Abstract
Cry toxins produced by Bacillus thuringiensis have been recognized as pore-forming toxins whose primary action is to lyse midgut epithelial cells in their target insect. In the case of the Cry1A toxins, a prepore oligomeric intermediate is formed after interaction with cadherin receptor. The Cry1A oligomer then interacts with glycosylphosphatidylinositol-anchored receptors. Two Manduca sexta glycosylphosphatidylinositol-anchored proteins, aminopeptidase (APN) and alkaline phosphatase (ALP), have been shown to bind Cry1Ab, although their role in toxicity remains to be determined. Detection of Cry1Ab binding proteins by ligand blot assay revealed that ALP is preferentially expressed earlier during insect development, because it was found in the first larval instars, whereas APN is induced later after the third larval instar. The binding of Cry1Ab oligomer to pure preparations of APN and ALP showed that this toxin structure interacts with both receptors with high affinity (apparent Kd = 0.6 nm), whereas the monomer showed weaker binding (apparent Kd = 101.6 and 267.3 nm for APN and ALP, respectively). Several Cry1Ab nontoxic mutants located in the exposed loop 2 of domain II or in β-16 of domain III were affected in binding to APN and ALP, depending on their oligomeric state. In particular monomers of the nontoxic domain III, the L511A mutant did not bind ALP but retained APN binding, suggesting that initial interaction with ALP is critical for toxicity. Our data suggest that APN and ALP fulfill two roles. First APN and ALP are initial receptors promoting the localization of toxin monomers in the midgut microvilli before interaction with cadherin. Then APN and ALP function as secondary receptors mediating oligomer insertion into the membrane. However, the expression pattern of these receptors and the phenotype of L511A mutant suggest that ALP may have a predominant role in toxin action because Cry toxins are highly effective against the neonate larvae that is the target for pest control programs.
Keywords: Bacterial Toxins, Insect, Membrane Proteins, Phosphatase, Receptor Structure-Function, Alkaline Phosphatase, Aminopeptidase, Bacillus thuringiensis, Cry1Ab Toxin, Receptor
Introduction
The Cry toxins produced by Bacillus thuringiensis are used worldwide as effective biological control agents for many species of insects including agricultural and forest pests and several vectors of human and animal diseases. Its insecticidal property results from crystalline inclusions produced during sporulation that are formed by δ-endotoxins known as Cry toxins (1).
The Cry protein family is composed of more than 54 groups, among which the three-domain Cry family forms the major group, having members that show toxicity to different insect orders and to nematodes. The crystal structure has been solved for six three-domain Cry toxins and one protoxin that display different insect specificities (2). The activated Cry toxins are globular molecules consisting of a bundle of seven α-helices (domain I), a three-β-sheet prism (domain II), and a β-sandwich (domain III) (3, 4). The N-terminal domain I is involved in oligomer formation, membrane insertion, and pore formation (5–8). Domain II and III are involved in the recognition and binding interaction with receptors in midgut cells (9–11).
The mode of action of Cry toxins has been extensively studied in lepidopteran insects and to some extent in coleopteran, dipteran, and nematodes. In the case of lepidopteran insects, a sequential binding of Cry1A toxins with at least two receptor molecules located in the midgut epithelium cells was proposed that resulted in toxin insertion into the membrane, pore formation, and cell death (12, 13). Susceptible larvae ingest Cry1A crystals formed of 130-kDa protoxins that after solubilization in the gut lumen are subject to proteolysis by midgut proteases, resulting in a 60-kDa toxic fragment composed of the three-domain structure described above. Cry1A monomers then bind to the primary receptor that has been identified in several species as a cadherin protein that is located in the microvilli of columnar midgut cells. This interaction provokes a conformational change of the toxin that facilitates further proteolytic cleavage of the domain I N-terminal helix α-1, resulting in toxin oligomerization (14, 15). In favor of this model, engineered modified Cry1Ab and Cry1Ac toxins lacking helix α-1 have been shown to retain toxicity to resistant insects that have mutations affecting the cadherin gene (16). The oligomer gains binding affinity to a second receptor that is anchored to the membrane by GPI2 and located in lipid rafts (13). Two proteins have been identified as secondary receptors, APN and ALP (17). Binding of the oligomeric toxin to the GPI-anchored receptors results in the insertion of the oligomer into the membrane and formation of pores, resulting in osmotic shock and cell death (13, 18).
Although APN was the first Cry1Ac-binding molecule identified in Manduca sexta (19, 20), its role as functional receptor is still controversial. Incorporation of M. sexta APN-1 in black lipid bilayers enhanced the Cry1Aa pore formation activity (21). Also, the transgenic expression of M. sexta APN-1 in Drosophila melanogaster resulted in sensitivity to Cry1Ac toxin (22). Nevertheless, Cry1Ac mutants in domain III that are affected in binding to APN-1 had a marginal effect on toxicity against M. sexta larvae (23). Also, in the case of Bombyx mori, it was shown that anti-cadherin antibodies protected detached midgut cells from the toxic effects of Cry1Aa, in contrast to anti-APN antibodies that had no effect on the toxicity of Cry1Aa (24). These apparently contradictory data could be explained if an additional secondary receptor could play the same role as APN in M. sexta midgut cells. In this regard, a GPI-anchored ALP was also identified as a binding protein of Cry1Ac toxin in M. sexta, although its role as a Cry1A receptor has not been analyzed until now (25, 26).
