Abstract
Urokinase plasminogen activator receptor (u-PAR) binds urokinase plasminogen activator (u-PA) and participates in plasminogen activation in addition to modulating several cellular processes such as adhesion, proliferation, and migration. u-PAR is susceptible to proteolysis by its cognate ligand and several other proteases. To elucidate the biological significance of receptor cleavage by u-PA, we engineered and expressed a two-chain urokinase plasminogen activator (tcu-PA) cleavage-resistant u-PAR (cr-u-PAR). This mutated receptor was similar to wild-type u-PAR in binding u-PA and initiating plasminogen activation. However, cr-u-PAR exhibited accelerated internalization and resurfacing due to direct association with the endocytic receptor α2-macroglobulin receptor/low density lipoprotein receptor-related protein in the absence of the enzyme·inhibitor complex of tcu-PA and plasminogen activator inhibitor-1 (tcu-PA·PAI-1). cr-u-PAR-expressing cells had enhanced migration compared with wild-type u-PAR-expressing cells, and cr-u-PAR was less sensitive to chymotrypsin cleavage as compared with wt u-PAR. Our studies suggest that these mutations in the linker region result in a rearrangement within the cr-u-PAR structure that makes it resemble its ligand-bound form. This constitutively active variant may mimic highly glycosylated cleavage-resistant u-PAR expressed in certain highly malignant cancer-cells.
Keywords: Protein/Cell Surface, Receptors/Recycling, Receptors/Structure-Function, Receptor Recycling, Receptors, LRP, Urokinase Receptor, Internalization, Linker Region, Vitronectin
Introduction
Urokinase plasminogen activator receptor (u-PAR),2 is a cell-surface multifunctional glycosylphosphatidylinositol-anchored protein that is heavily and heterogeneously glycosylated, with a molecular mass ranging between 55 and 65 kDa (1–3). Binding of its primary physiologic ligand, urokinase plasminogen activator (u-PA), to u-PAR supports cell-surface plasminogen (Pg) activation and promotes cellular processes such as migration and proliferation (4–8). The region between domains 1 and 2 (D1 and D2) of the three-domain structure of u-PAR is sensitive to proteolysis by several proteases, including its activated ligand two-chain u-PA (tcu-PA), plasmin (Pn), and chymotrypsin (9–11). This region also contains the chemotactic epitope, 88SRSRY92, which is exposed in the presence of u-PA either by conformational change to this area or cleavage of the D1–D2 linker region (12–14). Full-length u-PAR is required for high affinity binding of its cognate ligand, u-PA (9, 15).
One role of intact u-PAR is the regulation of the Pg activation cascade by co-localizing u-PA and Pg on the plasma membrane (16, 17). Receptor-bound u-PA is inhibited by its cognate serine protease inhibitor, plasminogen activator inhibitor-1 (PAI-1). This receptor-bound complex is cleared from the cell surface by a specific endocytic receptor, α2-macroglobulin receptor/low density lipoprotein receptor-related protein (LRP). The rapid internalization provides an efficient mechanism for immediate down-regulation of the cell-surface proteolysis mediated by the u-PA·u-PAR complex. Receptor internalization via binding to tcu-PA·PAI-1 complexes and association with LRP is also thought to modulate cell migration, because intact u-PAR is recycled and redistributed on the cell surface, changing its spatial relationships with proteins in the extracellular matrix and on the cell membrane (18). This is important, because u-PA binding to u-PAR leads to a conformational change in the receptor that enhances the affinity between u-PAR and vitronectin (Vtn) (19) in the matrix. Additionally, bound tcu-PA may be down-regulating functions by cleaving off D1 of a proximal u-PAR at two sites (Arg83 and Arg89), generating the cleaved form of u-PAR (D2D3) that is unable to bind u-PA (9), internalize tcu-PA·PAI-1 complexes, or efficiently bind Vtn and other matrix constituents (11), suggesting that the ligand that strengthens the interaction between u-PAR and the matrix also has the potential to abolish such activities. Even so, the role of receptor cleavage on regulation of u-PAR-associated cellular processes remains poorly understood.
We report here the development and characterization of a u-PAR mutant resistant to proteolysis by tcu-PA, to identify the importance of regulating receptor activity, especially in roles where the functions of active and cleaved u-PAR have been difficult to distinguish. This mutant is different from the previously generated u-PAR mutant, because this tcu-PA cleavage-resistant u-PAR (cr-u-PAR) was engineered with substitutions only in the u-PA cleavage sites (20–22), while avoiding disruption of residues important for binding to other proteins such as Vtn (23–25). Cells expressing this cleavage-resistant mutant compared with the wild-type receptor exhibited identical ability to promote cell-surface Pg activation but demonstrated increased internalization of the receptor in the absence of tcu-PA·PAI-1 complexes, increased receptor recycling, as well as tcu-PA-independent heightened cell migration. We hypothesize that the faster internalization of cr-u-PAR is related to a propensity for pre-assembly with LRP. The results suggest that the cleavage-resistant u-PAR possesses certain functions associated with the conformationally active liganded u-PAR. Whether cleavage resistance of this form confers certain advantages to cells that are similar to those present in heavily glycosylated cleavage-resistant u-PAR in cancer cells remains to be studied (11, 26, 27).
EXPERIMENTAL PROCEDURES
Materials
Single-chain u-PA was a kind gift from Dr. Jack Henkin (Abbot Laboratories, Rockford, IL). Active tcu-PA was generated by incubating single-chain u-PA with Pn-Sepharose beads, as previously described (16). Pg, Pn, and chymotrypsin were purchased from EMD Bioscience (San Diego, CA). Soluble u-PAR (su-PAR) was a kind gift from Dr. Andrew Mazar (Attenuon, San Diego, CA). The monoclonal antibody against the C terminus of LRP, 11H4, and receptor-associated protein (RAP) were kind gifts from Dr. Dudley Strickland (University of Maryland, MD). The somatomedin B (SMB) domain of Vtn was a kind gift from Dr. Michael Ploug (Finsen Laboratory, Denmark). Glu-Gly-Arg chloromethyl ketone (CMK) (EMD Bioscience, San Diego, CA) was used to generate inactive tcu-PA (CMK·u-PA) (28). CMK·u-PA was iodinated using 125I (PerkinElmer Life Sciences) and IODO-GEN-coated tubes as previously described (29). Active wt PAI-1 and the stable mutant PAI-114–1B (30) were kind gifts from Dr. Daniel Lawrence (University of Michigan, Ann Arbor, MI). PAI-114–1B was biotinylated using sulfo-NHS-LC-biotin (Pierce) following the manufacturer's suggestions. Rabbit α-human u-PAR polyclonal antibody was obtained by immunizing rabbits with human su-PAR. IgG fraction was immunopurified from sera using protein A-Sepharose beads (31). Antibody selectivity was determined via Coomassie staining and immunoblotting against human su-PAR. Pre-immunized serum was used as a control for nonspecific reactivity. For immunoblotting, ECL substrate was from Pierce, goat α-rabbit HRP and streptavidin-HRP were purchased from Jackson ImmunoResearch (West Grove, PA). G418 sulfate (Gemini Bio, West Sacramento, CA), Hanks' balanced saline solution (Invitrogen, Carlsbad, CA), non-enzymatic dissociation buffer (Invitrogen), and cycloheximide (CHX) (Sigma) were of highest quality available. Detergents Triton X-100 and deoxycholate were from Sigma. Immobilized Protein G, sulfo-NHS-S-S-LC-biotin, and streptavidin-agarose beads were from Pierce.
