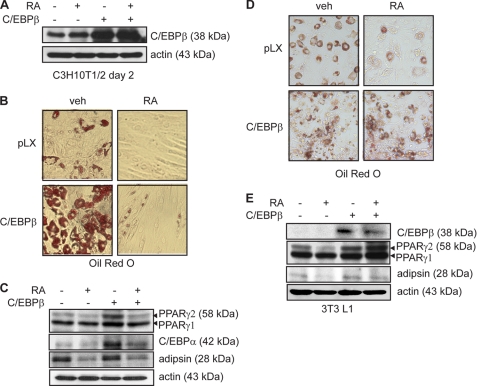
The work of Schwarz et al. (18) suggested that the retinoic acid-induced block of adipogenesis in preadipocytes occurred downstream of C/EBPβ induction, and because RA has no effect on C/EBPβ induction or protein expression, it likely involves a crippling of C/EBPβ transactivation activity. Consistent with this study, overexpression of C/EBPβ in C3H10T1/2 mesenchymal stem cells by retroviral transduction resulted in a robust stimulation of adipogenesis following induction to differentiate with induction mixture (data not shown). This strong induction of differentiation by C/EBPβ expression prevented the direct comparison of adipocyte marker expression in control and C/EBPβ cells. To overcome this difficulty, we induced differentiation of empty virus control (pLX) and C/EBPβ-expressing cells with the DNA demethyltransferase inhibitor 5-azacytidine. 5-Azacytidine treatment promotes the differentiation of C3H10T1/2 cells into all possible fates, although adipocytes are the first to appear in culture. Induction of differentiation with 5-azacytidine stimulated modest adipocyte differentiation in empty virus control cells (pLX) as evidenced by increased lipid accumulation (Fig. 2B) and adipocyte marker expression (Fig. 2C). Addition of RA to the induction and growth media of control cultures inhibited lipid accumulation completely (Fig. 2B) and markedly reduced adipsin and PPARγ2 expression (Fig. 2C).
FIGURE 2.
C/EBPβ overexpression prevents the inhibition of adipogenesis by RA. A, Western analysis of C/EBPβ expression in C3H10T1/2 cells retrovirally transduced to express C/EBPβ or with empty virus (pLX) and induced to differentiate with a 2-day 5-azacytidine treatment in the presence or absence of RA. Actin serves as a loading control. B, Oil red O micrographs of cells transduced and induced to differentiate as in A in the presence or absence of RA for 14 days. veh, vehicle. C, Western analysis of adipocyte marker expression in cells transduced and induced as in A. Actin serves as a loading control. D, Oil red O micrographs of 3T3-L1 cells retrovirally transduced to express C/EBPβ or with empty virus (pLX) and induced to differentiate with insulin, MIX, and dexamethasone for 2 days in the presence or absence of RA. Following induction, cells were refed every 2 days with growth media containing insulin and vehicle or RA as indicated for a total of 10 days. E, Western analysis of C/EBPβ and adipocyte marker expression in cells transduced and induced as in D. Actin serves as a loading control.
Differentiation of C/EBPβ-expressing C3H10T1/2 cultures resulted in a large increase in the number of lipid-laden cells (Fig. 2B) and robust expression of PPARγ2 and C/EBPα (Fig. 2C) over control cells. Similar to control cells, cotreatment with RA resulted in the inhibition of both lipid accumulation and adipocyte marker expression. However, although treatment with RA potently inhibited differentiation of both control and C/EBPβ-expressing cells, overexpression of C/EBPβ offered partial protection from the effects of RA, as evidenced by the presence of neutral fat (Fig. 2B) and increased levels of both C/EBPα and PPARγ2 expression as compared with control cells treated with RA (Fig. 2C). When C/EBPβ was overexpressed in 3T3-L1 cells, these cells began to differentiate in the absence of hormonal stimulation. As expected, the ectopic expression of C/EBPβ in these cells potentiated both the development of the adipocyte phenotype and adipocyte marker expression following hormonal induction (Fig. 2, D and E). Addition of RA to the media blocked the differentiation of the control cells and reduced their adipocyte marker expression (Fig. 2, D and E). In contrast to the C3H10T1/2 model, in C/EBPβ-expressing 3T3-L1 cells, RA was no longer able to block differentiation, as evidenced by the robust adipocyte marker expression comparable with that of cells not treated with RA (Fig. 2, D and E). These results suggest that overexpression of C/EBPβ can overcome the inhibitory effects of RA during adipocyte differentiation of preadipocytes. Indeed, we have observed that as little as a 2-fold overexpression of C/EBPβ is sufficient to block RA-induced osteoblastogenesis in C3H10T1/2 cells (12).
Retinoic Acid Decreases C/EBPβ Occupancy of the Cebpa Promoter
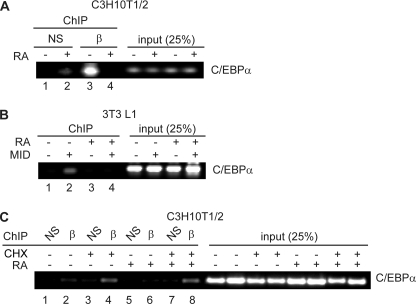
We have observed previously that treatment of mesenchymal stem cells with RA promoted osteoblastogenesis by reducing C/EBPβ occupancy of the Runx2 promoter, where it acts as an inhibitor (13). Because C/EBPβ is an important positive-acting transcription factor during adipogenesis, and the inhibitory effects of RA can be blocked by C/EBPβ overexpression in preadipocytes, we asked whether RA treatment resulted in decreased occupancy of C/EBPβ at adipogenic promoters, notably the Cebpa promoter. Indeed, in both C3H10T1/2 mesenchymal stem cells and 3T3-L1 preadipocytes, RA treatment resulted in a decrease in C/EBPβ occupancy of the Cebpa promoter, a principal C/EBPβ target gene (Fig. 3). In C3H10T1/2 cells induced to differentiate for 48 h with adipogenic induction mixture, C/EBPβ could be observed occupying the Cebpa promoter in the absence of RA treatment as measured by chromatin immunoprecipitation (Fig. 3A, lane 3). Cotreatment with RA resulted in a dramatic decrease in C/EBPβ occupancy of the Cebpa promoter without affecting C/EBPβ protein expression (Fig. 3A, lane 4, and Fig. 1C). This evidence in addition to the RA-evoked loss of C/EBPβ occupancy of the Runx2 promoter during osteoblastogenesis suggests that RA acts to decrease C/EBPβ occupancy in multiple promoter contexts (12).
