Abstract
Sirtuin 1 (SIRT1) is a NAD-dependent deacetylase that is critically involved in diverse cellular processes including metabolic disease, cancer, and possibly aging. Despite extensive studies on SIRT1 function, how SIRT1 levels are regulated remains relatively unknown. Here, we report that the nuclear bile acid receptor farnesoid X receptor (FXR) inhibits microRNA-34a (miR-34a) in the liver, which results in a positive regulation of SIRT1 levels. Activation of FXR by the synthetic agonist GW4064 decreases hepatic miR-34a levels in normal mice, and consistently, hepatic miR-34a levels are elevated in FXR-null mice. FXR induces expression of small heterodimer partner (SHP), an orphan nuclear receptor and transcriptional corepressor, which in turn results in repression of p53, a key activator of the miR-34a gene, by inhibiting p53 occupancy at the promoter. MiR-34a decreased SIRT1 levels by binding to the 3′-untranslated region of SIRT1 mRNA, and adenovirus-mediated overexpression of miR-34a substantially decreased SIRT1 protein levels in mouse liver. Remarkably, miR-34a levels were elevated, and SIRT1 protein levels were reduced in diet-induced obese mice, and FXR activation in these mice reversed the miR-34a and SIRT1 levels, indicating an intriguing link among FXR activation, decreased miR-34a, and subsequently, increased SIRT1 levels. Our study demonstrates an unexpected role of the FXR/SHP pathway in controlling SIRT1 levels via miR-34a inhibition and that elevated miR-34a levels in obese mice contribute to decreased SIRT1 levels. Manipulation of this regulatory network may be useful for treating diseases of aging, such as metabolic disease and cancer.
Keywords: Adenoviruses, Bile Acid, Metabolic Diseases, MicroRNA, p53, Transcription Factors, Transcription Regulation, SHP, SIRT1
Introduction
The NAD+-dependent Sirtuin 1 (SIRT1)3 deacetylase plays a critical role in cellular metabolism, the stress response, and possibly aging, by modulating the activity of its target proteins via protein deacetylation (1–4). Recent studies demonstrate that SIRT1 plays an important role in maintaining metabolic homeostasis in response to hormonal and nutritional fluctuations by modulating the activity of PGC-1α, a master metabolic regulator (5, 6). During nutritional deprivation, SIRT1 promotes fat mobilization and suppresses adipogenesis and regulates hepatic glucose and lipid metabolism by activating key metabolic regulators, including PGC-1α (5, 6). Activation of SIRT1 by natural or synthetic SIRT1 activators reduced acetylation levels of PGC-1α and protected against diet-induced obesity and insulin resistance by promoting mitochondrial function (7–9). SIRT1 levels are dynamically regulated in response to fasting and feeding under physiological conditions but markedly reduced in diet-induced obese mice (10). However, the molecular basis by which SIRT1 levels are regulated under normal conditions and why they are substantially reduced in metabolic disease states remains largely unknown.
The nuclear receptor, farnesoid X receptor (FXR), is the primary biosensor for endogenous bile acids and regulates expression of numerous genes involved in lipid and glucose metabolism (11–15). FXR indirectly inhibits its metabolic target genes by inducing the expression of small heterodimer partner (SHP), an orphan nuclear receptor and transcriptional corepressor (16, 17). It is well established that the FXR/SHP pathway plays an important role in maintaining bile acid and cholesterol levels by inhibiting the transcription of cholesterol cytochrome P450 7A1 hydroxylase, a key enzyme in hepatic bile acid biosynthesis (16, 17), and has been also shown to regulate fatty acid metabolism (18). Interestingly, FXR activation by the synthetic agonist, GW4064, or hepatic overexpression of constitutively active FXR using adenoviral delivery significantly improved hyperglycemia and hyperlipidemia in diabetic obese db/db mice (11). Although both SIRT1 and FXR are critical for hepatic metabolism and activation of both proteins improves metabolic outcomes (7, 8, 11), it remains largely unknown whether the expression and activity of these two proteins are coordinately regulated.
MicroRNAs (miRs) are recently discovered small (21–23-nucleotide) noncoding RNAs that inhibit translation and/or destabilize target mRNAs by binding to their 3′-untranslated regions (3′-UTRs) with partial base pairing (19). MiRs play important roles in cellular metabolism under normal and metabolic stress conditions (20, 21), and aberrant expression of miRs has been observed in human diseases, such as cancer and metabolic disorders (20–23). Here, we show that the FXR/SHP regulatory pathway inhibits expression of miR-34a, which results in a positive regulation of hepatic SIRT1 levels under physiological conditions. We further show this regulatory network is altered in diet-induced obese mice, resulting in elevated miR-34a levels and subsequently reduced SIRT1 levels in the liver.
