Abstract
The AIRE protein plays a remarkable role as a regulator of central tolerance by controlling the promiscuous expression of tissue-specific antigens in thymic medullary epithelial cells. Defects in the AIRE gene cause the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, a rare disease frequent in Iranian Jews, Finns, and Sardinian population. To this day, the precise function of the AIRE protein in regulating transcription and its interacting proteins has yet to be entirely clarified. The knowledge of novel AIRE interactors and their precise role will improve our knowledge of its biological activity and address some of the foremost autoimmunity-related questions. In this study, we have used a yeast two-hybrid system to identify AIRE-interacting proteins. This approach led us to the discovery of a new AIRE-interacting protein called DAXX. The protein is known to be a multifunctional adaptor with functions both in apoptosis and in transcription regulation pathways. The interaction between AIRE and DAXX has been validated by in vivo coimmunoprecipitation analysis and colocalization study in mammalian cells. The interaction has been further confirmed by showing in transactivation assays that DAXX exerts a strong repressive role on the transcriptional activity of AIRE.
Keywords: Protein/Protein-Protein Interactions, Protein/Repressor, Protein/Zinc Finger, Transcription/Regulation, Transcription/Repressor, AIRE Autoimmune Regulator, DAXX Protein
Introduction
Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (MIM 240300) is a rare recessive disease characterized by a variable combination of autoimmune endocrine tissues and liver destruction, mucocutaneous candidiasis, and ectodermal dystrophies (1–3). The defective gene, called AIRE (autoimmune regulator; MIM 607358), is a transcriptional regulator that coordinates the expression of a set of tissue-specific antigens in medullary thymic epithelial cells where self-reactive T cells experience negative selection. The absence of thymic AIRE expression leads to the escape of autoreactive T cells and results in manifest autoimmunity (4–7). Therefore, AIRE is a key molecule in the establishment of immunological tolerance. AIRE protein consists of multiple structural domains, conserved in the mouse homologue, which are indicative of a role as transcriptional regulator (8). AIRE is detected in the nucleus, where it is localized in the nuclear bodies associated with the nuclear matrix fraction of the cells (9, 10). Indeed, AIRE contains a potential bipartite nuclear localization signal, consisting of amino acids 110–114 and 131–133, even though only the latter part constitutes a functional nuclear localization signal (11). AIRE shares several domains with members of the Sp100 family of proteins, such as HSR, SAND, and PHD.3 The Sp100 family of proteins is a group of transcriptional regulators involved in both transcriptional activation and repression. The AIRE N-terminal HSR domain is an integral domain that drives homodimerization, subcellular localization, and protein-protein interactions (9, 12). Recently, the HSR domain function has been better defined by alignment and homology modeling studies, and for this reason the motif has been renamed the CARD domain (13). The CARD domain is a functional structure required for the correct function of signaling machineries that trigger apoptosis, inflammation, and innate immune recognition. The SAND domain is characteristic of proteins involved in chromatin-dependent transcriptional regulation and contains a conserved KDWD motif essential for DNA recognition (14). In addition to the DNA binding property, the SAND domain cooperates with the HSR/CARD domain in the homodimerization and nuclear localization function of AIRE (9, 12). AIRE contains two PHD zinc finger-type motifs that are known to be chromatin remodeling factors, indicating again that AIRE operates as a transcriptional regulator (15). AIRE PHDs are multifunctional domains, with transactivation and repression activity (16–18). In addition, it is debated whether the AIRE PHD behaves as an E3 ubiquitin ligase (19, 20). Interestingly, it has been recently shown by nuclear magnetic resonance solution structure that AIRE PHD1 binds unmethylated histone H3K4me0, an usual target of repressor factors involved in keeping chromatin in the inactive state (21–23). However, AIRE binding to H3-K4me0 is associated with methylation of Lys-4 leading to activation instead of repression of the adjacent chromatin (24). Finally, the AIRE protein includes four LXXLL motifs that mediate the binding of several coactivators to the nuclear receptors. The last LXXLL motif and the flanking PXXPXP sequence are essential for the transactivation capacity of AIRE (18). This finding is supported by the presence in this region of an autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy causing mutation (25). Previous studies have shown that AIRE is present in soluble high molecular weight complexes, and this evidence suggests the existence of several interacting proteins (9). A very recent work has just identified, via AIRE-targeted coimmunoprecipitation, a wide set of interactors that belong to many classes such as nuclear transport, chromatin binding/structure, transcription, and pre-mRNA processing (26). However, to this day, just a few AIRE protein partners have been functionally characterized, and those needed to be mentioned are as follows: 1) a heterotrimeric complex of DNA-dependent protein kinase, consisting of Ku70, Ku80, and the DNA protein kinase catalytic subunit responsible for the phosphorylation of the AIRE protein at Thr-68 and Ser-156 that modulate AIRE transactivation ability (27); 2) PIAS1 (protein inhibitor of activated STAT) protein that functionally interacts with AIRE to regulate the activity of AIRE target genes (28); 3) p63 protein, which interacts with the SAND domain of AIRE to regulate human leukocyte antigen class II genes (29); and 4) CREB-binding protein (CBP), a general transcriptional coregulator with histone acetyltransferase properties. CBP has been reported to bind AIRE directly in vitro. CBP colocalizes with AIRE into the nuclear bodies in monocytic cell cultures, and when coexpressed with AIRE, it enhances the transcription of reporter genes (17, 31). Despite the great amount of work on the AIRE protein, its precise mechanism of action still remains unsolved. Therefore, to clarify the functional role of AIRE and identify potential partners, we performed a yeast two-hybrid screen and isolated DAXX, a protein involved in apoptosis and in transcription regulation. The interaction between AIRE and DAXX was confirmed in vivo by coimmunoprecipitation and colocalization studies. Furthermore, by transactivation analysis, we established that DAXX protein acts as a repressor of AIRE.