FIGURE 3.
Retinoic acid treatment reduces C/EBPβ occupancy of the Cebpa promoter in vivo. A, ChIP analysis of C/EBPβ occupancy of the Cebpa promoter in C3H10T1/2 cells following treatment with vehicle or RA for 48 h as indicated. Isolated chromatin was immunoprecipitated using anti-C/EBPβ antibody (β) or with a nonspecific type-matched antibody (NS). B, ChIP analysis of C/EBPβ occupancy of the Cebpa promoter in 3T3-L1 cells following induction to differentiate with adipogenic mixture (MIX, insulin, dexamethasone = MID) and treated with vehicle or RA for 48 h as indicated. Unlike C3H10T1/2 cells, in the absence of adipogenic mixture, C/EBPβ protein levels are low and resulting in no occupancy of the Cebpa promoter. Pull down from unstimulated cells with anti-C/EBPβ antibody (lane 1) serves as a negative control for the ChIP. C, ChIP analysis of C/EBPβ occupancy (β) of the Cebpa promoter in C3H10T1/2 cells treated with RA and cycloheximide (CHX) as indicated for 48 h. Input represents 25% of the material used for immunoprecipitation. NS, nonspecific type-matched antibody.
Unlike C3H10T1/2 cells, unstimulated 3T3-L1 preadipocytes express only very low levels of C/EBPβ. In these cells, C/EBPβ cannot be detected interacting with the Cebpa promoter in the absence of treatment with induction mixture following immunoprecipitation with anti-C/EBPβ antibody (Fig. 3B, lane 1) (8). This condition therefore serves as a negative control for the ChIP. Following a 48-h treatment with adipogenic induction mixture (MID is MIX, insulin, and dexamethasone), C/EBPβ could be seen occupying the Cebpa promoter (Fig. 3B, lane 2). As was observed in MSCs, addition of RA to the medium of stimulated cells decreased C/EBPβ occupancy (Fig. 3B, lane 4) at this promoter.
The long RA treatments (48 h) required to elicit loss of C/EBPβ occupancy at target promoters suggest that the actions of RA are not direct. Rather, the time frame permits the retinoic acid receptor to act on other promoter targets and stimulate the up-regulation of a second factor that in turn could act on C/EBPβ DNA binding. To determine whether de novo protein synthesis is required for the RA-induced loss of C/EBPβ occupancy of the Cebpa promoter, we treated C3H10T1/2 cells with RA and cycloheximide, a protein synthesis inhibitor. This experiment was not possible in 3T3-L1 preadipocytes, as the treatment with cycloheximide inhibited the up-regulation of C/EBPβ. In untreated cells and those treated with cycloheximide, C/EBPβ can be seen occupying the Cebpa promoter (Fig. 3C, lane 2). As demonstrated in Fig. 3A, treatment with RA resulted in loss of C/EBPβ occupancy at this promoter. This effect of RA was abrogated by cotreatment with RA and cycloheximide, suggesting that the RA-induced reduction in C/EBPβ occupancy of the Cebpa promoter requires de novo protein synthesis (Fig. 3C, lane 8).
Smad3 Is a Novel Retinoic Acid Target Gene in Preadipocytes
Based on the kinetics of C/EBPβ displacement, the lack of protein-protein interaction between C/EBPβ and retinoic acid receptors, and the cycloheximide sensitivity (13), we hypothesized that RA induces the expression of a second protein that acts to modify C/EBPβ DNA affinity. We thus undertook the task of identifying a protein target that was both up-regulated by RA treatment and was able to modulate C/EBPβ DNA-binding affinity. Of interest, CHOP, a C/EBP family member and known negative regulator of C/EBPβ during adipogenesis has been shown to be up-regulated by RA in the blood (35). However, in both 3T3-L1 preadipocytes and C3H10T1/2 mesenchymal stem cells, RA failed to stimulate CHOP expression (data not shown). Given that TGFβ signaling can also inhibit adipogenesis, that Smad3/4 have been shown to interfere with C/EBPβ-mediated transcriptional responses, and that Smad4 can disrupt C/EBPβ binding to the Hp (haptoglobin) promoter in vitro (36), we hypothesized that a member of the Smad family of transcription factors may be responsible for transducing the effects of retinoic acid in our system. Smad3 has been shown to inhibit adipogenesis of 3T3 F442A preadipocytes, but it remains unclear whether Smad3 can itself inhibit C/EBPβ-dependent adipogenesis of 3T3-L1 preadipocytes or mesenchymal stem cells. Furthermore, Smad3 has not been described as an RA target gene in either of these systems, although recently Smad3 expression has been shown to be induced by RA treatment in CD4+ T cells (37).
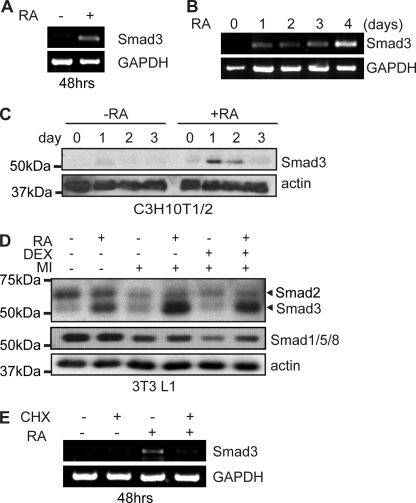
Following a 48-h treatment with RA, we observed a robust induction of Smad3 mRNA expression as measured by semi-quantitative RT-PCR in C3H10T1/2 cells (Fig. 4A). Further time course analysis revealed that although Smad3 mRNA is up-regulated by a shorter (24 h) RA treatment, mRNA continues to accumulate and reaches greater levels after 4 days of continuous RA treatment (Fig. 4B).