To determine whether the GPI-anchored ALP could act as a functional receptor of Cry1Ab toxin in M. sexta, we compared the binding interaction of monomeric and oligomeric forms of Cry1Ab toxin to pure preparations of both APN and ALP enzymes. Also, we analyzed the binding of Cry1Ab mutants located in regions of domain II and domain III previously characterized as APN-binding epitopes (27, 28). Our data suggest that both APN and ALP are functional receptor molecules that bind the oligomeric structure of Cry1Ab toxin with high affinity, facilitating membrane insertion and pore formation, and we also found that ALP may a have a predominant and more important role in Cry1Ab toxicity at earlier stages of larval development.
EXPERIMENTAL PROCEDURES
M. sexta Midgut BBMV Purification and Solubilization
Larvae of M. sexta were from a laboratory colony maintained on an artificial diet. BBMV were prepared from midgut tissue from each of the different larval instar by the magnesium precipitation method (29). The BBMV were centrifuged at 70,000 rpm for 40 min at 4 °C and suspended at 5 mg/ml in 20 mm Tris-HCl pH 8.5 buffer containing 100 mm NaCl, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 1% CHAPS for detergent solubilization. After 2 h at 4 °C, the solution was centrifuged at 70,000 rpm for 40 min. Protein content in the supernatant was measured by the DC protein dye method (Bio-Rad) using bovine serum albumin as a standard (Pierce).
Purification of Aminopeptidase-N
The APN protein was purified from fourth instar BBMV. The BBMV proteins that were solubilized with CHAPS as stated above were concentrated by Amicon YM-50 ultrafiltration, and a 2-ml aliquot was applied to a HR 5/10 Mono Q (GE Healthcare) column equilibrated with 20 mm Tris-HCl pH 8.5 buffer containing 2 mm MgCl2, 2 mm KCl (28, 30). The column was eluted with a gradient of NaCl (0.5–1 m) with a flow rate of 1 ml/min for 40 min. Fractions containing APN activity were pooled and concentrated by Amicon YM-50 ultrafiltration. The final sample was analyzed by SDS-PAGE and silver staining using the SilverSNAP II stain kit (Pierce) following the manufacturer's instructions.
Purification of Alkaline Phosphatase
ALP was purified by affinity chromatography using l-histidyl-diazo-benzylphosphonic acid agarose (Sigma) (31, 32). CHAPS-solubilized proteins from BBMV of third instar larvae were concentrated by Amicon YM-30 ultrafiltration, and a 1-ml aliquot was loaded into the column equilibrated with 20 mm Tris-HCl pH 8.5 buffer containing 1 mm MgCl2. After a washing step with the same buffer, ALP was eluted using 0.5 m KH2PO4. The final sample was analyzed by SDS-PAGE and silver staining as mentioned above.
Enzyme Assays
APN activity was assayed using l-leucine-p-nitroanilide as substrate, and ALP activity was assayed using p-nitrophenyl phosphate as substrate (33). Protein content was measured by the DC protein dye method (Bio-Rad) using bovine serum albumin as a standard (Pierce). The initial rate at 405 nm (Ultrospec II spectrophotometer; GE Healthcare) was used to calculate specific enzymatic activity of both enzymes. The absorption coefficient of p-nitroanilide used was 9.9 × 10−3 mol 1−1. One unit of specific APN activity was defined as the amount of enzyme catalyzing the hydrolysis of 1 μmol of l-leucine-p-nitroanilide min−1 mg of protein−11 at 25 °C. One unit of specific ALP activity was defined as the amount of enzyme producing 1 μmol of nitrophenol min−11 mg of protein−11 at 25 °C. Nitrophenol concentration was calculated by using a standard curve of 4-nitrophenol in 0.5 mm MgCl2, 100 mm Tris, pH 9.5.
Construction of Cry1Ab Mutants
Substitutions R368A/R369A, F371A, and L511A were produced by site-directed mutagenesis (QuikChange; Stratagene) using the pHT315Ab plasmid harboring the cry1Ab gene as template and the corresponding mutagenic primers: R368A/R369A, 5′-CAT TAT CGT CCA CTT TAT ATG CAG CAC CTT TTA ATA TAG GGA TAA ATA ATC-3′; F371A, 5′-CCA CTT TAT ATA GAA GAC CTG CTA ATA TAG GGA TAA ATA ATC-3′; and L511A, 5′-GGC CAG ATT TCA ACC GCG AGA GTA AAT ATT ACT GCA-3′. Automated DNA sequencing at facilities of the Instituto de Biotecnología-Universidad Nacional Autónoma de México verified the single-point mutations. Acrystalliferous B. thuringiensis strain 407 was transformed with the recombinant plasmids containing the mutated cry1Ab genes as reported (34) and selected in Luria broth at 30 °C supplemented with 10 μg ml−1 of erythromycin.
