Abstract
Skin hyperpigmentation disorders due to abnormal melanin production induced by ultraviolet (UV) irradiation are both a clinical and cosmetic problem. UV irradiation stimulates melanin production in melanocytes by increasing intracellular cAMP. Expression of heat shock proteins (HSPs), especially HSP70, is induced by various stressors, including UV irradiation, to provide cellular resistance to such stressors. In this study we examined the effect of expression of HSP70 on melanin production both in vitro and in vivo. 3-Isobutyl-1-methylxanthine (IBMX), a cAMP-elevating agent, stimulated melanin production in cultured mouse melanoma cells, and this stimulation was suppressed in cells overexpressing HSP70. IBMX-dependent transcriptional activation of the tyrosinase gene was also suppressed in HSP70-overexpressing cells. Expression of microphthalmia-associated transcription factor (MITF), which positively regulates transcription of the tyrosinase gene, was up-regulated by IBMX; however, this up-regulation was not suppressed in HSP70-overexpressing cells. On the other hand, immunoprecipitation and immunostaining analyses revealed a physical interaction between and co-localization of MITF and HSP70, respectively. Furthermore, the transcription of tyrosinase gene in nuclear extract was inhibited by HSP70. In vivo, UV irradiation of wild-type mice increased the amount of melanin in the basal layer of the epidermis, and this increase was suppressed in transgenic mice expressing HSP70. This study provides the first evidence of an inhibitory effect of HSP70 on melanin production both in vitro and in vivo. This effect seems to be mediated by modulation of MITF activity through a direct interaction between HSP70 and MITF.
Keywords: Aging, Chaperone Chaperonin, Heat Shock Protein, Skin, Transcription
Introduction
The skin can structurally be divided into several layers including the most apical layer, the epidermis, consisting of keratinocytes (1). In addition to changes with aging, the skin is damaged by various environmental stressors, especially by solar ultraviolet irradiation (photo-aging). UV light can be separated, based on the wavelength, into UVA (320–400 nm), UVB (290–320 nm), and UVC (100–290 nm) (2). Of these, most UVC can be absorbed by the ozone layer. Although the cell-damaging effect of UVA is relatively weak, UVA seems to play an important role in photo-aging because its content in solar UV is higher than UVB and UVC (3–5). Furthermore, UVB seems to also play the central role in photo-aging (6).
UV-induced skin hyperpigmentation disorders due to abnormal melanin production cause clinical and cosmetic problems. UV-dependent delayed pigmentation (induction of melanin production and distribution) plays a central role in the hyperpigmentation disorders. The induction of melanin production is mediated by various signal pathways (7–9). Of these pathways, a cAMP-dependent pathway seems to play a central role in UV-dependent stimulation of melanin production (7). In this pathway, exposure of keratinocytes to UV stimulates the release of signal molecules, such as α-melanocyte-stimulating hormone (α-MSH),2 prostaglandin E2, adenocorticotropic hormone, and endotheline-1, all of which stimulate melanin production in melanocytes through elevation of the level of intracellular cAMP. For example, the binding of α-MSH or adenocorticotropic hormone to melanocortin 1 receptor on melanocytes induces the expression of tyrosinase and other melanogenesis-related proteins through activation of adenylate cyclase, an increase in the intracellular cAMP level, activation of protein kinase A, activation of the cAMP response element-binding protein (CREB), and induction of expression of microphthalmia-associated transcription factor (MITF) that specifically binds to the promoter of the tyrosinase gene to promote its transcription (7). Tyrosinase is a rate-limiting enzyme in melanin synthesis, and an increase in the activity and expression of tyrosinase was observed in sites of UV-induced hyperpigmentation (10, 11); therefore, chemicals and natural products that suppress the activity and/or expression of tyrosinase could be pharmaceutically and cosmetically beneficial as hypopigmenting agents.
On the other hand, UV-induced modest melanin production plays an important role in protection of the skin against UV-dependent damage, including DNA damage (12). This protection is particularly important for the prevention of UV-induced development of melanoma and non-melanoma skin cancer (13). Synthesized melanin in the melanosomes in melanocytes is distributed and transported to the surrounding keratinocytes where it forms a melanin cap that acts as a filter to limit the penetration of UV into the epidermis and dermis (14). Melanin also acts as a scavenger of UV-produced reactive oxygen species that are also responsible for UV-dependent skin damage and development of skin cancer (15). Thus, identification of a mechanism that not only suppresses melanin production but also protects the skin from UV-induced damage is important for developing hypopigmenting agents (skin whitening agents) without worsening UV-induced skin damage.
When cells are exposed to stressors, a number of so-called stress proteins are induced to confer protection against such stressors. HSPs are representative of these stress proteins, and their cellular up-regulation of expression, especially that of HSP70, provides resistance as the HSPs re-fold or degrade denatured proteins produced by stressors such as reactive oxygen species (16). Because stressor-induced tissue damage is involved in various diseases, HSPs and HSP inducers have received much attention for their therapeutic potential. For example, we have shown using transgenic mice that HSP70 protects the gastrointestinal tract from development of gastric and small intestinal lesions and inflammatory bowel disease (17–20). Interestingly, geranylgeranylacetone, a leading anti-ulcer drug on the Japanese market, has been reported to be a non-toxic HSP-inducer, up-regulating various HSPs not only in cultured gastric mucosal cells but also in various tissues, including the gastric mucosa in vivo (21). It was recently reported that geranylgeranylacetone suppresses inflammatory bowel disease-related experimental colitis and lesion of small intestine (19, 22, 23). Based on these results, it is expected that non-toxic HSP inducers, including geranylgeranylacetone, will be therapeutically beneficial for various types of diseases.
It is known that various HSPs are constitutively expressed in the skin, and their expression, especially that of HSP70, is up-regulated by stressors such as heat treatment (24, 25). UV irradiation of keratinocytes induces the expression of HSPs not only in vitro but also in vivo (25–29). Furthermore, artificial expression of HSP70 in keratinocytes and melanocytes confers protection against UV not only in vitro (24, 29–32) but also in vivo; a sensitive phenotype of HSP70-null mice to UV-induced epidermal and dermal damage has been reported (33). Furthermore, protection of the skin against UV by expression of HSP70 has been suggested to occur in human skin (34). Therefore, if HSP70 can suppress melanin production, non-toxic HSP70 inducers should be beneficial as hypopigmenting agents because they can suppress melanin production while simultaneously protecting the skin against UV. It was recently reported that heat treatment of cultured melanoma cells suppresses melanin production; however, the contribution of HSPs to this suppression was not tested (35, 36). In this study we reproduced this suppression in another mouse melanoma cell line (B16) and found that in these cells artificial overexpression of HSP70 also suppresses melanin production. We also found that UVB irradiation-induced production of melanin in the epidermis was suppressed in transgenic mice expressing HSP70. Based on these results, we propose that non-toxic HSP70 inducers will be pharmaceutically and cosmetically beneficial as hypopigmenting agents.