Mutagenesis
Wild-type human u-PAR cDNA was obtained by reverse transcription-PCR of mRNA from U937 lymphoma cells (CRL-1593.2, ATCC, Manassas, VA). Variant u-PAR was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers for generating cr-u-PAR mutations, Arg83 → Ala and Arg89 → Ala, were 5′-GCA ACT CTG GCG CAG CTG TCA CCT ATT CCG CAA GCC GTT ACC-3′, and its reverse and complementary primer (IDT Inc., Coralville, IA). All cDNA sequences were verified (High-throughput Sequencing and Genotyping Unit, University of Illinois-Urbana, IL), and the cDNA were inserted into pcDNA3.1(+) (Invitrogen) via standard subcloning procedures using restriction digest.
Transfection of u-PAR Variants
The following cell lines HEK-293 (293, CRL-1573,) and U937 were purchased from ATCC. Stable transfection of pcDNA3.1-(+) carrying the wt u-PAR or cr-u-PAR genes into 293 cells was accomplished using the SuperFect transfection reagent (Qiagen, Valencia, CA).
u-PAR Detection and Cleavage Assay
1 × 106 non-transfected 293, 293 wt u-PAR, or 293 cr-u-PAR cells were dissociated using non-enzymatic dissociation buffer and resuspended in DMEM with 0.1% BSA (Calbiochem, San Diego, CA). Cells were acid-washed to remove surface-bound proteins as previously described (16). Samples were incubated with 100 nm tcu-PA for 20 h at 37 °C. Cell membrane proteins were solubilized for analysis with radioimmune precipitation assay buffer (32) with complete protease inhibitor mixture (Roche Applied Science). 5 μg of cell lysate was subjected to SDS-PAGE, and equivalent loading of proteins was achieved by quantitating samples using a BCA assay (Pierce). Samples were immunoblotted using polyclonal rabbit α-u-PAR and goat α-rabbit HRP. In some instances, purified su-PAR and endogenous u-PAR (5 × 106 3-day PMA-stimulated U937 cells prepared similarly for PAGE) (33) were used for comparison.
Cell-surface Competition Binding Assay
As previously described in Manchanda, et al. (16) CMK·u-PA was iodinated followed by a quenching step using 5 mm saturated tyrosine and 6 mm KI solution. 2.5 × 104 cells were harvested overnight in DMEM, washed, and incubated in serum-free DMEM, 0.1% BSA, 10 mm Hepes (pH 7.4), and 10 μg/ml CHX with increasing concentration of 125I-CMK·u-PA for 4 h at 4 °C in the presence or absence of 100-fold excess unlabeled CMK·u-PA. Cells were washed, lysed using 0.1 n NaOH and 1% SDS, and analyzed in a Wallac Wizard Gamma Counter (Ramsey, MN). Samples were normalized by total protein concentration.
Cell-surface u-PA·u-PAR in Pg Activation and Inhibition by PAI-1
5 × 104 cells were seeded overnight. Cells were washed with Hanks' balanced saline solution before the addition of reaction buffer (DMEM with 0.1% BSA), preincubation with CMK-tcu-PA, tcu-PA, or chymotrypsin, as shown, and incubated with 10 nm tcu-PA for 30 min at 37 °C. Real-time cell-surface generation was monitored at 37 °C on a SpectroMax Gemini XS microplate spectrofluorometer (Molecular Devices, Sunnyvale, CA) essentially as described (7), except using 56 nm Pg and 600 μm of the Pn substrate, H-d-Ala-Leu-Lys-7-amino-4-methylcoumarin (Bachem, Torrance, CA) in phosphate-buffered saline and 0.1% BSA. Excitation and emission wavelengths were 360 and 460 nm, respectively. The rates of Pn generation were determined from parabolic plots of relative fluorescence unit versus time, fitting to a second-order polynomial, and then conversion to nanomolar Pn by reference to the amidolytic activity of purified Pn using the same fluorogenic substrate (34, 35).
Biotinylated Protein Labeling and Internalization Assays
1.5 × 105 cells were harvested 18 h prior to labeling. tcu-PA·PAI-1 complex was generated by incubating for 30 min at 37 °C in 0.1 m Tris, pH 7.4, and full inhibition was determined using the chromogenic substrate, Spectrozyme UK (American Diagnostica, Stamford, CT). Cell-surface u-PAR was biotinylated using 200 μm sulfo-NHS-SS-LC-biotin as previously described (36). Cells were incubated without or with 10 nm tcu-PA·PAI-1 complex for 30 min at 4 °C and exposed to DMEM with 0.1% BSA prewarmed to 37 °C to initiate internalization. At specified times, cells were treated with 100 mm dithiothreitol for 3 min at 37 °C, to reduce cell-surface biotin label but not cytoplasmic biotin, followed by lysis in radioimmune precipitation assay buffer. Biotinylated membrane proteins were recovered from lysates via affinity-precipitation with streptavidin-agarose, followed by reduction and denaturation. 10 μl of total cell lysate were reduced to quantify the total amount of u-PAR found in the samples. Samples were subjected to SDS-PAGE and u-PAR detected using our polyclonal rabbit α-u-PAR antibody.
Alternatively, 293 cells expressing u-PAR were incubated with 10 nm of biotinylated PAI-114–1B·tcu-PA complex for 30 min at 4 °C. Cells were exposed to 37 °C to initiate internalization. At specified times, cells were acid-washed followed by lysis with radioimmune precipitation assay buffer. All samples were subjected to immunoblotting using streptavidin-HRP. Blots were analyzed by densitometry using the Kodak 1-D system. Samples were normalized to background. Control samples were prepared as described above and incubated with biotinylated PAI-114–1B·tcu-PA complex for 30 min at 37 °C except that 500 nm RAP was added to each incubation solution.
Receptor Recycling Assay
5 × 104 u-PAR-expressing cells were seeded overnight. Cells were incubated with tcu-PA·PAI-1 complex for 30 min at 4 °C. Unbound complex was removed, and the cells were incubated at 37 °C. At specified times, 10 nm tcu-PA was added and the amount of cell-surface u-PAR was measured via the Pg activation assay as described above. Cell surface-associated tcu-PA activity was normalized to the maximal amount of tcu-PA activity in the absence of tcu-PA·PAI-1 complexes. In sets of control samples, 500 nm RAP was added to all incubation solutions for the allotted times.
Co-immunoprecipitation of u-PAR and LRP
Immunoprecipitation experiments were performed as previously described (37). Suspensions of 1 × 106 cells (293 wt u-PAR or cr-u-PAR), were incubated in DMEM with 0.1% BSA only or with additional 10 nm tcu-PA·PAI-1 complex for 30 min at 4 °C. Parallel samples of cells were exposed to buffer or incubated with either 500 nm RAP or 4 μm SMB. LRP-containing complexes were immunoprecipitated from cell lysates using the α-LRP monoclonal antibody, 11H4, and protein G-conjugated agarose beads as described by Czekay et al. (37), and co-precipitated u-PAR was analyzed via immunoblotting as described above. 50 μl of total cell lysate were analyzed similarly to quantify the total amount of u-PAR found in the lysates. Blots were analyzed as described.
Chymotrypsin Cleavage Assay
Suspended 293 wt u-PAR and cr-u-PAR cells in DMEM, 0.1% BSA, and 100 μg/ml CHX were incubated with in the absence or presence of 100 nm CMK·u-PA for 30 min at 37 °C, followed by 100 nm chymotrypsin at 37 °C for the times shown (13). Cells were then washed and lysed using radioimmune precipitation assay buffer. 10 μg of total cellular protein lysate was analyzed via immunoblotting for u-PAR as described.