Cry1Ab Toxin Purification
Crystal inclusions of Cry1Ab toxin and their mutants were produced in B. thuringiensis strains described above. The cells were grown for 3 days at 30 °C in nutrient broth sporulation medium supplemented with erythromycin at 10 μg ml−1. After complete sporulation, the crystal inclusions were purified using discontinuous sucrose gradients, and the protein concentration was determined by the protein dye method of Bradford (35).
The monomeric toxins were obtained by trypsin activation in a mass ratio of 1:50 (1 h at 37 °C). Phenylmethylsulfonyl fluoride (final concentration, 1 mm) was added to stop proteolysis. The oligomeric Cry1Ab structure was produced by incubation of the crystals for 1 h with cadherin fragment CR7-CR12 (residues Met810–Ala1485) in a mass ratio 1:1 and digestion with midgut juice (5%) for 1 h at 37 °C in extraction buffer (50 mm Na2CO3 at pH 10.5, 0.02% mercaptoethanol). Phenylmethylsulfonyl fluoride (1 mm) was added to stop the reaction (36). Purification of monomeric and oligomeric structures of Cry1Ab toxin was achieved by size exclusion chromatography using a Superdex 200 HR 10/30 column on an AKTA Explorer (GE Healthcare) as reported previously (15).
Western Blots of Cry1Ab Toxin
Fractions of the purified toxin oligomers were separated in SDS-PAGE and transferred onto nitrocellulose membrane. The proteins were detected with polyclonal anti-Cry1Ab (1/80,000; 1 h) and a goat anti-rabbit secondary antibody coupled with horseradish peroxidase (Amersham Biosciences) (1/10,000; 1 h), followed by Super Signal chemiluminescence substrate (Pierce) as indicated by the manufacturer.
Ligand Blot
Cry1Ab toxin was biotinylated using biotinamidocaproate N-hydroxysuccinamide ester (Amersham Biosciences) according to the manufacturer's instructions. Samples of 5 μg of BBMV were separated by SDS-PAGE (9%) and transferred to polyvinylidene difluoride; after renaturation and blocking, the blots were incubated for 2 h with 10 nm of biotinylated Cry1Ab toxin in washing buffer (0.05% Tween 20 in PBS) at room temperature. Unbound toxin was removed by washing three times for 10 min in washing buffer, and bound toxin was identified by incubating the blots in PBS containing streptavidin-peroxidasa conjugate (1:5000 in PBS) for 1 h. The excess of streptavidin was removed by washing in three changes of washing buffer, and the membrane-bound complex was visualized by Super Signal chemiluminescence substrate (Pierce) as indicated by the manufacturer.
Toxicity Assays
Bioassays were performed with M. sexta neonate larvae by a surface contamination method. Toxin solution was poured on the diet surface and allowed to dry. Neonate M. sexta larvae were placed on the dried surface, and the mortality was monitored after 7 days. The lethal concentration required to kill 50% of the larvae (LC50) was estimated by Probit analysis (Polo-PC LeOra Software).
ELISA Binding Assays
One ng/well of purified APN or ALP was suspended in PBS buffer and used to coat a 96-well ELISA plate. After overnight incubation at 4 °C, 200 μl of PBS containing 2% skim milk was added to each well and incubated at 37 °C for 2 h. Then each well was washed three times with PBS buffer followed by incubation with different Cry1Ab toxin dilutions in PBS buffer (monomeric or oligomeric structures) at 37 °C. Excess toxin was removed by washing, and each well was incubated with anti-Cry1Ab antibody (1/10,000) for 1 h at 37 °C. After washing, peroxidase-conjugated rabbit antibody was added and incubated again at 37 °C for 1 h. After washing, substrate solution was added, and absorbance at 490 nm was measured. The data were analyzed using SigmaPlot version 10.
RESULTS
Expression of Aminopeptidase and Alkaline Phosphatase Proteins in M. sexta Larvae
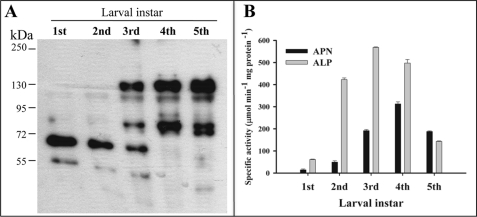
The expression of both APN and ALP in the different states of larval development was determined by ligand blot assays using biotinylated Cry1Ab toxin. The BBMV were isolated from midgut tissue of insects at different larval instar. Fig. 1A shows that Cry1Ab bound preferentially to a 65-kDa protein, previously identified as ALP (25) in BBMV isolated from first and second instar larvae. In contrast, these assays revealed that at the latter stages of larval development, such as the fourth and fifth instar, only the 120-kDa corresponding to APN protein but not the 65-kDa band was observed. However, it is important to mention that both proteins, APN and ALP, are present during all instar stages, although at different levels. Overexposure of the ligand blots revealed detectable low levels of APN in the first and second instar larvae and detectable low levels of ALP in the fourth and fifth instar larvae (data not shown). APN and ALP specific activities were determined in BBMV obtained from larvae in the different instars showing higher ALP enzymatic activity than APN during the first instars, whereas APN activity increased after the third instar, showing higher APN activities than ALP in the fifth instar (Fig. 1B).