EXPERIMENTAL PROCEDURES
Materials and Animals
Dulbecco's modified Eagle's medium was obtained from Nissui Pharmaceutical Co. [α-32P]GTP (6000 Ci/mmol) was from MP Biomedical. The RNeasy kit and HiPerFect transfection reagent were from Qiagen. PrimeScript® 1st strand cDNA synthesis kit was purchased from TAKARA Bio, and iQ SYBR Green Supermix was from Bio-Rad. Fetal bovine serum), melanin, 3-isobutyl-1-methylxanthine (IBMX), and α-MSH were from Sigma. Dynabeads Protein G, Lipofectamine (TM2000), Alexa Fluor 488 goat anti-mouse immunoglobulin G, and Alexa Fluor 594 goat anti-rabbit immunoglobulin G were purchased from Invitrogen. Antibodies against tyrosinase and actin were obtained from Santa Cruz. Antibodies against HSP70, HSP25, HSP47, HSP60, and HSP90 were from Stressgen. An antibody against MITF was obtained from Thermo Scientific. l-DOPA was from Nacalai, and the Dual Luciferase Assay System and NTP mixture were from Promega. Transgenic mice expressing HSP70 and their wild-type counterparts (8–10 weeks old, male) were gifts from Drs. C. E. Angelidis and G. N. Pagoulatos (University of Ioannina, Greece) and were prepared as described previously (17). Homozygotic male transgenic mice were used in the experiments. The experiments and procedures described here were carried out in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the National Institute of Health and were approved by the Animal Care Committee of Kumamoto University.
Cell Culture
B16 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere of 95% air with 5% CO2 at 37 °C. Transfection of B16 cells with pcDNA3.1 containing the hsp70 gene (37) was carried out using Lipofectamine (TM2000) according to the manufacturer's protocol. The stable transfectants expressing HSP70 were selected by immunoblotting and real-time RT-PCR analyses. Positive clones were maintained in the presence of 200 μg/ml G418.
Immunoblotting Analysis
Whole cell extracts were prepared as described previously (38). The protein concentration of each sample was determined by the Bradford method (39). Samples were applied to polyacrylamide SDS gels and subjected to electrophoresis after which proteins were immunoblotted with each antibody.
Determination of Melanin Content in Vitro
Melanin content was determined as described previously (40, 41) with some modifications. Cells were homogenized with 1 n NaOH. The melanin content of the cell extracts and the culture medium was determined by measuring the absorbance at 405 nm with a plate reader (Fluostar Galaxy).
Tyrosinase Activity Assay
Tyrosinase activity was assayed as described previously (42) with some modifications. Cells were washed with phosphate-buffered saline and homogenized with 20 mm Tris/HCl (pH 7.5) buffer containing 0.1% Triton X-100. Tyrosinase activity (oxidation of l-DOPA to DOPAchrome) was monitored as follows. Cell extracts (50 μl) were mixed with 100 μl of freshly prepared substrate solution (0.1% l-DOPA in phosphate-buffered saline) and incubated at 37 °C. The production of DOPAchrome was monitored by measuring the absorbance at 475 nm with a plate reader (Fluostar Galaxy) and corrected for auto-oxidation of l-DOPA.
Real-time RT-PCR Analysis
Real-time RT-PCR was performed as previously described (43) with some modifications. Total RNA was extracted from cells using an RNeasy kit according to the manufacturer's protocol. Samples (2.5 μg of RNA) were reverse-transcribed using a first-strand cDNA synthesis kit. Synthesized cDNA was used in real-time RT-PCR (Chromo 4 instrument (Bio-Rad)) experiments using iQ SYBR Green Supermix and analyzed with Opticon Monitor Software. Specificity was confirmed by electrophoretic analysis of the reaction products and by inclusion of template- or reverse transcriptase-free controls. To normalize the amount of total RNA present in each reaction, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was used as an internal standard.
Primers were designed using the Primer3 website. The primers used were (forward and reverse primers, respectively): tyrosinase, 5′-cctcctggcagatcatttgt-3′ and 5′-ggttttggctttgtcatggt-3′; gapdh, 5′-aactttggcattgtggaagg-3′ and 5′-acacattgggggtaggaaca-3′; mitf, 5′-ctagagcgcatggactttcc-3′ and 5′-acaagttcctggctgcagtt-3′.
siRNA Targeting of Genes
We used siRNA with the sequences of 5′-agcaguaccuuucuaccacdTdT-3′ and 5′-gugguagaaagguacugcudTdT-3′ as annealed oligonucleotides for repressing MITF expression. Cells were transfected with siRNA using HiPerFect transfection reagent according to the manufacturer's instructions. Non-silencing siRNA (5′-uucuccgaacgugucacgudTdT-3′ and 5′-acgugacacguucggagaadTdT-3′) was used as a negative control.
Luciferase Reporter Assay
The plasmid, pGL4-tyrosinase-luc (44), was a gift from Dr. M. Funaba (Kyoto University). The luciferase assay was performed as described previously (44, 45). Cells were transfected with 1 μg of each of the Photinus pyralis luciferase reporter plasmids (pGL4-tyrosinase-luc) and 0.125 μg of an internal standard plasmid bearing the Renilla reniformis luciferase reporter (pRL-SV40). P. pyralis luciferase activity in the cell extract was measured using the Dual Luciferase Assay System and then normalized for Renilla reniformis luciferase activity.
Co-immunoprecipitation Assay
Immunoprecipitation was carried out as described previously (46) with some modifications. Cells were harvested, lysed, and centrifuged. The antibody against HSP70 or MITF was added to the supernatant, and the samples were incubated for 12 h at 4 °C with rotation. Dynabeads Protein G was added and incubated for 2 h at 4 °C with rotation. Beads were washed four times, and proteins were eluted by boiling in SDS sample buffer.
Immunostaining Microscopy
After fixation with 4% paraformaldehyde, cells were incubated with antibody against HSP70 or MITF for overnight. Samples were further incubated with the respective secondary antibody. We acquired images with a confocal fluorescence microscope (Olympus FV500).
Chromatin Immunoprecipitation Assay
A chromatin immunoprecipitation assay was done as described previously (47) with some modifications. Cells were cross-linked with 1% formaldehyde for 10 min at 25 °C. After the addition of 125 mm (final concentration) glycine, cells were harvested and suspended in the lysis buffer (10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm dithiothreitol, 0.1% Nonidet P-40, and protease inhibitors). After 10 min of incubation on ice, cells were centrifuged to pellet the nuclei. Nuclei were then suspended in the nuclei lysis buffer (50 mm Tris/HCl (pH 8.1), 10 mm EDTA, 1% SDS, and protease inhibitors). Samples were sonicated 30 times for 10 s (to achieve an average fragment size of 0.5–1 kb). Immunoprecipitation was performed with magnetic beads that were coated with protein G and antibody against MITF or HSP70. Precipitates were washed, processed for DNA purification, and subjected to real time RT-PCR.
The primers used were (forward and reverse primers, respectively): tyrosinase promoter, 5′-gtctacttatgatctctaaatacaacaggcttg-3′ and 5′-tcatacaaaatctgcaccaataggttaatgagtg-3′; gapdh, 5′-acccagaagactgtggatgg-3′ and 5′-cacattgggggtaggaacac-3′.