FIGURE 4.
Retinoic acid treatment triggers up-regulation of Smad3 protein expression. A, semi-quantitative RT-PCR analysis of Smad3 mRNA expression in C3H10T1/2 cells treated with retinoic acid or vehicle for 48 h. Amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is used as a loading control. B, semi-quantitative RT-PCR analysis of Smad3 mRNA expression in C3H10T1/2 cells treated with RA for the indicated times. Amplification of glyceraldehyde-3-phosphate dehydrogenase is used as a loading control. C, Western analysis of Smad3 expression in C3H10T1/2 cells treated with vehicle or RA for the indicated time period in days. Actin serves as a loading control. D, Western analysis of Smad expression in 2-day post-confluent 3T3L1 cells induced to differentiate with the cAMP phosphodiesterase inhibitor MIX and insulin (+MI) in the presence or absence of the synthetic glucocorticoid dexamethasone (DEX) and RA as indicated for 48 h. Actin serves as a loading control. E, C3H10T1/2 cells treated with RA and cycloheximide (CHX) as indicated for 48 h were harvested for RNA isolation and semi-quantitative RT-PCR for Smad3 and glyceraldehyde-3-phosphate dehydrogenase.
Treatment with retinoic acid also resulted in a robust but transient increase in Smad3 protein expression in these same cells (Fig. 4C). We observed an increase in Smad3 expression after 24 h of RA treatment (Fig. 4C). Despite continued incubation with RA, Smad3 levels returned to base line after 3 days of treatment (Fig. 4C). Interestingly, the time frame of Smad3 up-regulation correlates tightly with the window of C/EBPβ activity during adipogenesis (7, 8). In 3T3-L1 preadipocytes, a 48-h RA treatment induced Smad3 protein expression in 3T3-L1 cells irrespective of cotreatment with the adipogenic mixture to induce C/EBPβ expression and differentiation (Fig. 4D). RA treatment did not stimulate the expression of Smad2, which has a greater molecular weight and is recognized by a Smad2/3 antibody. Rather, Smad2 levels are down-regulated by treatment with MIX and insulin, but not further affected by the addition of the synthetic glucocorticoid dexamethasone. Furthermore, the levels of other receptor-associated Smads (Smad1/5/8) were also down-regulated by adipogenic induction media and unchanged by RA treatment (Fig. 4D). Taken together, these results suggest that Smad3 is positively regulated by RA treatment in both mesenchymal stem cells and preadipocytes.
Although both protein and mRNA levels of Smad3 are rapidly induced by RA treatment, mRNA levels continue to rise while protein levels are down-regulated, suggesting a more complex mechanism for regulation by RA. To establish whether de novo protein synthesis is required for the induction of Smad3 mRNA expression, we treated C3H10T1/2 cells with the protein synthesis inhibitor cycloheximide and evaluated Smad3 mRNA levels following RA treatment by semi-quantitative RT-PCR. Although Smad3 mRNA is induced following a 48-h RA treatment, cotreatment with cycloheximide blocked this induction (Fig. 4E), indicating that de novo protein synthesis is required for the induction of Smad3 mRNA by RA and suggesting that the regulation of Smad3 expression by RA occurs via an indirect mechanism.
C/EBPβ DNA Occupancy Is Abrogated by Smad3
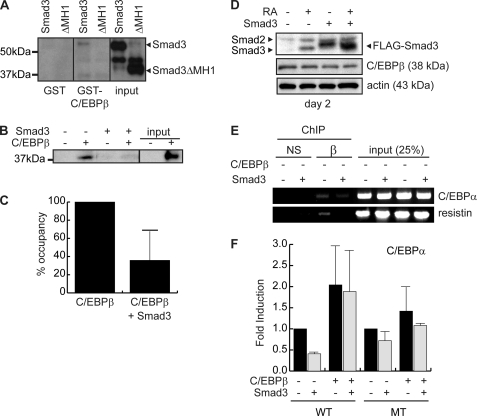
In HepG2 cells, the association of C/EBPβ with Smad4 is thought to provoke a decrease in occupancy of the Hp promoter in vitro (36). During both osteoblastogenesis and adipogenesis, we have observed that RA treatment results in decreased DNA occupancy by C/EBPβ (Fig. 3), which may be attributed to an association of C/EBPβ with Smad factors (13). Using a GST pulldown experiment, we evaluated the interaction of GST-C/EBPβ with in vitro translated Smad3 (Fig. 5A). Whereas GST alone did not interact with Smad3, we found a weak interaction between GST-C/EBPβ and Smad3 consistent with a previous report (38). Further analysis using an N-terminally truncated Smad3, which lacks the Smad MH1 domain (ΔMH1), revealed that this domain was necessary for the interaction of Smad3 with GST-C/EBPβ as the truncated Smad3 was unable to interact with GST-C/EBPβ (Fig. 5A). Previous studies concentrating on C/EBPδ have demonstrated that Smad3 interacts with this factor via its bZIP domain, a highly conserved region responsible for heterodimerization and DNA binding (38). Interaction of Smad3 with the C/EBPβ bZIP domain could conceivably inhibit DNA binding through steric interference.
FIGURE 5.