MATERIALS AND METHODS
Plasmids
Plasmids Used in Yeast Two-hybrid Assay
The AIRE cDNA (aa 1–517) and deletion fragments of AIRE (1–413, 1–161, 1–100, 12–517, and 68–517) were amplified by PCR using primers that carry NcoI and EcoRI restriction sites. The fragments were then cloned into pGBKT7-DNA-BD vector (Clontech). The full-length cDNA of DAXX protein was cloned into the pGAD expression vector (Clontech) using EcoRI and NotI restriction sites. pGAD-DAXX cod629stop and pGBKT7-AIRE1–517-W78R were obtained by site-specific mutagenesis.
Plasmids Used in Luciferase Assay
Full-length AIRE was cloned into EcoRI and NotI cloning sites of pEF5HA plasmid that was kindly provided by Nunzio Bottini (La Jolla Institute for Allergy and Immunology, La Jolla, CA). pEFHA vector allows the expression of constructs in fusion with an N-terminal HA tag and under control of the EFα promoter. The human full-length DAXX cDNA cloned into pCMV-XL5 plasmid was purchased from OriGene (Rockville, MD). The insulin promoter (pMG3-insulin/Luc) reporter plasmid (kindly provided by Paolo Moi, University of Cagliari, Italy) carries a single copy of the insulin promoter region that is cloned upstream of the luciferase reporter gene. Control pGL4 plasmid, driving constitutive expression of Renilla luciferase, was purchased from Promega.
Plasmids Used in Colocalization Assay
The full-length AIRE cDNA was amplified and cloned in Monster green fluorescent protein expression vector (Promega) carrying EcoRV restriction site. pCMV-XL5-DAXX was purchased from OriGene (Rockville, MD).
Plasmids Used in Coimmunoprecipitation Assay
The full-length AIRE cDNA was amplified and cloned into pEF5HA using primers that carry EcoRI-NotI restriction sites and into the EcoRI site of pcDNA3.1 V5/C (Invitrogen).
The AIRE constructs lacking 28, 132, 384, and 445 C-terminal amino acids were obtained by site-specific mutagenesis performed on pEF5HA AIRE full-length construct. The constructs lacking 11 and 67 N-terminal amino acids were obtained by PCR and cloned in pEF5HA vector. pCDNA 3.1 Xpress DAXX was prepared by PCR amplification with primers carrying EcoRI-Xba-I sites, and pCMV-XL5-DAXX was purchased from OriGene (Rockville, MD).
Plasmids Used in GST-Pulldown Assay
The pGEX-2T-AIRE aa 1–161 was created by site-specific mutagenesis that inserted a stop codon at position 162 in the pGEX-2T-AIRE full-length construct.
Antibodies
Polyclonal antibody against DAXX was purchased from Cell Signaling Technology (Boston, MA); anti-V5 monoclonal antibody was from Invitrogen; anti-HA monoclonal antibody (clone 16B12) was from Covance (Berkeley, CA); polyclonal anti-HDAC1 and HDAC2 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-rabbit Ig Texas Red secondary antibody, anti-mouse, and anti-rabbit secondary antibody were from GE Healthcare. Anti-actin antibody was purchased from Sigma.
Two-hybrid Assay Screens
Yeast two-hybrid assays were performed using GAL4 Matchmaker Two-hybrid System 3 (Clontech). A human thymus Matchmaker cDNA library in the pACT2 prey plasmid (Clontech) was screened for proteins that interact with AIRE (aa 1–517) in the pGBKT7-DNA-BD bait plasmid using the AH109 yeast reporter strain. In this study we used an AIRE construct lacking 28 C-terminal amino acids (AIRE aa 1–517) to eliminate the autoactivation phenomenon of the full-length AIRE. The yeast cells were transformed by the lithium acetate method and were plated on synthetic dropout (SD) medium lacking leucine and tryptophan (low stringency). The growing colonies were selected on synthetic dropout (SD) medium lacking leucine tryptophan, histidine, and adenine (high stringency) allowing the production of α-galactosidase. Positive clones were selected, and plasmid DNA was isolated and fully sequenced. Seven clones were found to encode the DAXX protein and precisely its C-terminal region (aa 629–740).
