Abstract
PCSK9 binds to the low density lipoprotein receptor (LDLR) and leads to LDLR degradation and inhibition of plasma LDL cholesterol clearance. Consequently, the role of PCSK9 in modulating circulating LDL makes it a promising therapeutic target for treating hypercholesterolemia and coronary heart disease. Although the C-terminal domain of PCSK9 is not involved in LDLR binding, the location of several naturally occurring mutations within this region suggests that it has an important role for PCSK9 function. Using a phage display library, we identified an anti-PCSK9 Fab (fragment antigen binding), 1G08, with subnanomolar affinity for PCSK9. In an assay measuring LDL uptake in HEK293 and HepG2 cells, 1G08 Fab reduced 50% the PCSK9-dependent inhibitory effects on LDL uptake. Importantly, we found that 1G08 did not affect the PCSK9-LDLR interaction but inhibited the internalization of PCSK9 in these cells. Furthermore, proteolysis and site-directed mutagenesis studies demonstrated that 1G08 Fab binds a region of β-strands encompassing Arg-549, Arg-580, Arg-582, Glu-607, Lys-609, and Glu-612 in the PCSK9 C-terminal domain. Consistent with these results, 1G08 fails to bind PCSK9ΔC, a truncated form of PCSK9 lacking the C-terminal domain. Additional studies revealed that lack of the C-terminal domain compromised the ability of PCSK9 to internalize into cells, and to inhibit LDL uptake. Together, the present study demonstrate that the PCSK9 C-terminal domain contribute to its inhibition of LDLR function mainly through its role in the cellular uptake of PCSK9 and LDLR complex. 1G08 Fab represents a useful new tool for delineating the mechanism of PCSK9 uptake and LDLR degradation.
Keywords: Annexin, Antibodies, Hepatocyte, Low Density Lipoprotein (LDL), Receptors, Fab, PCSK9
Introduction
Proprotein convertase subtilisin-like/kexin type 9 (PCSK9)4 is a key regulator of plasma low density lipoprotein (LDL) cholesterol and has emerged as a promising target for prevention and treatment of coronary heart disease. A strong link between PCSK9, LDL, cholesterol, and coronary heart disease has been established by multiple laboratories. Human genetic studies demonstrated remarkable correlations between several nonsense or missense PCSK9 mutations with plasma LDL cholesterol levels and the risk of coronary heart disease. Thus, putative gain- or loss-of-function mutants were found to correlate with increased or reduced plasma LDL levels and cardiovascular risk, respectively (1–7). A recent genome-wide association study further bolstered the importance of PCSK9 by establishing a linkage between a single nucleotide polymorphism at a locus near PCSK9 with early-onset myocardial infarction (8).
There is extensive evidence that plasma PCSK9 raises LDL cholesterol levels by binding to cell surface LDLR and targeting the receptor to lysosomes for degradation (9–13). Accordingly, inhibition of PCSK9 by recombinant LDLR fragments (14–16) or by mono- or polyclonal antibodies (17, 18) restored LDL cholesterol uptake in cells. Moreover, either RNAi targeting liver PCSK9 (19) or intravenous injection of a monoclonal antibody disrupting the PCSK9-LDLR interaction (17) were found to lower plasma LDL cholesterol in mice and in non-human primates. Collectively, these data strongly support PCSK9 as an attractive and viable target for therapeutic intervention against hypercholesterolemia.
PCSK9 is formed by an N-terminal prodomain, a subtilisin-like catalytic domain and a C-terminal cysteine/histidine-rich domain (CHRD). Recently, PCSK9 has been crystallized in complex with the epidermal growth factor-like repeat A (EGF-A) domain of the LDLR at both acidic and neutral pH values (14, 15, 20). Importantly, the LDLR EGF-A domain binds exclusively to the catalytic domain of PCSK9, and makes no contact with either the prodomain or the C-terminal domain.
The C-terminal domain of PCSK9 is composed of ∼230 residues with three symmetry-related repeats resembling a resistin homotrimer. This region contains 9 disulfide bonds (3 in each repeat) and has been suggested to mediate the colocalization of PCSK9 with LDLR at the cell surface (21). The C-terminal domain is not required for PCSK9 binding to LDLR (9, 14), but is involved in PCSK9-mediated LDLR degradation by a currently unknown mechanism (9, 21). Indeed, individuals expressing forms of PCSK9 with mutations in C-terminal domain have been shown to have either hyper- or hypocholesterolemia, thus strengthening the idea that this domain plays an important functional role. Here, we report the identification and characterization of a human antibody antigen binding fragment (Fab), 1G08, which binds to the C-terminal domain of PCSK9 and partially inhibits its effect on LDL uptake in vitro. In addition, we find that 1G08 binding to PCSK9 does not affect the PCSK9-LDLR interaction but inhibits PCSK9 internalization. Consistent with these results, PCSK9ΔC, which lacks the entire C-terminal domain, fails to bind 1G08 and has a reduced ability internalize and to inhibit cellular LDL uptake. Taken together, these results demonstrate the PCSK9 C-terminal domain contribute to its inhibition of LDLR function mainly through its role in the cellular uptake of PCSK9 and LDLR complex.
EXPERIMENTAL PROCEDURES
Expression Plasmids and Protein Purification
Full-length human PCSK9-V5-His protein was expressed and purified in stably transfected HEK293 cells as described previously (10). PCSK9 point mutations were generated by SeqWrite DNA Technology Service, and the mutant proteins were expressed and purified similarly to the full-length PCSK9. The truncated PCSK9ΔC (Gln-31–Ala-451) proteins with a C-terminal His tag were expressed and purified in Escherichia coli BL21 cells (14). The human LDLR ectodomain was purchased from R&D Systems. Full-length wild-type Annexin A2 (residues Met-1-Asp-339, bearing an N-terminal His6 tag followed by a linker with a TEV cleavage site), was expressed and readily purified to homogeneity by Ni affinity chromatography. The N-terminal tag was removed by overnight incubation with TEV protease. When studied by analytical size-exclusion chromatography, the purified Annexin A2 exhibited an elution profile corresponding to that of a monodisperse monomer of ∼40 kDa.
Isolation of Anti-PCSK9 Fab 1G08
The human combinatorial antibody HuCAL GOLD phage display libraries (22) were panned against recombinant human PCSK9-V5-His protein immobilized on Nunc Maxisorp plates. Three rounds of panning against human PCSK9-V5-His were carried out as described (22), and the XbaI-EcoRI inserts from the output of the third round were subcloned into Fab expression vector pMORPH_x9_MH (23), and individual chloramphenicol-resistant transformant colonies were picked and placed into 96-well plates for growth and screening for Fab expression.