Migration Assay
293 cells were resuspended as described above. 2 × 105 cells were incubated in the presence or absence of 0.5 nm tcu-PA in serum-free DMEM for 15 min at 37 °C and then placed on the top portion of Transwell chambers (Corning, Corning, NY) coated previously with 10 μg/ml purified Vtn (5). DMEM with 10% fetal calf serum was added to the bottom chamber, and the cells were incubated for 2 h at 37 °C. Migration was quantified using crystal violet staining as described previously (38). The absorption of the eluted stain was read at 506 nm on a SpectroMax Plus 384 microplate spectrophotometer.
RESULTS
Generation of u-PAR Resistant to Cleavage by tcu-PA
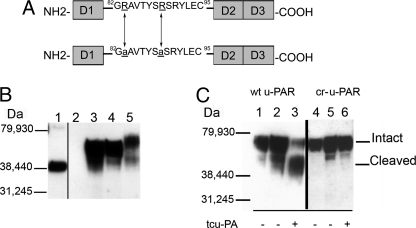
To study the effects of receptor cleavage, we made mutations in the linker region of u-PAR (12, 14, 39, 40) at two sites, Arg83 and Arg89 (Fig. 1A), which confer tcu-PA cleavage resistance. The arginine in position 91 was left unchanged, because this residue is essential for Vtn binding (23, 24). In Fig. 1B, expression of wt u-PAR and cr-u-PAR were detected by immunoblotting. However, non-transfected 293 cells lack a band for any u-PAR form as described by other investigators (41). In samples from cells expressing wt u-PAR and cr-u-PAR, there is an appearance of a band at ∼55 kDa that can be attributed to the intact receptor. The approximate 40-kDa band in these two u-PAR variants might be an underglycosylated u-PAR or partially degraded u-PAR. Cell lysates from PMA-stimulated U937 cells were positive controls (Fig. 1B). PMA stimulation increased u-PAR expression and increased glycosylation, leading to a higher molecular weight than 293-associated u-PAR (42). The expression of u-PAR in the 293 cells was confirmed by ligand-binding assays, wherein wt u-PAR was found to be expressed at a wide range of 0.7–2.2 × 106 receptors per cell among different clones, whereas cr-u-PAR was expressed at 1.5–5.5 × 106 receptors per cell among clones (Table 1).
FIGURE 1.
Domain structure, expression, and cleavage products of u-PAR. A, a diagram of wt u-PAR (top) shows the three domains designated D1–3 and a linker region. D3 contains the glycosylphosphatidylinositol anchor. The linker region contains the chemotactic epitope (59). A schematic of cr-u-PAR (bottom) shows the two mutated sites as underlined and in lowercase. B, immunoblotting using polyclonal rabbit α-u-PAR was used to detect total u-PAR. Samples were 125 ng of purified su-PAR as a positive control (lane 1) and lysates from non-transfected 293 cells (lane 2), 293 wt u-PAR cells (lane 3), 293 cr-u-PAR cells (lane 4), and 3-day PMA-stimulated U937 cells (lane 5). C, u-PAR cleavage products post-exposure to tcu-PA. Cells expressing wt u-PAR (lanes 1–3) or cr-u-PAR (lanes 4–6) were incubated in the absence of tcu-PA for 0 h (lanes 1 and 4) or 20 h (lanes 2 and 5) or in the presence of 100 nm tcu-PA for 20 h (lanes 3 and 6) followed by lysis, SDS-PAGE, and immunoblotting for total u-PAR.
TABLE 1.
Specific binding of CMK-u-PA to 293 cells
Cells were incubated with doses of radiolabeled CMK·u-PA in the presence of excess unlabeled CMK·u-PA, and cell-associated radiolabeled CMK·u-PA was measured as described under “Experimental Procedures.” Shown are the mean ± S.E. of three independent experiments.
| 293 wt u-PAR |
293 cr-u-PAR |
|||||
|---|---|---|---|---|---|---|
| Clone 2 | Clone 11 | Clone 13 | Clone 4 | Clone 7 | Clone 8 | |
| KD (nm) | 1.3 ± 0.6 | 1.1 ± 0.4 | 1.1 ± 0.4 | 1.4 ± 0.4 | 1.5 ± 0.3 | 1.5 ± 0.2 |
| Sites/cell (106) | 2.0 ± 0.2 | 2.2 ± 0.6 | 0.7 ± 0.1 | 1.5 ± 0.1 | 3.8 ± 0.3 | 5.5 ± 0.2 |
Receptor cleavability was tested by the prolonged incubation of u-PAR-expressing 293 cells with a high concentration of tcu-PA (Fig. 1C). Exposure to tcu-PA converted 70% of wt u-PAR into the cleaved form. In contrast, when treated similarly cr-u-PAR cells show a single predominant band of intact u-PAR. The overnight incubation alone did not lead to robust u-PAR cleavage in either cell type. These results suggest that substitution of Arg83 and Arg89 led to a u-PAR mutant unable to be proteolyzed by tcu-PA.
Dissociation constants for the binding of radiolabeled CMK·u-PA to three independent clones of cells expressing either wt u-PAR or cr-u-PAR ranged between 1.1 and 1.5 nm (Table 1), in close agreement with previous reports (43–46), suggesting that the expressed receptors were folded correctly and were adequately post-translationally modified. Non-transfected 293 cells showed no specific binding of CMK·u-PA in the ligand concentration range wherein u-PAR-expressing cells displayed saturable binding (data not shown). Based on the similar levels of receptor expression and affinities for u-PA, 293 wt u-PAR clone 2 and 293 cr-u-PAR clone 4 were chosen for further studies.
Effects of u-PAR Variants on Cell-surface Pg Activation
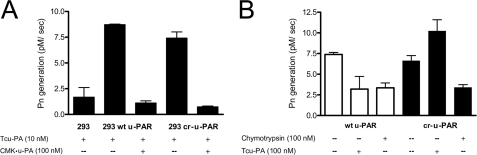
Recombinant wt u-PAR and cr-u-PAR were able to support the initiation of Pg activation in a specific manner (Fig. 2A). Fig. 2A shows that, although wt u-PAR and cr-u-PAR receptor-expressing cells bound tcu-PA and activated Pg, CMK·u-PA pretreatment prevented this binding and subsequent Pn generation in both cell types.
FIGURE 2.
Cr-u-PAR supports u-PA-dependent Pg activation that cannot be down-regulated by receptor cleavage. A, specific binding of tcu-PA to u-PAR leads to Pn generation that can be blocked by preincubation with excess CMK·u-PA. Cells were first incubated with or without 100 nm of CMK·u-PA for 30 min at 37 °C followed by 10 nm of tcu-PA. Cell-surface u-PA activity was measured via a Pg activation assay as described under “Experimental Procedures.” B, proteolysis of u-PAR results in a decrease in cell-surface Pn generation. Cells expressing wt u-PAR (white bars) or cr-u-PAR (black bars) were subjected to limited proteolysis with 10 nm chymotrypsin or 100 nm tcu-PA, and residual intact u-PAR was detected by incubation with 10 nm of tcu-PA followed by a Pg activation assay. Data represent the mean and S.D. of three individual replicates.
u-PAR cleavage should decrease the amount of available intact wild-type receptors that may bind tcu-PA, slowing Pg activation. To test this hypothesis, cells expressing wt u-PAR or cr-u-PAR were treated with tcu-PA or chymotrypsin to remove D1, followed by u-PA binding and assaying subsequent Pg activation as described. Fig. 2B shows that incubation of wt u-PAR cells with either excess tcu-PA or chymotrypsin resulted in a 55% decrease in Pn generation compared with non-treated cells, in agreement with previous findings (9, 47). In contrast, the addition of tcu-PA to cr-u-PAR cells led to a 38% increase in Pg activation over non-treated cells, which we attribute to the lack of receptor cleavage coupled with higher saturation of available uncleaved cr-u-PAR sites by the molar excess of tcu-PA. As with wt u-PAR cells, exposure of cr-u-PAR cells to chymotrypsin resulted in a similar 55% decrease in Pn generation compared with non-treated cells, demonstrating that the chymotrypsin cleavage site remains intact. PAI-1 was able to inactivate tcu-PA bound to either u-PAR variant (data not shown). These data suggest that initiation and inhibition of Pg activation occur similarly on cells bearing wt u-PAR or cr-u-PAR.