Smad3 interferes with C/EBPβ occupancy of target promoters. A, interaction of in vitro translated full-length Smad3 and a truncated Smad3 lacking the MH1 domain (ΔMH1) with GST and GST-C/EBPβ. Following binding, precipitated Smad3 was revealed by Western blotting. B, avidin biotin-conjugated DNA assay evaluating the interaction of endogenous C/EBPβ from 3T3-L1 cells induced to differentiate for 24 h, with a double-stranded oligonucleotide coding for four repeats of a C/EBP consensus motif in the presence or absence of recombinant Smad3. Input represents 10% of the material used in the binding reaction. C, quantification of the interaction of C/EBPβ with a consensus DNA motif as in A by phosphorimager analysis. Data represent results from three experiments. Error bars represent the mean ± S.E. D, Western analysis of Smad and C/EBPβ expression in 3T3-L1 cells retrovirally transduced to express Smad3 or with empty virus and induced to differentiate into adipocytes with standard mixture for 48 h in the presence or absence of RA as indicated. Note that retrovirally expressed Smad3 is FLAG-tagged and thus migrates higher than endogenous Smad3. Endogenous Smad3 is indicated as the lower band observed in RA-treated empty virus control cells. The larger Smad2 is indicated by an arrowhead. Actin is used as a loading control. E, ChIP analysis of C/EBPβ occupancy of the Cebpa (C/EBPα) promoter and the Retn (resistin) promoter in 3T3-L1 cells retrovirally transduced to express Smad3 or with empty virus and induced to differentiate for 48 h. Chromatin was immunoprecipitated using anti-C/EBPβ antibody (β) or a type-matched nonspecific antibody (NS). Inputs represent 25% of the material used for precipitation. F, transient transcription assay in C3H10T1/2 cells measuring activation of the mouse Cebpa promoter by C/EBPβ and Smad3. Both the wild type (WT) promoter and a mutant reporter (MT) with the C/EBP-response element abolished were used. Data are reported as fold induction over the activity of the respective promoters in the absence of C/EBPβ and Smad3. Luciferase activity was corrected with β-galactosidase activity from a cotransfected reporter to correct for transfection efficiency. Data represent the means of two independent experiments performed in duplicate. Error bars represent the mean ± S.E.
To measure the impact of Smad3 expression on C/EBPβ DNA binding ability, we used an in vitro avidin biotin-conjugated DNA assay, which measures the occupancy of endogenous C/EBPβ on a short double-stranded oligonucleotide encoding four consensus C/EBP elements. Although C/EBPβ interacts efficiently with this element in the absence of Smad3, addition of GST-Smad3 reduced interaction with the promoter element by greater than 60% as compared with addition of GST alone (Fig. 5, B and C). This is the first study implicating Smad3 in the modulation of C/EBPβ DNA binding activity, although Smad4-mediated inhibition of DNA binding has been described in vitro (36).
To test the effects of ectopic Smad3 expression on C/EBPβ DNA binding in vivo, we produced pooled stable 3T3-L1 cells lines expressing FLAG-tagged Smad3 by retroviral transduction (Fig. 5D). The expression of Smad3 in our stable cell line was ∼5 times greater than the levels of Smad3 in control cells following induction with RA (Fig. 5D), and this level of expression did not impact on the C/EBPβ expression levels as demonstrated by Western blotting (Fig. 5D).
To evaluate the effect of ectopic Smad3 expression on C/EBPβ promoter occupancy in vivo, we used chromatin immunoprecipitation to assess C/EBPβ occupancy of the C/EBPα and the Retn (resistin) promoter (Fig. 5F). In cells overexpressing Smad3, the occupancy of C/EBPβ at both the Cebpa promoter and the Retn promoter was reduced as compared with the occupancy observed in empty vector controls (Fig. 5E). However, occupancy of the Cebpa promoter, although reduced, was still observed in Smad3-expressing cells, suggesting that ectopic expression of Smad3 alone cannot completely recapitulate the effects of RA in this system. Thus, although Smad3 can act to interfere with C/EBPβ DNA binding in vitro, ectopic expression of Smad3 in itself is not sufficient for this effect in vivo. Despite the modest reduction of C/EBPβ occupancy in vivo following Smad3 overexpression, we observed a reduction of ∼50% in the C/EBPβ-mediated transactivation from the Cebpa promoter in a transient transcription assay in C3H10T1/2 cells (Fig. 5F). This inhibition of transcription by ectopic Smad3 is lost upon overexpression of C/EBPβ in these cells. Furthermore, Smad3 was unable to inhibit transcription from a mutant Cebpa promoter, which has the C/EBP element abolished by site-directed mutagenesis, suggesting that the repressive actions of Smad3 at this promoter are mediated through the C/EBP element (Fig. 5F).
Retinoic Acid Treatment Induces Nuclear Accumulation and Activation of Smad3
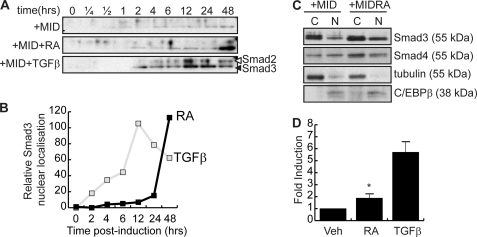
Smad3 is a TGFβ receptor-associated transcription factor that, following TGFβ binding to its receptor, is phosphorylated leading to its heterodimerization with Smad4 and its nuclear accumulation (39). Although this mechanism appears to be the primary mode of nuclear accumulation for Smad3, Smad3 nuclear localization has been described in the absence of Smad4 (40). Because C/EBPβ is a nuclear protein and RA treatment does not affect its subcellular localization (data not shown), Smad3 must gain access to the nucleus in the absence of TGFβ signaling to interfere with C/EBPβ DNA binding activity. To assess the subcellular localization of Smad3 following retinoic acid treatment, 3T3-L1 cells were treated with the adipogenic induction mixture, including vehicle, TFGβ, or RA for 48 h, and nuclear Smad3 was evaluated by subcellular fractionation and Western blotting (Fig. 6). Western analysis of differentiating 3T3-L1 cell nuclear extracts revealed that although adipogenic induction mixtures had little effect on Smad3 nuclear accumulation, TGFβ treatment resulted in nuclear accumulation of both Smad3 and Smad2 (Fig. 6A). Treatment with RA, which we have demonstrated to increase cellular Smad3 levels, resulted in an increase in nuclear Smad3 but not Smad2. Peak levels of nuclear Smad3 were observed with 48 h of RA treatment, corresponding to the peak of Smad3 protein expression (Fig. 6, A and B, and Fig. 4, C and D). For TGFβ-treated cells, maximal nuclear accumulation of Smad3 was observed between 12 and 24 h of treatment (Fig. 6D), whereas levels of nuclear Smad3 were highest following 48 h of RA treatment. The kinetics of Smad3 nuclear accumulation is in concordance with our previous observations that noted that the displacement of C/EBPβ by RA required at least 48 h to be produced, with shorter hormone treatments being ineffective (13).