EXPERIMENTAL PROCEDURES
Reagents and Materials
Anti-miR-34a, scrambled RNA, and primers for reverse transcription-quantitative PCR (qRT-PCR) for measuring miRNAs including miR-34a were purchased from Applied Biosystems. Antibodies for FXR (sc-1204, sc-13063), SHP (sc-30169), p53 (sc-6243), HNF-4 (sc-8987), lamin (sc-20680), tubulin (sc-8085), actin (sc-1616), and green fluorescent protein (sc-8334) were purchased from Santa Cruz Biotechnology. M2 antibody was from Sigma, and antibodies for SIRT1 and acetylated histone K9/K14 were purchased from Upstate.
MicroRNA Gene Profiling
Total RNA was isolated from livers from three male wild type mice or FXR-null mice using the mirVanaTM miRNA isolation kit (Ambion, Austin, TX). MicroRNA microarray studies including labeling, hybridization, image scanning, and data analysis were carried out as described previously (24).
qRT-PCR Quantification of miRs and mRNAs
RNA was isolated from liver or cultured cells. The levels of mRNA or miR were determined by qRT-PCR according to the manufacturer's protocol (Applied Biosystems).
Cell Cultures and Transfection Reporter Assay
HepG2 cells (ATCC HB8065) were maintained in Dulbecco's modified Eagle's medium/F12 (1:1) medium. COS-1 cells were maintained in Dulbecco's modified Eagle's medium. Cells were transfected with luciferase reporters along with β-galatosidase expression plasmid and further infected with adenoviral vectors as described previously (25, 26). Consistent results were observed in at least two independent triplicate assays in each experiment.
Construction of Adenoviral MiR-34a
For construction of Ad-miR-34a, the XbaI/Not1 fragment encoding miR-34a from pCDH-miR-34a was inserted into pCI. Then the Kpn1/Not1 fragment from this resulting plasmid was inserted into Kpn1/Not1-digested Ad-Track. Positive clones were identified by DNA sequencing. Ad-empty, Ad-3FLAG-FXR, and Ad-FLAG-SHP have been described previously (27, 28).
In Vivo Experiments
FXR-null mice, ob/ob mice, and congenic CJ57BL mice were purchased from the Jackson Laboratory. BALB/c male mice were fed normal chow or high fat and high calorie Western-style chow for 16–20 weeks. Recombinant adenoviral vectors (0.5–1.0 × 109 active viral particle in 200 μl of phosphate-buffered saline) were injected via the tail vein of mice as described previously (25, 28). A synthetic FXR agonist, GW4064 (30 mg/kg in corn oil), or vehicle was administered by intraperitoneal injection, and mice were killed after 1 h. Adenoviral-mediated overexpression of FLAG-FXR was performed as described previously (28). All of the animal use and adenoviral protocols were approved by the Institutional Animal Care and Use and Institutional Biosafety Committees at University of Illinois at Urbana-Champaign and were in accordance with National Institutes of Health guidelines.
Chromatin Immunoprecipitation (ChIP) Assay, Glutathione S-Transferase (GST) Pulldown, and Coimmunoprecipitation (CoIP) Assay
ChIP assays in mouse liver and HepG2 cells were carried out essentially as described (25, 27, 29). For ChIP assays along with siRNA experiments, HepG2 cells were either infected with Ad-siSHP or control Ad-empty, or transiently transfected with siRNA for HNF-4 or control siRNA. Two days later, cells were treated with 100 nm GW4064 for 1 h and collected for ChIP assays. The linearity of the PCRs was established using different number of cycles and different amounts of template. Each ChIP experiment was repeated at least three times with similar reproducible results. Sequences of the ChIP primers for mouse and human miR-34a promoters are available upon request. GST pulldown and CoIP studies were performed as described previously (25–27, 29).
RESULTS
FXR Down-regulates Hepatic MiR-34a
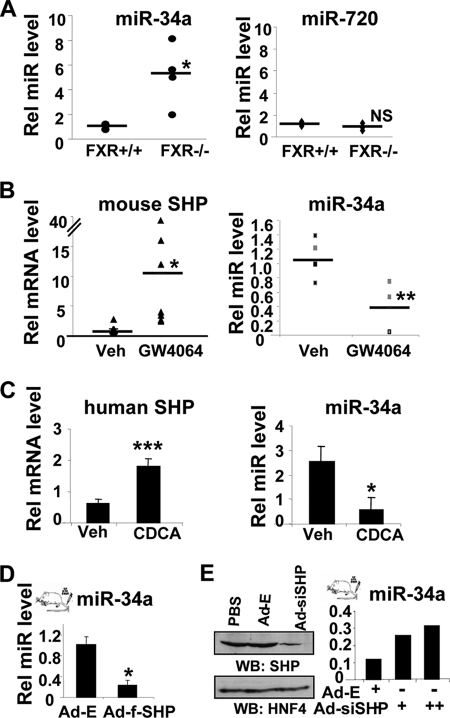
FXR is an important regulator of many metabolic genes in liver (13–15), and it is possible that miRs, as metabolic regulators, might mediate some FXR actions in hepatocyes. MiRs regulated by FXR were detected by miR microarray analysis with hepatic RNA of wild type or FXR-null mice. Of the miRs tested, the levels of miR-34a were up-regulated the most in FXR-null mice (supplemental Fig. S1). These results were confirmed by q-RTPCR studies in which control miR-720 levels were unchanged and miR-34a levels were elevated about 5-fold although miR-34a levels were not substantially increased in one of four mice which resulted in a large standard error (Fig. 1A). These results suggest that FXR usually down-regulates hepatic expression of miR-34a. To test this possibility, normal mice were treated with a synthetic FXR agonist, GW4064. As expected, mRNA levels of a known FXR target SHP (16, 17) were increased whereas miR-34a levels were decreased in mice treated with GW4064 (Fig. 1B). Similar results were observed in HepG2 cells treated with a natural FXR bile acid agonist, chenodeoxycholic acid (Fig. 1C). These results demonstrate that FXR normally down-regulates hepatic expression of miR-34a and, further, raise the possibility that the orphan nuclear receptor and transcriptional corepressor SHP may mediate FXR inhibition of miR-34a gene expression.