FIGURE 1.
Expression of aminopeptidase N and alkaline phosphatase during larval development. A, ligand blot analysis of biotin-labeled Cry1Ab toxin to BBMV proteins from M. sexta larvae isolated from each instar larvae (first through fifth indicated). B, specific enzymatic activities of APN (black bars) and ALP (gray bars) determined in BBMV from each larval instar.
Analysis of Binding of Cry1Ab Oligomer and Monomer to APN and ALP
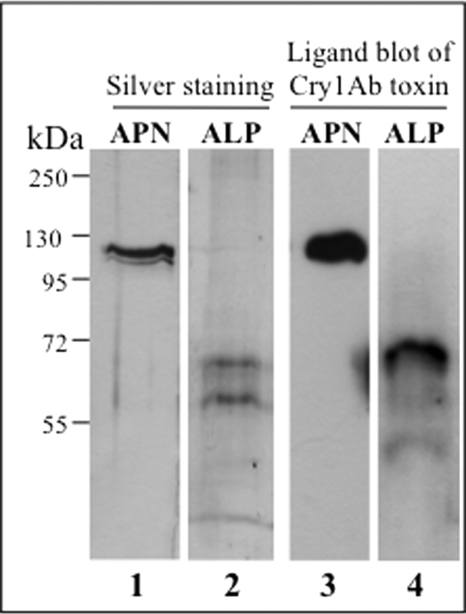
To determine the role of M. sexta ALP in Cry1Ab toxicity, we decided to analyze the binding of Cry1Ab oligomer to pure ALP preparations and compare it with the binding to pure APN samples. APN was purified from fourth instar larvae to avoid ALP contamination, and ALP was purified from third instar larvae because first and second instar larvae are too small. The BBMV were solubilized with detergent, and APN and ALP were purified by ion exchange and affinity chromatography, respectively, as described under “Experimental Procedures.” The purified fractions containing only APN or ALP activity were examined by SDS-PAGE and silver staining to assess their quality (Fig. 2, lanes 1 and 2). APN appears as a single band at ∼120 kDa, whereas ALP appeared as two bands of ∼65 and 60 kDa. The purity of both receptors was further examined by a Cry1Ab toxin ligand blot showing that both the 120-kDa APN and the 65-kDa ALP bound Cry1Ab (Fig. 2, lanes 3 and 4). Tables 1 and 2 show the results of representative purification experiments of APN and ALP, respectively. These data also indicate that the 65- and 120-kDa bands previously identified by Cry1Ab binding in BBMV samples (Fig. 1A) correspond to ALP and APN proteins, respectively. The identity of the 65-kDa band as ALP protein was confirmed by mass spectrometry analysis (data not shown).
FIGURE 2.
Purification of M. sexta APN and ALP proteins. Silver stain 10% SDS-PAGE of APN (lane 1) and ALP (lane 2) pure fractions from Mono Q (Table 1) and affinity chromatography (Table 2), respectively, are shown. Ligand blots with biotin-labeled Cry1Ab toxin of APN (lane 3) and ALP (lane 4) fractions are shown. Molecular mass markers are shown at the left of the figure.
TABLE 1.
Purification of aminopeptidase-N from midgut brush border membrane vesicles of fourth instar M. sexta larvae
| Fraction | Total activity | Total protein | Specific activity | Recovery |
|---|---|---|---|---|
| units | mg | units/mg | % | |
| BBMV | 87.3 | 20 | 4.3 | 100 |
| BBMV/CHAPS | 49.3 | 5.4 | 9.1 | 56.4 |
| Mono Qa | 16.6 | 0.212 | 78.2 | 19.01 |
a Fig. 2 (lane 1) shows the protein profile of purified APN.
TABLE 2.
Purification of alkaline phosphatase from midgut brush border membrane vesicles of third instar M. sexta larvae
| Fraction | Total activity | Total protein | Specific activity | Recovery |
|---|---|---|---|---|
| units | mg | units/mg | % | |
| BBMV | 35.6 | 10 | 3.56 | 100 |
| BBMV/CHAPS | 21.6 | 4.5 | 4.8 | 60.6 |
| Affinity columna | 1.135 | 0.1 | 11.35 | 3.18 |
a Fig. 2 (lane 2) shows the protein profile of purified ALP.
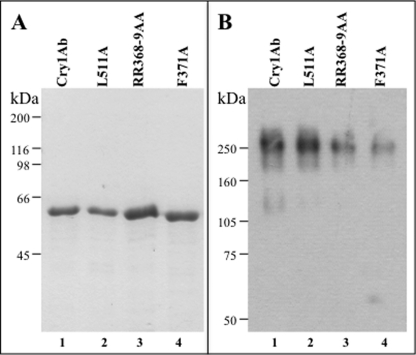
We then analyzed the interaction of the two Cry1Ab structures, corresponding to the oligomeric or monomeric Cry1Ab structures, with both receptors, APN or ALP. The Cry1Ab oligomers were purified by size exclusion chromatography from Cry1Ab protoxin samples activated with M. sexta midgut juice in the presence of a cadherin fragment that contains cadherin repeats (CR) 7–12, whereas monomeric Cry1Ab was prepared by activating Cry1Ab protoxin with trypsin as described under “Experimental Procedures.” Fig. 3 shows the Cry1Ab trypsin-activated 60-kDa monomer and the 250-kDa oligomer after purification (Fig. 3, A and B, respectively, lanes 1).