Cultures of transformant colonies were isopropyl-1-thio-β-d-galactopyranoside (IPTG)-induced and grown overnight in 96-well plates for Fab expression. Culture supernatants were incubated with purified human PCSK9-V5-His protein immobilized in 96-well Nunc Maxisorp plates, washed with 0.1% TweenTM 20 in phosphate-buffered saline using a plate washer, incubated with horseradish peroxidase (HRP)-coupled anti-Fab antibody, and washed again with phosphate-buffered saline/TweenTM 20. Bound HRP was detected by addition of TMP substrate, and A450 values of wells were read with a plate reader.
Fabs from ELISA-positive clones were expressed by IPTG-induction in E. coli TG1F− cells. Cultures were lysed, the His-tagged Fabs were purified by nickel-nitrilotriacetic acid chromatography (Qiagen) and exchanged into a buffer of 25 mm HEPES pH 7.3–150 mm NaCl by centrifugal diafiltration. Proteins were analyzed by electrophoresis on Caliper Lab-Chip 90 and by conventional SDS-PAGE and quantified by BCA protein assay (Pierce). Purified Fab protein was re-assayed by ELISA in serial dilutions to confirm activity of purified Fab. Fab 1G08 was identified as ELISA-positive against human PCSK9.
Surface Plasmon Resonance (SPR)
All SPR experiments were performed using Biacore instruments at 25 °C. For PCSK9-1G08 binding studies, a purified full-length C-terminally biotinylated PCSK9 was noncovalently immobilized on a streptavidin-coated sensor surface (SA chip, GE Healthcare). Running buffers contained 150 mm NaCl, 1 mm CaCl2, 0.005% (v/v) P-20 surfactant, and 25 mm Hepes, pH 7.4. Binding constants were obtained from a series of 1G08-Fab injections. Following injections of 1G08-Fab, sensor chip surfaces were regenerated with a 5-s injection of 30 mm NaOH. Data were analyzed using BIAevaluation software, with reference sensorgrams subtracted from experimental sensorgrams to yield curves representing specific binding. Steady-state analysis was used to plot equilibrium binding response (Req) against analyte concentration to obtain thermodynamic dissociation constants (KD).
For competition studies, the purified full-length LDLR was covalently immobilized by amine coupling to a carboxymethylated dextran sensor surface (CM5 chip, GE Healthcare). First, the purified full-length PCSK9 protein was injected, followed by injection of the 1G08 Fab during the initial dissociation phase of the PCSK9-LDLR complex.
LDLR and PCSK9 Interaction Studies by Time-resolved Fluorescence Resonance Energy Transfer (TR-FRET) Assay
TR-FRET experiments were performed as previously described (10) using 4 nm Eu3+-labeled LDLR ectodomain and 10 nm Alexa Fluor 647-labeled PCSK9. Reactions were performed at 20 °C in 50 μl of buffer containing 10 mm Hepes, pH 7.4, 150 mm NaCl, 0.1 mm CaCl2, and 0.05% (w/v) bovine serum albumin. Disruption of the PCSK9-LDLR interaction was assessed in titrations in which putative competitor proteins, typically in concentration ranges of 0.1 nm to 2.5 μm, were added to the reaction mixture with incubations for 16 h prior to fluorescence measurements. IC50 values were determined from triplicate experiments by fitting data to a sigmoidal dose-response curve using nonlinear regression (GraphPad Software Inc.).
1G08 and PCSK9 Binding TR-FRET Assay
1G08 and PCSK9 binding was studied using TR-FRET assay similarly to that described above. 2–4 nm europium Eu3+-labeled 1G08-IgG and 8 to 16 nm Alexa Fluor 647-labeled PCSK9 were used in 50 μl of reaction buffer containing 10 mm Hepes, pH 7.4, 150 mm NaCl, 0.1 mm CaCl2, and 0.05% (w/v) bovine serum albumin at 20 °C. Disruption of the 1G08-PCSK9 binding was assessed in titrations in which unlabeled recombinant wild-type or mutant PCSK9 proteins, or Annexin A2 were added to the reaction mixture with incubations for 2–4 h prior to fluorescence measurements typically in concentration ranges of 0.03–1000 nm.
Isolation of LDL and Labeling of LDL and LDLR and PCSK9
LDL was isolated from healthy human volunteers as previously described (10). Labeling of PCSK9 with Alexa Fluor 647, LDL with Alexa Fluor 546 and LDLR with Eu3+(8044)-DTA (Perkin-Elmer, Waltham, MA) were performed as previously reported (24).
LDL Uptake and PCSK9 Uptake
Cellular uptake of Alexa Fluor 546-labeled LDL or Alexa Fluor647-labeled PCSK9 was studied in HEK293 cells as described previously (10), with the exception that Alexa Fluor647-labeled PCSK9 were incubated with cells in fibronectin plates for 5 h before imaging.
Size-exclusion Chromatography (SEC)
SEC was used to assess the size of 1G08 Fab, PCSK9, PCSK9ΔC, and the complex of 1G08 Fab and PCSK9. Equimolar amounts of purified PCSK9 (0.18 mm, MW 75,979, ϵ280 = 57,030 m−1 cm−1) or purified PCSK9ΔC (0.18 mm, MW 45,708, ϵ280 = 28,270 m−1 cm−1) and 1G08-Fab (0.18 mm, MW 48,853, ϵ280 = 79,050 m−1 cm−1) were mixed and incubated for 2 h at 4 °C in 40 mm Tris-HCl buffer, pH 7.5, containing 100 mm NaCl, 5% glycerol, and 20 μm CaCl2. Samples were loaded on a Superdex 200 10/30 column (Amersham Biosciences) equilibrated with the buffer described above. Elution was performed at 4 °C, and the flow rate was held at 0.50 ml/min. The column was calibrated using markers of known molecular weights (Bio-Rad).
Proteolysis
PCSK9-1G08-Fab complex in 25 mm Hepes pH 7.5, 100 mm NaCl was digested at room temperature with two different enzymes: trypsin (Promega) and GluC (Roche). PCSK9 or 1G08-Fab (alone) samples was digested in the same conditions and used as controls. The PCSK9 to endoprotease ratio used (w/w) was 1000/1 and 500/1 for trypsin and GluC, respectively. The cleavage sites on PCSK9 and PCSK9/1G08-Fab complex at 5, 15, 30, 60, 120, 240, and 360 min digestions were followed by MALDI-TOF mass spectrometry before and after dithiothreitol reduction.
MALDI-TOF MS Analysis
MALDI-TOF spectra of positive ions were acquired in linear mode on a MALDI-TOF mass spectrometer (Voyager-STR, Applied Biosystems, Inc Framingham, MA) equipped with a delayed extraction device and a nitrogen laser (λ = 337 nm). A saturated solution of sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid, Aldrich) in 50% acetonitrile and 1% trifluoroacetic acid was used to obtain the MALDI crystals. 500 shots were averaged for each spectrum acquired. The spectra were externally calibrated using des-Arg1-Bradikinin, Angiotensin I, Glu1-Fibrinopeptide B, ACTH fragments, insulin, thioredoxin, and apomyoglobin according to the mixture provided with the SequazymeTM Peptide Mass Standards kit (Applied Biosystems). The difference between the calculated average mass and the experimental mass measured (0.05% to 0.1%) was consistent with the accuracy of MALDI-TOF MS in linear mode.