Cells Expressing cr-u-PAR Internalize tcu-PA·PAI-1 Complexes More Rapidly than Those Cells Expressing wt u-PAR
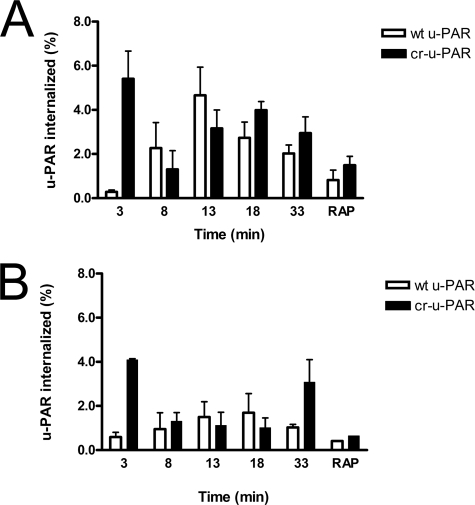
We examined the effect of u-PAR mutations on the ability of cells to rapidly endocytose tcu-PA·PAI-1 complexes. Upon pulse-labeling a pool of cell-surface receptors with biotin and addition of tcu-PA·PAI-1 complex, we detected u-PAR in the cytoplasmic fraction. Consistent with previous reports, cytoplasmic wt u-PAR increases in a gradual time-dependent manner to a maximum internalized amount in 13 min followed by a gradual decline (Fig. 3A) (48). In contrast, cr-u-PAR bound to tcu-PA·PAI-1 complex was internalized with an initial peak that was detected at the first studied time point, followed by a secondary peak that resembles the endocytic profile of wt u-PAR, thus showing a shift in time frame. RAP binding to LRP inhibits endocytosis mediated by this cell-surface receptor (49). Fig. 3A depicts RAP inhibiting LRP-mediated endocytosis of u-PAR·tcu-PA·PAI-1 complexes to 10% of peak values seen in the absence of RAP. These data suggest that, although a similar process may mediate the internalization of both types of receptors, cr-u-PAR undergoes this process at a much faster rate compared with wt-u-PAR, this process may also regulate the fraction of cell-surface receptor internalized and is likely limited by the LRP expression levels (50–52).
FIGURE 3.
cr-u-PAR is internalized rapidly compared with wt u-PAR. Cells expressing wt u-PAR (white bars) and cr-u-PAR (black bars) were briefly surface-labeled with reducible biotin and incubated with 10 nm tcu-PA·PAI-1 complex (A) or buffer (B) at 4 °C. Internalization was initiated by exposure to 37 °C and terminated via cell-surface reduction with dithiothreitol and cell lysis, with time points representing the duration of incubation at 37 °C plus the dithiothreitol exposure. 500 nm RAP was added to the control reaction with every new incubation. Internalized non-reduced u-PAR was isolated and detected as described. Total u-PAR was detected by subjecting 20 μl of total cell lysate to SDS-PAGE and immunoblotting. Data represent the amount of non-reduced u-PAR (internalized) as a percentage of total u-PAR in each sample, and shown are the means ± S.E. of at least three independent replicates.
We evaluated the internalization of wt u-PAR and cr-u-PAR in the absence of tcu-PA·PAI-1 complexes to identify if the initial burst of cr-u-PAR endocytosis was due to a mutant-specific response toward tcu-PA·PAI-1 exposure or due to a new intrinsic property of unliganded cr-u-PAR (Fig. 3B). cr-u-PAR was rapidly internalized at 3 min in a manner similar to that seen in the presence of tcu-PA·PAI-1 complex (Fig. 3A). A secondary internalization peak was also present at 33 min similar to Fig. 3A, consistent with a basal endocytosis cycle of about 30 min. Although wt u-PAR internalization remained under 2% of total labeled receptor, internalization of cr-u-PAR reached twice as high (Fig. 3B). These data suggest that the initial rapid cr-u-PAR internalization event can occur independently of the binding of tcu-PA·PAI-1 complexes.
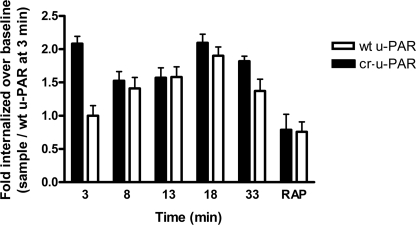
To determine if biotinylation of u-PAR affected its internalization profile, we performed a similar experiment utilizing biotinylated-PAI-114–1B in complex with tcu-PA. Fig. 4 shows that labeled tcu-PA·PAI-114–1B complex exhibited a similar endocytosis profile as the biotinylated u-PAR. Importantly, we found an initial burst of complex internalization similar to the burst of labeled cr-u-PAR endocytosis in Fig. 3. Additionally, lower molecular weight biotin-positive species appeared as the amount of cytoplasmic biotinylated tcu-PA·PAI-1 complex diminished over time, consistent with lysosomal degradation of the endocytosed complex (data not shown). RAP inhibited tcu-PA·PAI-1 complex internalization, indicating involvement of LRP in this endocytic process.
FIGURE 4.
cr-u-PAR promotes biotinylated-PAI-114–1B·tcu-PA complex internalization. 293 wt u-PAR (white bars) and cr-u-PAR (black bars) cells were incubated with 10 nm biotinylated-PAI-114–1B·tcu-PA complex at 4 °C, and internalization was initiated at 37 °C and terminated as described earlier. Cells were acid-washed to remove non-internalized cell-surface-bound biotinlyated-PAI-I14–1B·tcu-PA complex. Time points represent the duration of the 37 °C incubation plus the acid wash. Internalized biotinylated complex was detected as described with streptavidin-HRP, and the amount of complex internalized by wt u-PAR cells at 3 min was set as baseline. As a control, RAP (500 nm) was incubated with tcu-PA·PAI-1 with continuous addition of RAP following incubation for 30 min at 37 °C. Data represent the mean ± S.E. of minimum three independent replicates.