In Vitro Transcription Assay
In vitro transcription assay was done as described (48) with some modifications. Nuclear extracts (5 μg protein) were incubated with DNA fragments containing the tyrosinase promoter and transcribed region (−270/+59) or the glucose-regulated protein (grp78) promoter and transcribed region (−304/+253) in the buffer containing 20 mm HEPES/KOH (pH 7.9), 100 mm KCI, 0.5 mm dithiothreitol, 1 mm EDTA, 20% glycerol, 0.4 mm ATP, 0.4 mm CTP, 0.4 mm UTP, 0.016 mm GTP, [α-32P]GTP, and 4 mm MgC12 for 60 min at 30 °C. Isolated RNAs were resolved by electrophoresis on 15% polyacrylamide gel containing 7 m urea. The gel was dried and autoradiographed.
Assay for Melanin Production in Vivo
Animals were exposed to UVB irradiation with a double bank of UVB lamps (peak emission at 312 nm, VL-215LM lamp, Vilber Lourmat). The energy of the UV was monitored by radiometer sensor (UVX-31, UV Products). Skin reflective colorimetric measurements were assessed with a narrow-band simple reflectance meter (Mexameter MX18, Courage-Khazaka). The measurement was done for four parts of the skin, and the mean was calculated. The measurement area was 5 mm in diameter, and the instrument was calibrated using black and white calibration plates. Skin biopsies were harvested and processed for Fontana-Masson staining, as described (49).
Statistical Analysis
All values are expressed as the mean ± S.D. or S.E. Two-way analysis of variance followed by the Tukey test was used to evaluate differences between more than three groups. Differences were considered to be significant for values of p < 0.05.
RESULTS
Inhibition of Melanin Production by Expression of HSP70
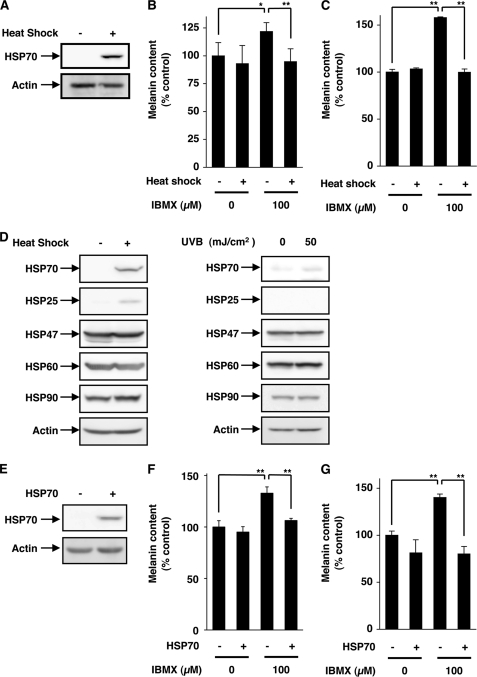
As mentioned above, it has been reported that heat treatment of mouse melanoma cells (Mel-Ab) suppresses melanin production (35, 36). In this study we tested whether similar results are observed for a different mouse melanoma cell line (B16). Because UVB irradiation of B16 cells did not stimulate the melanin production (supplemental Fig. S1, A and B), we used either α-MSH or IBMX (which is a cAMP-elevating agent that acts through inhibition of phosphodiesterase) to mimic UV-stimulated melanin production in vivo and measured the amount of melanin in both the culture medium and cell extract. We confirmed that heat treatment (42 °C, 1.5 h) induced expression of HSP70 (Fig. 1A). As shown in Fig. 1, B and C, treatment of B16 cells with IBMX increased the melanin content in both fractions, and pretreatment of the cells with the heat shock suppressed this increase significantly, suggesting that the heat treatment inhibited IBMX-stimulated production of melanin. We also examined the effect of heat shock on melanin production in cells in which the production had been already activated by IBMX and found that the heat treatment inhibited the melanin production even under these conditions (supplemental Fig. S1, C and D). Similar results were observed for α-MSH-stimulated production of melanin (supplemental Fig. S1, E and F). As shown in Fig. 1D, this heat treatment also induced the expression of HSP25 weakly but did not induce the expression of other HSPs. We also found that UVB irradiation of B16 cells did not clearly affect the expression of HSPs (Fig. 1D).
FIGURE 1.
Effect of heat treatment or overexpression of HSP70 on IBMX-stimulated melanin production. B16 cells were incubated for 1.5 h at 42 °C (Heat Shock +) or 37 °C (Heat Shock −) then for 6 h at 37 °C and finally for 72 h at 37 °C with or without 100 μm IBMX (A–C). B16 cells were incubated for 1.5 h at 42 °C (Heat Shock +) or 37 °C (Heat Shock −) or irradiated with or without indicated doses of UVB and cultured for 24 h (D). HSP70-overexpressing B16 cells (HSP70 +) and mock transfectant control cells (HSP70 −) were incubated for 72 h with or without 100 μm IBMX (E–G). Whole cell extracts were analyzed by immunoblotting (A, D, and E). The amounts of melanin in the conditioned medium (B and F) or cell extract (C and G) were determined as described under “Experimental Procedures” and are expressed relative to the control. Values are given as the mean ± S.D. (n = 3). **, p < 0.01; *, p < 0.05.
We established a clone of B16 cells that stably overexpresses HSP70. The extent of expression of HSP70 in this clone is shown (Fig. 1E). We confirmed that this overexpression of HSP70 did not affect the cell growth (data not shown). IBMX- and α-MSH-dependent increases in the amount of melanin in the culture medium and cell extract were observed in the control clone but were not so apparent in the clone overexpressing HSP70 (Fig. 1, F and G, supplemental Fig. S1, G and H), showing that expression of HSP70 somehow inhibits the synthesis of melanin in B16 cells.
We also examined the effect of heat shock and/or UVB irradiation on melanin production with or without IBMX. As shown in supplemental Fig. S1, I and J, irradiation with UVB did not affect the melanin production with or without IBMX. UVB did not affect the melanin production even with simultaneous heat treatment (supplemental Fig. S1, I and J).
Mechanism for Inhibition of Melanin Production by Expression of HSP70
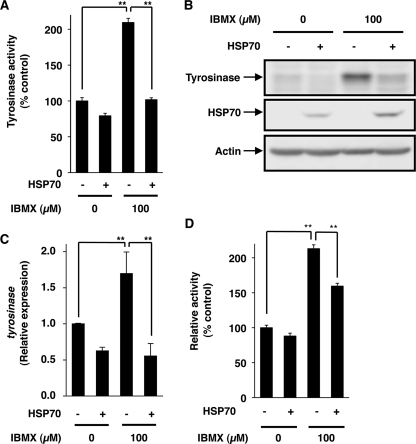
To understand the mechanisms governing inhibition of melanin production by expression of HSP70, we first examined tyrosinase activity. As shown in Fig. 2A, treatment of control cells with IBMX increased the tyrosinase activity in the cell extract as described previously (42), and this increase was not observed for cells overexpressing HSP70. We also examined the effect of IBMX and/or overexpression of HSP70 on the expression of tyrosinase. Treatment of cells with IBMX increased the level of tyrosinase, and this increase was suppressed in HSP70-overexpressing cells (Fig. 2B). Similar results were observed at the mRNA level, which was monitored by real-time RT-PCR (Fig. 2C), suggesting that expression of HSP70 inhibits transcription of the tyrosinase gene. To confirm this notion, we performed a luciferase reporter assay using a reporter plasmid where the promoter of the tyrosinase gene is inserted upstream of the luciferase gene. As shown in Fig. 2D, treatment of cells with IBMX increased luciferase activity in the cell extract, and the activity was significant lower in IBMX-treated HSP70-overexpressing cells than in the control cells, supporting the notion that expression of HSP70 inhibits the expression of tyrosinase at the level of transcription. Results similar to those in Fig. 2 were observed when heat treatment was used for induction of expression of HSP70 (supplemental Fig. S2, A–D).