RESULTS
1G08 Binds PCSK9 and Inhibits Effects of PCSK9 on Cellular LDL Uptake
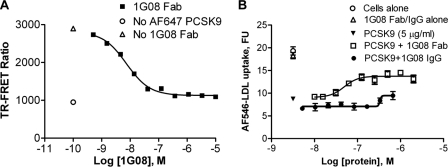
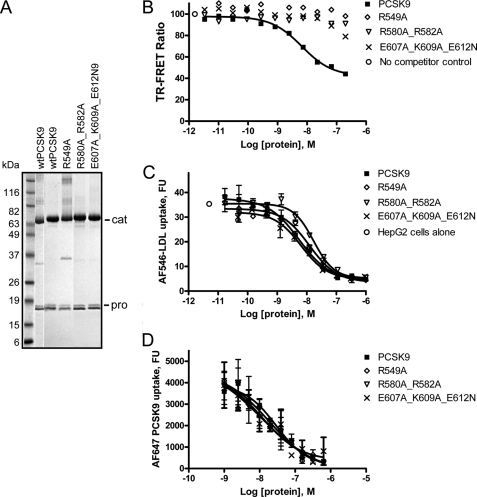
Using phage library panning and a PCSK9 binding ELISA, we identified a human Fab, 1G08, which binds purified recombinant human PCSK9 with an affinity (KD) of 550 pm as measured by surface plasmon resonance (SPR). 1G08 IgG (IgG2) binds PCSK9 with similar affinity. A TR-FRET assay confirmed the high binding affinity of 1G08 for PCSK9. In this assay, unlabeled 1G08 Fab was used to compete with Eu3+-labeled 1G08 for the binding to Alexa Fluor (AF) 647 labeled PCSK9 (Fig. 1A). The apparent inhibition constant (IC50) was 4.0 ± 0.3 nm (mean ± S.E.). It is possible that the IC50 value is an overestimation, as it is close to the IC50 detection limit of this assay (∼4 nm). 1G08 Fab and IgG do not bind mouse PCSK9 (data not shown).
FIGURE 1.
1G08 binds PCSK9 and inhibits PCSK9 cellular function. A, 1G08-PCSK9 binding TR-FRET assay. A preformed complex of AF647-labeled PCSK9 (16 nm) and Eu3+-labeled1G08-IgG (4 nm) was combined with increasing amounts of unlabeled PCSK9 protein at pH 7.4. No treatment control and assay background (No AF647-PCSK9) are shown as indicated. B, effect of 1G08 Fab or IgG on PCSK9-mediated inhibition of LDL uptake in HEK293 cells. AF546-LDL uptake were measured in cells incubated with PCSK9 (5 μg/ml) and increasing concentrations of 1G08 Fab or IgG. 1G08 Fab dose-dependently decreases AF546-labeled LDL uptake. The IC50 is ∼50 nm, maximal inhibition is ∼50%.
Next, we assessed whether binding of 1G08 Fab to PCSK9 alters cellular LDL uptake. As reported earlier (10, 24), addition of purified recombinant human PCSK9 to HEK293 cell culture medium significantly decreased cellular uptake of AF546-labeled LDL. Addition of 1G08 Fab partially blocked (∼50–60%) PCSK9 inhibition of LDL uptake in HEK293 cells, with an IC50 of ∼50 nm (Fig. 1B). A similar inhibitory effect was also observed in HepG2 cells with an IC50 of ∼150 nm (data not shown). Of note, 1G08 Fab had no effect on cellular LDL uptake in the absence of exogenously added PCSK9 (Fig. 1B), which is likely due to the low levels of endogenous PCSK9 secreted by HEK293 or HepG2 cells (<1 nm). Interestingly, unlike 1G08 Fab, even at the highest concentration tested (1 μm), 1G08 IgG2 had very little effect on PCSK9 cellular function (Fig. 1B).
1G08 Does Not Affect the PCSK9-LDLR Interaction
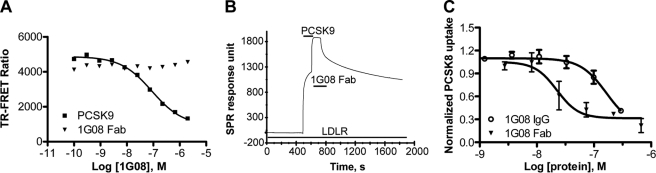
To test whether 1G08 Fab inhibits PCSK9 function by altering the interaction between PCSK9 and the LDLR, we used an in vitro TR-FRET assay that monitors PCSK9-LDLR binding. In this assay, unlabeled PCSK9 dose-dependently inhibited PCSK9-LDLR binding; in contrast, 1G08 Fab did not affect PCSK9 binding to the receptor (Fig. 2A).
FIGURE 2.
1G08 does not affect PCSK9 and LDL receptor interaction but inhibits cellular PCSK9 uptake. A, PCSK9-LDLR interaction TR-FRET assay. Increasing concentrations of unlabeled PCSK9 or 1G08-Fab was titrated into reactions containing a preformed complex of AF647-PCSK9 (10 nm) and Eu3+-labeled recombinant LDLR (4 nm) at pH 7.4. Data are means ± S.D. of triplicates. Unlabeled PCSK9, but not 1G08-Fab, disrupted the complex between Eu3+-labeled soluble LDLR and wild-type PCSK9. B, Biacore sensorgram showed the binding of 1G08-Fab (5 μm) to PCSK9 bound to immobilized LDLR ectodomain. The sensorgram shows first an increase in RU because of the association of PCSK9 with LDLR and then a second increase due to the association of 1G08 Fab with the PCSK9-LDLR complex. Line bars show application of LDLR, PCSK9, or 1G08 as indicated. In separate SPR experiments, the 1G08 Fab did not bind to the immobilized LDL receptor. C, 1G08 Fab and IgG impairs PCSK9 internalization. HepG2 cells were incubated (37 °C, 5 h) with 25 nm AF647-labeled PCSK9 in the presence of 1G08 Fab or IgG at concentration as indicated. The background fluorescence level was measured by incubating cells with AF647-PCSK9 in the presence of 50-fold unlabeled PCSK9.
Consistent with these results, SPR studies showed that addition of 1G08 Fab following PCSK9 to immobilized LDLR further increase the response signal, suggesting 1G08 Fab was able to bind LDLR-bound PCSK9 without disrupting the complex (Fig. 2B), suggesting that the binding sites of 1G08 and LDLR on PCSK9 do not overlap.