Cr-u-PAR Recycling Is Increased following Internalization of u-PAR·tcu-PA·PAI-1 Complexes
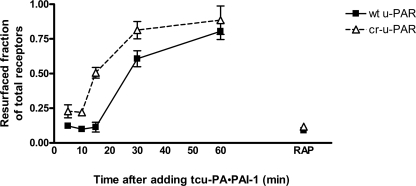
u-PAR and LRP internalization by cells is subsequently followed by recycling of these receptors to the cell surface, spatially redistributing the receptors (53). Having observed an initial rapid internalization of cr-u-PAR, we sought to determine if the endocytosis of tcu-PA·PAI-1-bound cr-u-PAR is followed by recycling/resurfacing of unliganded receptor (Fig. 5). Receptor recycling was studied by saturating cell-surface u-PAR with tcu-PA·PAI-1 complexes, inducing internalization as described, and detecting resurfaced unoccupied u-PAR via binding of active tcu-PA in a Pg activation assay. Fig. 5 shows that cells expressing cr-u-PAR recycle the receptor faster than their wild-type receptor-expressing counterparts. Approximately one-half of internalized cr-u-PAR had reappeared on the cell surface by 15 min after the induction of endocytosis, whereas internalized wt u-PAR required nearly twice the time (30 min) to reach a similar level of resurfacing. For the entire course of the experiments, synthesis of new receptor was prevented by the addition of CHX. Exposure of cells to RAP before addition of tcu-PA·PAI-1 complexes resulted in marked inhibition of receptor resurfacing, suggesting that LRP-mediated endocytosis is required for the appearance of unoccupied receptor on the cell surface and that unoccupied receptor does not appear via simple dissociation of the initially formed u-PAR·tcu-PA·PAI-1 complex.
FIGURE 5.
Cells expressing cr-u-PAR resurface recently unoccupied receptor faster than wt u-PAR-expressing cells. wt u-PAR (■) and cr-u-PAR (Δ)-expressing cells were incubated with pre-assembled tcu-PA·PAI-1 complexes at 4 °C. Cells were washed and incubated at 37 °C for the times shown, and the amount of resurfaced unoccupied receptor was assessed via a Pg activation assay. Pg activating activities at each time point are shown relative to samples of cells not incubated with tcu-PA·PAI-1 complex, leaving a full complement of unoccupied u-PAR. As a control, 500 nm RAP was added to each incubation step. Data represent the means ± S.E. of four independent replicates.
cr-u-PAR Binds LRP in the Absence of tcu-PA·PAI-1 Complex
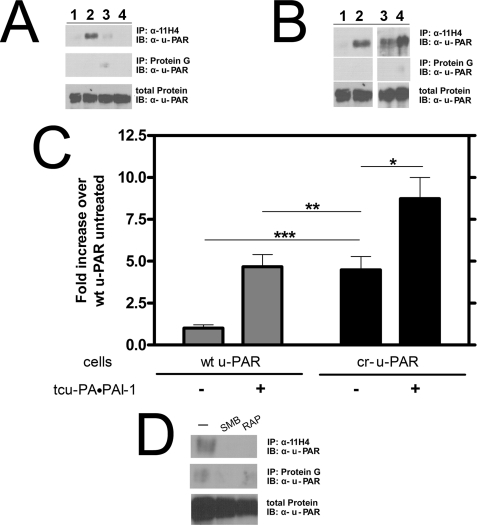
LRP associates with u-PAR-bound tcu-PA·PAI-1 complexes (54). This interaction is primarily between the tcu-PA·PAI-1 complex and LRP, although a direct u-PAR-LRP interaction may play a minor role (37). Because cr-u-PAR has an altered LRP-mediated internalization profile, which occurs even in the absence of tcu-PA·PAI-1 complex, we determined whether cr-u-PAR and LRP are closely associated by detecting u-PAR that was co-immunoprecipitated with the monoclonal anti-LRP antibody, 11H4. In Fig. 6 (A and B), a trace amount wt u-PAR was detected using 11H4 in the absence of tcu-PA·PAI-1 complex. In the presence of complex, the amount of immunoprecipitated wt u-PAR was enhanced, an effect that was blocked by RAP (Fig. 6A). In contrast, cr-u-PAR alone co-immunoprecipitated with LRP, an association that was enhanced by the addition of tcu-PA·PAI-1 complex (Fig. 6, B–D). The amount of cr-u-PAR precipitated with 11H4 in the absence of tcu-PA·PAI-1 complex was similar to the amount of wt u-PAR detected in the presence of complex.
FIGURE 6.
cr-u-PAR co-immunoprecipitates with LRP in the absence of tcu-PA·PAI-1 complexes. A, cells expressing wt u-PAR were incubated with buffer (lane 1), 10 nm tcu-PA·PAI-1 (lane 2), 500 nm RAP (lane 3), or 500 nm RAP, and 10 nm tcu-PA·PAI-1 complex (lane 4). u-PAR was affinity-precipitated with the monoclonal α-LRP antibody, 11H4, and detected using polyclonal α-u-PAR antibody. Nonspecific binding of u-PAR to protein G beads was minimal. B, cells expressing wt u-PAR (lanes 1 and 2) or cr-u-PAR (lanes 3 and 4) were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 10 nm tcu-PA·PAI-1 complex and u-PAR affinity-precipitated as described under “Experimental Procedures.” The band intensities are shown (C), with data representing the mean ± S.D. of three individual replicates (t test; *, p < 0.03; **, not significant; and ***, p < 0.0005). D, cr-u-PAR-expressing cells were treated with buffer only (lane 1), 4 μm SMB (lane 2), or 500 nm RAP (lane 3) for 30 min before lysis and immunoprecipitation with α-LRP antibody followed by immunoblotting with α-u-PAR antibody.
To disrupt the close association of unliganded cr-u-PAR to LRP, we selected molecules that bind specifically to u-PAR or to LRP. The SMB domain of Vtn directly interacts with u-PAR in the linker region N-terminal to our mutated sites, whereas RAP prevents binding of several ligands to LRP. Fig. 6D shows that both SMB and RAP inhibited the affinity precipitation of cr-u-PAR by 11H4. Disruption of the cr-u-PAR-LRP interaction suggests that the receptors can directly interact or may be bridged by other unidentified molecules that specifically bind both receptors.
Chymotrypsin Cleaves cr-u-PAR at a Different Rate than wt u-PAR
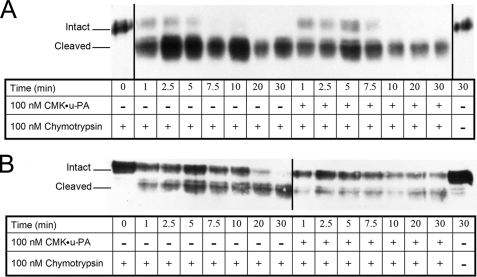
u-PA bound to u-PAR exposes the chemotactic epitope situated between residues 88 and 92 in the linker region (12, 14, 39). X-ray crystallographic evidence suggests this u-PA-dependent conformational change in the receptor involves the rotation of Tyr87 away from the bulk solvent, thus decreasing its availability to serve as the P1 residue for the chymotrypsin active site (14, 40). To investigate if the linker region of cr-u-PAR is conformationally altered compared with that of wt u-PAR, cells expressing wt u-PAR and cr-u-PAR were exposed to chymotrypsin in the presence or absence of CMK·u-PA (Fig. 7). wt u-PAR alone was rapidly cleaved by chymotrypsin, with nearly all of the receptor converted to the D2D3 form by the first time point (1 min) and no detectable intact receptor remaining after ∼5 min. The presence of CMK·u-PA slightly reduced the cleavage of wt u-PAR. In contrast, cr-u-PAR cleavage by chymotrypsin was delayed, with intact receptor detectable until ∼20 min into the reaction. CMK·u-PA further delayed the cleavage of cr-u-PAR by chymotrypsin, with roughly half of the intact receptor still being detected by the final time point (30 min). Thus the addition of CMK·u-PA to cr-u-PAR-expressing cells reduced chymotrypsin-associated receptor cleavage to a much greater extent than seen in similarly treated wt u-PAR-expressing cells. From our previous assays, we know that u-PA can bind with high affinity to both u-PARs (Table 1), indicating that the differential protection of CMK·u-PA between wt u-PAR and cr-u-PAR against chymotrypsin is not due to dissimilar u-PA-binding. These data indicate that there is a significant conformational difference between the linker regions of wt u-PAR and cr-u-PAR that may correlate with different degrees of receptor behavior, such as receptor internalization and LRP-association.