FIGURE 2.
Effect of overexpression of HSP70 on IBMX-stimulated activity and expression of tyrosinase. HSP70-overexpressing B16 cells (HSP70 +) and mock transfectant control cells (HSP70 −) were incubated for 48 h with or without 100 μm IBMX (A–C). Tyrosinase activity was determined as described under “Experimental Procedures” and expressed relative to the control (A). Whole cell extracts were analyzed by immunoblotting as described in the legend of Fig. 1B. Total RNA was extracted and subjected to real-time RT-PCR using a specific primer for the tyrosinase gene. Values were normalized to gapdh gene expression and expressed relative to the control sample (C). HSP70-expressing B16 cells (HSP70 +) and mock transfectant control cells (HSP70 −) were transiently transfected with pRL-SV40 (internal control plasmid carrying the R. reniformis luciferase gene) and pGL4-tyrosinase-luc. After 24 h, the cells were incubated for 24 h with or without 100 μm IBMX. P. pyralis luciferase activity was measured and normalized for R. reniformis luciferase activity (D). Values are given as the mean ± S.D. (n = 3). **, p < 0.01.
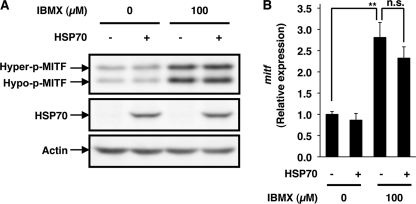
As described in the introduction, MITF plays a central role in UV-induced expression of tyrosinase through its specific binding to the promoter of the tyrosinase gene (50). We examined the effect of IBMX and/or expression of HSP70 on the expression of MITF. As described previously (51), treatment of cells with IBMX increased the level of MITF (Fig. 3A). Surprisingly, expression of HSP70 did not affect the level of MITF irrespective of the presence of IBMX (Fig. 3A). Similar results were observed for mRNA expression (Fig. 3B). These results suggest that the inhibitory effect of HSP70 expression on the promoter activity of the tyrosinase gene is not mediated by alterations to the expression of MITF. It is known that MITF is activated by phosphorylation (50); however, the results in Fig. 3A also suggest that the level of phosphorylation (the ratio of the hyperphosphorylated form to the hypophosphorylated form of MITF) is not affected by expression of HSP70. We also examined the effect of heat shock on the level of MITF. As shown in supplemental Fig. S2, E and F, different from the case of overexpression of HSP70 (Fig. 3), heat treatment decreased the level of MITF and mitf mRNA.
FIGURE 3.
Effect of overexpression of HSP70 on IBMX-stimulated expression of MITF. HSP70-overexpressing B16 cells (HSP70 +) or mock transfectant control cells (HSP70 −) were incubated for 3 h with or without 100 μm IBMX (A and B). Whole cell extracts were analyzed by immunoblotting as described in the legend of Fig. 1. The bands representing the hyperphosphorylated (Hyper-p) and hypophosphorylated (Hypo-p) forms of MITF are shown (A). mitf mRNA expression was monitored as described in the legend of Fig. 2B. Values are given as the mean ± S.D. (n = 3). **, p < 0.01; n.s., not significant.
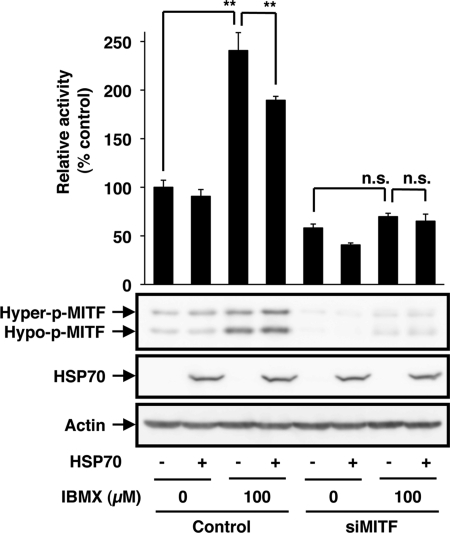
To test whether MITF is required for HSP70-dependent regulation of the promoter activity of the tyrosinase gene, we examined the effect of siRNA for MITF on the promoter activity of the tyrosinase gene. As shown in Fig. 4, transfection with siRNA clearly inhibited the expression of MITF irrespective of the presence of IBMX and HSP70 overexpression. The siRNA suppressed IBMX-dependent activation of the promoter activity of the tyrosinase gene, and HSP70 overexpression did not affect the promoter activity in cells transfected with the siRNA (Fig. 4), suggesting that MITF plays an important role in the inhibitory effect of HSP70 on the promoter activity of the tyrosinase gene.
FIGURE 4.
Effect of siRNA for MITF on the IBMX-stimulated promoter activity of the tyrosinase gene. HSP70-overexpressing B16 cells (HSP70 +) or mock transfectant control cells (HSP70 −) were transiently transfected with not only pRL-SV40 and pGL4-tyrosinase-luc but also with siRNA for MITF (siMITF) or non-silencing siRNA (Control). After 24 h, the cells were incubated for 24 h with or without 100 μm IBMX. Luciferase activity was measured and expressed as described in the legend of Fig. 2. Values are given as the mean ± S.D. (n = 3). **, p < 0.01; n.s., not significant. Whole cell extracts were analyzed by immunoblotting as described in the legend of Fig. 3.
We also examined the expression of MITF-regulated genes other than the tyrosinase gene, such as tyrosinase-related protein 1 (Tyrp1), dopachrome tautomerase (Dct), and protein phosphatase methylesterase 1 (PmeI). Expression of these genes was enhanced by IBMX and this enhancement was suppressed in HSP70-overexpressing cells (supplemental Fig. S2, G–I).
Involvement of an Interaction between HSP70 and MITF in the Inhibitory Effect of HSP70 on Melanin Production
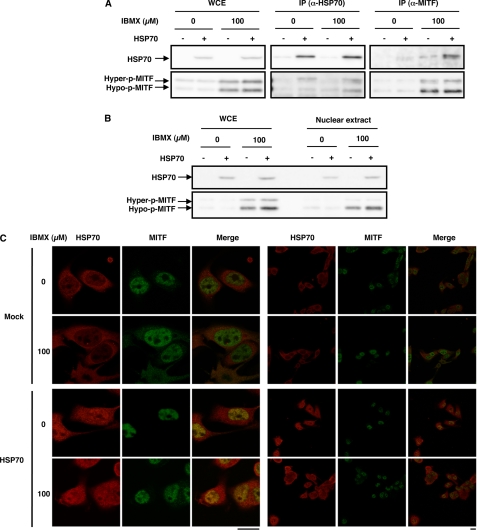
Based on the results mentioned above and the fact that HSP70 is a molecular chaperone modifying the activity of other proteins by its direct binding, we hypothesized that HSP70 interacts with MITF to modulate its effect on transcription of the tyrosinase gene. At first, we examined the physical interaction between HSP70 and MITF by a co-immunoprecipitation assay; we immunoprecipitated HSP70 and looked for the presence of MITF. Efficient precipitation of HSP70 was observed in a manner that was dependent on both the overexpression of HSP70 (Fig. 5A) and the specific antibody (data not shown). MITF was co-immunoprecipitated, and this co-immunoprecipitation was stimulated by overexpression of HSP70 (Fig. 5A). We also performed a reciprocal co-immunoprecipitation assay; we immunoprecipitated MITF and looked for the presence of HSP70. Efficient precipitation of MITF was observed in a manner that was dependent on the specific antibody (data not shown), and HSP70 was co-immunoprecipitated (Fig. 5A). These results suggest that HSP70 can physically interact with MITF.