FIGURE 1.
FXR down-regulates miR-34a. A, hepatic miR-34a and control miR-720 levels from wild type (+/+) and FXR-null mice (−/−) were measured by qRT-PCR, and values from each individual mouse were plotted. Bars indicate average values from three or four animals. B, normal mice were treated with GW4064 or vehicle (Veh) for 1 h, and livers were collected for qRT-PCR, and values from each individual mouse were plotted. Bars indicate average values from five to seven animals. C, HepG2 cells were treated with 50 μm chenodeoxycholic acid (CDCA) overnight and collected for qRT-PCR. Error bars, S.D. D and E, normal mice were injected via the tail vein with adenoviral vectors as indicated, and 5 days later, hepatic miR-34a levels were measured by qRT-PCR and SHP protein levels were measured by Western blot (WB) analysis. Statistical significance was measured using Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, statistically not significant, S.E. (n = 5).
SHP May Mediate FXR Inhibition of MiR-34a
To determine whether SHP may mediate FXR inhibition of miR-34a gene expression, either siRNA for SHP or exogenous FLAG-SHP was expressed in livers in vivo by infection with adenoviral vectors. MiR-34a levels were substantially reduced by overexpression of FLAG-SHP (Fig. 1D) but were markedly increased by down-regulation of endogenous SHP with siRNA (Fig. 1E). These results strongly suggest that SHP mediates inhibition of miR-34a by FXR in mouse liver in vivo.
SHP Interacts with p53, a Key Activator of the MiR-34a Gene
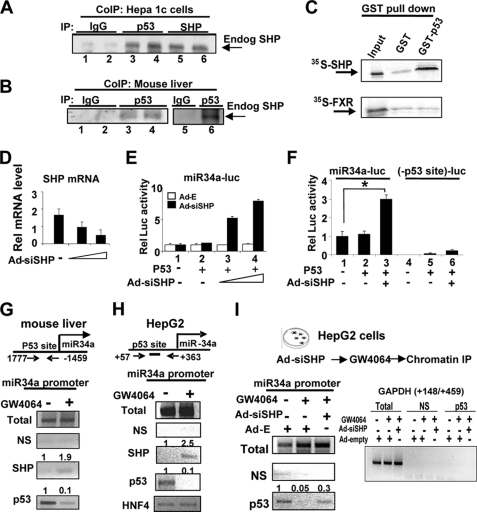
SHP has been shown to interact with and inhibit numerous DNA-binding transcription factors (26, 29, 30). Because p53 is a key activator of the miR-34a gene (23), we asked whether SHP could inhibit hepatic p53 activity. By CoIP assays, interaction between SHP and p53 in cells overexpressing these proteins was detected (supplemental Fig. S2). Endogenous SHP also interacted with endogenous p53 in Hepa1c1c7 cells (Fig. 2A) and in liver extracts (Fig. 2B). In in vitro GST pulldown assays, SHP interacted with GST-p53, whereas FXR did not (Fig. 2C). These results indicate that SHP directly interacts with p53 in hepatic cells and mouse liver.
FIGURE 2.
SHP inhibits expression of the miR-34a gene by inhibiting p53 binding to the promoter. A and B, CoIP assays were done using mouse Hepa1c1c7 cell (A) and mouse liver (B) extracts to detect interaction between endogenous p53 and SHP. C, 35S-SHP or 35S-FXR was synthesized, and binding to GST or GST-p53 was determined by GST pulldown assays. D–F, HepG2 cells were cotransfected with expression plasmids as indicated and with the miR34a promoter (−1402 to +578)-luc or a mutant reporter and then infected with Ad-siSHP. Two days later, cells were collected for qRT-PCR (D) and reporter assays (E and F). Error bars, S.D. G and H, mouse (G) or HepG2 cells (H) were treated with 100 nm GW4064 for 1 h, and samples were collected for ChIP assays. Band intensities were measured, and the intensities relative to samples treated with vehicle (−) are indicated at the top of each band. I, HepG2 cells infected as indicated were treated with GW4064 and collected for ChIP assays.