1G08 Binding Inhibits PCSK9 Internalization
The binding of 1G08 Fab to PCSK9 does not affect the PCSK9-LDLR interaction, yet, it inhibits PCSK9-mediated inhibition on cellular LDL uptake. To delineate the underlying mechanisms, we tested whether 1G08 affects PCSK9 internalization. AF647-labeled PCSK9 uptake was measured by fluorescence imaging in HEK293 cells cultured in Dulbecco's modified Eagle's medium containing 10% lipoprotein-deficient serum (10). 1G08 Fab dose-dependently inhibited the uptake of AF647-PCSK9 in these cells (Fig. 2C). Consistent with its partially inhibitory effect on LDL uptake, 1G08 Fab did not completely inhibit AF647-PCSK9 internalization even at the highest concentration tested (2 μm). Similarly, 1G08 IgG is also a partial inhibitor of AF-647-PCSK9 internalization, although with much less efficiency. The IC50 values of 1G08 Fab and IgG is 22 nm and great than 170 nm, respectively (Fig. 3). A control Fab that does not bind PCSK9 had no effect on AF647-PCSK9 internalization. These results indicate that the inhibitory effect of 1G08 on PCSK9 cellular function is mediated, at least in part, through attenuation of cellular PCSK9 uptake.
FIGURE 3.
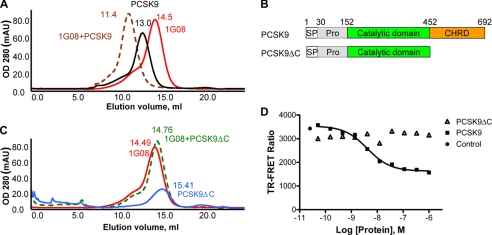
1G08 does not bind PCSK9 C-terminal truncation mutant. A, size-exclusion chromatography of PCSK9-1G08-Fab complex (brown-dotted color, 50 μl, 180 μm), PCSK9 (black color, 50 μl, 180 μm), 1G08-Fab (red color, 50 μl, 180 μm). B, schematic illustration of the functional domain organization of full-length PCSK9 and the C-terminal-truncated PCSK9ΔC protein. C, size-exclusion chromatography of PCSK9ΔC-1G08-Fab co-elution (green-dotted color, 50 μl, 180 μm), PCSK9ΔC (blue color, 50 μl, 180 μm), 1G08-Fab (red color, 50 μl, 180 μm). The retention volume (ml) is indicated on top of each peak. D, 1G08-PCSK9 binding TR-FRET assay showing that unlabeled PCSK9, but not unlabeled PCSK9ΔC disrupted the preformed 1G08-Fab and PCSK9 complex. Fluorescence level in the control reaction (no competitor) was also shown. Abbreviations: Pro, inhibitory prodomain; SP, signal peptide. CHRD, cysteine/histidine-rich domain.
In Vitro Characterization of the PCSK9-1G08 Fab Complex
Previous studies have shown that the C-terminal domain of PCSK9 plays a role in mediating the colocalization of PCSK9 with LDLR at the cell surface and PCSK9 trafficking (21). To further characterize the PCSK9/1G08-Fab interaction, we initially studied these proteins using SEC. When PCSK9 and 1G08-Fab proteins were loaded onto the column separately (Fig. 3A, peaks in black and red colors), PCSK9 eluted as a single peak at retention volume 13.0 ml corresponding to a protein with an apparent molecular mass (MW) of 76 kDa, which accounts for one PCSK9 monomer (PCSK9 MW 75,979 Da); while 1G08-Fab eluted as a single peak at 14.5 ml, which corresponds to 49 kDa, in agreement with the molecular weight expected for a Fab. When equimolar concentrations of PCSK9 and 1G08-Fab were applied to a SEC column, a single, near symmetrical peak was eluted at a retention volume (Ve) −11.4 ml (Fig. 3A, peak in brown color), with no evidence of aggregation or proteolytic degradation. The retention volume of the complex peak corresponds to a 130-kDa protein, which closely matches a heterodimer formed by one PCSK9 molecule and one 1G08-Fab molecule.
Interestingly, 1G08-Fab did not form a complex with PCSK9ΔC (amino acids 31–451), a truncated form of PCSK9 lacking the C-terminal domain but capable to bind LDLR like full-length PCSK9 (14) (Fig. 3B). When equimolar concentrations of PCSK9ΔC and 1G08-Fab were loaded into the SEC column, the resulting elution profile was consistent with that of the two individual free proteins co-eluting separately. The retention volume of the co-eluting PCSK9ΔC and 1G08-Fab is 14.8 ml (Fig. 3C, peak in green color), very close to that of the individual proteins (15.4 and 14.8 ml, respectively. Fig. 3C, peak in blue and magenta color). The size-exclusion chromatography behavior of the proteins was unaffected by lowering or increasing the protein concentrations (0.05–0.5 mm), suggesting that the proteins did not undergo concentration-dependent aggregation and that there is no appreciable binding of 1G08 and PCSK9ΔC even in the high μm to low mm concentration range.
In agreement with these results, 1G08 also failed to bind PCSK9ΔC in the TR-FRET assay (Fig. 3D). In contrast to unlabeled full-length PCSK9, which completely blocked the binding of 1G08-Fab to fluorescent-labeled PCSK9, PCSK9ΔC had no effect. Taken together, these results clearly suggest that 1G08-Fab binds at the C-terminal domain of PCSK9 (Fig. 3D).
Mapping 1G08 Binding Site on PCSK9 C-terminal Domain
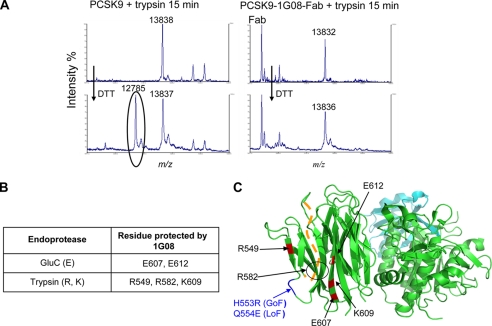
To further define the binding site of 1G08-Fab on PCSK9, we subjected PCSK9 to proteolysis in the presence or absence of 1G08-Fab, then followed by mass spectrometry determination of the PCSK9 residues protected by 1G08 from proteolysis. PCSK9-1G08 complex was digested with endoproteinase trypsin or GluC (Staphylococcus aureus protease V8), serine proteinases, which selectively cleaves peptide bonds C-terminal domain to arginine and lysine (R, K) or glutamic acid (E) residues, respectively. PCSK9 or 1G08 samples were digested separately under the same conditions and used as controls. The cleavage sites on PCSK9 and PCSK9-1G08-Fab complex were identified by MALDI-TOF mass spectrometry at 5, 15, 30, 60, 120, 240, and 360 min sample digestion. Following proteolysis with trypsin and DTT reduction, the mass spectrum of PCSK9 revealed a peak at 12785.0 m/z that corresponds to peptide Gly-583—Arg-705. This peak was absent from the spectrum derived from the PCSK9-1G08 complex, demonstrating that 1G08 protected PCSK9 residue Arg-582 from digestion by trypsin (Fig. 4A). Similarly, 1G08-Fab protected PCSK9 residues Arg-549, Lys-609, and Glu-607, Glu-612 by trypsin and GluC, respectively (Fig. 4B). All residues protected by 1G08-Fab are located in the C-terminal of PCSK9 (highlighted in red in Fig. 4C), in agreement with the SEC study results which indicate that 1G08 binds PCSK9 in this region.