FIGURE 5.
Physical interaction between MITF and HSP70 and their intracellular co-localization. HSP70-overexpressing B16 cells (HSP70 +) or mock transfectant control cells (HSP70 −) were incubated for 3 h with or without 100 μm IBMX (A–C), and whole cell extracts (WCE) were prepared (A and B). WCE were immunoprecipitated with antibodies against HSP70 (IP (α-HSP70)) or those against MITF (IP (α-MITF)) (A), or nuclear extracts were prepared from whole cell extracts (B). Each fraction was analyzed by immunoblotting as described in the legend of Fig. 1 (A and B). After fixation, samples were incubated with antibody against HSP70 or MITF. After incubation with the respective secondary antibody, cells were inspected using fluorescence microscopy. The left three panels are the magnified image of the right three panels. Scale bar, 20 μm (C).
We also examined the interaction using purified proteins. After incubation of purified glutathione S-transferase (GST)-fused MITF and HSP70, we precipitated GST-MITF and looked for the presence of HSP70. As shown in supplemental Fig. S3, precipitation of HSP70 was observed with full-length GST-MITF (GST-MITF-1) but not with GST alone, suggesting the direct interaction between MITF and HSP70. We also constructed a series of deletion mutants of GST-MITF to identify the domain of MITF responsible for its interaction with HSP70. Deletion of the N-terminal region of MITF (1–99 amino acid residues) diminished the interaction with HSP70, and the N-terminal fragment of MITF (1–99 amino acid residues) interacted with HSP70, suggesting that this region is responsible for the interaction (supplemental Fig. S3).
We then tested the co-localization of HSP70 and MITF by immunoblotting and immunostaining assays. As shown in Fig. 5B, MITF was detected in the nuclear extract irrespective of whether HSP70 was overexpressed, and HSP70 was also detected in the nuclear extract from cells overexpressing HSP70 but not in extract from control cells. These observations were confirmed by an immunostaining assay; MITF localized in the nucleus irrespective of the overexpression of HSP70, and HSP70 localized in the nucleus in a manner that was dependent on its overexpression (Fig. 5C). As a result, co-localization of HSP70 and MITF in the nucleus was observed in HSP70-overexpressing cells (see the Merge panel in Fig. 5C).
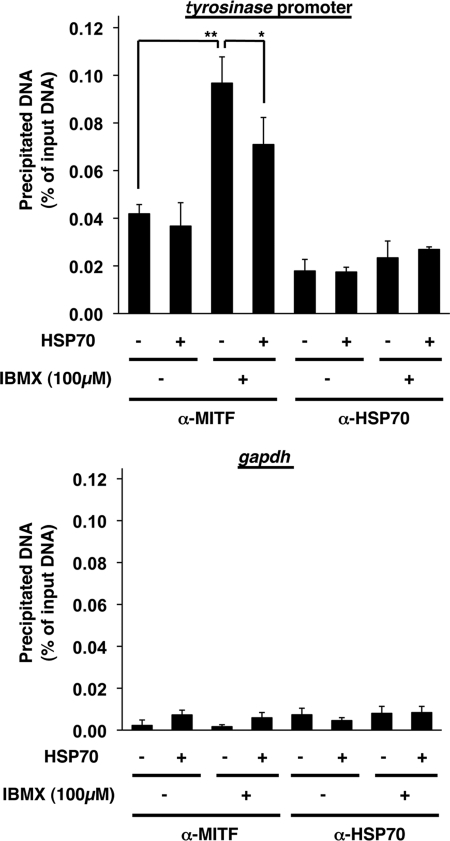
We then examined the effect of HSP70 overexpression on the specific binding of MITF to the promoter of the tyrosinase gene by chromatin immunoprecipitation assay. As shown in Fig. 6, DNA fragments containing the promoter of tyrosinase gene were precipitated with antibody against MITF more efficiently than control DNA fragments, suggesting that MITF specifically binds to the promoter of the tyrosinase gene in cells. We also found that this binding was stimulated by treatment of cells with IBMX, and HSP70 overexpression significantly inhibited this binding (Fig. 6). Results in Fig. 6 also suggest that HSP70 does not bind to the promoter of the tyrosinase gene so apparently in cells.
FIGURE 6.
The binding of MITF to the promoter of the tyrosinase gene in cells. HSP70-overexpressing B16 cells (HSP70 +) and mock transfectant control cells (HSP70 −) were incubated for 24 h with or without 100 μm IBMX. Whole cell extracts were immunoprecipitated with an antibody against MITF or HSP70. DNA fractions were prepared from the immunoprecipitated samples, and whole cell extracts and subjected to real time RT-PCR with specific primer sets for the tyrosinase promoter (upper panel) and gapdh gene (lower panel). Values are expressed relative to the input DNA (whole cell extracts) and are given as mean ± S.D. (n = 3). **, p < 0.01; *, p < 0.05.
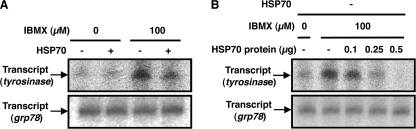
Finally, we examined the effect of HSP70 on the transcription of the tyrosinase gene in nuclear extract. DNA fragments containing the promoter region of the tyrosinase or grp78 (control) were incubated with nuclear extract to promote the transcription, and transcripts were detected by autoradiography after separation on polyacrylamide gel electrophoresis. The band with the expected size was detected depending on the template DNA and nuclear extract (data not shown), showing that the band corresponds to the transcript of tyrosinase or grp78. The intensity of bands corresponding to the transcript of tyrosinase but not that of grp78 increased by treatment of cells with IBMX (Fig. 7A). As shown in Fig. 7A, the intensity of band corresponding to the transcript of tyrosinase was lower with extracts prepared from HSP70-overexpressing cells treated with IBMX than those from control cells treated with IBMX. Such effect was not observed for the band corresponding to the transcript of grp78 (Fig. 7A). Furthermore, the addition of purified HSP70 to extract prepared from control cells decrease the intensity of band corresponding to the transcript of tyrosinase but not of grp78 (Fig. 7B). These results suggest that HSP70 directly suppresses the transcription of the tyrosinase.
FIGURE 7.
Effect of HSP70 on the transcription of the tyrosinase gene in nuclear extract. HSP70-overepressing B16 cells (HSP70 +) or mock transfectant control cells (HSP70 −) were incubated for 3 h with or without 100 μm IBMX. Nuclear extracts were prepared and incubated with DNA fragments containing the tyrosinase or grp78 promoter region in the presence (B) or absence (A) of indicated amounts of purified HSP70 (B). Isolated RNAs were electrophoresed on a 15% polyacrylamide gel and autoradiographed.
