Abstract
The influenza A virus PB1-F2 protein, encoded by an alternative reading frame in the PB1 polymerase gene, displays a high sequence polymorphism and is reported to contribute to viral pathogenesis in a sequence-specific manner. To gain insights into the functions of PB1-F2, the molecular structure of several PB1-F2 variants produced in Escherichia coli was investigated in different environments. Circular dichroism spectroscopy shows that all variants have a random coil secondary structure in aqueous solution. When incubated in trifluoroethanol polar solvent, all PB1-F2 variants adopt an α-helix-rich structure, whereas incubated in acetonitrile, a solvent of medium polarity mimicking the membrane environment, they display β-sheet secondary structures. Incubated with asolectin liposomes and SDS micelles, PB1-F2 variants also acquire a β-sheet structure. Dynamic light scattering revealed that the presence of β-sheets is correlated with an oligomerization/aggregation of PB1-F2. Electron microscopy showed that PB1-F2 forms amorphous aggregates in acetonitrile. In contrast, at low concentrations of SDS, PB1-F2 variants exhibited various abilities to form fibers that were evidenced as amyloid fibers in a thioflavin T assay. Using a recombinant virus and its PB1-F2 knock-out mutant, we show that PB1-F2 also forms amyloid structures in infected cells. Functional membrane permeabilization assays revealed that the PB1-F2 variants can perforate membranes at nanomolar concentrations but with activities found to be sequence-dependent and not obviously correlated with their differential ability to form amyloid fibers. All of these observations suggest that PB1-F2 could be involved in physiological processes through different pathways, permeabilization of cellular membranes, and amyloid fiber formation.
Keywords: Diseases/Amyloid, Protein/Viral, Protein/Folding, Protein/Structure, Protein/Synthesis, Viruses/Negative-strand RNA, Influenza A Virus, PB1-F2 Protein
Introduction
Influenza A virus (IAV)3 is a major pathogen of humans and animals responsible for three main pandemics of the last century: 1957, 1968, and the most severe in 1918 with 20–40 million deaths reported (1). Every year, influenza epidemics cause considerable illness and death. Since 2003, one of the greatest worldwide pandemic threats remains the highly pathogenic H5N1 avian viruses that display a high lethality rate (60% of the 438 cases resulted in death) (2, 3). Since its identification in April 2009, a novel swine origin influenza A virus H1N1 is currently causing a pandemic as declared by the WHO in June 2009 (around 1% of the 300,000 cases resulted in death) (4). IAV belongs to the Orthomyxoviridae viral family, and its genome is composed of eight negative-strand RNA gene segments encoding 11 proteins (5). The virulence of IAV is complex and can be influenced by each of the eight viral segments and notably the HA, NA, and PB1 segments (6).
PB1-F2, the 11th discovered IAV protein, is translated from an alternative reading frame in the PB1 gene (+1 reading frame of PB1, PB1-Frame 2) (7). PB1-F2 is expressed by many IAV isolates from a wide range of hosts and in nearly all avian and human strains (8–10). Recent studies have described the influence of PB1-F2 on viral pathogenesis in mouse models (11–14). Loss of PB1-F2 expression results in attenuation of the virus and in a more rapid clearance of the virus from the lungs of infected mice. Insertion of a single N66S mutation present in the PB1-F2 of the 1918 pandemic strain is sufficient to increase virulence of a low pathogenic IAV in mice (12). Moreover, a mouse model of secondary Streptococcus pneumoniae pneumonia following A/PR/834 virus infection shows greater weight loss of infected mice, more severe pneumonia, and a higher lethality with the virus expressing PB1-F2 compared with the PB1-F2 knock-out virus (13, 14). In addition, contemporary H1N1 strains that no longer express PB1-F2 seem to be less virulent. PB1-F2 is considered as one of the factors that contribute to IAV virulence (15).
Compared with the other IAV proteins, PB1-F2 shows several distinct features. PB1-F2 is expressed differently among infected cells and independently of the expression level of other viral proteins. PB1-F2 has a short half-life and is rapidly degraded (7). PB1-F2 is generally described as a proapoptotic factor that could facilitate evasion of host defenses (with NS1), allowing the virus to escape from the immune system by inducing apoptosis in macrophages and monocytes (1, 7, 16, 17). Thus, PB1-F2 is thought to compromise the ability of the host to mobilize adaptive immune responses (18). Another specific feature is the mitochondrial tropism of PB1-F2, which is only partial because PB1-F2 can also be present in the nucleus and cytoplasm of infected cells, depending on the cell type (7, 19). The carboxyl-terminal domain of the protein, containing the mitochondrial targeting signal, is capable by itself to interfere with mitochondrial function and cellular viability (20, 21). Studies of the interaction of a synthetic form of PB1-F2 with membrane showed that PB1-F2 was able to create pores of variable size in planar lipid membranes, suggesting that PB1-F2 could oligomerize to permeabilize membranes (22). PB1-F2 was also shown to interact with two proteins implicated in the formation of the mitochondrial permeability transition pore complex, ANT-3 (adenine nucleotide translocator 3), and VDAC-1 (voltage-dependent ion channel 1) (17).
Although new insights have been gained in understanding the function of PB1-F2, very little structural information is available. Bruns and co-workers (23, 24) produced a full-length synthetic PB1-F2 and performed a study by CD and 1H NMR (25). The stability of the structured regions of PB1-F2 was shown to be dependent on the hydrophobicity of the solvent: random coil in aqueous solution and α-helical structure upon the addition of trifluoroethanol (TFE). This α-helical structure present in the positively charged C-terminal domain of PB1-F2 was described as not a true amphipathic helix and is more compact than previously predicted. In an effort to obtain deeper insight into the structure-function of PB1-F2 and to better understand its role in the virus cycle, we produced and compared PB1-F2 originating from seven IAV isolates, including the highly pathogenic strain H5N1, the 1918 Brevig Mission strain, and two apathogenic avian strains, in biochemical and biophysical assays. We showed that PB1-F2 can be classified in the newly described group of intrinsically disordered proteins, capable of conformation changes switching from a random to an α-helical or β-sheet secondary structure. PB1-F2 is able to oligomerize to form amyloid fibers upon different environmental conditions. The presence of amyloid PB1-F2 structures was also evidenced in infected cells. Moreover, functional tests showed that the seven variants of PB1-F2 efficiently permeabilized membranes.
EXPERIMENTAL PROCEDURES
Constructions and Protein Expression
PB1-F2 genes were cloned in the Escherichia coli expression vector pET-22b+ (Novagen) between the NdeI and XhoI restriction sites, leading to the expression of C-terminal His6 fusion proteins. Genes coding for PB1-F2 A/WSN/33 (H1N1), A/HK/156/1997 (H5N1), and A/Swan/FR/06299/2006 (H5N1) strains were amplified with gene-specific primers. cDNAs for PB1-F2 A/PR/8/1934 (H1N1), A/Udorn/1972 (H3N2), and A/Tu/Mass/3740/1965 (H6N2) strains were obtained by reverse transcription using U12 universal primer (26) from total RNA of infected Madin-Darby canine kidney cells with the different viruses and extracted with the RNeasy minikit (Qiagen) according to the manufacturer's instructions. PB1-F2 A/BrevigMission/1/1918 (H1N1) strain was artificially synthesized (GeneCust) and amplified with gene-specific primers. The sequence of each cloned gene was confirmed by nucleotide sequencing. All primers and sequences are available upon request. Competent BL-21 Rosetta cells (Stratagene) transformed with the different pET22-PB1-F2-HisTag plasmids were cultured to an A600 of 0.6 in L medium. To express PB1-F2, the transformed cells were incubated for 4 h at 37 °C in 1 mm isopropyl 1-thio-β-d-galactopyranoside with agitation.