SHP Inhibits p53 Transactivation of the MiR-34a Promoter
To test whether SHP inhibits p53 transactivation ability on the miR-34a promoter, we next examined the effects of down-regulation of SHP on miR-34a promoter activity. SHP mRNA and protein levels were efficiently decreased in cells infected with Ad-siSHP (Fig. 2D and supplemental Fig. S3), and miR-34a promoter activity was increased in a dose-dependent manner, but not by infection with control Ad-empty (Fig. 2E). Further, mutation of the p53 binding site nearly abolished the promoter activity (Fig. 2F, lanes 2 and 5), indicating that p53 is the dominant regulator of miR-34a expression. These results, taken together, indicate that SHP inhibits transactivation of p53, a key activator of miR-34a promoter activity.
In addition, although activity was nearly abolished by mutation of the p53 binding site, small increases were observed in cells expressing SHP siRNA (Fig. 2F, lanes 5 and 6) compared with wild type cells (lanes 2 and 3), which suggests that SHP may also interact with and inhibit transcription factor(s) other than p53 at the miR-34a promoter. A potential DNA binding site for HNF-4, a known SHP-interacting protein (30), is present at upstream of the p53 binding site in the miR-34a promoter. Overexpression of HNF-4, but not other nuclear receptors including FXR, enhanced p53 transactivation of the miR-34a promoter activity (supplemental Fig. S4). Similar transactivation of the miR-34a promoter by HNF-4 was observed with a promoter lacking the p53 binding site, indicating that HNF-4 is a minor contributor to miR-34a activation in a p53-independent manner (supplemental Fig. S5).
SHP Inhibits p53 Binding to the MiR-34a Promoter
To delineate mechanisms by which SHP inhibits p53transactivation of miR-34a, we performed ChIP assays. Occupancy of p53 was substantially decreased, whereas that of SHP was increased after GW4064 treatment (Fig. 2G), and similar results were observed in HepG2 cells (Fig. 2H). These results suggest that activated FXR signaling results in dissociation of p53 and recruitment of SHP at the miR-34a promoter.
To determine directly whether SHP is involved in decreased occupancy of p53 at the miR-34a promoter, SHP was down-regulated with siRNA. The inhibition of p53 binding to the miR-34a promoter by GW4064 treatment was markedly reversed by expression of SHP siRNA, whereas little binding to the control GAPDH gene was observed (Fig. 2I). These results demonstrate SHP-dependent inhibition of p53 binding to the miR-34a promoter. Because SHP has been shown to interact directly with HNF-4 (30), we also examined whether HNF-4 plays a role in the recruitment of SHP to the miR-34a promoter. Occupancy of HNF-4 at the miR-34a promoter was decreased in cells expressing siRNA for HNF-4 as expected, and the association of SHP at the promoter was decreased (supplemental Fig. S6). These results suggest that HNF-4 may also play a role in the recruitment of SHP to the miR-34a promoter.
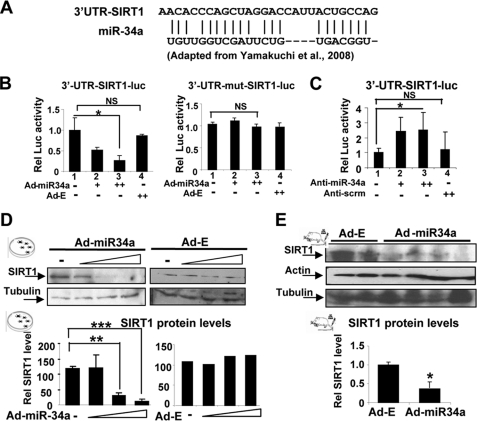
MiR-34a Binds to the 3′-UTR of SIRT1 Transcript
MiR-34a was shown to regulate cell proliferation and apoptosis by imperfect base-pairing to the 3′-UTR of SIRT1 mRNA (Fig. 3A) in human colon cancer cells (22). To test whether miR-34a inhibits SIRT1 activity in hepatic cells, reporter assays were performed using gain- and loss-of-function experiments. Overexpression of miR-34a decreased the 3-UTR-SIRT1-luc activity in a dose-dependent manner (Fig. 3B, left panel, and supplemental Fig. S7), and these effects were blunted in 3′-UTR SIRT1-luc in which miR-34a binding sites are mutated (Fig. 3B, right panel). Overexpression of anti-miR-34a significantly increased the reporter activity (Fig. 3C, lanes 1 and 3, and supplemental Fig. S8), whereas control scrambled RNA did not (lanes 1 and 4). These results confirm the previous findings that SIRT1 is a target of miR-34a (22) and show that this inhibition occurs in hepatic cells.
FIGURE 3.
SIRT1 is a target of miR-34a in hepatocytes. A, the miR-34a sequence and miR-34a binding site in the 3′-UTR of the SIRT1 transcript are shown. B, HepG2 cells were transfected with plasmids as indicated and then infected with Ad-miR-34a or control Ad-empty and collected for reporter assays. C, COS-1 cells were transfected with reporter plasmids as indicated along with anti-miR-34a or control scrambled RNA (Anti-scrm) and then collected for reporter assays. D, HepG2 cells were infected with Ad-miR-34a or control Ad-empty, and SIRT1 and control tubulin levels were detected. E, normal mice were injected in the tail vein with Ad-miR-34a or control Ad-empty, and 5 days later, hepatic SIRT1 and control actin and tubulin levels were measured. In the lower panels of D and E, band intensities were measured, and the intensities relative to those of tubulin were plotted. Statistical significance was determined by the Student's t test (S.E., n = 3). **, p < 0.01; ***, p < 0.001.