FIGURE 7.
Chymotrypsin cleaves cr-u-PAR less efficiently compared with wt u-PAR. Suspension of cells expressing wt u-PAR (A) or cr-u-PAR (B) were incubated with buffer or 100 nm CMK·u-PA as shown at 37 °C, followed by additional incubation with 100 nm chymotrypsin at 37 °C for the times shown. 10 μg of total cell lysate was analyzed by immunoblotting using polyclonal rabbit α-u-PAR. Blots were imaged using the Kodak 1D system. Shown are images representative of four independent replicates.
cr-u-PAR Expression Enhances Migration in 293 Cells
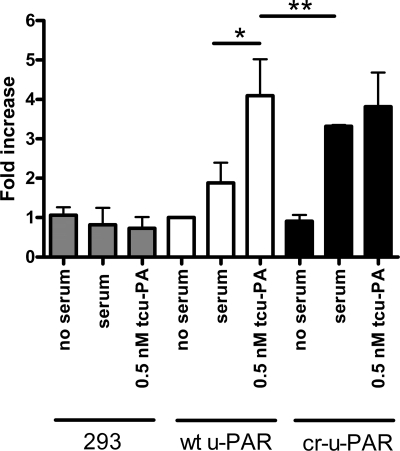
The altered endocytosis profile, ligand-free close association with LRP, and resistance to chymotrypsin-associated cleavage exhibited by cr-u-PAR indicated that this form may represent a constitutively active receptor. u-PAR-expressing cells can undergo u-PA-dependent migration, and importantly, this has been previously shown in 293 cells expressing recombinant u-PAR (55). We thus tested whether cells expressing cr-u-PAR behaved similarly as cells expressing wt u-PAR in the presence of such an activating ligand in a well described u-PA-dependent process, cellular migration. Fig. 8 depicts a lack of migration for non-transfected 293 cells on a Vtn-coated matrix when exposed to serum or to u-PA. Serum-stimulated wt u-PAR-expressing cells demonstrated a 2.7-fold increase in migration over serum-free treatment. This effect was further enhanced in the presence of 0.5 nm tcu-PA leading to a 4.5-fold increase in migration. However, cr-u-PAR-expressing cells showed enhanced migration in the presence of serum that was similar to u-PA-stimulated migration (Fig. 8). These effects were not due to changes in random chemotaxis as cells expressing either u-PAR variant exhibited little directed migration in the absence of serum or with serum in both top and bottom chambers (data not shown). Thus, although tcu-PA is able to enhance wt u-PAR-associated migration, this additional effect was not seen in cr-u-PAR-expressing cells, which already migrate strongly in the absence of ligand. These data further support our hypothesis that cr-u-PAR is a receptor that may be in an activated state.
FIGURE 8.
293 cr-u-PAR-expressing cells promote u-PA-independent migration. Non-transfected 293 (gray bars)-, 293 wt u-PAR (white bars)-, and 293 cr-u-PAR (black bars)-expressing cells were serum-starved and suspended in serum-free DMEM. Cells were either pre-treated with serum-free DMEM and allowed to migrate over Vtn-coated chambers with no serum gradient (no serum), pre-treated with serum-free DMEM and allowed to migrate toward 10% fetal calf serum (serum), or pre-treated with 0.5 nm tcu-PA and allowed to migrate toward 10% fetal calf serum (0.5 nm tcu-PA) for 2 h at 37 °C. Samples were analyzed as described under “Experimental Procedures.” Data represent the means ± S.D. of at least three individual replicates and were analyzed using a Student's t test (*, p < 0.0001 and **, not significant).
DISCUSSION
Models regarding the functions of u-PAR are complex and involve roles for the unoccupied receptor, u-PA-bound receptor, and tcu-PA-cleaved receptor. Functions of u-PAR in any one of these states may differ depending on the other receptors expressed on the same cell, the composition of the cellular environment, and the type of matrix in contact with the cell. Further confounding matters is the fact that the primary physiologic ligand of u-PAR, u-PA, is also a potent source of cleaved receptor (10, 56). In a simplified model, u-PA binding is proposed to enhance the association of u-PAR with Vtn (47) and integrins (19), whereas tcu-PA-dependent release of D1 abolishes affinity for these two proteins and promotes binding to and signaling via G-protein-coupled receptors such as formyl peptide receptor like-1 (FPRL-1) (57). Signaling pathways affected by these different interactions lead to changes in cellular migration, proliferation, and adhesion, via what may be simultaneous down-regulation of some pathways, such as those involving integrins, and up-regulation of others, such as those involving G-protein-coupled receptors (58). However, in a mixed population of receptors, the contributions of each state of u-PAR are difficult to distinguish and have not yet been delineated.
In a previous attempt to address this problem, a cleavage-resistant u-PAR variant, hcr-u-PAR, was created, in which several mutations were made to the cleavage sites in the linker region for u-PA, plasmin, matrix metalloproteinases, and chymotrypsin (20–22). Although cells expressing wt u-PAR were shown to exhibit u-PA-dependent ERK activation, those expressing hcr-u-PAR did not, suggesting that cleaved and intact u-PAR may access alternate signaling pathways (20). Additionally, disruption of fibroblast to myofibroblast differentiation by hcr-u-PAR led to the conclusion that u-PAR cleavage may play an important role in cell differentiation (22). However, because two of five residues in the chemotactic epitope (Arg89 and Arg91) were altered in hcr-u-PAR, including a residue essential to Vtn binding (Arg91), it is unclear whether the functional differences observed between intact wt u-PAR and hcr-u-PAR are solely related to cleavability (23–25).
In this study, we engineered a u-PAR mutant, cr-u-PAR, which is resistant to cleavage by u-PA but alters only one residue, Arg89, in the chemotactic epitope and preserves the ability of the cells expressing the receptor to interact with a Vtn-rich matrix to promote migration. Through a systematic characterization of cr-u-PAR, we found that the mutant receptor bound to u-PA and promoted Pg activation indistinguishably from wt u-PAR, implying that cr-u-PAR is properly folded and oriented on the cell surface to allow u-PA and cell-surface bound Pg to interact. We also found evidence of a close association between cr-u-PAR and LRP, a result that was unanticipated given that the current paradigm holds that tight interaction of these two receptors requires the bridging effect of the tcu-PA·PAI-1 complex. Nevertheless, this hypothesis is strongly supported by the findings that unliganded cr-u-PAR is constitutively endocytosed in an LRP-dependent manner, that cr-u-PAR and LRP can be affinity co-precipitated from cell lysate, and that this co-precipitation can be specifically blocked by RAP and SMB. These data may be explained by direct binding between cr-u-PAR and LRP, by the two molecules indirectly co-associating within a larger cell-surface complex, or by the two receptors being co-localized in the same plasma membrane micro-domains. Although we cannot yet discriminate among these possibilities, the results suggest an intriguing implication, that cr-u-PAR may represent a receptor variant that is constitutively active in the absence of ligand.