FIGURE 4.
Proteolysis and mass spectrometry study of 1G08 binding site on PCSK9 C-terminal domain. A, 1G08 Fab binding site analysis. Following limited proteolysis with trypsin (R, K) and dithiothreitol reduction, the mass spectrum of PCSK9 reveals a peak at 12785.0 m/z (circled) that corresponds to peptide Gly-583—Arg-705, this peak is absent from the spectrum derived from the trypsin-treated PCSK9-1G08-Fab complex, demonstrating that 1G08-Fab protected PCSK9 residue Arg-582 from digestion by trypsin. B, summary of PCSK9 residues protected by 1G08 Fab binding in proteolysis experiments. C, ribbon representation of PCSK9 and a summary of the data obtained by the MS analyses. The PCSK9 residues protected by 1G08-Fab after limited proteolysis with trypsin and GluC are shown in red and are all located in the PCSK9 C-terminal domain. The orange dashed line denotes residues Gly-572 to Pro-585, which form a disordered loop not resolved in the x-ray structure.
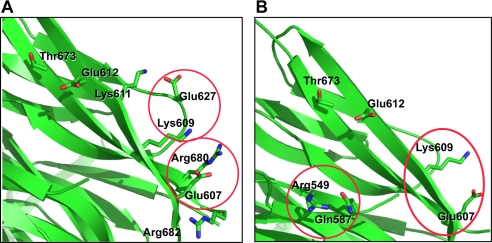
Molecular modeling analysis revealed the spatial relations among the residues involved in the putative 1G08-Fab binding epitope. Based on structural analyses, Glu-607 interacts with Lys-609 and Arg-549 interacts with Gln-587 when PCSK9 is crystallized alone (25) (Fig. 5A); whereas Glu-607 and Lys-609 interact with Arg-680 and Glu-627, respectively, via salt bridges in PCSK9 when it was co-crystallized with LDLR EGF-A (20) (Fig. 5B). This suggests that this region is structurally flexible, and the residues involved may therefore be able to change interaction partners from the residues within PCSK9 to those of 1G08-Fab. Mutation of those residues to alanine would be expected to weaken the inter- and intrasalt bridge interactions as indicated from the mutation study shown in Table 1.
FIGURE 5.
Spatial relations among the residues in the putative 1G08-Fab binding epitope. A, interaction points of the mutated residues from the crystal structure of PCSK9-LDLR complex (PDB ID core: 3 bps). Glu-607 and Lys-609 interact with Arg-680 and Glu-627, respectively, via salt bridge. B, interaction points of the mutated residues from the crystal structure of PCSK9 (PDB ID core: 2p4e). Arg-549 and Lys-609 interact with Gln-587 and Gln-607, respectively. This region is more ordered in free PCSK9 than in PCSK9-LDLR complex.
TABLE 1.
IC50 values of PCSK9 C terminus mutants in 1G08 binding
| Protein name | IC50 for binding to 1G08 |
|---|---|
| nm | |
| wt PCSK9 | 4.8 ± 1.2 (n = 12) |
| R549A | ≫1000 (n = 2) |
| R580A_R582A | ≫1000 (n = 2) |
| E607A_K609A_E612N | ≫1000 (n = 2) |
Mutation Analysis of 1G08 Binding Site on PCSK9 C-terminal Domain
To confirm that residues Arg-549, Arg-582, Glu-607, Lys-609, and Arg-612 are part of the 1G08 binding epitope, we made three mutants: R549A, R580A_R582A, and E607A_K609A_E612N. Both Arg-580 and Arg-582 are located in a region that is disordered in all the PCSK9 crystal structures (shown as an orange dashed line in Fig. 4C). Arg-580, being followed by a proline in the primary sequence of PCSK9, was not subjected to trypsin cleavage. As a consequence, its accessibility/protection information could not be assessed by trypsin digestion. Therefore, the R580A/R582A double mutant was generated even though Arg-580 was not shown to be protected by 1G08 in the proteolysis studies. SDS-PAGE analysis of the wild-type PCSK9 and all three PCSK9 mutant proteins suggested that the mutations at these sites did not affect the overall integrity of the PCSK9 proteins (Fig. 6A). Remarkably, in the PCSK9-1G08 TR-FRET assay, the IC50 values of these three PCSK9 mutants were more than 1000-fold higher than that of unlabeled wild-type PCSK9 (Fig. 6B and Table 1), which demonstrate that, indeed, mutations at these sites impair 1G08-PCSK9 binding. However, these mutations had only modest impact on the potency of PCSK9: the IC50 of the wild-type PCSK9 on LDL uptake was 5.5 ± 1.6 nm (n = 10); the IC50 of the triple mutant E607A_K609A_E612N was 5.9 ± 0.8 nm (n = 2); the IC50 values of R549A and R580A_R582A were 13.1 ± 0.8 nm (n = 3) and 16.5 ± 0.1 nm (n = 2), respectively (Fig. 6C). Further, the mutant PCSK9 proteins showed similar cellular uptake potencies as the wild-type PCSK9 protein (Fig. 6D). In agreement with these results, 1G08 Fab did not inhibit the effect of R549A on LDL uptake (data not shown).
FIGURE 6.
Mutation analysis of 1G08 binding site on PCSK9 C-terminal domain. A, Coomassie Blue-stained SDS-PAGE gel of purified wild-type and PCSK9 C-terminal domain mutants. Cat and Pro denote the catalytic and C-terminal domains and the prodomain, respectively. B, 1G08-PCSK9 binding TR-FRET assay showing that R549A, R580A_R582A, and E607A_K609A_E612N mutants have significantly reduced ability to disrupt the 1G08-PCSK9 complex. C, competition study showing the inhibition of AF546-LDL uptake by recombinant wild-type, R549A, R580A_R582A, and E607A_K609A_E612N mutant PCSK9 proteins in HepG2 cells. D, competition study showing the inhibition of AF647-PCSK9 uptake by recombinant wild-type, R549A, R580A_R582A, and E607A_K609A_E612N mutant PCSK9 proteins in HepG2 cells.