Protein Purification
Following incubation in isopropyl 1-thio-β-d-galactopyranoside, the transformed cells were pelleted by centrifugation for 15 min at 5000 × g. All of the recombinant PB1-F2-HisTag proteins accumulated in insoluble cytoplasmic inclusion bodies. After cell lysis by sonication in 50 mm Tris-HCl, pH 7.5, 10 mm EDTA buffer containing 0.1% Triton X-100, the crude lysate was centrifuged for 30 min at 10,000 × g at 4 °C. The pellets, containing the inclusion bodies were solubilized overnight under agitation at 4 °C with 20 mm Tris-HCl, pH 8, 0.5 m NaCl, 5 mm imidazole, 8 m urea buffer (solubilization buffer). The lysate was clarified by centrifugation for 30 min at 10,000 × g at 4 °C, and the supernatant was collected and loaded on a Hitrap-IMAC column using the AKTA Purifier-100 FPLC chromatographic system (GE Healthcare). The column was equilibrated with solubilization buffer. After sample injection to the column, the flow-through fraction was recovered, the column was washed with low imidazole buffer, and a 10-min linear gradient of 5–800 mm imidazole in the same buffer was applied to elute the column-bound proteins at a flow rate of 1 ml/min. One-ml fractions were collected during the gradient application. Column fractions were analyzed by 12.5% SDS-PAGE and Coomassie Blue staining. The PB1-F2-HisTag-containing fractions were pooled and further purified by size exclusion chromatography on a Sepharose S200 column equilibrated with 20 mm Tris-HCl, pH 8, 0.5 m NaCl, 8 m urea buffer. The S200-obtained fractions were subjected to 12.5% SDS-PAGE analysis. Urea was removed from the pool of S200 PB1-F2-HisTag-containing fractions on a G25 desalting column equilibrated with 10 mm ammonium acetate, pH 5, buffer. 5–10 mg of purified PB1-F2 were obtained per liter of bacterial culture. The resulting purified proteins were subjected to 12.5% SDS-PAGE analysis (0.1% SDS), and proteins were detected by Coomassie Blue staining. Prosieve color protein markers (Lonza) were used. Matrix-assisted laser desorption ionization mass spectrometry analysis was performed to confirm the molecular weight and integrity of the recombinant PB1-F2 proteins. PB1-F2-HisTag proteins were freeze-dried to be further used at the desired concentration and in different buffers.
Circular Dichroism Spectroscopy
CD measurements were performed on a JASCO J-810 spectropolarimeter equipped with a thermostatic cell holder, using a quartz cell of 1.0-mm path length. Spectra were collected over the wavelength range of 180–250 nm with a bandwidth of 1.0 nm and corrected for the contribution of the buffer. CD spectra of the PB1-F2 variants were recorded at 20 °C and at a final concentration of 50 μm. Each spectrum was an average of 16 scans. For estimation of secondary structure content, CD spectra curves obtained in TFE, acetonitrile, SDS, and asolectin were analyzed and quantified using the DICROPROT 2000 software (27).
Size Measurements by Dynamic Light Scattering (DLS)
The size measurements with the Zetasizer Nano serie (Malvern), based on the principle of dynamic light scattering, were made at 20 °C using a helium-neon laser wavelength of 633 nm and detection angle of 173°. The results were presented as size distribution calculated from the Malvern software.
Thioflavin T Fluorescence Measurements
Thioflavin T (ThT) binding assays were performed by adding freshly prepared 10 μm stock solution of ThT in 10 mm sodium acetate buffer, pH 5, to PB1-F2-HisTag samples at equimolar ratios. Fluorescence measurements were performed on a Jasco FP-6200 spectrofluorimeter at 20 °C. PB1-F2 protein samples at a final concentration ranging from 0.25 to 5 μm were preincubated at room temperature for 40 min before measurements. Fluorescence intensities were measured using an excitation wavelength of 435 nm and emission wavelength of 485 nm at a 0.2-cm path length after 5 min of incubation with ThT (28).
Liposome Preparation
An appropriate amount of soybean asolectin (Fluka) in chloroform solution was evaporated to dryness under a stream of nitrogen, and traces of solvent were removed by evacuation under a SpeedVac. The lipid residues were hydrated in sodium acetate, pH 5.0, buffer containing 80 mm calcein and gently vortexed and sonicated for a few minutes. Then liposomes were freeze-thawed three times in liquid nitrogen. Dye-loaded liposomes were separated from free calcein by passage through a Sephadex G-25 column using a sodium acetate elution buffer, pH 5.0. The liposome suspension obtained was finally extruded through a polycarbonate membrane with a pore diameter of 100 nm (Osmonics) to obtain calibrated liposomes. The size of asolectin vesicles was checked with DLS. Prepared liposome solution of typically 2.5 mg/ml were stored at 4 °C and were stable for several days. Liposomes used in a leakage assay analyzed by electron microscopy and used for CD analyses were prepared by the same method without the addition of calcein.
Assay of Liposome Permeabilization
The ability of the different PB1-F2 variants to permeabilize liposomes was checked by monitoring the fluorescence decrease of the encapsulated calcein when complexed with Co2+ contained in the buffer by quenching reaction. After the addition of peptide solution at the desired peptide/lipid ratio, the mixtures were excited at 492 nm, and their emission of fluorescence was recorded at 520 nm using a TECAN microplate reader. The WSN PB1-F2 fibers used in the assay presented in the supplemental material were obtained after incubation in the presence of 0.01% SDS at room temperature for 30 min (supplemental Fig. S2). The total release of calcein was achieved with the addition of Triton X-100 at a final concentration of 0.1% (v/v) to determine the maximum of fluorescence intensity (100%).
Electron Microscopy
Electron micrographs were acquired using a transmission electron microscope at a 80-kV excitation voltage. Samples of PB1-F2 variants at a concentration of 50 μm in 10 mm sodium acetate, pH 5, buffer were incubated in the presence of 0.01% SDS at room temperature for 30 min. A 10-μl sample was placed onto Formvar/carbon-coated 200-mesh copper grids (Agar Scientific). To investigate the interaction between lipid vesicles and PB1-F2 variants, asolectin liposomes were prepared in 10 mm sodium acetate, pH 5, buffer at a final concentration of 0.1 mg/ml in the presence or absence of a 50 μm concentration of the different PB1-F2 variants and incubated at room temperature for 5 min. Based on the permeabilization data, these conditions are suitable to observe a significant interaction between PB1-F2 and membranes. After the incubation, 10 μl of the lipid-protein sample were adsorbed onto Formvar/carbon-coated 200-mesh copper grids. The grids were blotted and, after drying, negatively stained by floating on 10-μl drops of 2% (w/v) uranyl acetate (Sigma).
Cell Culture and Viral Infections
293T cells in Dulbecco's minimum essential medium (Lonza) and Madin-Darby canine kidney cells in Eagle's minimum essential medium (Lonza), supplemented with 10% fetal calf serum (Perbio), 2 mm l-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin, were grown as monolayers. The human leukemic monocyte lymphoma (U937) cell line was maintained in RPMI 1640 medium (Lonza) supplemented with 10% fetal calf serum, 2 mm l-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin. All cultures were maintained at 37 °C in a 5% CO2 incubator. For infection, growing cells were seeded at 8 × 105 cells/plate and allowed to attach overnight before infection. Cells were washed with fetal calf serum-free medium and incubated with virus at a multiplicity of infection of 0.1 for 1 h at 37 °C. Infected cells were covered with complete fetal calf serum-free culture medium and incubated at 37 °C until collection.