Several lines of evidence suggest that cr-u-PAR not only exhibits cleavage resistance, but also displays properties of an activated receptor. In several functional aspects, free cr-u-PAR behaved like liganded wt u-PAR. The peak amount of wt u-PAR endocytosed in the presence of tcu-PA·PAI-1 complex is similar to the peak amount of cr-u-PAR internalized in the absence of complex. Similarly, the amount of cr-u-PAR that was affinity-precipitated with an anti-LRP antibody in the absence of tcu-PA·PAI-1 complex was identical to the amount of wt u-PAR captured in the presence of complex. Furthermore, cells expressing cr-u-PAR when stimulated with only serum migrated as much as those expressing wt u-PAR stimulated by serum and u-PA. It should be noted that, although there may be bovine-derived u-PA from the serum included in some of these experiments, the binding of u-PA to human u-PAR is species-specific. In particular, regions known to mediate this specificity do not involve the residues mutated in cr-u-PAR, and a residue in human u-PA, Trp30, critical to this specificity is not conserved in bovine u-PA (12). Therefore, it is highly probable that, without the addition of exogenous human u-PA, cr-u-PAR is unoccupied yet retains some important functions of the u-PA-bound wt u-PAR. Additionally, the behavior of cr-u-PAR as an active receptor is evident in other cellular events as well, including proliferation and adhesion.3 The mechanism of this activation may involve local changes to the conformation of the linker region and chemotactic epitope, as evidenced by the differential susceptibilities of wt u-PAR and cr-u-PAR to partial proteolysis in this area by chymotrypsin. This structural change may in turn affect the inter-domain relationships in the receptor, subtly altering its global conformation as well. Indeed, crystal structures of u-PAR in complex with various ligands, including an antagonist peptide, the amino-terminal fragment of u-PA, and SMB, demonstrate just such differences in inter-domain distances and inter-domain loop conformations (12, 14, 39, 40). Although more biophysical means of characterization are needed to ascertain whether unliganded cr-u-PAR is conformationally similar to u-PA-bound wt u-PAR, it is clear that cr-u-PAR is a functionally activated receptor in the absence of bound ligand. Other changes in receptor function and their effects on cell behavior remain to be studied.
The precise role of u-PAR cleavage in cancer biology remains elusive. Soluble and cell-surface-cleaved u-PAR appear to be promising markers in the early detection, prognosis, and response to treatment of cancers of the prostate, ovary, breast, and bone marrow (25). Because cleaved u-PAR can induce u-PA-independent cell migration via G-protein-coupled receptors (26), receptor cleavage may promote metastasis. However, in a highly invasive anaplastic thyroid carcinoma, u-PAR glycosylation prevents cleavage (27). Less aggressive forms of thyroid carcinoma as well as thyroid adenoma that do not possess great invasive potential express cleavable receptor, suggesting that, in thyroid cancers, u-PAR cleavage may down-regulate invasiveness (27). Thus cr-u-PAR is potentially an important tool in the dissection of the roles of u-PAR in cancer, and further investigation into its constitutive activity may shed light on the ligand-induced conformational activation of the intact wild-type receptor.
Acknowledgments
Many thanks to Dr. Bradford Schwartz, Dr. Daniel Lawrence, and Dr. Dudley Strickland for donating proteins used in this report. We thank Lucienne Burrus for excellent technical assistance. Also, many thanks to Dr. Shih-Hon Li for his input and editing.
E. C. Nieves and N. Manchanda, manuscript in preparation.
- u-PAR
- urokinase plasminogen activator receptor
- u-PA
- urokinase plasminogen activator
- Pg
- plasminogen
- tcu-PA
- two-chain u-PA
- Pn
- plasmin
- PAI-1
- plasminogen activator inhibitor-1
- LRP
- α2-macroglobulin receptor/low density lipoprotein receptor-related protein
- Vtn
- vitronectin
- cr-u-PAR
- tcu-PA cleavage-resistant u-PAR
- su-PAR
- soluble u-PAR
- CMK
- chloromethyl ketone
- CMK·u-PA
- inactive CMK-treated u-PA
- CHX
- cycloheximide
- RAP
- receptor-associated protein
- SMB
- somatomedin B domain of vitronectin
- wt
- wild type
- HRP
- horseradish peroxidase
- DMEM
- Dulbecco's modified Eagle's medium
- BSA
- bovine serum albumin
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1.Ploug M., Rahbek-Nielsen H., Nielsen P. F., Roepstorff P., Dano K. (1998) J. Biol. Chem. 273, 13933–13943 [DOI] [PubMed] [Google Scholar]
- 2.Blasi F., Stoppelli M. P., Cubellis M. V. (1986) J. Cell. Biochem. 32, 179–186 [DOI] [PubMed] [Google Scholar]
- 3.Møller L. B., Pöllänen J., Rønne E., Pedersen N., Blasi F. (1993) J. Biol. Chem. 268, 11152–11159 [PubMed] [Google Scholar]
- 4.Chapman H. A., Wei Y., Simon D. I., Waltz D. A. (1999) Thromb. Haemost. 82, 291–297 [PubMed] [Google Scholar]
- 5.Nguyen D. H., Hussaini I. M., Gonias S. L. (1998) J. Biol. Chem. 273, 8502–8507 [DOI] [PubMed] [Google Scholar]
- 6.Blasi F., Behrendt N., Cubellis M. V., Ellis V., Lund L. R., Masucci M. T., Møller L. B., Olson D. P., Pedersen N., Ploug M., et al. (1990) Cell Differ. Dev. 32, 247–253 [DOI] [PubMed] [Google Scholar]
- 7.Ellis V., Behrendt N., Danø K. (1991) J. Biol. Chem. 266, 12752–12758 [PubMed] [Google Scholar]
- 8.Ossowski L., Aguirre-Ghiso J. A. (2000) Curr. Opin. Cell Biol. 12, 613–620 [DOI] [PubMed] [Google Scholar]
- 9.Høyer-Hansen G., Ronne E., Solberg H., Behrendt N., Ploug M., Lund L. R., Ellis V., Danø K. (1992) J. Biol. Chem. 267, 18224–18229 [PubMed] [Google Scholar]
- 10.Høyer-Hansen G., Ploug M., Behrendt N., Rønne E., Danø K. (1997) Eur. J. Biochem. 243, 21–26 [DOI] [PubMed] [Google Scholar]
- 11.Montuori N., Rossi G., Ragno P. (1999) FEBS Lett. 460, 32–36 [DOI] [PubMed] [Google Scholar]
- 12.Huai Q., Mazar A. P., Kuo A., Parry G. C., Shaw D. E., Callahan J., Li Y., Yuan C., Bian C., Chen L., Furie B., Furie B. C., Cines D. B., Huang M. (2006) Science 311, 656–659 [DOI] [PubMed] [Google Scholar]
- 13.Fazioli F., Resnati M., Sidenius N., Higashimoto Y., Appella E., Blasi F. (1997) EMBO J. 16, 7279–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huai Q., Zhou A., Lin L., Mazar A. P., Parry G. C., Callahan J., Shaw D. E., Furie B., Furie B. C., Huang M. (2008) Nat. Struct. Mol. Biol. 15, 422–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behrendt N., Ronne E., Dano K. (1996) J. Biol. Chem. 271, 22885–22894 [DOI] [PubMed] [Google Scholar]