Effect of Expression of HSP70 on UVB-induced Melanin Production in Vivo
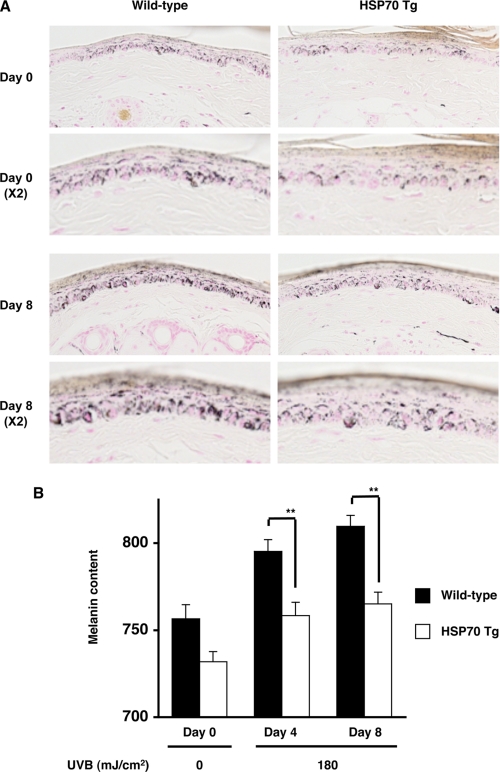
Finally, we tested the in vivo relevance of our in vitro results using transgenic mice expressing HSP70. The transgenic mice and wild-type mice were exposed to UVB irradiation for 8 days, and the melanin content was estimated by Fontana-Masson staining of sections or by a narrow-band simple reflectance meter (Mexameter). For this we used the tail skin because murine tail skin resembles human skin, as epidermal melanocytes are present and UV-dependent melanin production has been observed (52). We confirmed the overexpression of HSP70 in the skin of the transgenic mice by an immunoblotting assay (data not shown). As shown in Fig. 8A, an increase in melanin staining at the basal layer of the epidermis (the dermal/epidermal border) was observed in the wild-type mice after UVB irradiation, but this increase was not so obvious in sections prepared from the transgenic mice expressing HSP70. Measurement of melanin content by a Mexameter also showed that the melanin content was lower in the UVB-treated transgenic mice expressing HSP70 than in the wild-type controls (Fig. 8B). We also found that heat treatment of tail skin caused overexpression of HSP70 and a lower level of melanin content after irradiation with UVB (supplemental Fig. S4).
FIGURE 8.
Effect of overexpression of HSP70 on UVB-induced melanin production in the skin in vivo. Transgenic mice expressing HSP70 (HSP70 Tg) and wild-type mice (C57/BL6) were irradiated with 180 mJ/cm2 UVB once per 2 days for 8 days (totally 4 times) (A and B). Sections were prepared from the tail skin and subjected to Fontana-Masson staining (A). The amount of melanin in the tail skin was measured as described under “Experimental Procedures.” Values are given as the mean ± S.E. (n = 17). **, p < 0.01 (B).
DISCUSSION
It has been reported that heat treatment of mouse melanoma cells (Mel-Ab) suppresses melanin production; however, it is not clear whether up-regulation of expression of HSPs is involved in this phenomenon because an HSP-independent mechanism such as activation of extracellular signal-regulated kinase (ERK) and inhibition of p38 mitogen-activated protein kinase (p38 MAPK) has been proposed to be responsible for this phenomenon (35, 36). In this study we confirmed the inhibitory effect of heat treatment on melanin production in another type of mouse melanoma (B16) and showed that artificial overexpression of HSP70 also suppresses melanin production, suggesting that up-regulation of HSP70 expression is involved in the inhibitory effect of heat treatment on melanin production. Other mechanisms, such as activation of ERK and inhibition of p38 MAPK, may also contribute to the inhibitory effect of heat treatment on melanin production (35, 36). Furthermore, a decrease in the level of MITF after heat treatment (supplemental Fig. S2, E and F) should be involved in the inhibitory effect of heat treatment on melanin production.
Because HSPs are known to affect the intracellular traffic of vesicles, it is possible that expression of HSP70 affects the intracellular traffic of melanosomes, resulting in alterations to the amount of melanin in the culture medium. However, as well as heat treatment, artificial overexpression of HSP70 decreased the amount of melanin not only in the culture medium but also in cell extracts. This suggests that the decrease in the amount of melanin in the culture medium cannot be simply explained by the alteration of intracellular traffic of melanosomes and that synthesis of melanin by melanosomes is suppressed by overexpression of HSP70. In fact, we found that the activity and expression of tyrosinase (a rate-limiting enzyme in the synthesis of melanin) are suppressed in cells overexpressing HSP70. It is well known that the activity of tyrosinase is regulated mainly at the level of transcription, and we found by real-time RT-PCR analysis and luciferase reporter assay that the transcriptional activity of the tyrosinase gene is suppressed in cells overexpressing HSP70. Furthermore, in cells transfected with siRNA specific for MITF (a key transcription factor regulating the transcription of the tyrosinase gene), overexpression of HSP70 did not affect the promoter activity of the tyrosinase gene, suggesting that MITF plays an important role in HSP70-dependent regulation of the transcription of the tyrosinase gene.
The pathway of activation of adenylate cyclase followed by an increase in the intracellular cAMP level and activation of protein kinase A/cAMP response element-binding protein has been proposed to play an important role in the UV-induced expression of tyrosinase mainly through induction of MITF expression (7). In this study we have confirmed that IBMX up-regulates the expression of MITF. However, surprisingly, overexpression of HSP70 did not affect this up-regulation. Thus, we propose an alternative mechanism for the inhibitory effect of HSP70 on expression of the tyrosinase gene; HSP70 directly binds to MITF in the nucleus and inhibits its specific binding to the promoter of the tyrosinase gene. This is based on the following results; HSP70 can physically interact with MITF (co-immunoprecipitation assay), overexpressed HSP70 co-localized with MITF in the nucleus (co-immunostaining assay), overexpression of HSP70 inhibited the specific binding of MITF to the promoter of the tyrosinase gene (chromatin immunoprecipitation assay), and HSP70 inhibited the transcription of the tyrosinase gene in nuclear extract.
On the other hand, it has been reported that heat treatment suppresses the promoter activity of the mitf gene and suppresses the expression of MITF through inhibition of protein phosphatase 2A and ERK activation (36). Furthermore, it was suggested that heat treatment suppresses the activity of tyrosinase through p38 MAPK suppression (35). Thus, the inhibitory effect of heat treatment on melanin production would be mediated not only by expression of HSP70 but also by ERK activation and p38 MAPK suppression.