MiR-34a Down-regulates Hepatic SIRT1 Protein Levels
Exogenous expression of miR-34a also significantly reduced SIRT1 protein levels in a dose-dependent manner, but SIRT1 mRNA levels were only modestly, and not statistically significantly, reduced (Fig. 3D and supplemental Fig. S9). These results indicate that miR-34a down-regulates SIRT1 protein levels in hepatic cells. In in vivo studies, infection of mice with either Ad-miR-34a or control Ad-empty resulted in similar infection efficiency, detected by green fluorescent protein expression, and elevated miR-34a levels in mice infected with Ad-miR-34a (supplemental Fig. S10). Consistent with the HepG2 cell studies, overexpression of miR-34a in mouse liver significantly decreased SIRT1 protein levels (Fig. 3E). These results demonstrate that SIRT1 is a target of miR-34a in mouse liver in vivo.
Elevated Hepatic MiR-34a and Reduced SIRT1 Levels in FXR-null Mice
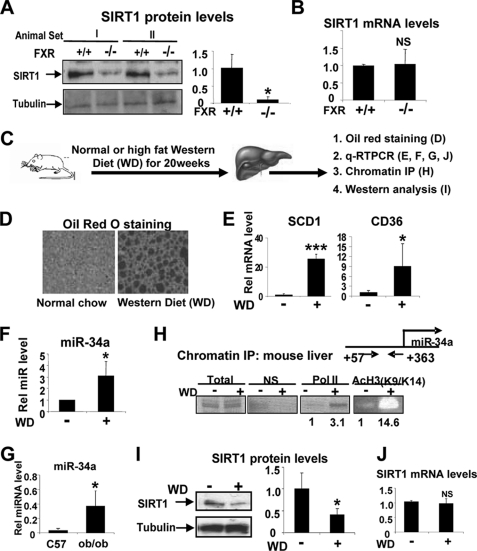
The results above suggest that activation of a regulatory pathway involving FXR and SHP inhibits miR-34a expression resulting in increased SIRT1 levels. Because miR-34a levels were highly elevated in FXR-null mice (Fig. 1) and miR-34a targets SIRT1 (Fig. 3), we next asked whether SIRT1 levels are decreased in FXR-null mice. Consistent with the results that miR-34a levels were increased in FXR-null mice (Fig. 1A), SIRT1 protein levels were indeed substantially decreased (Fig. 4A), whereas SIRT1 mRNA levels were not changed (Fig. 4B).
FIGURE 4.
Elevated miR-34a and decreased SIRT1 levels in diet-induced obese mice. A and B, hepatic SIRT1 protein levels from wild type or FXR-null mice were detected by Western blot analysis (A) and qRT-PCR (B). A, right panel, band intensities relative to those of tubulin were plotted. Statistical significance was determined by the Student's t test (S.E., n = 4). *, p < 0.05; NS, statistically not significant. C–J, C, experimental outline. Livers from mice fed normal chow (−) or a high fat Western-style diet (WD) for 20 weeks were collected for oil red o staining (D), qRT-PCR (E, F, G, and J), ChIP assays (H), and Western blot analysis (I). I, band intensities relative to those of tubulin were plotted. Statistical significance was determined by the Student's t test (S.E., n = 5). *, p < 0.05.
Deregulation of Hepatic MiR-34a and SIRT1 Levels in Diet-induced Obese Mice
FXR-null mice exhibit proartherogenic lipid profiles and elevated lipid and glucose levels (11, 13, 31), and activation of FXR in metabolic disease mice resulted in improved metabolic outcomes (11). Therefore, to examine whether the FXR targets miR-34a and whether SIRT1 levels are altered in metabolic disease, we utilized high fat Western-style diet (WD)-induced obese mice (Fig. 4C). Hepatic lipid levels and mRNA levels of lipogenic FAS and CD36 genes were substantially elevated in WD mice (Fig. 4, D and E). Interestingly, miR-34a levels were significantly elevated in diet-induced obese mice (Fig. 4F) and in leptin-deficient ob/ob mice (Fig. 4G). Consistent with increased miR-34a expression in diet-induced obese mice (Fig. 4F), association of RNA polymerase II, detected by ChIP assay, was increased at the miR-34a promoter, and acetylated histone 3 K9/K14, a gene activation histone code (32), was dramatically elevated (Fig. 4H). Consistent with elevated miR-34a levels, reduced SIRT1 protein, but not mRNA, levels were detected in liver (Fig. 4, I and J).