- 16.Manchanda N., Schwartz B. S. (1991) J. Biol. Chem. 266, 14580–14584 [PubMed] [Google Scholar]
- 17.Ellis V., Whawell S. A., Werner F., Deadman J. J. (1999) Biochemistry 38, 651–659 [DOI] [PubMed] [Google Scholar]
- 18.Cao C., Lawrence D. A., Li Y., Von Arnim C. A., Herz J., Su E. J., Makarova A., Hyman B. T., Strickland D. K., Zhang L. (2006) EMBO J. 25, 1860–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman H. A., Wei Y. (2001) Thromb. Haemost. 86, 124–129 [PubMed] [Google Scholar]
- 20.Mazzieri R., D'Alessio S., Kenmoe R. K., Ossowski L., Blasi F. (2006) Mol. Biol. Cell 17, 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D., Aguirre Ghiso J., Estrada Y., Ossowski L. (2002) Cancer Cell 1, 445–457 [DOI] [PubMed] [Google Scholar]
- 22.Bernstein A. M., Twining S. S., Warejcka D. J., Tall E., Masur S. K. (2007) Mol. Biol. Cell 18, 2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madsen C. D., Ferraris G. M., Andolfo A., Cunningham O., Sidenius N. (2007) J. Cell Biol. 177, 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gårdsvoll H., Ploug M. (2007) J. Biol. Chem. 282, 13561–13572 [DOI] [PubMed] [Google Scholar]
- 25.Rasch M. G., Lund I. K., Almasi C. E., Hoyer-Hansen G. (2008) Front. Biosci. 13, 6752–6762 [DOI] [PubMed] [Google Scholar]
- 26.Montuori N., Carriero M. V., Salzano S., Rossi G., Ragno P. (2002) J. Biol. Chem. 277, 46932–46939 [DOI] [PubMed] [Google Scholar]
- 27.Ragno P., Montuori N., Covelli B., Hoyer-Hansen G., Rossi G. (1998) Cancer Res. 58, 1315–1319 [PubMed] [Google Scholar]
- 28.Kettner C., Shaw E. (1979) Biochim. Biophys. Acta 569, 31–40 [DOI] [PubMed] [Google Scholar]
- 29.Schwartz B. S., Monroe M. C., Levin E. G. (1988) Blood 71, 734–741 [PubMed] [Google Scholar]
- 30.Berkenpas M. B., Lawrence D. A., Ginsburg D. (1995) EMBO J. 14, 2969–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrew S. M., Titus J. A. (2001) Curr. Protoc. Cell Biol. Chapter 16, Unit 16-13 [DOI] [PubMed] [Google Scholar]
- 32.Wei Y., Lukashev M., Simon D. I., Bodary S. C., Rosenberg S., Doyle M. V., Chapman H. A. (1996) Science 273, 1551–1555 [DOI] [PubMed] [Google Scholar]
- 33.Picone R., Kajtaniak E. L., Nielsen L. S., Behrendt N., Mastronicola M. R., Cubellis M. V., Stoppelli M. P., Pedersen S., Danø K., Blasi F. (1989) J. Cell Biol. 108, 693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiore M. M., Neuenschwander P. F., Morrissey J. H. (1992) Blood 80, 3127–3134 [PubMed] [Google Scholar]
- 35.Griffith M. J., Breitkreutz L., Trapp H., Briet E., Noyes C. M., Lundblad R. L., Roberts H. R. (1985) J. Clin. Invest. 75, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L., Gonias S. L. (2005) J. Cell. Biochem. 96, 1021–1033 [DOI] [PubMed] [Google Scholar]
- 37.Czekay R. P., Kuemmel T. A., Orlando R. A., Farquhar M. G. (2001) Mol. Biol. Cell 12, 1467–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen D. H., Catling A. D., Webb D. J., Sankovic M., Walker L. A., Somlyo A. V., Weber M. J., Gonias S. L. (1999) J. Cell Biol. 146, 149–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llinas P., Le Du M. H., Gårdsvoll H., Danø K., Ploug M., Gilquin B., Stura E. A., Ménez A. (2005) EMBO J. 24, 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barinka C., Parry G., Callahan J., Shaw D. E., Kuo A., Bdeir K., Cines D. B., Mazar A., Lubkowski J. (2006) J. Mol. Biol. 363, 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montuori N., Salzano S., Rossi G., Ragno P. (2000) FEBS Lett. 476, 166–170 [DOI] [PubMed] [Google Scholar]
- 42.Behrendt N., Rønne E., Ploug M., Petri T., Løber D., Nielsen L. S., Schleuning W. D., Blasi F., Appella E., Danø K. (1990) J. Biol. Chem. 265, 6453–6460 [PubMed] [Google Scholar]
- 43.Stoppelli M. P., Corti A., Soffientini A., Cassani G., Blasi F., Assoian R. K. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4939–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plow E. F., Freaney D. E., Plescia J., Miles L. A. (1986) J. Cell Biol. 103, 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Estreicher A., Wohlwend A., Belin D., Schleuning W. D., Vassalli J. D. (1989) J. Biol. Chem. 264, 1180–1189 [PubMed] [Google Scholar]
- 46.Beaufort N., Leduc D., Rousselle J. C., Magdolen V., Luther T., Namane A., Chignard M., Pidard D. (2004) J. Immunol. 172, 540–549 [DOI] [PubMed] [Google Scholar]
- 47.Høyer-Hansen G., Behrendt N., Ploug M., Danø K., Preissner K. T. (1997) FEBS Lett. 420, 79–85 [DOI] [PubMed] [Google Scholar]
- 48.Cortese K., Sahores M., Madsen C. D., Tacchetti C., Blasi F. (2008) PLoS One 3, e3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conese M., Nykjaer A., Petersen C. M., Cremona O., Pardi R., Andreasen P. A., Gliemann J., Christensen E. I., Blasi F. (1995) J. Cell Biol. 131, 1609–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Kuo A., Kochan J., Strickland D., Kariko K., Barnathan E. S., Cines D. B. (1994) J. Biol. Chem. 269, 8153–8158 [PubMed] [Google Scholar]
- 51.Henic E., Sixt M., Hansson S., Høyer-Hansen G., Casslén B. (2006) Gynecol. Oncol. 101, 28–39 [DOI] [PubMed] [Google Scholar]
- 52.Nykjaer A., Kjøller L., Cohen R. L., Lawrence D. A., Garni-Wagner B. A., Todd R. F., 3rd, van Zonneveld A. J., Gliemann J., Andreasen P. A. (1994) J. Biol. Chem. 269, 25668–25676 [PubMed] [Google Scholar]
- 53.Nykjaer A., Conese M., Christensen E. I., Olson D., Cremona O., Gliemann J., Blasi F. (1997) EMBO J. 16, 2610–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andreasen P. A., Sottrup-Jensen L., Kjøller L., Nykjaer A., Moestrup S. K., Petersen C. M., Gliemann J. (1994) FEBS Lett. 338, 239–245 [DOI] [PubMed] [Google Scholar]
- 55.Jo M., Takimoto S., Montel V., Gonias S. L. (2009) Am. J. Pathol. 175, 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Høyer-Hansen G., Pessara U., Holm A., Pass J., Weidle U., Danø K., Behrendt N. (2001) Biochem. J. 358, 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Resnati M., Pallavicini I., Wang J. M., Oppenheim J., Serhan C. N., Romano M., Blasi F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1359–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blasi F., Carmeliet P. (2002) Nat. Rev. Mol. Cell Biol. 3, 932–943 [DOI] [PubMed] [Google Scholar]
- 59.Mannari C., Santi S., Migliori M., Filippi C., Origlia N., Sansò M., Boldrini E., Giovannini L. (2008) Int. J. Immunopathol. Pharmacol. 21, 651–658 [DOI] [PubMed] [Google Scholar]