Generation of Recombinant Knocked Out PB1-F2 Mutant Influenza Virus
The 12-plasmid reverse genetic system was used to generate recombinant viruses as described previously (29). Briefly, 293T cells were transfected with eight plasmids encoding the individual vRNA segments of influenza A/WSN/33 (H1N1) virus from a truncated human polymerase I promoter and four expression plasmids encoding the corresponding subunits of the viral polymerase and the nucleocapsid protein. The generated wild type (WT) and knocked out for PB1-F2 expression (ΔPB1-F2) viruses were amplified on Madin-Darby canine kidney cells and titrated by plaque assays. Generation of the PB1 plasmid knocked out for the expression of PB1-F2 was achieved using the QuikChange mutagenesis kit (Stratagene) at nucleotides coding for the four initiation codons changing ATG to ACG, resulting in silent mutation in the PB1 open reading frame. The mutations introduced were confirmed by reverse transcription-PCR and sequencing of the PB1 segment of the mutated generated viruses (supplemental Fig. S1).
Western Blot Analysis
U937 cells were infected with the recombinant viruses WT and ΔPB1-F2 and lysed at 12 h postinfection in a buffer containing 500 mm NaCl, 50 mm Tris-HCl, pH 8, 2% Triton X-100, and protease inhibitors (Roche Applied Science). Samples were loaded and separated on a 12.5% polyacrylamide gel. Gels were transferred onto Immobilon-P membranes (Millipore). The lack of PB1-F2 expression in ΔPB1-F2-infected cells was confirmed by Western blot analysis using rabbit polyclonal anti-PB1-F2 antibody (produced by immunization of rabbits with recombinant WSN PB1-F2). The relative infectivity of each recombinant virus was confirmed by Western blot analysis using a monoclonal anti-NS1 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Proteins were visualized using relevant peroxidase-labeled secondary antibodies by chemiluminescence (Amersham Biosciences).
Surface Plasmon Resonanse (SPR)
A Biacore 3000 SPR biosensor system equipped with control and evaluation software purchased from Biacore (GE Healthcare) was used in the study. PB1-F2 in cell homogenates was detected using rabbit hybridoma monoclonal anti-PB1-F2 antibodies. For this, an anti-rabbit phosphatase antibody was first immobilized on a CM5 chip using amine-coupling chemistry. The surface of the chip was activated for 7 min with a mixture of 0.05 m N-hydroxysuccinimide and 0.2 m 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide, in Hepes-buffered saline buffer, pH 7.4. Anti-rabbit phosphatase antibody diluted in 10 mm sodium acetate buffer, pH 5, was then covalently linked to the surface, giving up to 7000 resonance units. Ethanolamine (1 m, pH 8.5) was injected for 7 min to block the remaining activated groups. Monoclonal anti-PB1-F2 antibody was captured to the surface. Afterward, a linear calibration curve was obtained with recombinant WSN PB1-F2 in a concentration range from 10 nm to 1 μm (5 mm sodium acetate buffer, pH 5). These binding interactions were performed at an injection flow of 10 μl/min until stabilization of the immobilization level (2 min) was achieved. Report points were recorded before and after each injection, and the relative response units were determined. Regeneration of the anti-rabbit antibody was achieved by a 2-min injection of glycine buffer (pH 2.2). The same experimental parameters were used to probe cell lysates. The sensorgram observed with infected cell lysates was corrected by subtracting the response observed with non-infected cells or cells infected by the virus invalided for the PB1-F2 gene (reference surface) and usually normalized to a base line of 0 resonance units. Concentration of PB1-F2 in infected cells was determined relative to the calibration curve. All measurements were performed in triplicate at 20 °C.
Thioflavin S Staining
At 12 h postinfection, U937 cells were resuspended, centrifuged at low speed, and fixed in 3.2% paraformaldehyde for 10 min at room temperature. Fixed cells were rinsed in phosphate-buffered saline and stained using a 0.01% solution of thioflavin S (ThS) in 40% EtOH for 3 min at room temperature. Finally, stained cells were differentiated in 50% EtOH in phosphate-buffered saline for 10 min, rinsed in phosphate-buffered saline, and observed using a Leica DMR fluorescence microscope equipped with a Olympus DP71 camera and using Cell-F software (Soft Imaging System, Olympus).
RESULTS
PB1-F2 Variants Present a Soluble Disordered Structure in Solution
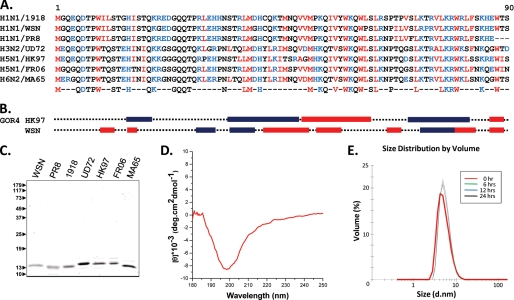
Fig. 1A shows a sequence alignment of seven PB1-F2 variants selected to represent its sequence variability among IAVs. This alignment includes the sequences of three highly pathogenic viruses, the H1N1 virus A/BrevigMission/1/1918 and two H5N1 viruses (A/Swan/Fr/06299/06 and A/HK/156/97). All variants are 90-amino acid-long proteins with the exception of A/PR/8/34, which is 87 amino acids long. The analysis revealed 41% amino acid identity in the PB1-F2 proteins, including six conserved glutamine residues, 12 hydrophobic residues (five tryptophans), and nine charged residues. Clusters of conserved amino acids were found over all of the sequences. Whereas the extreme N-terminal domain is negatively charged, the other part of the molecule is positively charged with conserved histidine, arginine, and lysine residues.
FIGURE 1.
A, multialignment of the seven PB1-F2 variants included in this study. Strains are abbreviated as follows. 1918, A/Brevig Mission/1/1918 (H1N1); WSN, A/WSN/1933 (H1N1); PR8, A/PR/8/1934 (H1N1); UD72, A/Udorn/1972 (H3N2); HK97, A/HK/156/1997 (H5N1); FR06, A/Swan/FR/06299/2006 (H5N1); MA65, A/Tu/Mass/3740/1965 (H6N2). Conserved residues for the seven sequences are indicated on the bottom line (blue, charged residues; red, hydrophobic residues). B, secondary structure predictions (blue boxes, α-helix; red boxes, β-turn) were obtained with the software GOR-IV (available on the World Wide Web). C, the left panel shows SDS-PAGE analysis and Coomassie Blue staining of purified PB1-F2 proteins. The relative molecular weights (shown in thousands) were determined by reference to marker proteins. D, far-UV CD spectra for WSN PB1-F2 recorded in 10 mm sodium acetate, pH 5, buffer. E, the right panel shows the evolution in time (24 h) of the mean RH of WSN PB1-F2 (50 μm) in 10 mm sodium acetate, pH 5, buffer measured by DLS at 20 °C.
All of these variants were produced in the E. coli and found to form bacterial inclusion bodies. Purification was carried out after solubilization using a chaotropic agent by His tag chromatography and gel filtration (Fig. 1C). To obtain the native refolded form of the PB1-F2 variants, only dialysis performed in acidic conditions (pH 5) prevents their aggregation. The secondary structure of the refolded PB1-F2 variants was then estimated using CD spectroscopy. All of the recorded spectra showed very similar shape characteristics of a random coil conformation with no evidence of stable secondary structures (Fig. 1D). Furthermore, DLS analyses were carried out to measure the mean hydrodynamic radius (RH) of the PB1-F2 in this condition. As exemplified with the WSN variant in Fig. 1E, the RH values of the different PB1-F2s were found ranging from 3 to 10 nm as expected for a disorder protein of 11 kDa (30). These data suggest that all of the PB1-F2 variants adopted a soluble disordered structure in acidic solution.