FIGURE 3.
Cry1Ab domain II loop 2 and domain III mutants are structurally stable. A, SDS-PAGE electrophoresis pattern of trypsin activated Cry1Ab (lane 1), L511A (lane 2), R368A/R369A (lane 3), and F371A (lane 4) toxins. B, Western blot of pure oligomer samples obtained after size exclusion chromatography of toxin samples activated in the presence of cadherin fragment CR7-CR12. Cry1Ab (lane 1), L511A (lane 2), R368A/R369A (lane 3), and F371A (lane 4) toxins are shown. Molecular mass markers are shown at the left of the figure.
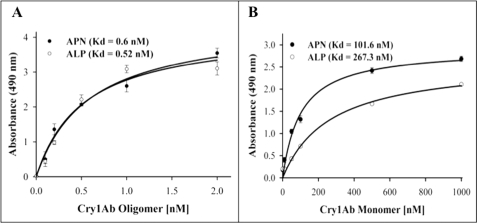
Specific binding of Cry1Ab oligomer to both receptors, APN and ALP, proved to be concentration-dependent and saturable. Oligomer binding to APN was saturated at 1 nm (Fig. 4A), whereas saturable binding of Cry1Ab monomer was observed with a much higher toxin concentration, 400 nm (Fig. 4B). Determination of binding parameters indicated that the oligomer structure bound APN and ALP with an apparent binding affinity (Kd) in the range of 0.5–0.6 nm, whereas the monomer bound APN or ALP molecules with an apparent binding affinity of 100 and 260 nm, respectively. These results suggest that the Cry1Ab oligomeric structure gains at least 200-fold higher affinity to both APN and ALP proteins in comparison with the Cry1Ab monomeric structure.
FIGURE 4.
Binding of Cry1Ab oligomer and monomer structure to APN and ALP proteins. The purified APN (Table 1) and ALP (Table 2) were immobilized on ELISA plates and incubated with Cry1Ab oligomer (A, 0–2 nm) or monomer (B, 0–1000 nm). Kd values obtained by Scatchard analysis are indicated inside the graphs.
Cry1Ab-binding Regions Involved in APN and ALP Interaction
Domain II loop 2 and 3 mutants were previously shown to be affected in APN binding (27, 37, 38). Also, a scFv antibody that bound the domain III β16-β22 region competed the binding of Cry1Ab to APN (10). However, the role of these binding regions in the interaction of Cry1Ab oligomer with APN has not been analyzed, nor whether the same regions are involved in the interaction with ALP. To gain insight on the role of these Cry1Ab epitopes in the binding interaction of Cry1Ab oligomer with both APN and ALP, we characterized the binding of monomeric and oligomeric structures of three Cry1Ab mutants to APN and ALP. Two mutants were located in loop 2 of domain II (R368A/R369A and F371A), and the third mutant was located in the β-16 of domain III (L511A). Toxicity assays showed that the three mutants were not toxic to M. sexta (Table 3). The protoxins of the three Cry1Ab mutants characterized produced a 60-kDa protein after trypsin activation, indicating no major structural constraints for these mutants (Fig. 3A). Although the loop 2 mutants produced lower amounts of oligomer after proteolytic activation in the presence of the cadherin fragment CR7-CR12, enough oligomer was obtained to perform binding assays (Fig. 3B).
TABLE 3.
Toxicity of Cry1Ab and mutant toxins to M. sexta larvae
| Toxin | LC50a | Relative toxicityb |
|---|---|---|
| ng/cm2 | ||
| Cry1Ab | 3.9 (2.2–6.6) | 1 |
| R368A/R369A | >1000 | >260 |
| F371A | >1000 | >260 |
| L511A | >1000 | >260 |
a LC50 indicates the concentration required to kill 50% of the tested larvae. The numbers in parentheses are 95% confidence limits.
b LC50 mutant/LC50 wild type.
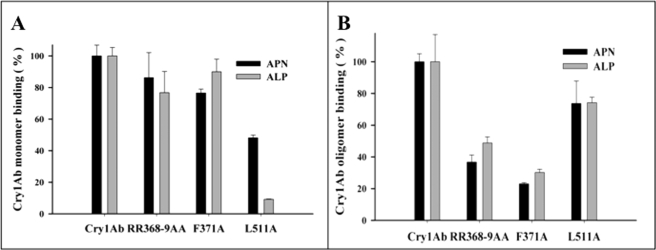
ELISA binding assays of monomeric (25 nm) or oligomeric (0.1 nm) structures of Cry1Ab mutants to APN and ALP were performed under nonsaturated conditions. These studies showed that the monomeric structure of the L511A mutant, located in domain III, was severely affected in binding to ALP protein (Fig. 5A). In contrast, it is clear in this figure that this mutant retained significant binding to APN (Fig. 5A). Fig. 5A also shows that monomeric structures of the domain II loop 2 mutants, F371A and R368A/R369A, were not affected in their binding interaction with both APN and ALP. The binding of oligomeric structures of the Cry1Ab mutants to both APN and ALP revealed that domain II loop 2 mutations are affected on binding to both receptors, whereas the domain III mutant L511A had just a marginal effect in its interaction with both receptors (Fig. 5B).