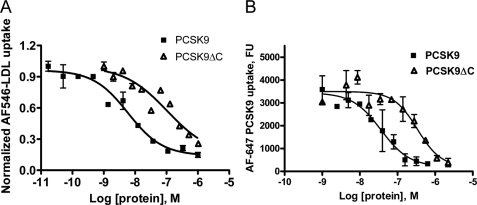
The C-terminal domain of PCSK9 has been reported to affect the ability of PCSK9 to down-regulate cell surface LDLR levels, although the mechanism was not well understood (9). We confirmed the role of the PCSK9 C-terminal domain in the modulation of LDLR levels by assessing the functional activity of the PCSK9ΔC protein in the LDL uptake assay. As shown in Fig. 7A, the IC50 of PCSK9ΔC (128.5 ± 31.8 nm, n = 2) was ∼20 fold higher than that of the wild-type PCSK9 (6.9 ± 4.2 nm, n = 4) in HepG2 cells, despite the fact that PCSK9ΔC and PCSK9 bind the LDLR with similar affinity in the TR-FRET assay (14). Similar results were also obtained in HEK293 cells (data not shown). Consistent with these results, relative to full-length PCSK9, PCSK9ΔC was also less potent inhibitor for AF-647 PCSK9 uptake, the IC50 was also ∼10 fold higher. Together these data point to a critical role of the PCSK9 C-terminal domain for cellular PCSK9 uptake and PCSK9-dependent LDLR degradation.
FIGURE 7.
The PCS9 C terminus is important for its internalization and its effect on LDL uptake. The dose-dependent inhibition of AF546-LDL (A) or AF647-PCSK9 (B) uptake by recombinant wild-type and PCSK9ΔC proteins is shown.
Effect of Serum and Annexin A2
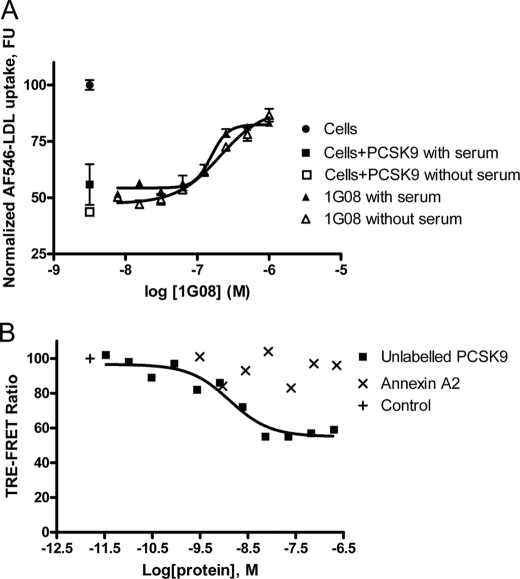
It is possible that the binding of 1G08 to PCSK9 inhibited the action of a serum protein required for PCSK9 function, as 10% lipoprotein-deficient serum is present during the reaction. We therefore tested the effect of 1G08 Fab in the absence of serum. As shown in Fig. 8A, 1G08 Fab acted similarly in the absence or presence of serum, suggesting that a serum accessory protein is not necessary for its effect.
FIGURE 8.
Effect of serum and Annexin A2. A, effect of 1G08 Fab on PCSK9-mediated inhibition of LDL uptake in HEK293 cells. AF546-LDL uptake were measured in HepG2 cells incubated with PCSK9 (1 μg/ml) and increasing concentrations of 1G08 Fab in the presence or absence of 10% lipoprotein-deficient serum. B, Annexin A2 does not affect 1G08 binding to PCSK9. Eu3+-G08 IgG 4 nm and AF647-PCSK9 16 nm were used to in the 1G08-PCSK9 interaction assay. Annexin A2, up to a concentration of 19 μm did not affect 1G08-PCSK9 binding.
A recent study has shown that a membrane associated protein Annexin A2 binds PCSK9 at the C-terminal domain and impairs PCSK9-mediated LDLR degradation (26). We tested whether 1G08 and Annexin A2 binds the PCSK9 at the same site. As shown in Fig. 8B, Annexin A2 does not affect 1G08-PCSK9 binding, suggesting that the binding sites of Annexin A2 and 1G08 do not overlap.
DISCUSSION
Studies have shown that the PCSK9 C-terminal domain is not directly involved in PCSK9-LDLR binding (9, 14). Yet, several gain- (H553R) or loss- (S462P and Q554E) of function point mutations within the C-terminal domain have been shown to result in hyper- or hypocholesterolemia, respectively (1, 27–29), which pointing to the functional significance of this region. The mechanism by which the C-terminal domain modulates the cellular function of PCSK9 is not well understood. Here, we report that 1G08, an anti-PCSK9 Fab, binds PCSK9 with a high affinity and impairs the PCSK9-mediated inhibitory effect on LDL uptake mainly through its inhibition of PCSK9 internalization. These studies demonstrated that 1G08 Fab is a useful new tool for delineating mechanism of PCSK9 uptake and LDLR degradation.
Proteolysis and mass spectrometry analysis, coupled with biochemical and site-directed mutagenesis studies revealed that 1G08 binds to the C-terminal domain of PCSK9. Importantly, 1G08 did not affect PCSK9-LDLR binding. Rather, it partially inhibited PCSK9 internalization. Consistent with these findings, 1G08 failed to bind PCSK9ΔC, a truncated form of PCSK9 lacking the C-terminal domain (i.e. after Ala-451). These results are in good agreement with those from a recent study which showed that a similar C-terminally truncated PCSK9 variant (amino acids 1–454) was able to interact with LDLR, but was unable to induce LDLR degradation when overexpressed in hepatoma HuH-7 cells (9). Further, the present study also demonstrate that relative to full-length PCSK9, PCSK9ΔC had greatly reduced ability to inhibit cellular uptake of fluorescence labeled PCSK9 and LDL uptake in hepatocytes and in HEK293 cells, even though its binding affinity for LDLR is unaltered (14). Together, these results demonstrate that the PCSK9 C-terminal domain contribute to its inhibition of LDLR function mainly through its impact on the cellular uptake of PCSK9 and LDLR complex. Substitution of Arg-549, Arg-580, Arg-582, Glu-607, Lys-609, and Glu-612 (residues involved in 1G08 binding) with alanines have very little effect in PCSK9 internalization; suggesting these residues are outside domains directly impacting PCSK9 internalization. Further studies are needed to identify the C-terminal residues involved in PCSK9 internalization.