The Conformational Diversity of the PB1-F2 Variants in Different Environments
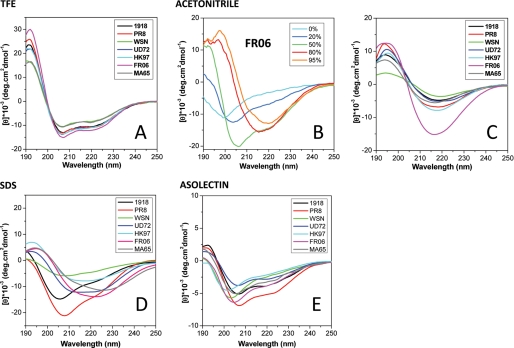
In a polar environment, the C-terminal domain of the PB1-F2 PR8 variant was shown to form an amphipathic α-helix with positively charged residues on one side of the helix and hydrophobic residues on the other side (25). The seven sequences used in our study were subjected to GOR-IV (31) secondary structure prediction analysis (Fig. 1B). Sequences of HK97 and PR8 PB1-F2 are predicted to form α-helices on 33.3 and 29.9% of their length, respectively, in contrast to the five other PB1-F2 variants that only displayed scores around 18%. All sequences are predicted to form β-sheets around one-third of their sequences (31.03–37.78%) except for HK97, which has a score of 18%. These observations suggest that, due to its sequence variability, PB1-F2 variants present alternative folding pathways. To investigate the conformational diversity of PB1-F2, the effect of different solvents on the PB1-F2 structure was then analyzed. First, CD spectra of the seven variants were recorded in the presence of TFE or acetonitrile. As previously reported in the presence of 50% TFE with the PR8 PB1-F2 variant (25), the CD spectra were typical of α-helical folding, with minima at 208 and 222 nm and a maximum at 192 nm (Fig. 2A). Deconvolution of the data obtained at a different TFE/water ratio showed that PB1-F2 was mainly helical upon the addition of 20% TFE to reach a maximum ranging from 35 to 50% α-helicity at 50% TFE. The propensity to form α-helix in the presence of TFE appears to be a common property of the seven PB1-F2 variants. In contrast, the spectra obtained upon increasing concentrations of acetonitrile showed a transition from a random coil conformation to β-sheets (exemplified in Fig. 2B for the FR06 variant) with the characteristic signature of a minimum at 217 nm and a maximum at 195 nm. Fig. 2C shows that all variants form β-sheets in 80% acetonitrile, indicating the presence of 20% (PR8 variant) to 50% of β-sheet content (FR06 variant) upon deconvolution.
FIGURE 2.
Secondary structure formation of the seven PB1-F2 variants in different environments. Far-UV CD spectra were recorded in 10 mm sodium acetate, pH 5, buffer complemented with solvants (50% TFE (A) or 80% acetonitrile (C)), detergent (0.01% SDS (D)), or 0.1 mg/ml asolectin liposomes (E). B, the secondary structural switch from random coil to β-sheet conformation while increasing acetonitrile percentage in the buffer for FRO6 PB1-F2.
To further investigate the structure of PB1-F2 in membrane-mimicking conditions, CD spectra were recorded in an ionic detergent SDS solution or in the presence of asolectin liposomes. In SDS solution (Fig. 2D), the spectra were indicative of the presence of a mix of random coil conformations and β-sheets. Deconvolution data showed variable percentages of β-sheet content: 22% for PR8 variant to 75–80% for FR06 and several other variants. For asolectin (Fig. 2E), the curves suggest the presence of a mix of random and β-sheet structures. The curves could reflect some distortion of the CD spectra due to light scattering on membrane vesicles. Taken together, the CD data showed that the secondary structure of PB1-F2 is strongly dependent on the environment. Although PB1-F2 is not structured in aqueous conditions, PB1-F2 forms α-helices in TFE or β-sheets in acetonitrile as well as in membrane-mimicking detergent or in the presence of liposomes. The intrinsic propensity of the PB1-F2 variants to form β-sheets appears to be sequence-specific.
Dynamic Light Scattering Analysis Reveals the Formation of Oligomeric or Aggregated Structures in Different Environments
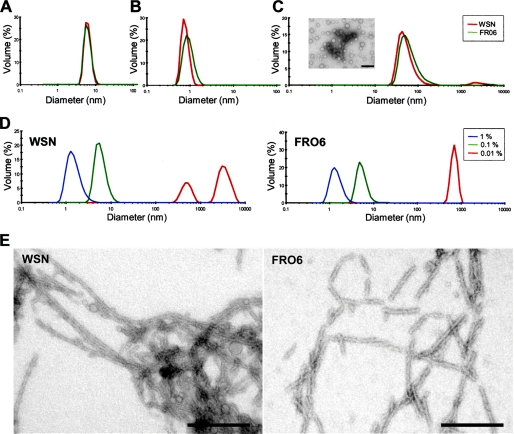
To characterize the consequences of the acquisition of an ordered secondary structure by the PB1-F2 variants on their three-dimensional structure, we first carried out DLS experiments in the environmental conditions described above. Fig. 3 shows representative results obtained with two PB1-F2 variants (WSN and FR06). As illustrated in Fig. 3B, incubation in 50% TFE allows detection of objects with a diameter of about 1 nm, a size compatible with a monomeric and structured form of an 11-kDa protein. It should be noted that the HK97 variant is the only one that precipitates in these conditions. In contrast, incubation in acetonitrile results in the formation of oligomers/aggregates with sizes ranging from 20 to 1000 nm (Fig. 3C). Incubation with lower concentrations of acetonitrile allows the detection of smaller oligomers/aggregates (data not shown). Fig. 3D shows the behavior of the two PB1-F2 variants in the presence of different concentrations of SDS. In 1% SDS, both variants are more compact (RH of ∼1 nm). When the SDS concentration was lowered to 0.1%, PB1-F2 was found to form small oligomers with RH of <10 nm. Interestingly, the variants were found to form oligomers/aggregates of variable and larger sizes in 0.01% SDS. Whereas the FR06 PB1-F2 forms oligomers/aggregates with an RH of ∼1000 nm (like PR8, UD72, and HK97), the WSN, 1918, and MA65 PB1-F2 form oligomers from 2 to 8 μm. Electron microscopy of negatively stained preparations indicates that PB1-F2 in 0.01% SDS forms fibers that can vary in length from 100 nm to 1 μm and with a diameter of around 10–15 nm (Fig. 3E).
FIGURE 3.
DLS size distributions of the mean RH, of different PB1-F2 variants at a concentration of 50 μm in 10 mm sodium acetate, pH 5, buffer alone (A) or complemented with 50% TFE (B) or 80% acetonitrile (C). The inset in C shows aggregates formed by WSN PB1-F2 in acetonitrile by electron microscopy. D, evolution of the RH of WSN and FRO6 PB1-F2 in the presence of 0.01% (red), 0.1% (green), or 1% (blue) SDS. Observation by electron microscopy of the fibers (E) formed by WSN (left) and FRO6 (right) PB1-F2 variants in the presence of 0.01% SDS. Bars, 200 nm.