FIGURE 6.
RA treatment increases nuclear Smad3. A, time course of Western analysis of nuclear Smad3 in 3T3-L1 cells induced to differentiate with MIX, insulin, and dexamethasone (DEX) (+MID) and treated with vehicle, RA, or TGFβ for the times indicated. B, quantification of nuclear Smad3 following RA or TGFβ treatment as in A. C, nuclear (N) and cytoplasmic (C) localization of Smad3 and Smad4 in 3T3-L1 cells induced to differentiate for 48 h in the presence or absence of RA. Tubulin is a cytoplasmic marker, whereas C/EBPβ is used to evaluate the integrity of the nuclear compartment. D, transient transcription assay measuring the activation of a synthetic Smad-responsive reporter construct by a 48-h RA treatment in NIH 3T3 cells. Relative light units were corrected for β-galactosidase activity from a cotransfected constitutively active reporter construct to correct for transfection efficiency. Data represent three independent experiments, and error bars represent the mean ± S.E. (*, p < 0.05). Veh, vehicle.
Given that cellular Smad3 levels increase following RA treatment, the increase in nuclear Smad3 may be a result of higher cellular levels of this factor. However, the molecular weight of Smad3 precludes the passive accumulation into the cell nucleus. Despite this, many different modes of Smad3 transport into the nucleus have been described. The classic mode of Smad3 nuclear transport involves heterodimerization with Smad4. In RA-treated cells, we were unable to correlate Smad4 nuclear accumulation with the presence of nuclear Smad3 (Fig. 6C). Nuclear and cytoplasmic extracts prepared from differentiating 3T3-L1 preadipocytes cotreated with RA or vehicle for 48 h indicate that although RA treatment stimulates the up-regulation of Smad3 expression in both the cytoplasm and the nucleus, Smad4 is not redistributed to the nucleus accordingly, suggesting that the increased nuclear Smad3 occurs independently of Smad4 (Fig. 6C). Tubulin and C/EBPβ are shown as markers of the cytoplasmic and nuclear fractions, respectively. Furthermore, despite the reported ability of RA to stimulate Smad3 phosphorylation via MAPK activation, we were unable to correlate changes in phosphorylation with RA-induced nuclear accumulation of Smad3 (data not shown) (41). Taken together, these results suggest that RA may act at least in part to induce an increase in nuclear Smad3 in a Smad4-independent fashion.
Despite the novel mechanics of Smad3 nuclear accumulation in retinoic acid-treated cells, RA-induced Smad3 was able to drive Smad-mediated transcription, suggesting that this population of Smad3 is functional (Fig. 6D). Transient transcription experiments in NIH 3T3 cells revealed that treatment with RA in the absence of ectopically expressed Smad3 was sufficient to induce a 2-fold activation of a synthetic Smad-responsive promoter (Fig. 6D). NIH 3T3 cells were chosen for this experiment because they have the same origin as 3T3-L1 cells but are easier to transfect. In contrast to the RA-treated cells, ectopic expression of Smad3 and treatment with TGFβ resulted in a 5-fold activation of this same promoter. Taken together, these results strongly suggest that treatment of cells with RA results in the up-regulation of Smad3 expression such that transcription from Smad-responsive genes can be induced.
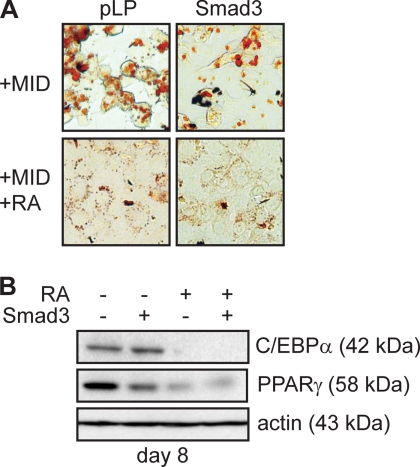
Smad3 Overexpression Is Not Sufficient to Inhibit Adipogenesis
To directly investigate the role of Smad3 in the inhibition of adipogenesis by RA, we used the pooled stable 3T3-L1 cell lines created by retroviral transduction described in Fig. 5. These cells express ∼5 times the Smad3 as control cells (Fig. 5D). When these cells were induced to differentiate for 8 days, we observed a significant reduction in Oil red O-positive cells (Fig. 7A) in cells overexpressing Smad3 as compared with empty vector controls. However, adipogenesis was more potently inhibited by the addition of RA to the adipogenic mixture in both Smad3-expressing and empty vector control cell lines (Fig. 7A). Despite a significant reduction in the number of lipid-containing cells in cultures overexpressing Smad3, Western analysis of adipogenic marker expression indicated that whereas RA potently inhibited the expression of both C/EBPα and PPARγ2, ectopic expression of Smad3 alone was unable to entirely recapitulate these effects. Although C/EBPα levels were unaffected by Smad3 expression, PPARγ2 levels were reduced 2-fold in Smad3-expressing cells stimulated to differentiate for 8 days (Fig. 7B). Taken together with the ChIP data that indicates that Smad3 overexpression is insufficient to prevent C/EBPβ DNA binding in vivo, these results suggest that in 3T3-L1 preadipocytes, expression of Smad3 alone was not sufficient to inhibit adipocyte differentiation in the absence of RA treatment (7, 8). Indeed, if RA acts to actively promote Smad3 nuclear accumulation in addition to stimulating its expression, ectopic expression of Smad3 alone would not be expected to exert its effects on a nuclear target such as C/EBPβ in the absence of RA.
FIGURE 7.