The cysteine/histidine-rich C-terminal domain of PCSK9 consists of a cylindrically shaped domain with quasi 3-fold internal symmetry. Each of the three subdomains is a sandwich made up of six antiparallel β-strands. This domain resembles the structure of a resistin homotrimer (28) and is composed of three β-sheet modules each held together by three internal disulfide bonds, for a total of nine disulfide bonds within the C-terminal domain (30). Structural studies have shown that several regions in the PCSK9 C-terminal domain are structurally flexible suggesting that they may be involved in binding to a protein other than LDLR (25, 28, 30). Our studies showed that the 1G08-Fab binding epitope on PCSK9 is comprised of three adjacent β-strands containing Arg-549, Arg-580, Arg-582, Glu-607, Lys-609, and Glu-612 in the C-terminal domain. These residues are close to an exposed loop 550VHCHQQGH557, which contains two natural gain- and loss-of-function mutations H553R and Q554E, respectively (20, 28). Because of the lack of readily cleavable residues in this region, we were unable to resolve whether this region was protected by 1G08 Fab in the proteolysis study. It is possible that this loop may also be part of the 1G08 binding epitope. Although the residues identified in the PCSK9 C-terminal domain (Arg-549, Arg-580, Arg-582, Glu-607, Lys-609, and Glu-612) are crucial for the binding of 1G08 Fab; they do not appear to be important for PCSK internalization or inhibition of LDL uptake, as mutation at these sites do not affect cellular uptake of the PCSK9 or inhibition of LDL uptake.
We have determined that the inhibition of 1G08 Fab on PCSK9 cellular function does not require any serum soluble protein. Another plausible mechanism for 1G08 Fab inhibition of PCSK9 internalization is by preventing PCSK9 interaction with a currently unidentified cell surface molecule required for cellular uptake of PCSK9. This hypothesis may explain the discrepancy between the IC50 value of wild-type PCSK9 in inhibiting LDL uptake in our cell-based assay (∼60 nm) and the lower affinity of PCSK9 for LDLR measured by SPR (KD = 600–800 nm) (10). Binding of PCSK9 to LDLR and to a cell surface coreceptor may allow the formation of a tighter complex and a more efficient internalization of the PCSK9-LDLR complex. In addition, the need for a tissue-specific cell surface cofactor for PCSK9 function could also explain the preferential down-modulation of liver LDLR by PCSK9 in mice (12). Consistently, a recent study showed that a membrane-associated protein Annexin A2 binds PCSK9 C-terminal domain and impairs PCSK9-mediated LDLR degradation (26). The finding that 1G08 Fab and Annexin A2 bind distinctive epitopes suggests that multiple site of modulation exist in the PCSK9 C-terminal.
Interestingly, relative to 1G08 Fab, 1G08 IgG is less effective for inhibiting PCSK9 uptake and for restoring cellular LDL uptake. The reason for this disparity is unclear. It is conceivable that the smaller Fab is easier to access its epitope on the C-terminal domain and prevent the binding of a cell surface coreceptor. PCSK9 inhibitors are highly desirable because of their promise as new LDL-lowering agents. Recent reports demonstrated that anti-PCSK9 antibodies targeting the catalytic domain were able to restore LDL uptake in HepG2 cells overexpressing PCSK9 or treated with exogenous recombinant PCSK9 protein, and to reduce serum cholesterol levels in mice and in non-human primates (17, 18). To our knowledge, this is the first report of a human antibody antigen binding fragment against the C-terminal domain of PCSK9 that partially restores LDL uptake through inhibition of PCSK9 internalization. The 1G08 antibody antigen binding fragment will serve as a useful tool for delineating mechanism of PCSK9 uptake and LDLR degradation; and for identifying cofactors, which may bind the C-terminal domain of PCSK9 and regulate its function. Further, the work presented herein provides a novel strategy for inhibiting PCSK9 function.
Acknowledgment
We thank Vijayalakshmi Agnani for production of full-length PCSK9.
Footnotes
- PCSK9
- proprotein convertase subtilisin-like/kexin type 9
- AF
- Alexa Fluor
- Fab
- fragment antigen binding
- EGF-A
- epidermal growth factor-like repeat A
- LDL
- low density lipoprotein
- LDLR
- LDL receptor
- RFU
- relative fluorescence units
- SEC
- size-exclusion chromatography
- SPR
- surface plasmon resonance
- TR-FRET
- time-resolved fluorescence resonance energy transfer
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight.
REFERENCES
- 1.Abifadel M., Varret M., Rabès J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derré A., Villéger L., Farnier M., Beucler I., Bruckert E., Chambaz J., Chanu B., Lecerf J. M., Luc G., Moulin P., Weissenbach J., Prat A., Krempf M., Junien C., Seidah N. G., Boileau C. (2003) Nat. Genet. 34, 154–156 [DOI] [PubMed] [Google Scholar]
- 2.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. (2005) Nat. Genet. 37, 161–165 [DOI] [PubMed] [Google Scholar]
- 3.Berge K. E., Ose L., Leren T. P. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1094–1100 [DOI] [PubMed] [Google Scholar]
- 4.Kotowski I. K., Pertsemlidis A., Luke A., Cooper R. S., Vega G. L., Cohen J. C., Hobbs H. H. (2006) Am. J. Hum. Genet. 78, 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J. C., Boerwinkle E., Mosley T. H., Jr., Hobbs H. H. (2006) N. Engl. J. Med. 354, 1264–1272 [DOI] [PubMed] [Google Scholar]
- 6.Timms K. M., Wagner S., Samuels M. E., Forbey K., Goldfine H., Jammulapati S., Skolnick M. H., Hopkins P. N., Hunt S. C., Shattuck D. M. (2004) Hum Genet 114, 349–353 [DOI] [PubMed] [Google Scholar]
- 7.Abifadel M., Rabes J. P., Devillers M., Munnich A., Erlich D., Junien C., Varret M., Boileau C. (2009) Hum. Mutat. 30, 520–529 [DOI] [PubMed] [Google Scholar]