We also suggest that the expression of HSP70 suppresses UVB-induced synthesis of melanin in vivo; transgenic mice expressing HSP70 showed a smaller UV-dependent increase in the melanin content of the skin than wild-type mice. We have considered that the mechanism that is suggested by our in vitro studies is responsible for this in vivo phenomenon. Because hypopigmenting reagents are useful as drugs and cosmetics, a number of compounds that inhibit tyrosinase and/or MITF have been discovered; however, most of their cosmetic and pharmaceutical applications have not been successful due to skin irritation and permanent depigmentation (53). This seems to be due to the fact that UV-induced melanogenesis has a protective role against UV-induced skin damage. Based on the results of this study, we propose that non-toxic HSP70-inducers could be cosmetically and pharmaceutically beneficial because HSP70 protects keratinocytes from UV-induced cell damage both in vitro and in vivo (24, 29–33). Furthermore, human HSP70 has been reported to stimulate deoxyribonucleic acid base excision repair by its direct binding to human AP endonuclease and uracil DNA glycosylase (54). In E. coli, an HSP70 homologue (DnaK) stimulates the nucleotide excision repair of damaged DNA by maintaining repair proteins in their properly folded state (55), suggesting that human HSP70 may also stimulate nucleotide excision repair. We recently found that expression of HSP70 suppresses UV-induced DNA damage and stimulates its repair process at the skin in mice (56). Other beneficial effects of HSP70, such as stimulation of wound-healing at the skin and anti-aging activity have also been suggested (57–60). Furthermore, because recent reports suggest that MITF is an oncogene and its activation is involved in the progression of melanoma (61), the inhibitory effect of HSP70 on MITF would be beneficial for the prevention of UV-induced melanoma. We have already screened for non-toxic HSP70 inducers from natural products and found that some of their HSP-inducing activities were more potent than geranylgeranylacetone (62). We hope to develop some of these as whitening cosmetics or drugs for melanin-related diseases.
Supplementary Material
Acknowledgments
We thank Drs. C. E. Angelidis and G. N. Pagoulatos (University of Ioannina) or Dr. M. Funaba (Kyoto University) for generously providing transgenic mice expressing HSP70 or a plasmids, respectively.
This work was supported by grants-in-aid for scientific research from the Ministry of Health, Labour, and Welfare of Japan as well as the Japan Science and Technology Agency and grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- α-MSH
- α-melanocyte-stimulating hormone
- ERK
- extracellular signal-regulated kinase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GRP
- glucose-regulated protein
- HSP
- heat shock protein
- IBMX
- 3-isobutyl-1-methylxanthine
- MITF
- microphthalmia-associated transcription factor
- p38 MAPK
- p38 mitogen-activated protein kinase
- siRNA
- small interfering RNA
- RT
- reverse transcription
- GST
- glutathione S-transferase.
REFERENCES
- 1.Rabe J. H., Mamelak A. J., McElgunn P. J., Morison W. L., Sauder D. N. (2006) J. Am. Acad. Dermatol. 55, 1–19 [DOI] [PubMed] [Google Scholar]
- 2.Svobodova A., Walterova D., Vostalova J. (2006) Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 150, 25–38 [DOI] [PubMed] [Google Scholar]
- 3.McMillan T. J., Leatherman E., Ridley A., Shorrocks J., Tobi S. E., Whiteside J. R. (2008) J. Pharm. Pharmacol. 60, 969–976 [DOI] [PubMed] [Google Scholar]
- 4.Shorrocks J., Paul N. D., McMillan T. J. (2008) J. Invest. Dermatol. 128, 685–693 [DOI] [PubMed] [Google Scholar]
- 5.Douki T., Reynaud-Angelin A., Cadet J., Sage E. (2003) Biochemistry 42, 9221–9226 [DOI] [PubMed] [Google Scholar]
- 6.Matsumura Y., Ananthaswamy H. N. (2004) Toxicol. Appl. Pharmacol. 195, 298–308 [DOI] [PubMed] [Google Scholar]
- 7.Buscà R., Ballotti R. (2000) Pigment Cell Res. 13, 60–69 [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi Y., Brenner M., Hearing V. J. (2007) J. Biol. Chem. 282, 27557–27561 [DOI] [PubMed] [Google Scholar]
- 9.D'Orazio J. A., Nobuhisa T., Cui R., Arya M., Spry M., Wakamatsu K., Igras V., Kunisada T., Granter S. R., Nishimura E. K., Ito S., Fisher D. E. (2006) Nature 443, 340–344 [DOI] [PubMed] [Google Scholar]
- 10.Costin G. E., Hearing V. J. (2007) FASEB J. 21, 976–994 [DOI] [PubMed] [Google Scholar]
- 11.Miyamura Y., Coelho S. G., Wolber R., Miller S. A., Wakamatsu K., Zmudzka B. Z., Ito S., Smuda C., Passeron T., Choi W., Batzer J., Yamaguchi Y., Beer J. Z., Hearing V. J. (2007) Pigment Cell Res. 20, 2–13 [DOI] [PubMed] [Google Scholar]
- 12.Agar N., Young A. R. (2005) Mutat. Res. 571, 121–132 [DOI] [PubMed] [Google Scholar]
- 13.Sturm R. A., Duffy D. L., Box N. F., Chen W., Smit D. J., Brown D. L., Stow J. L., Leonard J. H., Martin N. G. (2003) Pigment Cell Res. 16, 266–272 [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi N., Nakagawa A., Muramatsu T., Yamashina Y., Shirai T., Hashimoto M. W., Ishigaki Y., Ohnishi T., Mori T. (1998) J. Invest. Dermatol. 110, 806–810 [DOI] [PubMed] [Google Scholar]
- 15.Bustamante J., Bredeston L., Malanga G., Mordoh J. (1993) Pigment Cell Res. 6, 348–353 [DOI] [PubMed] [Google Scholar]
- 16.Morimoto R. I., Santoro M. G. (1998) Nat. Biotechnol. 16, 833–838 [DOI] [PubMed] [Google Scholar]
- 17.Tanaka K., Namba T., Arai Y., Fujimoto M., Adachi H., Sobue G., Takeuchi K., Nakai A., Mizushima T. (2007) J. Biol. Chem. 282, 23240–23252 [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K., Tsutsumi S., Arai Y., Hoshino T., Suzuki K., Takaki E., Ito T., Takeuchi K., Nakai A., Mizushima T. (2007) Mol. Pharmacol. 71, 985–993 [DOI] [PubMed] [Google Scholar]
- 19.Asano T., Tanaka K., Yamakawa N., Adachi H., Sobue G., Goto H., Takeuchi K., Mizushima T. (2009) J. Pharmacol. Exp. Ther. 330, 458–467 [DOI] [PubMed] [Google Scholar]