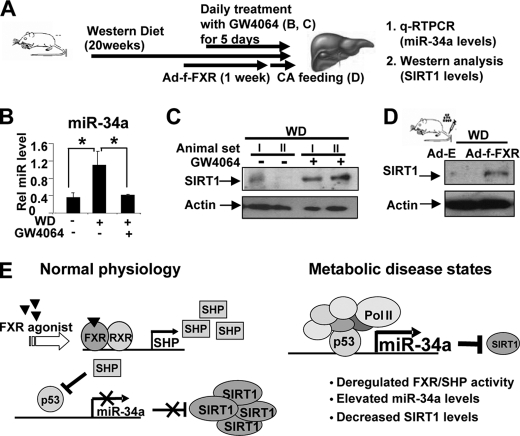
FXR Activation Decreases Hepatic MiR-34a Levels and Increases SIRT1 Levels in Obese Mice
It was demonstrated that activation of FXR in db/db mice improved metabolic profiles by decreasing serum glucose and lipid levels (11). We, therefore, asked whether activation of FXR in diet-induced obese mice could alter miR-34a levels and subsequently, SIRT1 protein levels in these mice (Fig. 5A). Treatment with the FXR activator, GW4064, daily for 5 days substantially reduced the miR-34a levels in diet-induced obese mice compared with those in normal mice (Fig. 5B). Remarkably, hepatic SIRT1 protein levels were substantially increased in obese mice treated with GW4064 (Fig. 5C). In control experiments, treatment with GW4064 increased mRNA levels of SHP and decreased mRNA and protein levels of p53 in diet-induced obese mice (supplemental Fig. S11). Overexpression of FXR in mouse liver using tail vein injection of Ad-3FLAG-FXR also resulted in substantial increases in SIRT1 protein levels in diet-induced obese mice (Fig. 5D). These results demonstrate an intriguing link between FXR activation, decreased miR-34a levels, and subsequently, increased hepatic SIRT1 protein levels in obese mice.
FIGURE 5.
Activation of FXR decreased miR-34a levels and increased SIRT1 protein levels in diet-induced obese mice. A, experimental outline is shown. B and C, mice fed WD chow were treated daily with GW4064 for 5 days, and hepatic miR-34a levels (B) and SIRT1 protein levels (C) were measured. D, mice fed WD chow were injected with Ad-FLAG-FXR, and 1 week later, mice were fed 0.5% cholic acid-chow for 6 h; hepatic SIRT1 protein levels were measured. E, a proposed model for FXR/SHP/miR-34a regulatory network controlling hepatic SIRT1 levels is shown. In this model, miR-34a inhibits translation of hepatic SIRT1 by binding to the 3′-UTR of SIRT1 transcript with partial base pairing. In normal mice, activation of FXR induces expression of SHP, which suppresses transcription of the miR-34a gene by inhibiting p53 binding to the miR-34a promoter. In contrast, in diet-induced obese mice, the FXR/SHP pathway is defective, which results in elevated miR-34a levels and subsequently decreased SIRT1 protein levels in the liver.
DISCUSSION
Despite recent advances in understanding the biological functions of SIRT1, the molecular basis by which SIRT1 levels are regulated under normal conditions and why they are substantially reduced in metabolic disease states remains unclear. Our studies, at least in part, provide an explanation by showing that the FXR/SHP pathway plays a role in controlling hepatic SIRT1 levels via miR-34a inhibition and that elevated miR-34a levels in obese mice contribute to decreased SIRT1 protein levels.
As summarized in Fig. 5E, in normal mice, hepatic miR-34a levels are regulated via a cascade pathway involving the nuclear bile acid receptor FXR and an orphan nuclear receptor and transcription corepressor SHP. Activation of FXR signaling inhibits the expression of hepatic miR-34a gene through the induction of SHP. SHP suppresses transcription of the miR-34a gene by inhibiting the promoter occupancy of p53, the key activator of the miR-34a gene. In contrast, in diet-induced obese mice, the FXR/SHP pathway is deregulated so that miR-34a levels are highly elevated, which contributes to reduced SIRT1 protein levels. Remarkably, activation of FXR in these obese mice by daily treatment with GW4064 for 5 days restores SIRT1 levels and decreases miR-34a levels. Edwards and colleagues (11) demonstrated previously that daily treatment of diabetic obese db/db mice with GW4064 for 5 days improved metabolic profiles by decreasing serum glucose and lipid levels. Therefore, our findings, together with these previous studies, indicate an intriguing link among FXR activation, decreased miR-34a levels, increased hepatic SIRT1 levels, and beneficial metabolic outcomes.
The present studies demonstrate that FXR positively regulates hepatic SIRT1 levels through miR-34a inhibition. Interestingly, SIRT1 may positively regulate hepatic FXR as well. PGC-1α was shown to enhance FXR activity in the regulation of triglyceride metabolism during fasting by increasing expression of the FXR gene and also by coactivating FXR transactivation (33). Because SIRT1 deacetylates and increases PGC-1α activity (1, 6), SIRT1 should increase FXR activity via modulation of PGC-1α activity. Further, we recently reported that SIRT1 increases FXR transactivation ability by dynamic deacetylation of FXR (28). FXR acetylation inhibits its transactivation potential by inhibiting interaction of FXR with RXR and subsequently DNA binding (28). Although FXR acetylation is dynamically modulated by p300 and SIRT1, FXR acetylation levels are highly elevated in ob/ob mice and diet-induced obese mice (28). These previous studies, along with current findings, suggest that an intriguing positive regulatory loop between FXR and SIRT1 is operating in hepatocytes in normal conditions. In obese mice, however, the FXR/SHP signaling may be deregulated because of highly elevated FXR acetylation levels, which results in elevated miR-34a levels.