- 8.Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., Erdmann J., Reilly M. P., Rader D. J., Morgan T., Spertus J. A., Stoll M., Girelli D., McKeown P. P., Patterson C. C., Siscovick D. S., O'Donnell C. J., Elosua R., Peltonen L., Salomaa V., Schwartz S. M., Melander O., Altshuler D., Merlini P. A., Berzuini C., Bernardinelli L., Peyvandi F., Tubaro M., Celli P., Ferrario M., Fetiveau R., Marziliano N., Casari G., Galli M., Ribichini F., Rossi M., Bernardi F., Zonzin P., Piazza A., Yee J., Friedlander Y., Marrugat J., Lucas G., Subirana I., Sala J., Ramos R., Meigs J. B., Williams G., Nathan D. M., Macrae C. A., Havulinna A. S., Berglund G., Hirschhorn J. N., Asselta R., Duga S., Spreafico M., Daly M. J., Nemesh J., Korn J. M., McCarroll S. A., Surti A., Guiducci C., Gianniny L., Mirel D., Parkin M., Burtt N., Gabriel S. B., Thompson J. R., Braund P. S., Wright B. J., Balmforth A. J., Ball S. G., Hall A. S., Linsel-Nitschke P., Lieb W., Ziegler A., Konig I. R., Hengstenberg C., Fischer M., Stark K., Grosshennig A., Preuss M., Wichmann H. E., Schreiber S., Ouwehand W., Deloukas P., Scholz M., Cambien F., Li M., Chen Z., Wilensky R., Matthai W., Qasim A., Hakonarson H. H., Devaney J., Burnett M. S., Pichard A. D., Kent K. M., Satler L., Lindsay J. M., Waksman R., Epstein S. E., Scheffold T., Berger K., Huge A., Martinelli N., Olivieri O., Corrocher R., Holm H., Thorleifsson G., Thorsteinsdottir U., Stefansson K., Do R., Xie C., Siscovick D. (2009) Nat. Genet. 41, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13045–13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher T. S., Lo Surdo P., Pandit S., Mattu M., Santoro J. C., Wisniewski D., Cummings R. T., Calzetta A., Cubbon R. M., Fischer P. A., Tarachandani A., De Francesco R., Wright S. D., Sparrow C. P., Carfi A., Sitlani A. (2007) J. Biol. Chem. 282, 20502–20512 [DOI] [PubMed] [Google Scholar]
- 11.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. (2006) J. Clin. Invest. 116, 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grefhorst A., McNutt M. C., Lagace T. A., Horton J. D. (2008) J. Lipid Res. 49, 1303–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt R. J., Beyer T. P., Bensch W. R., Qian Y. W., Lin A., Kowala M., Alborn W. E., Konrad R. J., Cao G. (2008) Biochem. Biophys. Res. Commun. 370, 634–640 [DOI] [PubMed] [Google Scholar]
- 14.Bottomley M. J., Cirillo A., Orsatti L., Ruggeri L., Fisher T. S., Santoro J. C., Cummings R. T., Cubbon R. M., Lo Surdo P., Calzetta A., Noto A., Baysarowich J., Mattu M., Talamo F., De Francesco R., Sparrow C. P., Sitlani A., Carfí A. (2009) J. Biol. Chem. 284, 1313–1323 [DOI] [PubMed] [Google Scholar]
- 15.McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., Lagace T. A. (2009) J. Biol. Chem. 284, 10561–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan L., Pang L., Zhang R., Murgolo N. J., Lan H., Hedrick J. A. (2008) Biochem. Biophys. Res. Commun. 375, 69–73 [DOI] [PubMed] [Google Scholar]
- 17.Chan J. C., Piper D. E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., Di Y., Shetterly S., Arimura Z., Salomonis H., Romanow W. G., Thibault S. T., Zhang R., Cao P., Yang X. P., Yu T., Lu M., Retter M. W., Kwon G., Henne K., Pan O., Tsai M. M., Fuchslocher B., Yang E., Zhou L., Lee K. J., Daris M., Sheng J., Wang Y., Shen W. D., Yeh W. C., Emery M., Walker N. P., Shan B., Schwarz M., Jackson S. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9820–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duff C. J., Scott M. J., Kirby I. T., Hutchinson S. E., Martin S. L., Hooper N. M. (2009) Biochem. J. 419, 577–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank-Kamenetsky M., Grefhorst A., Anderson N. N., Racie T. S., Bramlage B., Akinc A., Butler D., Charisse K., Dorkin R., Fan Y., Gamba-Vitalo C., Hadwiger P., Jayaraman M., John M., Jayaprakash K. N., Maier M., Nechev L., Rajeev K. G., Read T., Röhl I., Soutschek J., Tan P., Wong J., Wang G., Zimmermann T., de Fougerolles A., Vornlocher H. P., Langer R., Anderson D. G., Manoharan M., Koteliansky V., Horton J. D., Fitzgerald K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11915–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon H. J., Lagace T. A., McNutt M. C., Horton J. D., Deisenhofer J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nassoury N., Blasiole D. A., Tebon Oler A., Benjannet S., Hamelin J., Poupon V., McPherson P. S., Attie A. D., Prat A., Seidah N. G. (2007) Traffic 8, 718–732 [DOI] [PubMed] [Google Scholar]
- 22.Rothe C., Urlinger S., Löhning C., Prassler J., Stark Y., Jäger U., Hubner B., Bardroff M., Pradel I., Boss M., Bittlingmaier R., Bataa T., Frisch C., Brocks B., Honegger A., Urban M. (2008) J. Mol. Biol. 376, 1182–1200 [DOI] [PubMed] [Google Scholar]
- 23.Rauchenberger R., Borges E., Thomassen-Wolf E., Rom E., Adar R., Yaniv Y., Malka M., Chumakov I., Kotzer S., Resnitzky D., Knappik A., Reiffert S., Prassler J., Jury K., Waldherr D., Bauer S., Kretzschmar T., Yayon A., Rothe C. (2003) J. Biol. Chem. 278, 38194–38205 [DOI] [PubMed] [Google Scholar]
- 24.Pandit S., Wisniewski D., Santoro J. C., Ha S., Ramakrishnan V., Cubbon R. M., Cummings R. T., Wright S. D., Sparrow C. P., Sitlani A., Fisher T. S. (2008) J. Lipid Res. 49, 1333–1343 [DOI] [PubMed] [Google Scholar]
- 25.Cunningham D., Danley D. E., Geoghegan K. F., Griffor M. C., Hawkins J. L., Subashi T. A., Varghese A. H., Ammirati M. J., Culp J. S., Hoth L. R., Mansour M. N., McGrath K. M., Seddon A. P., Shenolikar S., Stutzman-Engwall K. J., Warren L. C., Xia D., Qiu X. (2007) Nat. Struct. Mol. Biol. 14, 413–419 [DOI] [PubMed] [Google Scholar]
- 26.Mayer G., Poirier S., Seidah N. G. (2008) J. Biol. Chem. 283, 31791–31801 [DOI] [PubMed] [Google Scholar]
- 27.Costet P., Krempf M., Cariou B. (2008) Trends Biochem. Sci. 33, 426–434 [DOI] [PubMed] [Google Scholar]
- 28.Hampton E. N., Knuth M. W., Li J., Harris J. L., Lesley S. A., Spraggon G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14604–14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cameron J., Holla O. L., Laerdahl J. K., Kulseth M. A., Berge K. E., Leren T. P. (2009) Atherosclerosis 203, 161–165 [DOI] [PubMed] [Google Scholar]
- 30.Piper D. E., Jackson S., Liu Q., Romanow W. G., Shetterly S., Thibault S. T., Shan B., Walker N. P. (2007) Structure 15, 545–552 [DOI] [PubMed] [Google Scholar]