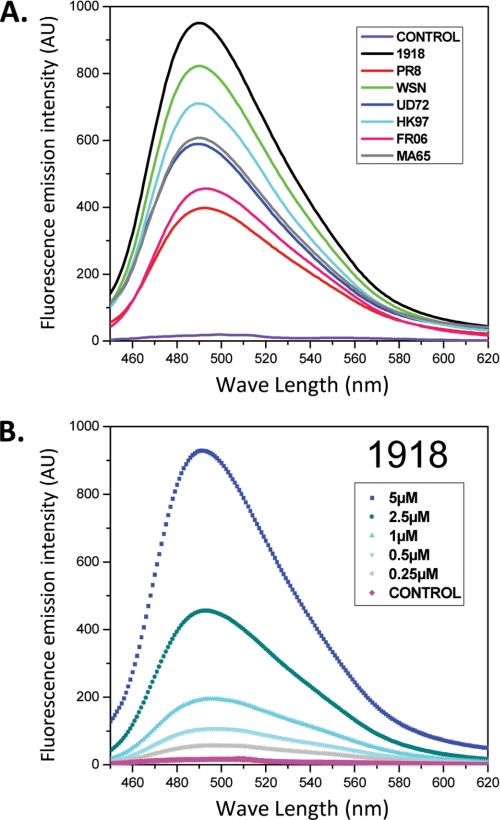
To characterize the nature of the PB1-F2 fibers formed in the presence of SDS, we incubated them in the presence of ThT, which specifically binds to amyloid fibrils (28). In the assay, the fluorescence signal emitted at 485 nm after an excitation at 435 nm reveals that the ThT binds to amyloid fibers. ThT was found to bind all of the PB1-F2 fibers formed in the presence of 0.01% SDS, thus revealing that all PB1-F2 variants form amyloid fibers in these experimental conditions. Although the WSN and the 1918 variants provided a strong emission signal, the FR06 and PR8 variants displayed a weak signal (Fig. 4A). The magnitude of the emission signal correlates well with the size of the fibers observed by electron microscopy, WSN and 1918 forming the longest fibers. The increase of fluorescence is concentration-dependent, as exemplified by the 1918 variant (Fig. 4B). The presence of fibers was detectable at a concentration as low as 0.25 μm. It should be noted that amyloid fibers were not detected when the PB1-F2s were incubated in aqueous solutions and in TFE/water or acetonitrile/water mixtures. In 0.1 and 1% SDS solutions, no amyloid fibers were detected.
FIGURE 4.
A, measure of the fluorescence increase of ThT after binding with amyloid formed by the seven PB1-F2 variants at a final concentration of 5 μm in 10 mm sodium acetate pH5 buffer containing 0.01% SDS. B, ThT binding by 1918 PB1-F2 at concentrations ranging from 0.25 to 5 μm. The control was obtained with PB1-F2 at the same concentration in sodium acetate buffer without SDS. AU, absorbance units.
PB1-F2 Proteins Permeabilize Liposomes in a Sequence-specific Manner
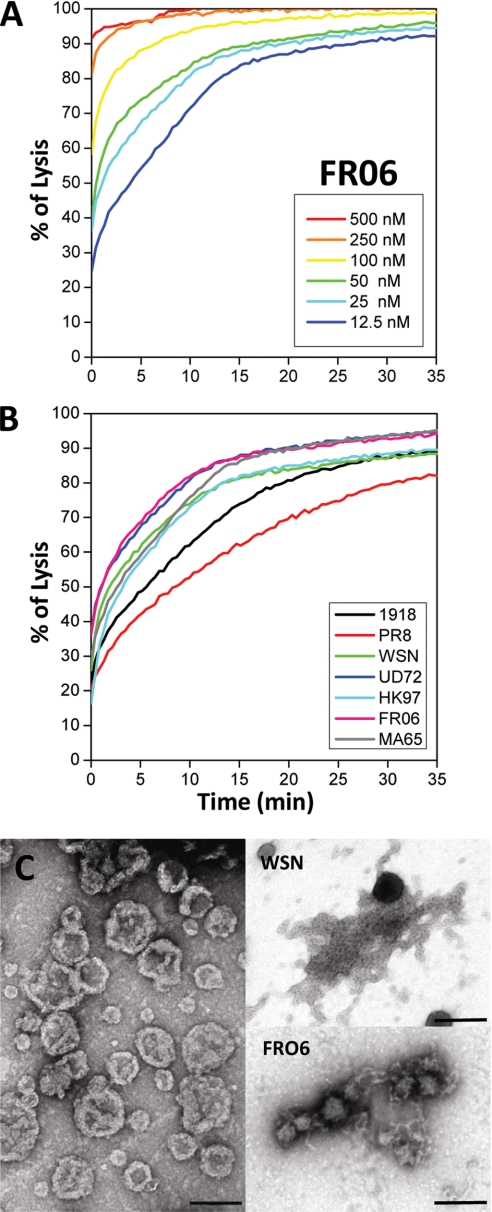
To determine if the conformational diversity of the PB1-F2 variants correlates with their intrinsic ability to form pores in membranes as was reported for amyloidogenic proteins (32, 33), permeabilization tests were carried out on synthetic membranes. PB1-F2 variants were incubated with liposomes containing a fluorescent soluble probe. The release of fluorescent calcein from perforated liposomes was revealed by a quenching reaction using cobalt present in the buffer and not inside the liposomes. Thus, a decrease of fluorescence corresponds to liposome lysis. Each PB1-F2 variant caused permeabilization of the liposomes in a time- and dose-dependent manner (see Fig. 5A for the FR06 variant). All of the variants were able to achieve a complete lysis at a concentration of 500 nm within a few seconds. However, at lower concentrations, the kinetic of the probe release was different between each variant, indicating that the sequence variability of PB1-F2 may affect its ability to interact with membrane and induce the lysis (Fig. 5B). No permeabilization was detected for any variant under a critical concentration of 5 nm. The physical interaction between synthetic liposomes and PB1-F2 variants was investigated by electron microscopy. The liposomes shown in Fig. 5C (left) had initially a spherical appearance with an apparent diameter range from 100 to 400 nm in sodium acetate buffer, pH 5. Incubation of asolectin vesicle suspensions with soluble WSN or FRO6 PB1-F2 leads to a spectacular liposome destruction (Fig. 5C, right). Both PB1-F2 variants assemble into fibrils with a high tendency to associate with membrane fragments. As observed with SDS, WSN PB1-F2 forms longer fibers than the FR06 PB1-F2 variant. The lysis activity of each variant was not found to correlate well with their ability to form long or short amyloid fibers.
FIGURE 5.
Membrane permeabilization by PB1-F2 variants. Asolectin liposomes were assayed for calcein release against nanomolar concentrations of PB1-F2 variants. A, permeabilization of liposomes upon the addition of 12.5–500 nm FRO6 PB1-F2 variant in 10 mm sodium acetate, pH 5. B, lysis activity of all seven PB1-F2 variants at a concentration of 25 nm in 10 mm sodium acetate, pH 5, buffer. C, images of liposomes in the presence of different PB1-F2 variants by electron microscopy. Left, untreated liposomes in sodium acetate buffer, pH 5; upper right, WSN PB1-F2; lower right, FRO6 PB1-F2. Bar, 200 nm.
To check whether the fibers were the active species causing liposome leakage, we carried out a permeabilization assay with preformed WSN PB1-F2 fibers. Interestingly, there was no pore formation with fibers. Supplemental Fig. S2 shows that no pore formation activity was evidenced with preformed fibers. A weak and similar increase of fluorescent signal was observed with liposomes treated with PB1-F2 fibers and/or 0.01% SDS. In contrast, monomeric PB1-F2 significantly permeabilized liposomes. This shows that prefibrillar aggregates and not the fibers are the species that possess a pore-forming activity.
In Vivo PB1-F2 Oligomerization
To address the physiological relevance of the PB1-F2 ability to form amyloidal fibers in vitro, we first estimated the PB1-F2 concentration in infected cells to determine if the PB1-F2 concentrations required for oligomerization and fiber assembly are of the same order of magnitude as the PB1-F2 intracellular concentration.