FIGURE 5.
Binding analysis of Cry1Ab and loop 2 and domain III mutants to APN or ALP. A, ELISA binding assays of 25 nm of monomeric structures of Cry1Ab and mutant toxins to APN (black bars) or ALP (gray bars). B, ELISA binding assays of 0.1 nm of oligomeric structures of Cry1Ab and mutant toxins to APN (black bars) or ALP (gray bars). Standard deviations from three replica plates were obtained.
DISCUSSION
In the pore forming model describing the mode of action of Cry1Ab toxin (33), binding to cadherin receptor results in a conformational change that allows further toxin proteolysis, this cleavage induces the formation of a toxin prepore structure. Then the GPI-anchored receptors, APN and ALP, have been proposed to bind the prepore oligomeric structure and drive it into lipid rafts where pore formation takes place (12, 13, 33, 39). The role of the prepore oligomeric structure in Cry1A toxicity was demonstrated by isolation of single-point mutations in helix α-3, which were affected in oligomerization and were severally affected in toxicity. Also, it was reported that Cry1Ab mutants in helix α-4 were affected in pore formation activity and were inactive. The mutants in helix α-4 are dominant-negative inhibitors of Cry1Ab toxin (5, 40).
We previously reported that the Cry1Ab oligomeric structure bound a protein extract enriched in M. sexta APN activity with an apparent binding affinity of 1 nm, in contrast with the monomeric toxin that bound this protein extract with 160 nm apparent binding affinity. The protein extract was enriched in GPI-anchored proteins because it was prepared by treatment of M. sexta BBMV with phospholipase-C that cleaves out GPI-anchored proteins (13). Nevertheless, we did not rule out the contribution of GPI-anchored ALP to the binding of the Cry1Ab oligomer to that protein extract. To directly determine the properties of toxin receptor binding interaction, the GPI-anchored APN and ALP receptors were purified, and their interaction with Cry1Ab oligomeric or monomeric structures was analyzed. We found that Cry1Ab oligomer bound to APN and ALP from M. sexta with similar high affinities, suggesting that both proteins could act as receptors for Cry1Ab toxin. These results suggest that both APN and ALP could have a similar role in the binding of Cry1Ab prepore oligomer and in mediating toxicity. However, it was also reported that APN and ALP are differentially expressed within M. sexta gut, because APN was preferentially expressed in the posterior part of the gut, whereas ALP was expressed at similar levels through out the gut (41). Interestingly, we show in this work that both APN and ALP have also a differential expression through larval development. ALP was expressed at higher levels than APN in the two first larval instars, whereas APN was the predominant GPI-anchored Cry1Ab binding protein in the fourth and fifth instar (Fig. 1A). The identities of both the 65- and 120-kDa proteins as ALP and APN were confirmed by purification of these proteins and determination of their enzymatic activities (Tables 1 and 2). In addition the 65-kDa band was confirmed to be ALP by mass spectrometry analysis (data not shown). Overall, these data suggest that ALP could have a predominant role in toxicity because toxicity bioassays are performed with neonate larvae where ALP is highly abundant and present in the anterior region of the gut. The susceptibility of M. sexta larvae to Cry1Ab is greatly diminished through larval development, showing lower susceptibility to this toxin in higher instars (42), and these data correlate with the developmental expression pattern of ALP protein (Fig. 1A).
We characterized two Cry1Ab mutants, located in domain II loop 2 (R368A/R369A and F371A), with respect to their binding interaction with APN and ALP. Accordingly, we found that the monomeric structures of these nontoxic mutants were not affected in their binding interaction with APN or ALP. However, these mutations had an important effect on the binding of their corresponding oligomeric structure to APN or ALP. These results show that domain II loop 2 of Cry1Ab oligomer is an important epitope in the interaction with APN and ALP receptors. It was previously reported that F371A mutation did not affect APN binding, but these studies were done only with the monomeric structure of the mutant toxin (37). Also it was proposed that this mutant was affected in membrane insertion, and this effect was used as an argument to say that domain II loop 2 may be participating in triggering membrane insertion (43). Here we show that F371A mutation greatly affects binding of the oligomeric structure to both APN and ALP. Because it was shown that binding of the Cry1A oligomer to APN facilitates insertion of the toxin into the membrane (18), the results presented here suggest that the previously observed effect of F371A mutation in regard to its membrane insertion is due to the severe effects of this mutation on the binding interaction of the oligomeric structure with APN and ALP.