In accordance with important roles of FXR in regulating miR-34a and SIRT1 protein levels, miR-34 levels were elevated, and SIRT1 protein levels are substantially decreased in FXR-null mice. However, it is unclear and counterintuitive why activation of the nuclear bile acid receptor FXR, which would presumably occur during the fed state due to high levels of circulating bile acids, positively regulates SIRT1, which mediates hepatic fasting responses. In part, this probably results from our lack of complete understanding of the complex interplay of regulatory pathways in hepatic metabolism, but it may also reflect an adaptive process related to the continuous cycles of feeding and fasting. Staels and his colleagues (34, 35) demonstrated that FXR plays an important role in adaptive responses during the transition from feeding to fasting and fasting-associated responses were markedly blunted in FXR-null mice. In line with these results, Auwerx and his colleagues (10) demonstrated that SIRT1 levels are dynamically regulated under fasting and feeding in normal mice. SIRT1 levels were modestly increased upon short term fasting (6 h), but prominent expression was still observed in the fed state in normal mice (10). We, therefore, envision that under normal conditions, FXR dynamically regulates metabolic pathways in response to fasting and feeding cycles, and activated FXR signaling during feeding would contribute to maintenance of basal levels of hepatic SIRT1 via miR-34a inhibition, which would allow for an efficient transition to the next fasting cycle. In contrast, this regulatory network is altered in metabolic disease states, resulting in constitutively elevated miR-34a levels and subsequently reduced SIRT1 levels in the liver.
Recent studies demonstrate that SIRT1 deacetylates and inhibits p53 activity, which results in decreased expression of miR-34a (3, 22, 36, 37). Consistent with this feedback regulatory loop, we also observed that overexpression of SIRT1 in liver of diet-induced obese mice using tail vein injection of Ad-FLAG-SIRT1 substantially decreased hepatic miR-34a levels (supplemental Fig. S12). Furthermore, daily treatment for 1 week with resveratrol, which activates SIRT1, also decreased miR-34a levels in obese mice (supplemental Fig. S12). Resveratrol indirectly activates SIRT1 by AMP-activated protein kinase activation (38, 39), so the possibility that resveratrol is acting in a SIRT1-independent manner cannot be ruled out. However, the SIRT1 expression studies, combined with the resveratrol results, further support the existence of the regulatory loop between miR-34a and SIRT1 in hepatocytes in which SIRT1 inhibits expression of its own inhibitor, miR-34a, thereby further enhancing SIRT1 expression.
MiRs are emerging as important cellular regulators critically involved in diverse biological pathways, including metabolic regulation, cell proliferation, and apoptosis (19, 37). Approximately 30% of all human genes are thought to be regulated by miRs, and elevated levels of miRs have been detected in diverse pathophysiological conditions (20, 21). Recent in vivo studies demonstrated that antisense miRs may have therapeutic value by down-regulating miRs in disease conditions (20, 40). It will be important to see whether down-regulation of elevated miR-34a in obese mouse liver using anti-miR-34a approaches would increase hepatic SIRT1 levels and improve metabolic outcomes in these mice. Modulation of SIRT1 levels by manipulation of the FXR-miR-34a-SIRT1 pathway, therefore, may provide a novel therapeutic target for treating aging-related diseases, such as metabolic disorders and cancer, in which SIRT1 plays an important role.
Supplementary Material
Acknowledgments
We thank Drs. J. Mendell for miR-34a promoter-luc plasmids, M. Yamakuchi for 3′-UTR SIRT1 plasmids, and W. Gu for GST-p53 and pcDNA3-FLAG-p53. We also thank B. Kemper for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK062777 and DK080032. This work was also supported by an American Diabetes Association Basic Science Award (to J. K. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12.
- SIRT1
- Sirtuin 1
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator α
- FXR
- farnesoid X receptor
- SHP
- small heterodimer partner
- miR
- microRNA
- 3′-UTR
- 3′-untranslated region
- HNF-4
- hepatic nuclear factor 4
- qRT-PCR
- reverse transcription-quantitative PCR
- Ad
- adenovirus
- ChIP
- chromatin immunoprecipitation
- GST
- glutathione S-transferase
- CoIP
- coimmunoprecipitation
- siRNA
- small interfering RNA
- luc
- luciferase
- WD
- western style diet.