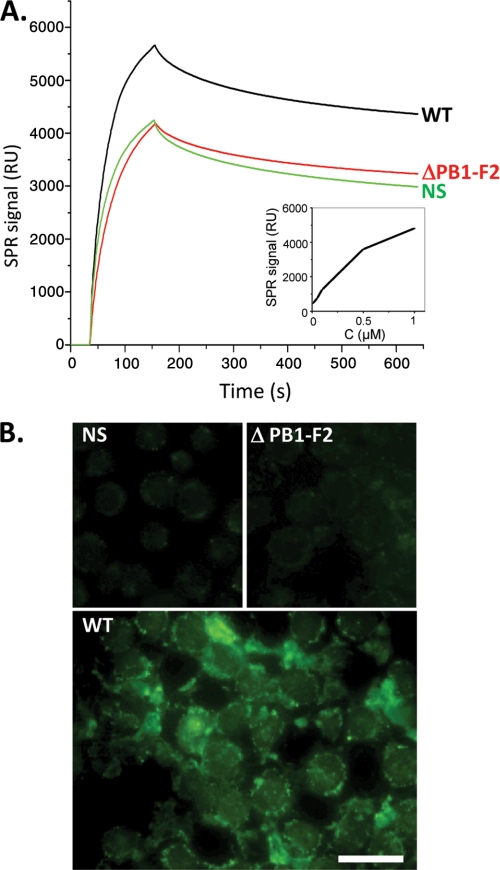
To this end, we produced a specific anti-PB1-F2 monoclonal antibody (to be described elsewhere). Cells were infected with a recombinant WT influenza virus A/WSN/1933 (H1N1) or with a mutant that was knocked out for the PB1-F2 gene (ΔPB1-F2). The amount of PB1-F2 in infected cells was quantified by a sensorchip (Biacore) coated with the anti-PB1-F2 antibody (Fig. 6A). The PB1-F2-specific SPR signal was calculated by subtracting the ΔPB1-F2 SPR signal obtained from the WT virus. By comparing this signal with those obtained using dilutions of recombinant PB1-F2 (Fig. 6A, inset), we estimated the intracellular concentration of PB1-F2 to be roughly 0.5–1 μm, a value of the same range as in the in vitro assays in which we evidenced by the thioflavin assay amyloid fibers at a 0.25 μm concentration.
FIGURE 6.
PB1-F2 expression and aggregation in influenza virus-infected cells. A, real-time kinetics of interaction between lysates obtained from U937-infected cells with wild type A/WSN/1933 (H1N1) virus (WT), virus knocked out for PB1-F2 expression (ΔPB1-F2), or non-infected cells with chip-immobilized monoclonal anti-PB1-F2 antibody. The sensorgrams are representative for three independent experiments. Inset, a calibration curve obtained for interaction of purified WSN PB1-F2 with the immobilized anti-PB1-F2 antibody. B, ThS staining of nonstimulated (NS), WT, and ΔPB1-F2 U937-infected cells was observed by fluorescence microscopy after excitation with UV light. Bar, 10 μm.
We next tried to evidence the presence of PB1-F2 amyloid fibers/aggregates in influenza virus infected cells using ThS staining (34, 35). ThS, like ThT, binds to amyloid fibers and was chosen because it resulted in an overall reduction of background cell autofluorescence compared with ThT staining (data not shown). Fig. 6B shows that in contrast to what was observed in non-infected and ΔPB1-F2-infected cells, WT-infected cells exhibit an intense fluorescent staining, thus revealing the presence of amyloid β-sheet structures. The ThS staining appeared to be diffuse in the cytoplasm with intense spots associated with membranes. This result demonstrates that PB1-F2 is associated or forms amyloid-like fibers in infected cells.
DISCUSSION
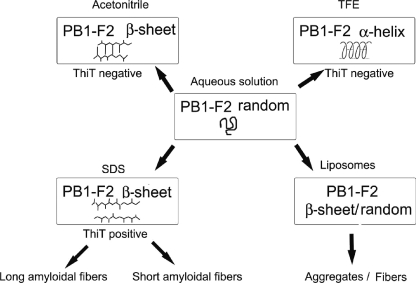
In the present work, we have investigated both secondary and oligomeric structures of seven PB1-F2 variants to gain insight into their structural and functional relationships. We show that PB1-F2 has distinct folding pathways leading to the formation of either disordered conformations or α-helices or oligomeric β-enriched structures, depending on physico-chemical conditions (Fig. 7). As previously observed, our results confirm the disordered structure of PB1-F2 in aqueous solutions (acidic) as well as the formation of an α-helical secondary structure in TFE (25). TFE is well known to induce α-helix folding by stabilizing hydrogen bonds in peptides (36, 37) even if they do not have an intrinsic propensity to adopt α-helical structure. In contrast, all PB1-F2 variants exhibit β-sheet secondary structures in the presence of acetonitrile. Compared with TFE, acetonitrile is less polar, and thus it is a weaker hydrogen bond donor (38). Moreover, the β-sheet structural conformational switch of PB1-F2 was also observed with the addition of SDS. This mild detergent is commonly used for probing conformation of small proteins due to its ability to induce and stabilize ordered conformations in peptides having structure-forming potential (39–41). Furthermore, SDS micelles have been used extensively for the structural investigations of membrane peptides because they provide an anionic, membrane-mimicking environment with a hydrophobic core and a polar head (42). Therefore, it clearly appears that the microenvironment surrounding PB1-F2 strongly affects its folding pathway. Finally, in the presence of small unilamellar asolectin liposomes of net negative charge, all PB1-F2 variants adopt a β-sheet conformation similar to those observed with acetonitrile and SDS.
FIGURE 7.
Model for the PB1-F2 folding pathways. The scheme represents the different folding pathways leading to the formation of either disorder conformations, α-helices, or oligomeric β-enriched structures, depending on the physical and chemical parameters of the environment. All PB1-F2 variants exhibit random structures in aqueous (acidic pH) solutions. PB1-F2 variants adopt an α-helical structure in TFE-containing solutions. Whereas ThT-negative β-sheet assemblies are evidenced in acetonitrile solutions, PB1-F2 variants generate ThT positive amyloidic assemblies in the presence of 0.01% SDS. The length of the fibers differs, depending on the PB1-F2 variants. β-Oligomers are also observed in the presence of liposomes.
PB1-F2 oligomerizes in all of the conditions that favor β-sheet-enriched structure formation. In most cases, protein fibers emerge from an equilibrium of folded protein states with local unfolded or marginally stable structures that can be self-assembled into β-sheet oligomers, leading finally to the irreversible formation of insoluble fiber (43). In contrast, oligomeric structures were not observed in TFE. The α-helical conformation evidenced with TFE may be considered as an end conversion pathway because only the PB1-F2 monomeric structured form was observed in this environmental condition. Fluorinated alcohols, such as TFE, have been shown to favor monomeric protein conformation by disrupting hydrophobic interaction in aggregated amyloid preparations (44). Only amorphous aggregates were observed upon the addition of acetonitrile. Amyloid fibers were generated in 0.01% SDS solution. Fibers were also observed in the presence of asolectin liposomes, as visualized by electronic microscopy. PB1-F2 variants form fibers of different lengths, from 100 nm to 1 μm. Because PB1-F2 variants exhibit differential abilities to form β-sheets and amyloid fibers, this process is likely to be sequence-specific. The structural switch leading to an amyloid pathway was observed for other viral proteins, such as the DNA binding domain of papillomavirus E2 proteins and EBNA-1 Epstein-Barr virus protein in vitro (45, 46).