Smad3 overexpression is not sufficient to inhibit adipocyte differentiation. A, Oil red O micrographs of 3T3-L1 cells retrovirally infected to express Smad3 or with empty vector (pLP) and induced to differentiate into adipocytes with standard mixture for 8 days in the presence or absence of RA. B, Western analysis of adipocyte marker expression in 3T3-L1 cells transduced and induced to differentiate as in A. Actin expression is shown as a loading control. MID, MIX, insulin, and dexamethasone.
Up-regulation of Smad3 by Retinoic Acid Is Necessary for Inhibition of Adipogenesis by RA
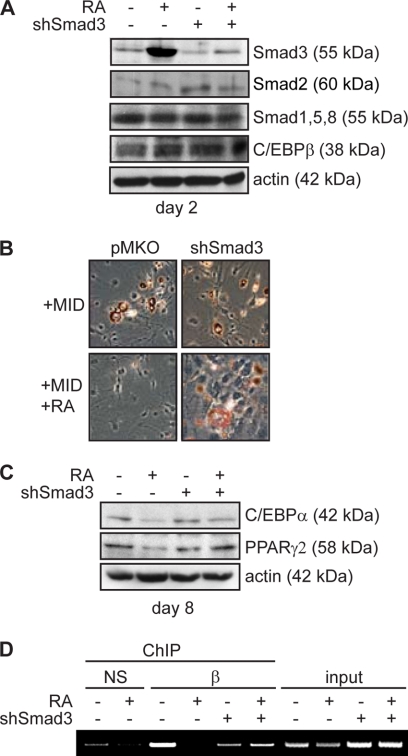
To determine whether the stimulation of Smad3 expression by RA is required for the inhibition of adipogenesis, we retrovirally transduced 3T3-L1 cells with virus to express a small hairpin RNA directed against Smad3 (shSmad3) or with empty virus. The shRNA was designed to a region specific to Smad3 and is not predicted to bind any other known Smad protein. This targeting sequence has been used with success by other groups without cross-reactivity (29). Expression of the shRNA reduced both the basal and RA-induced levels of Smad3 such that following RA treatment, the cells expressing shRNA did not up-regulate Smad3 expression (Fig. 8A). In addition, as predicted the shRNA did not affect Smad2 levels, the Smad1/5/8 levels, or C/EBPβ expression (Fig. 8A).
FIGURE 8.
Inhibition of adipogenesis by RA is abrogated by loss of Smad3 expression. A, Western analysis of Smad and C/EBPβ expression in 3T3-L1 cells retrovirally transduced to express a small hairpin RNA directed against Smad3 (shSmad3) or with empty vector (pMKO) and induced to differentiate into adipocytes with standard mixture for 48 h in the presence or absence of RA. B, Oil red O micrographs of 3T3-L1 cells retrovirally transduced as in A and induced to differentiate into adipocytes with induction mixture for 8 days. MID, MIX, insulin, and dexamethasone. C, Western analysis of adipocyte marker expression in 3T3-L1 cells transduced and induced to differentiate as in B. D, ChIP analysis of C/EBPβ occupancy of the Cebpa promoter in cells retrovirally transduced and induced to differentiate as in A. Chromatin was immunoprecipitated with anti-C/EBPβ antibody (β) or a type-matched nonspecific antibody (NS) as indicated. Input represents 25% of the material used for immunoprecipitation.
When these cells were induced to differentiate with standard adipogenic mixture and cultured for 8 days, no apparent differences in morphology or lipid accumulation were observed between shSmad3-expressing and empty vector controls (Fig. 8B). However, when cells were cotreated with RA, lipid accumulation was profoundly inhibited in empty vector control cells but largely unaffected in shSmad3-expressing cells (Fig. 8B). These results are supported by analysis of adipocyte marker expression (Fig. 8C). In the absence of RA, both empty vector control cells and shSmad3-expressing cells express similar levels of C/EBPα and PPARγ2. Following treatment with RA, although empty vector control cells display reduced levels of both C/EBPα and PPARγ2, shSmad3-expressing cells maintain high levels of these proteins (Fig. 8C).
Analysis of C/EBPβ occupancy of the Cebpa promoter by ChIP in shSmad3-expressing cells revealed that Smad3 was required for the modulation of C/EBPβ DNA binding ability in 3T3-L1 cells following RA treatment. Although RA treatment reduced C/EBPβ occupancy of the Cebpa promoter in empty vector control cells induced to differentiate for 48 h, C/EBPβ occupancy was unaffected by retinoic acid treatment in shSmad3-expressing cells. Taken together, these data suggest that the stimulation of Smad3 expression by retinoic acid is necessary for the inhibition of adipogenesis by this hormone and the reduction of C/EBPβ occupancy.
DISCUSSION
Despite a role for retinoids in the inhibition of adipocyte differentiation both in vivo and in vitro, the use of retinoids for weight loss is precluded by the varied toxic effects of these compounds and their teratogenicity. However, understanding how RA impacts on adipocyte differentiation reveals critical regulatory networks and may assist in the development of new pharmacological strategies to inhibit adipocyte hyperplasia.
Here, we provide compelling evidence that RA inhibits adipogenesis of both mesenchymal stem cells and preadipocytes through the up-regulation of Smad3 and the stimulation of its nuclear accumulation. Interestingly, although overexpression of C/EBPβ in MSCs only partially rescues inhibition of adipogenesis by RA, it completely blocks the inhibition in preadipocytes. This observation highlights the differences between the two models studied. Indeed, although both models express C/EBPβ, the actions of C/EBPβ, including the regulation of clonal expansion, appear to be more important in preadipocytes than MSCs. Notwithstanding these differences, RA is a potent inhibitor of adipocyte differentiation in both models. In MSCs, the effects of RA may extend to the promotion of alternative cell fates, making the study of the biological effects of C/EBPβ activity more complex in this model.
The activity of Smad3 in preadipocytes converges on the transcription factor C/EBPβ whose DNA binding activity on target promoters is decreased in vitro by Smad3 and in vivo by Smad3 and RA. Although the simple overexpression of Smad3 does not completely recapitulate the effects of RA in our models, this difference may be due to several factors. First, Smad3 expression is sustained in our system, whereas RA triggers only a transient rise in Smad3 levels, coinciding with the window of C/EBPβ activity. Because our stable cell lines maintain high levels of Smad3 expression throughout differentiation, this may influence downstream signaling pathways. Indeed, given that Smad3 has been shown to bind the C/EBPδ bZIP domain, it is possible that Smad3 expression could, in our overexpression system, impinge on C/EBPα transcriptional responses as well.