- 20.Suemasu S., Tanaka K., Namba T., Ishihara T., Katsu T., Fujimoto M., Adachi H., Sobue G., Takeuchi K., Nakai A., Mizushima T. (2009) J. Biol. Chem. 284, 19705–19715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirakawa T., Rokutan K., Nikawa T., Kishi K. (1996) Gastroenterology 111, 345–357 [DOI] [PubMed] [Google Scholar]
- 22.Ohkawara T., Nishihira J., Takeda H., Miyashita K., Kato K., Kato M., Sugiyama T., Asaka M. (2005) Scand. J. Gastroenterol. 40, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 23.Ohkawara T., Nishihira J., Takeda H., Katsurada T., Kato K., Yoshiki T., Sugiyama T., Asaka M. (2006) Int. J. Mol. Med. 17, 229–234 [PubMed] [Google Scholar]
- 24.Wilson N., McArdle A., Guerin D., Tasker H., Wareing P., Foster C. S., Jackson M. J., Rhodes L. E. (2000) J. Cutan. Pathol. 27, 176–182 [DOI] [PubMed] [Google Scholar]
- 25.Morris S. D. (2002) Clin. Exp. Dermatol. 27, 220–224 [DOI] [PubMed] [Google Scholar]
- 26.Jonak C., Klosner G., Trautinger F. (2006) Int. J. Cosmet. Sci. 28, 233–241 [DOI] [PubMed] [Google Scholar]
- 27.Trautinger F., Kokesch C., Klosner G., Knobler R. M., Kindas-Mügge I. (1999) Exp. Dermatol. 8, 187–192 [DOI] [PubMed] [Google Scholar]
- 28.Allanson M., Reeve V. E. (2004) J. Invest. Dermatol. 122, 1030–1036 [DOI] [PubMed] [Google Scholar]
- 29.Trautinger F. (2001) J. Photochem. Photobiol. B 63, 70–77 [DOI] [PubMed] [Google Scholar]
- 30.Trautinger F., Kindås-Mügge I., Barlan B., Neuner P., Knobler R. M. (1995) J. Invest. Dermatol. 105, 160–162 [DOI] [PubMed] [Google Scholar]
- 31.Park K. C., Kim D. S., Choi H. O., Kim K. H., Chung J. H., Eun H. C., Lee J. S., Seo J. S. (2000) Arch. Dermatol. Res. 292, 482–487 [DOI] [PubMed] [Google Scholar]
- 32.Maytin E. V., Wimberly J. M., Kane K. S. (1994) J. Invest. Dermatol. 103, 547–553 [DOI] [PubMed] [Google Scholar]
- 33.Kwon S. B., Young C., Kim D. S., Choi H. O., Kim K. H., Chung J. H., Eun H. C., Park K. C., Oh C. K., Seo J. S. (2002) J. Dermatol. Sci. 28, 144–151 [DOI] [PubMed] [Google Scholar]
- 34.Trautinger F., Knobler R. M., Hönigsmann H., Mayr W., Kindås-Mügge I. (1996) J. Invest. Dermatol. 107, 442–443 [DOI] [PubMed] [Google Scholar]
- 35.Kim D. S., Park S. H., Kwon S. B., Na J. I., Huh C. H., Park K. C. (2007) Arch. Pharm. Res. 30, 581–586 [DOI] [PubMed] [Google Scholar]
- 36.Kim D. S., Park S. H., Kwon S. B., Youn S. W., Park E. S., Park K. C. (2005) Cell. Signal. 17, 1023–1031 [DOI] [PubMed] [Google Scholar]
- 37.Fujimoto M., Takaki E., Hayashi T., Kitaura Y., Tanaka Y., Inouye S., Nakai A. (2005) J. Biol. Chem. 280, 34908–34916 [DOI] [PubMed] [Google Scholar]
- 38.Hoshino T., Tsutsumi S., Tomisato W., Hwang H. J., Tsuchiya T., Mizushima T. (2003) J. Biol. Chem. 278, 12752–12758 [DOI] [PubMed] [Google Scholar]
- 39.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 40.Kim K. S., Kim J. A., Eom S. Y., Lee S. H., Min K. R., Kim Y. (2006) Pigment Cell Res. 19, 90–98 [DOI] [PubMed] [Google Scholar]
- 41.Lei T. C., Virador V. M., Vieira W. D., Hearing V. J. (2002) Anal. Biochem. 305, 260–268 [DOI] [PubMed] [Google Scholar]
- 42.Yang J. Y., Koo J. H., Song Y. G., Kwon K. B., Lee J. H., Sohn H. S., Park B. H., Jhee E. C., Park J. W. (2006) Acta Pharmacol. Sin. 27, 1467–1473 [DOI] [PubMed] [Google Scholar]
- 43.Mima S., Tsutsumi S., Ushijima H., Takeda M., Fukuda I., Yokomizo K., Suzuki K., Sano K., Nakanishi T., Tomisato W., Tsuchiya T., Mizushima T. (2005) Cancer Res. 65, 1868–1876 [DOI] [PubMed] [Google Scholar]
- 44.Murakami M., Iwata Y., Funaba M. (2007) Mol. Cell Biochem. 303, 251–257 [DOI] [PubMed] [Google Scholar]
- 45.Namba T., Homan T., Nishimura T., Mima S., Hoshino T., Mizushima T. (2009) J. Biol. Chem. 284, 4158–4167 [DOI] [PubMed] [Google Scholar]
- 46.Hoshino T., Nakaya T., Araki W., Suzuki K., Suzuki T., Mizushima T. (2007) Biochem. J. 402, 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takehara M., Makise M., Takenaka H., Asano T., Mizushima T. (2008) Biochem. J. 413, 535–543 [DOI] [PubMed] [Google Scholar]
- 48.Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito T., Ito N., Saathoff M., Bettermann A., Takigawa M., Paus R. (2005) Br. J. Dermatol. 152, 623–631 [DOI] [PubMed] [Google Scholar]
- 50.Levy C., Khaled M., Fisher D. E. (2006) Trends Mol. Med. 12, 406–414 [DOI] [PubMed] [Google Scholar]
- 51.Park H. Y., Wu C., Yonemoto L., Murphy-Smith M., Wu H., Stachur C. M., Gilchrest B. A. (2006) Biochem. J. 395, 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui R., Widlund H. R., Feige E., Lin J. Y., Wilensky D. L., Igras V. E., D'Orazio J., Fung C. Y., Schanbacher C. F., Granter S. R., Fisher D. E. (2007) Cell 128, 853–864 [DOI] [PubMed] [Google Scholar]
- 53.Ando H., Kondoh H., Ichihashi M., Hearing V. J. (2007) J. Invest. Dermatol. 127, 751–761 [DOI] [PubMed] [Google Scholar]
- 54.Bases R. (2006) Cell Stress Chaperones 11, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou Y., Crowley D. J., Van Houten B. (1998) J. Biol. Chem. 273, 12887–12892 [DOI] [PubMed] [Google Scholar]
- 56.Matsuda M., Hoshino T., Yamashita Y., Tanaka K., Maji D., Sato K., Adachi H., Sobue G., Ihn H., Funasaka Y., Mizushima T. (2010) J. Biol. Chem. 285, 5848–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon M. M., Reikerstorfer A., Schwarz A., Krone C., Luger T. A., Jäättelä M., Schwarz T. (1995) J. Clin. Invest. 95, 926–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gutsmann-Conrad A., Heydari A. R., You S., Richardson A. (1998) Exp. Cell Res. 241, 404–413 [DOI] [PubMed] [Google Scholar]
- 59.Kimura K., Tanaka N., Nakamura N., Takano S., Ohkuma S. (2007) J. Biol. Chem. 282, 5910–5918 [DOI] [PubMed] [Google Scholar]
- 60.Rattan S. I. (1998) Biochem. Mol. Biol. Int. 45, 753–759 [DOI] [PubMed] [Google Scholar]
- 61.Garraway L. A., Widlund H. R., Rubin M. A., Getz G., Berger A. J., Ramaswamy S., Beroukhim R., Milner D. A., Granter S. R., Du J., Lee C., Wagner S. N., Li C., Golub T. R., Rimm D. L., Meyerson M. L., Fisher D. E., Sellers W. R. (2005) Nature 436, 117–122 [DOI] [PubMed] [Google Scholar]
- 62.Yamashita Y., Hoshino T., Matsuda M., Kobayashi C., Tominaga A., Nakamura Y., Nakashima K., Yokomizo K., Ikeda T., Mineda K., Maji D., Niwano Y., Mizushima T. (2010) Exp. Dermatol., in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.