We also checked whether the structural switch between the unfolded state and oligomeric β-sheet-enriched structures may constitute a critical element for the regulation of PB1-F2 activities. PB1-F2 was previously reported to form pores of variable sizes in planar membranes (22). Otherwise, PB1-F2 was shown to be cytotoxic when added to cell medium at a low concentration (7) and to promote loss of mitochondrial membrane potential by inducing morphological alterations (7, 17). We show here that all of the variants used in this study permeabilize asolectin liposomes very efficiently, even at 5 nm nanomolar range concentrations. The fact that PB1-F2 adopts a β-sheet structure in the presence of synthetic membranes suggests a possible pore formation activity, as observed for many amyloid-forming proteins (33). Furthermore, the net positive charge of PB1-F2 in acidic conditions may promote its incorporation into negatively charged asolectin liposomes by electrostatic interaction. Lysis assays showed that there is no direct correlation between the length of fibers and PB1-F2 activity. Interestingly, the addition of preformed fibers to liposomes did not lead to liposome permeabilization. This suggests that a prefibrillar protein but not amyloid may be the active lytic form. The ordered prefibrillar aggregates may form nonspecific pores perturbing cell ionic homeostasis. Frequently, prefibrillar aggregates display much higher toxicity than mature fibrils (47–49). The fast leakage kinetics observed here (within a few seconds) suggests the formation of β-amyloid pore structures rather than the generation of long fiber aggregates. The high activity of PB1-F2 on membranes (with pores formed at nanomolar concentrations) suggests that PB1-F2 within an infected cell may contribute to perturbation of the intracellular ionic homeostasis, leading ultimately to cell death.
Our results do not rule out the possibility that free or membrane-bound PB1-F2 interacts with cellular and/or viral proteins. Actually, PB1-F2 was shown to interact with the mitochondrial proteins VDAC-1 and ANT-3 and to enhance the opening of the permeability transition pore complex (17). It has also been recently proposed that PB1-F2 could be involved in interactions with members of the Bcl-2 family known to regulate mitochondrially mediated apoptosis (50). On the other hand, PB1-F2 was shown to regulate viral polymerase activity by interaction with the PB1 protein in the nucleus (19). The natively disordered state of PB1-F2 in aqueous solutions could allow interactions with all of these proteins and lipids. Proteins other than PB1-F2 have also been shown to require a high degree of structural disorder to fulfill their functions and need to undergo disorder-order transition during or prior to their biological function (51). Such proteins are frequently involved in some of the most important regulatory functions in the cell, and the lack of intrinsic structure in many cases is relieved when the protein binds to its target molecule (52).
A critical finding of our work is that PB1-F2 forms amyloid-like fibers in infected cells. Although amyloidosis is mainly implicated in neurodegenerative diseases, fibrillization of some viral or cellular proteins has been evidenced in viral infections. Recently, semen-derived amyloid fibrils were shown to drastically enhance human immunodeficiency virus infection (53). This capability is not restricted to retroviruses and suggests a possible role of amyloids in the transmission and pathogenesis of other enveloped viruses (54). Viral proteins by themselves can also initiate protein aggregation and regulate amyloid formation (55). Recent work suggests that highly pathogenic H5N1 influenza virus or other neurotropic influenza viruses could initiate central nervous system disorders, including Parkinson and Alzheimer diseases (56). Our observation that PB1-F2 is able to form amyloidal fibers in virus-infected cells may be related to the nervous system disorders observed with influenza neurotropic viruses and opens the way for further studies of the role of PB1-F2 in the pathogenesis of the influenza virus.
Overall, our results provide new insights into structural behavior of PB1-F2 able to switch from a disordered state to α-helix and to β-sheet structures in membranes. We demonstrate that PB1-F2 forms amyloid fibers in infected cells, an observation that explains the pathogenicity associated with this protein. These observations open the way for further studies to elucidate the role of PB1-F2 in the influenza virus cycle.
Supplementary Material
Acknowledgments
We thank Céline Henry (Plateau d'Analyse Protéomique par Séquençage et Spectrométrie de Masse (PAPSSM), Institut National de la Recherche Agronomique (INRA), Jouy-en-Josas, France) for the mass spectrometry analysis and Sophie Chat and Christine Longin (Génomique et Physiologie de la Lactation (GPL), INRA, Jouy-en-Josas, France) for electronic microscopy observations. We greatly appreciate the gift of PB1-F2 genes from Ervin Fodor (University of Oxford), Nadia Naffakh and Nicolas Escriou (Institut Pasteur, Paris, France), and Pascale Massin and Véronique Jestin (Agence Française de Sécurité Sanitaire des Ailments (AFSSA), Ploufragan, France). We thank George Browlee for providing the reverse genetic system for virus strain WSN. We thank Christophe Lemaire for the spectrofluorimeter facilities of Laboratoire de Génétique et Biologie Cellulaire (LGBC) (University of Versailles). We also thank Stéphane Biacchesi (Virologie et Immunologie Moléculaires, INRA, Jouy-en-Josas, France) and Rachel Young (Unité Mixte de Recherche de Génétique Animale et Biologie Intégrative (UMR GABI), INRA, Jouy-en-Josas, France) for critical reading of the manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- IAV
- influenza A virus
- TFE
- trifluoroethanol
- DLS
- dynamic light scattering
- ThT
- thioflavin T
- ThS
- thioflavin S
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Lamb R. A., Takeda M. (2001) Nat. Med. 7, 1286–1288 [DOI] [PubMed] [Google Scholar]
- 2.Lipatov A. S., Govorkova E. A., Webby R. J., Ozaki H., Peiris M., Guan Y., Poon L., Webster R. G. (2004) J. Virol. 78, 8951–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gambotto A., Barratt-Boyes S. M., de Jong M. D., Neumann G., Kawaoka Y. (2008) Lancet 371, 1464–1475 [DOI] [PubMed] [Google Scholar]
- 4.Garten R. J., Davis C. T., Russell C. A., Shu B., Lindstrom S., Balish A., Sessions W. M., Xu X., Skepner E., Deyde V., Okomo-Adhiambo M., Gubareva L., Barnes J., Smith C. B., Emery S. L., Hillman M. J., Rivailler P., Smagala J., de Graaf M., Burke D. F., Fouchier R. A., Pappas C., Alpuche-Aranda C. M., López-Gatell H., Olivera H., López I., Myers C. A., Faix D., Blair P. J., Yu C., Keene K. M., Dotson P. D., Jr., Boxrud D., Sambol A. R., Abid S. H., St George K., Bannerman T., Moore A. L., Stringer D. J., Blevins P., Demmler-Harrison G. J., Ginsberg M., Kriner P., Waterman S., Smole S., Guevara H. F., Belongia E. A., Clark P. A., Beatrice S. T., Donis R., Katz J., Finelli L., Bridges C. B., Shaw M., Jernigan D. B., Uyeki T. M., Smith D. J., Klimov A. I., Cox N. J. (2009) Science 325, 197–20119465683 [Google Scholar]