Second, ectopic expression of Smad3 alone may not permit the nuclear accumulation required to affect C/EBPβ-mediated transcriptional responses. RA may then act, in addition to up-regulating Smad3 expression, to stimulate Smad3 nuclear accumulation in a Smad4-independent fashion. Indeed, in our system, overexpression of Smad3 was unable to completely recapitulate the actions of RA, suggesting that Smad3 alone is not sufficient to inhibit adipogenesis. However, we demonstrate that the up-regulation of Smad3 by retinoic acid is a necessary step for the inhibition of adipogenesis by RA, and in the absence of Smad3, both inhibition of adipogenesis and the effect of C/EBPβ DNA binding are lost.
Interestingly, although Smad3 mRNA is rapidly up-regulated following RA treatment, its level continues to rise over a period of 4 days, a time frame that corresponds to the down-regulation of Smad3 protein expression. This apparent discrepancy may be attributed to multiple actions of RA. RA may act to stimulate the transcription from the Smad3 gene through a mechanism that requires de novo protein synthesis and results in the accumulation of Smad3 protein. With continued RA treatment, although the mRNA levels continue to rise, the protein synthesis of this message may be inhibited, or alternatively, the Smad3 protein stability may be altered resulting in a decrease in Smad3 protein expression despite high mRNA levels.
The observed up-regulation and nuclear accumulation of Smad3 occurs independently of TGFβ signaling. However, there have been numerous reports describing cross-talk, both positive and negative, between RA signaling and the TGFβ pathway. Our results may provide the link to explain the pleiotropic observed effects of retinoid treatment. Interestingly, during adipogenesis, TGFβ is also a potent inhibitor of differentiation, and this effect has been attributed to the actions of Smad3/4 (38). Although TGFβ does not affect C/EBPβ protein levels, it does impinge on its ability to drive transcription from a synthetic C/EBP-responsive promoter and the Lep (leptin) promoter in transient transcription assays (38). These results may be explained at least in part by the effects of Smad3 on C/EBPβ DNA binding ability but also on reported reduction of C/EBPβ transactivation activity following TGFβ treatment (38).
C/EBPβ is a ubiquitously expressed transcription factor whose activity is tightly controlled by multiple translational starts, post-translational modification, and homo- and heterodimerization with other bZIP transcription factors (7, 32, 42, 43). Because of the numerous levels of control, the transcriptional activity of C/EBPβ ranges the entire spectrum from repression to potent activation. Here, we show that retinoic acid treatment can prevent the interaction of C/EBPβ with its target promoters and thereby can interfere with C/EBPβ-mediated transcriptional responses. We expect that the importance of this mechanism extends beyond adipogenesis. Indeed, our own results indicate that C/EBPβ, a repressor of osteoblastogenesis, can be prevented from interacting with the RunX2 promoter by retinoic acid, thereby stimulating the osteoblast differentiation process (13). Interestingly, Smad3 is also a regulator of osteoblast differentiation; its expression stimulates early differentiation but represses late differentiation in a runx2-dependent fashion (44, 45). These results are consistent with our own observations that demonstrate that C/EBPβ expression potently represses early osteoblast differentiation but can act as an activator at later stages (13).
The RA/Smad3-mediated interference of C/EBPβ activity may also be important outside of mesenchymal lineage decisions. Indeed, whereas C/EBPβ is required for both liver regeneration and ductal morphogenesis of the mammary gland, retinoic acid has the opposite effect in both systems (10, 15, 43, 46–49). It is interesting to speculate that these effects may be mediated through the Smad3 pathway described herein.
Acknowledgments
We thank Drs. Rik Derynck and Bob Weinberg for providing plasmids.
This work was supported by Heart and Stroke Foundation of Ontario Grant NA 6375.
- MIX
- methylisobutylxanthine
- C/EBP
- CCAAT/enhancer-binding protein
- RA
- retinoic acid
- RAR
- retinoic acid receptor
- MH1
- Mad homology domain 1
- TGFβ
- Transforming growth factor β
- PPAR
- peroxisome proliferator-activated receptor
- MSC
- mesenchymal stem cell
- shRNA
- small hairpin
- GST
- glutathione S-transferase
- ChIP
- chromatin immunoprecipitation
- RT
- reverse transcription.