REFERENCES
- 1.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 2.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 3.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 4.Guarente L. (2007) Cold Spring Harb. Symp. Quant. Biol. 72, 483–488 [DOI] [PubMed] [Google Scholar]
- 5.Rodgers J. T., Puigserver P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodgers J. T., Lerin C., Gerhart-Hines Z., Puigserver P. (2008) FEBS Lett. 582, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. (2006) Cell 127, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 8.Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008) Cell Metab. 8, 347–358 [DOI] [PubMed] [Google Scholar]
- 10.Coste A., Louet J. F., Lagouge M., Lerin C., Antal M. C., Meziane H., Schoonjans K., Puigserver P., O'Malley B. W., Auwerx J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Lee F. Y., Barrera G., Lee H., Vales C., Gonzalez F. J., Willson T. M., Edwards P. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. (2000) Cell 102, 731–744 [DOI] [PubMed] [Google Scholar]
- 13.Lee F. Y., Lee H., Hubbert M. L., Edwards P. A., Zhang Y. (2006) Trends Biochem. Sci. 31, 572–580 [DOI] [PubMed] [Google Scholar]
- 14.Fiorucci S., Rizzo G., Donini A., Distrutti E., Santucci L. (2007) Trends Mol. Med. 13, 298–309 [DOI] [PubMed] [Google Scholar]
- 15.Cariou B., Staels B. (2007) Trends Pharmacol. Sci. 28, 236–243 [DOI] [PubMed] [Google Scholar]
- 16.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. (2000) Mol. Cell 6, 507–515 [DOI] [PubMed] [Google Scholar]
- 17.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., Maloney P. R., Willson T. M., Kliewer S. A. (2000) Mol Cell 6, 517–526 [DOI] [PubMed] [Google Scholar]
- 18.Watanabe M., Houten S. M., Wang L., Moschetta A., Mangelsdorf D. J., Heyman R. A., Moore D. D., Auwerx J. (2004) J. Clin. Invest. 113, 1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobert O. (2008) Science 319, 1785–1786 [DOI] [PubMed] [Google Scholar]
- 20.Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., Watts L., Booten S. L., Graham M., McKay R., Subramaniam A., Propp S., Lollo B. A., Freier S., Bennett C. F., Bhanot S., Monia B. P. (2006) Cell Metab. 3, 87–98 [DOI] [PubMed] [Google Scholar]
- 21.van Rooij E., Sutherland L. B., Qi X., Richardson J. A., Hill J., Olson E. N. (2007) Science 316, 575–579 [DOI] [PubMed] [Google Scholar]
- 22.Yamakuchi M., Ferlito M., Lowenstein C. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang T. C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Lee K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., Arking D. E., Beer M. A., Maitra A., Mendell J. T. (2007) Mol. Cell 26, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song G., Wang L. (2008) Nucleic Acids Res. 36, 5727–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao J., Xiao Z., Kanamaluru D., Min G., Yau P. M., Veenstra T. D., Ellis E., Strom S., Suino-Powell K., Xu H. E., Kemper J. K. (2009) Genes Dev. 23, 986–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemper J. K., Kim H., Miao J., Bhalla S., Bae Y. (2004) Mol. Cell. Biol. 24, 7707–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang S., Tsang S., Jones R., Ponugoti B., Yoon H., Wu S. Y., Chiang C. M., Willson T. M., Kemper J. K. (2008) J. Biol. Chem. 283, 35086–35095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemper J. K., Xiao Z., Ponugoti B., Miao J., Fang S., Kanamaluru D., Tsang S., Wu S. Y., Chiang C. M., Veenstra T. D. (2009) Cell Metab. 10, 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang S., Miao J., Xiang L., Ponugoti B., Treuter E., Kemper J. K. (2007) Mol. Cell. Biol. 27, 1407–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y. K., Dell H., Dowhan D. H., Hadzopoulou-Cladaras M., Moore D. D. (2000) Mol. Cell. Biol. 20, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma K., Saha P. K., Chan L., Moore D. D. (2006) J. Clin. Invest. 116, 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Castellani L. W., Sinal C. J., Gonzalez F. J., Edwards P. A. (2004) Genes Dev. 18, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duran-Sandoval D., Cariou B., Percevault F., Hennuyer N., Grefhorst A., van Dijk T. H., Gonzalez F. J., Fruchart J. C., Kuipers F., Staels B. (2005) J. Biol. Chem. 280, 29971–29979 [DOI] [PubMed] [Google Scholar]
- 35.Cariou B., van Harmelen K., Duran-Sandoval D., van Dijk T., Grefhorst A., Bouchaert E., Fruchart J. C., Gonzalez F. J., Kuipers F., Staels B. (2005) FEBS Lett. 579, 4076–4080 [DOI] [PubMed] [Google Scholar]
- 36.Yamakuchi M., Lowenstein C. J. (2009) Cell Cycle 8, 712–715 [DOI] [PubMed] [Google Scholar]
- 37.He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., Cohen R. A., Zang M. (2008) J. Biol. Chem. 283, 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krützfeldt J., Kuwajima S., Braich R., Rajeev K. G., Pena J., Tuschl T., Manoharan M., Stoffel M. (2007) Nucleic Acids Res. 35, 2885–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.