- 5.Palese P. (1977) Cell 10, 1–10 [DOI] [PubMed] [Google Scholar]
- 6.Pappas C., Aguilar P. V., Basler C. F., Solórzano A., Zeng H., Perrone L. A., Palese P., García-Sastre A., Katz J. M., Tumpey T. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3064–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W., Calvo P. A., Malide D., Gibbs J., Schubert U., Bacik I., Basta S., O'Neill R., Schickli J., Palese P., Henklein P., Bennink J. R., Yewdell J. W. (2001) Nat. Med. 7, 1306–1312 [DOI] [PubMed] [Google Scholar]
- 8.Pancuchárová H., Russ G. (2006) Acta Virol. 50, 269–272 [PubMed] [Google Scholar]
- 9.Zell R., Krumbholz A., Wutzler P. (2006) Emerg. Infect. Dis. 12, 1607–1608; author reply 1608–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zell R., Krumbholz A., Eitner A., Krieg R., Halbhuber K. J., Wutzler P. (2007) J. Gen Virol. 88, 536–546 [DOI] [PubMed] [Google Scholar]
- 11.Zamarin D., Ortigoza M. B., Palese P. (2006) J. Virol. 80, 7976–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conenello G. M., Zamarin D., Perrone L. A., Tumpey T., Palese P. (2007) PLoS Pathog. 3, 1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conenello G. M., Palese P. (2007) Cell Host Microbe 2, 207–209 [DOI] [PubMed] [Google Scholar]
- 14.McAuley J. L., Hornung F., Boyd K. L., Smith A. M., McKeon R., Bennink J., Yewdell J. W., McCullers J. A. (2007) Cell Host Microbe 2, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basler C. F., Aguilar P. V. (2008) Antiviral Res. 79, 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowy R. J. (2003) Int. Rev. Immunol. 22, 425–449 [DOI] [PubMed] [Google Scholar]
- 17.Zamarin D., García-Sastre A., Xiao X., Wang R., Palese P. (2005) PLoS Pathog. 1, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman J. R. (2007) Virol. J. 4, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazur I., Anhlan D., Mitzner D., Wixler L., Schubert U., Ludwig S. (2008) Cell Microbiol. 10, 1140–1152 [DOI] [PubMed] [Google Scholar]
- 20.Gibbs J. S., Malide D., Hornung F., Bennink J. R., Yewdell J. W. (2003) J. Virol. 77, 7214–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada H., Chounan R., Higashi Y., Kurihara N., Kido H. (2004) FEBS Lett. 578, 331–336 [DOI] [PubMed] [Google Scholar]
- 22.Chanturiya A. N., Basañez G., Schubert U., Henklein P., Yewdell J. W., Zimmerberg J. (2004) J. Virol. 78, 6304–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henklein P., Bruns K., Nimtz M., Wray V., Tessmer U., Schubert U. (2005) J. Pept. Sci. 11, 481–490 [DOI] [PubMed] [Google Scholar]
- 24.Röder R., Bruns K., Sharma A., Eissmann A., Hahn F., Studtrucker N., Fossen T., Wray V., Henklein P., Schubert U. (2008) J. Pept. Sci. 14, 954–962 [DOI] [PubMed] [Google Scholar]
- 25.Bruns K., Studtrucker N., Sharma A., Fossen T., Mitzner D., Eissmann A., Tessmer U., Röder R., Henklein P., Wray V., Schubert U. (2007) J. Biol. Chem. 282, 353–363 [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R. (2001) Arch. Virol. 146, 2275–2289 [DOI] [PubMed] [Google Scholar]
- 27.Deléage G., Geourjon C. (1993) Comput. Appl. Biosci. 9, 197–199 [DOI] [PubMed] [Google Scholar]
- 28.Naiki H., Higuchi K., Hosokawa M., Takeda T. (1989) Anal. Biochem. 177, 244–249 [DOI] [PubMed] [Google Scholar]
- 29.Fodor E., Devenish L., Engelhardt O. G., Palese P., Brownlee G. G., García-Sastre A. (1999) J. Virol. 73, 9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohn J. E., Millett I. S., Jacob J., Zagrovic B., Dillon T. M., Cingel N., Dothager R. S., Seifert S., Thiyagarajan P., Sosnick T. R., Hasan M. Z., Pande V. S., Ruczinski I., Doniach S., Plaxco K. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12491–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garnier J., Gibrat J. F., Robson B. (1996) Methods Enzymol. 266, 540–553 [DOI] [PubMed] [Google Scholar]
- 32.Engel M. F. (2009) Chem. Phys. Lipids 160, 1–10 [DOI] [PubMed] [Google Scholar]
- 33.Lashuel H. A. (2005) Sci. Aging Knowledge Environ. 2005, pe28. [DOI] [PubMed] [Google Scholar]
- 34.Sun A., Nguyen X. V., Bing G. (2002) J. Histochem. Cytochem. 50, 463–472 [DOI] [PubMed] [Google Scholar]
- 35.LeVine H., 3rd (1993) Protein Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buck M. (1998) Q. Rev. Biophys. 31, 297–355 [DOI] [PubMed] [Google Scholar]
- 37.Montserret R., Aubert-Foucher E., McLeish M. J., Hill J. M., Ficheux D., Jaquinod M., van der Rest M., Deléage G., Penin F. (1999) Biochemistry 38, 6479–6488 [DOI] [PubMed] [Google Scholar]
- 38.Kamlet M. J., Abboud J. L., Taft R. W. (1977) J. Am. Chem. Soc. 99, 6027–6037 [Google Scholar]
- 39.Chang D. K., Cheng S. F., Chien W. J. (1997) J. Virol. 71, 6593–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Najbar L. V., Craik D. J., Wade J. D., Salvatore D., McLeish M. J. (1997) Biochemistry 36, 11525–11533 [DOI] [PubMed] [Google Scholar]
- 41.Zhong L., Johnson W. C., Jr. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4462–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montserret R., McLeish M. J., Böckmann A., Geourjon C., Penin F. (2000) Biochemistry 39, 8362–8373 [DOI] [PubMed] [Google Scholar]
- 43.Dobson C. M. (2004) Semin. Cell Dev. Biol. 15, 3–16 [DOI] [PubMed] [Google Scholar]
- 44.Barrow C. J., Yasuda A., Kenny P. T., Zagorski M. G. (1992) J. Mol. Biol. 225, 1075–1093 [DOI] [PubMed] [Google Scholar]
- 45.Wetzler D. E., Castaño E. M., de Prat-Gay G. (2007) Protein Sci. 16, 744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freire E., Oddo C., Frappier L., de Prat-Gay G. (2008) Proteins 70, 450–461 [DOI] [PubMed] [Google Scholar]
- 47.Stefani M., Dobson C. M. (2003) J. Mol. Med. 81, 678–699 [DOI] [PubMed] [Google Scholar]
- 48.Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 49.Simoneau S., Rezaei H., Salès N., Kaiser-Schulz G., Lefebvre-Roque M., Vidal C., Fournier J. G., Comte J., Wopfner F., Grosclaude J., Schätzl H., Lasmézas C. I. (2007) PLoS Pathog. 3, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLean J. E., Datan E., Matassov D., Zakeri Z. F. (2009) J. Virol. 83, 8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uversky V. N. (2002) Protein Sci. 11, 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright P. E., Dyson H. J. (1999) J. Mol. Biol. 293, 321–331 [DOI] [PubMed] [Google Scholar]
- 53.Münch J., Rücker E., Ständker L., Adermann K., Goffinet C., Schindler M., Wildum S., Chinnadurai R., Rajan D., Specht A., Giménez-Gallego G., Sánchez P. C., Fowler D. M., Koulov A., Kelly J. W., Mothes W., Grivel J. C., Margolis L., Keppler O. T., Forssmann W. G., Kirchhoff F. (2007) Cell 131, 1059–1071 [DOI] [PubMed] [Google Scholar]
- 54.Wojtowicz W. M., Farzan M., Joyal J. L., Carter K., Babcock G. J., Israel D. I., Sodroski J., Mirzabekov T. (2002) J. Biol. Chem. 277, 35019–35024 [DOI] [PubMed] [Google Scholar]
- 55.Pulliam L. (2009) J. Neuroimmune Pharmacol. 4, 213–217 [DOI] [PubMed] [Google Scholar]
- 56.Jang H., Boltz D., Sturm-Ramirez K., Shepherd K. R., Jiao Y., Webster R., Smeyne R. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14063–14068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.