REFERENCES
- 1.Green H., Kehinde O. (1975) Cell 5, 19–27 [DOI] [PubMed] [Google Scholar]
- 2.Rubin C. S., Hirsch A., Fung C., Rosen O. M. (1978) J. Biol. Chem. 253, 7570–7578 [PubMed] [Google Scholar]
- 3.Cao Z., Umek R. M., McKnight S. L. (1991) Genes Dev. 5, 1538–1552 [DOI] [PubMed] [Google Scholar]
- 4.Lekstrom-Himes J., Xanthopoulos K. G. (1998) J. Biol. Chem. 273, 28545–28548 [DOI] [PubMed] [Google Scholar]
- 5.Wang L., Shao J., Muhlenkamp P., Liu S., Klepcyk P., Ren J., Friedman J. E. (2000) J. Biol. Chem. 275, 14173–14181 [DOI] [PubMed] [Google Scholar]
- 6.Tanaka T., Yoshida N., Kishimoto T., Akira S. (1997) EMBO J. 16, 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiper-Bergeron N., Salem H. A., Tomlinson J. J., Wu D., Haché R. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiper-Bergeron N., Wu D., Pope L., Schild-Poulter C., Haché R. J. (2003) EMBO J. 22, 2135–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z., Xie Y., Bucher N. L., Farmer S. R. (1995) Genes Dev. 9, 2350–2363 [DOI] [PubMed] [Google Scholar]
- 10.Greenbaum L. E., Li W., Cressman D. E., Peng Y., Ciliberto G., Poli V., Taub R. (1998) J. Clin. Invest. 102, 996–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darlington G. J., Ross S. E., MacDougald O. A. (1998) J. Biol. Chem. 273, 30057–30060 [DOI] [PubMed] [Google Scholar]
- 12.Schrem H., Klempnauer J., Borlak J. (2004) Pharmacol. Rev. 56, 291–330 [DOI] [PubMed] [Google Scholar]
- 13.Wiper-Bergeron N., St-Louis C., Lee J. M. (2007) Mol. Endocrinol. 21, 2124–2135 [DOI] [PubMed] [Google Scholar]
- 14.Robinson G. W., Johnson P. F., Hennighausen L., Sterneck E. (1998) Genes Dev. 12, 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seagroves T. N., Krnacik S., Raught B., Gay J., Burgess-Beusse B., Darlington G. J., Rosen J. M. (1998) Genes Dev. 12, 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sterneck E., Tessarollo L., Johnson P. F. (1997) Genes Dev. 11, 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landschulz W. H., Johnson P. F., McKnight S. L. (1989) Science 243, 1681–1688 [DOI] [PubMed] [Google Scholar]
- 18.Schwarz E. J., Reginato M. J., Shao D., Krakow S. L., Lazar M. A. (1997) Mol. Cell. Biol. 17, 1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue J. C., Schwarz E. J., Chawla A., Lazar M. A. (1996) Mol. Cell. Biol. 16, 1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeyakumar S. M., Vajreswari A., Giridharan N. V. (2006) Obesity 14, 52–59 [DOI] [PubMed] [Google Scholar]
- 21.Mercader J., Ribot J., Murano I., Felipe F., Cinti S., Bonet M. L., Palou A. (2006) Endocrinology 147, 5325–5332 [DOI] [PubMed] [Google Scholar]
- 22.Ribot J., Felipe F., Bonet M. L., Palou A. (2001) Obes. Res. 9, 500–509 [DOI] [PubMed] [Google Scholar]
- 23.Hathcock J. N., Hattan D. G., Jenkins M. Y., McDonald J. T., Sundaresan P. R., Wilkening V. L. (1990) Am. J. Clin. Nutr. 52, 183–202 [DOI] [PubMed] [Google Scholar]
- 24.Boyd A. S. (1989) Am. J. Med. 86, 568–574 [DOI] [PubMed] [Google Scholar]
- 25.Choy L., Skillington J., Derynck R. (2000) J. Cell Biol. 149, 667–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X., Sun Y., Constantinescu S. N., Karam E., Weinberg R. A., Lodish H. F. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10669–10674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Feng X., We R., Derynck R. (1996) Nature 383, 168–172 [DOI] [PubMed] [Google Scholar]
- 28.Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., Novina C. D. (2003) RNA 9, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard D. J. (2004) Mol. Endocrinol. 18, 606–623 [DOI] [PubMed] [Google Scholar]
- 30.Gregoire F. M., Smas C. M., Sul H. S. (1998) Physiol. Rev. 78, 783–809 [DOI] [PubMed] [Google Scholar]
- 31.Millward C. A., Heaney J. D., Sinasac D. S., Chu E. C., Bederman I. R., Gilge D. A., Previs S. F., Croniger C. M. (2007) Diabetes 56, 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramji D. P., Foka P. (2002) Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor S. M., Jones P. A. (1979) Cell 17, 771–779 [DOI] [PubMed] [Google Scholar]
- 34.Jefcoate C. R., Wang S., Liu X. (2008) Methods Mol. Biol. 456, 173–193 [DOI] [PubMed] [Google Scholar]
- 35.Gery S., Park D. J., Vuong P. T., Chih D. Y., Lemp N., Koeffler H. P. (2004) Blood 104, 3911–3917 [DOI] [PubMed] [Google Scholar]
- 36.Zauberman A., Lapter S., Zipori D. (2001) J. Biol. Chem. 276, 24719–24725 [DOI] [PubMed] [Google Scholar]
- 37.Xiao S., Jin H., Korn T., Liu S. M., Oukka M., Lim B., Kuchroo V. K. (2008) J. Immunol. 181, 2277–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choy L., Derynck R. (2003) J. Biol. Chem. 278, 9609–9619 [DOI] [PubMed] [Google Scholar]
- 39.Xu L. (2006) Biochim. Biophys. Acta 1759, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill C. S. (2009) Cell Res. 19, 36–46 [DOI] [PubMed] [Google Scholar]
- 41.Cao Z., Flanders K. C., Bertolette D., Lyakh L. A., Wurthner J. U., Parks W. T., Letterio J. J., Ruscetti F. W., Roberts A. B. (2003) Blood 101, 498–507 [DOI] [PubMed] [Google Scholar]
- 42.Trautwein C., van der Geer P., Karin M., Hunter T., Chojkier M. (1994) J. Clin. Invest. 93, 2554–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luedde T., Duderstadt M., Streetz K. L., Tacke F., Kubicka S., Manns M. P., Trautwein C. (2004) Hepatology 40, 356–365 [DOI] [PubMed] [Google Scholar]
- 44.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. (2001) EMBO J. 20, 2254–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonewald L. F., Dallas S. L. (1994) J. Cell. Biochem. 55, 350–357 [DOI] [PubMed] [Google Scholar]
- 46.Kistler A. (1986) Carcinogenesis 7, 1175–1182 [DOI] [PubMed] [Google Scholar]
- 47.Wang Y. A., Shen K., Wang Y., Brooks S. C. (2005) Dev. Dyn. 234, 892–899 [DOI] [PubMed] [Google Scholar]
- 48.Ledda-Columbano G. M., Pibiri M., Molotzu F., Cossu C., Sanna L., Simbula G., Perra A., Columbano A. (2004) Carcinogenesis 25, 2061–2066 [DOI] [PubMed] [Google Scholar]
- 49.Ozeki A., Tsukamoto I. (1999) Biochim. Biophys. Acta 1450, 308–319 [DOI] [PubMed] [Google Scholar]