Recently we proposed that APN fulfills two roles in the mode of action of Cry1Ab: first as a low affinity and highly abundant binding site that locates monomeric toxin in the vicinity of the microvilli before cadherin binding and second as a secondary high affinity receptor that mediates insertion of the oligomeric Cry1Ab structure into the membrane, suggesting a ping-pong binding mechanism (38). Interestingly, characterization of nontoxic domain II loop 3 mutants revealed an opposite phenotype when compared with the loop 2 mutations regarding the binding of monomeric or oligomeric Cry1Ab to APN. Domain II loop 3 mutants affected monomer binding to APN in contrast to the binding of the oligomeric structure that was not affected (38). Here we show that the monomeric structures of domain II loop 2 mutations had no effect on their binding to APN or ALP, whereas oligomer binding to both GPI-anchored molecules was greatly affected. These results show that domain II loops 2 and 3 mediate different steps of Cry1Ab interaction with GPI-anchored receptors depending on their oligomeric state.
In relation to the interaction of domain III with APN and ALP, previous work identified domain III β-16 of Cry1Ab as an important binding determinant to M. sexta APN (10). The nontoxic L511A mutant is located in this β16, and this mutant was specially affected in its interaction with ALP retaining significant binding to APN (Fig. 5A). The severe effect on toxicity shown by L511A mutant indicates an important role for ALP binding in vivo and suggests that ALP has a predominant role, in comparison with APN, on Cry1Ab toxicity. Nevertheless, the precise role of both GPI-anchored proteins on Cry1Ab toxicity should be determined in the future; probably toxicity assays of larvae that are specifically silenced by RNA interference for either APN or ALP could help in answering this specific question.3
Acknowledgments
We thank Josué Océlotl for isolation of Cry1Ab L511A mutant and Jorge Sánchez and Lizbeth Cabrera for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01 AI066014. This work was also supported by Consejo Nacional de Ciencia y Tecnología Grants 83135 and 81639, Dirección General de Asuntos del Personal Académico–UNAM Grants IN218608 and IN210208, and United States Department of Agriculture Grant 2207-35607-17780.
B. Flores-Escobar, A. Bravo, M. Soberón, and I. Gómez, personal communication.
- GPI
- glycosylphosphatidylinositol
- APN
- aminopeptidase-N
- ALP
- alkaline phosphatase
- BBMV
- brush border membrane vesicles
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- ELISA
- enzyme-linked immunosorbent assay
- BBMV
- brush border membrane vesicle(s)
- PBS
- phosphate-buffered saline
- CR
- cadherin repeat(s).
REFERENCES
- 1.Schnepf E., Crickmore N., Van Rie J., Lereclus D., Baum J., Feitelson J., Zeigler D. R., Dean D. H. (1998) Microbiol. Mol. Biol. Rev. 62, 775–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo S., Ye S., Liu Y., Wei L., Xue J., Wu H., Song F., Zhang J., Wu X., Huang D., Rao Z. (2009) J. Struct. Biol. 168, 259–266 [DOI] [PubMed] [Google Scholar]
- 3.de Maagd R. A., Bravo A., Berry C., Crickmore N., Schnepf H. E. (2003) Annu. Rev. Genet. 37, 409–433 [DOI] [PubMed] [Google Scholar]
- 4.Bravo A., Gill S. S., Soberón M. (2007) Toxicon 49, 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiménez-Juárez N., Muñoz-Garay C., Gómez I., Saab-Rincon G., Damian-Almazo J. Y., Gill S. S., Soberón M., Bravo A. (2007) J. Biol. Chem. 282, 21222–21229 [DOI] [PubMed] [Google Scholar]
- 6.Gazit E., La Rocca P., Sansom M. S., Shai Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12289–12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grochulski P., Masson L., Borisova S., Pusztai-Carey M., Schwartz J. L., Brousseau R., Cygler M. (1995) J. Mol. Biol. 254, 447–464 [DOI] [PubMed] [Google Scholar]
- 8.Rausell C., Pardo-López L., Sánchez J., Muñoz-Garay C., Morera C., Soberón M., Bravo A. (2004) J. Biol. Chem. 279, 55168–55175 [DOI] [PubMed] [Google Scholar]
- 9.Herrero S., González-Cabrera J., Ferré J., Bakker P. L., de Maagd R. A. (2004) Biochem. J. 384, 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gómez I., Arenas I., Benitez I., Miranda-Ríos J., Becerril B., Grande R., Almagro J. C., Bravo A., Soberón M. (2006) J. Biol. Chem. 281, 34032–34039 [DOI] [PubMed] [Google Scholar]
- 11.Rajamohan F., Lee M. K., Dean D. H. (1998) Prog. Nucleic Acid Res. Mol. Biol. 60, 1–27 [DOI] [PubMed] [Google Scholar]
- 12.Jurat-Fuentes J. L., Adang M. J. (2006) J. Invertebr. Pathol. 92, 166–171 [DOI] [PubMed] [Google Scholar]
- 13.Bravo A., Gómez I., Conde J., Muñoz-Garay C., Sánchez J., Miranda R., Zhuang M., Gill S. S., Soberón M. (2004) Biochim. Biophys. Acta 1667, 38–46 [DOI] [PubMed] [Google Scholar]
- 14.Gómez I., Pardo-López L., Muñoz-Garay C., Fernandez L. E., Pérez C., Sánchez J., Soberón M., Bravo A. (2007) Peptides 28, 169–173 [DOI] [PubMed] [Google Scholar]
- 15.Gómez I., Sánchez J., Miranda R., Bravo A., Soberón M. (2002) FEBS Lett. 513, 242–246 [DOI] [PubMed] [Google Scholar]
- 16.Soberón M., Pardo-López L., López I., Gómez I., Tabashnik B. E., Bravo A. (2007) Science 318, 1640–1642 [DOI] [PubMed] [Google Scholar]
- 17.Pigott C. R., Ellar D. J. (2007) Microbiol. Mol. Biol. Rev. 71, 255–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pardo-López L., Gómez I., Rausell C., Sanchez J., Soberón M., Bravo A. (2006) Biochemistry 45, 10329–10336 [DOI] [PubMed] [Google Scholar]
- 19.Knight P. J., Crickmore N., Ellar D. J. (1994) Mol. Microbiol. 11, 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sangadala S., Walters F. S., English L. H., Adang M. J. (1994) J. Biol. Chem. 269, 10088–10092 [PubMed] [Google Scholar]
- 21.Schwartz J. L., Lu Y. J., Söhnlein P., Brousseau R., Laprade R., Masson L., Adang M. J. (1997) FEBS Lett. 412, 270–276 [DOI] [PubMed] [Google Scholar]
- 22.Gill M., Ellar D. (2002) Insect Mol. Biol. 11, 619–625 [DOI] [PubMed] [Google Scholar]
- 23.Jenkins J. L., Lee M. K., Sangadala S., Adang M. J., Dean D. H. (1999) FEBS Lett. 462, 373–376 [DOI] [PubMed] [Google Scholar]
- 24.You T. H., Lee M. K., Jenkins J. L., Alzate O., Dean D. H. (2008) BMC Biochem. 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNall R. J., Adang M. J. (2003) Insect Biochem. Mol. Biol. 33, 999–1010 [DOI] [PubMed] [Google Scholar]
- 26.Krishnamoorthy M., Jurat-Fuentes J. L., McNall R. J., Andacht T., Adang M. J. (2007) Insect Biochem. Mol. Biol. 37, 189–201 [DOI] [PubMed] [Google Scholar]
- 27.Lee M. K., Rajamohan F., Jenkins J. L., Curtiss A. S., Dean D. H. (2000) Mol. Microbiol. 38, 289–298 [DOI] [PubMed] [Google Scholar]
- 28.Jenkins J. L., Lee M. K., Valaitis A. P., Curtiss A., Dean D. H. (2000) J. Biol. Chem. 275, 14423–14431 [DOI] [PubMed] [Google Scholar]
- 29.Wolfersberger M. G. (1993) Arch. Insect Biochem. Physiol. 24, 139–147 [DOI] [PubMed] [Google Scholar]
- 30.Valaitis A. P., Lee M. K., Rajamohan F., Dean D. H. (1995) Insect Biochem. Mol. Biol. 25, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 31.Landt M., Boltz S. C., Butler L. G. (1978) Biochemistry 17, 915–919 [DOI] [PubMed] [Google Scholar]
- 32.Asgeirsson B., Adalbjörnsson B. V., Gylfason G. A. (2007) Biochim. Biophys. Acta 1774, 679–687 [DOI] [PubMed] [Google Scholar]
- 33.Bravo A., Miranda R., Gómez I., Soberón M. (2002) Biochim. Biophys. Acta 1562, 63–69 [DOI] [PubMed] [Google Scholar]
- 34.Lereclus D., Arantès O., Chaufaux J., Lecadet M. (1989) FEMS Microbiol. Lett. 51, 211–217 [DOI] [PubMed] [Google Scholar]
- 35.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 36.Pacheco S., Gómez I., Gill S. S., Bravo A., Soberón M. (2009) Peptides 30, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins J. L., Dean D. H. (2000) in Genetic Engineering: Principles and Methods (Setlow J. K. ed) Vol. 22, pp. 33–54, Plenum Press, New York: [DOI] [PubMed] [Google Scholar]
- 38.Pacheco S., Gómez I., Arenas I., Saab-Rincon G., Rodríguez-Almazán C., Gill S. S., Bravo A., Soberón M. (2009) J. Biol. Chem. 284, 32750–32757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuang M., Oltean D. I., Gómez I., Pullikuth A. K., Soberón M., Bravo A., Gill S. S. (2002) J. Biol. Chem. 277, 13863–13872 [DOI] [PubMed] [Google Scholar]
- 40.Rodríguez-Almazán C., Zavala L. E., Muñoz-Garay C., Jiménez-Juárez N., Pacheco S., Masson L., Soberón M., Bravo A. (2009) PloS One 4, e5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J., Brown M. R., Hua G., Adang M. J. (2005) Cell Tissue Res. 321, 123–129 [DOI] [PubMed] [Google Scholar]
- 42.Griko N., Zhang X., Ibrahim M., Midboe E. G., Bulla L. A., Jr. (2008) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 151, 59–63 [DOI] [PubMed] [Google Scholar]
- 43.Nair M. S., Liu X. S., Dean D. H. (2008) Biochemistry 47, 5814–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]