To map the AIRE interaction domain, PCR fragments encoding deletion derivatives of AIRE were amplified and subcloned into pGBKT7 DNA-BD; the mutant form of pGBKT7-AIRE (pGBKT7-AIRE-(1–517)-W78R) was obtained by site-specific mutagenesis. All constructs were cotransformed with a full-length pGAD-AD-DAXX in AH109 yeast strain. To establish whether the Ser/Pro/Thr-rich region of DAXX protein is the only domain involved in AIRE interaction, we produced a pGAD-DAXX construct where a stop codon at position 629 was created by site-specific mutagenesis. This construct lacking 111 amino acids was cotransformed with pGBKT7AIRE (aa 1–517) and pGBKT7-AIRE (aa 1–161) in AH109 yeast strains.
α-Galactosidase Activity Assay
The quantitative α-galactosidase activity assay was performed using p-nitrophenyl α-d-galactopyranoside as substrate according to the Clontech manual with some minor modifications. Noninteracting proteins grown on synthetic dropout (SD) medium lacking leucine and tryptophan as well as interacting proteins grown on synthetic dropout (SD) medium lacking leucine, tryptophan, histidine, and adenine were inoculated in the respective culture medium from a single fresh yeast colony and incubated overnight at 30 °C. The supernatant was harvested by centrifugation at 13,000 × g for 2 min, and 400 μl of the supernatant was mixed with 600 μl of fresh prepared Assay Buffer (2:1 ratio of 0.5 m sodium acetate, pH 4.5, to 100 mm p-nitrophenyl α-d-galactopyranoside (Sigma)) and incubated at 30 °C for 4 h. The reaction was stopped with the addition of 200 μl of 1 m Na2CO3. The A410 nm of each reaction was recorded by a spectrophotometer. Yeast transformations were performed in duplicate, and each yeast colony was assayed in triplicate.
Cell Culture and Transient Transfections
HeLa and COS-1 cells were grown in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine (Invitrogen) at 37 °C and 5% CO2. All transfections were performed by Lipofectamine 2000 method according to the manufacturer's instructions (Invitrogen). Each experiment was performed at least three times, and within each experiment, transfections were performed in quadruplicate.
Proteasome Inhibition Assay
COS-1 cells (∼3 × 105) were transiently transfected with 1.5 μg of pEF5HA-AIRE aa 1–100. 24 h from transfection, cells were treated with 2 and 5 μm proteosome inhibitor benzyloxycarbonyl-Leu-Leu-Leu-H (MG132; Sigma) for 3 h or left untreated. Cells were than harvested and lysed in sample buffer, and the lysates were analyzed by anti-HA Western blotting.
Luciferase Assay
300 ng of pMG3-insulin/Luc reporter plasmid and 50 ng of control pGL4 plasmid were cotransfected in HeLa and COS-1 cells with the appropriate amount of HA-tagged full-length AIRE (2 μg) and variable amounts of DAXX construct ranging from 100 to 500 ng using the Lipofectamine 2000 method. After incubation for 24 h, cells were washed with 1× PBS and lysed. Luciferase assays were performed on cell lysates using the Dual-Luciferase reporter assay kit from Promega (Madison, WI) according to the manufacturer's instructions, and reading luminescence on MicroLumat LB 96P luminometer (Berthold). The ratio between firefly and Renilla luciferase activity was then plotted against the expression of constructs assessed by densitometric scanning of anti-HA and anti-DAXX blots of total lysates. For deacetylation inhibition studies, trichostatin A (TSA) (Sigma) was added 5 h following transfections at a final concentration of 100 ng/ml and incubated for 24 h.
Immunofluorescence
1.5 × 105 HeLa cells seeded on culture slides were transfected with 320 ng of GFP-AIRE and 320 ng of pXL5-DAXX. After 48 h of transfection, HeLa cells were fixed with methanol for 5 min on ice and with ethanol at room temperature, permeabilized with PBS containing 1% Triton X-100 for 10 min at room temperature, and blocked for 45 min with 8% bovine serum albumin in PBS. Anti-DAXX staining was performed by incubation for 1 h at 37 °C with anti-DAXX polyclonal antibody (Cell Signaling Technology) diluted 1:200 in PBS containing 3% bovine serum albumin. The antigen-primary antibody complex was detected by incubating for 20 min at 37 °C with anti-rabbit Ig Texas Red secondary antibody (GE Healthcare) diluted 1:50 in 3% bovine serum albumin in PBS. The nucleus were stained with 4′,6-diamidino-2-phenylindole diluted 1:1000 in PBS for 5 min at room temperature. The culture slides were mounted with a coverglass in Vectashield mounting medium (Vector Laboratories). Microscopic observation was performed under a confocal laser-scanning microscope LSM Leica equipped with fluorescein isothiocyanate and TRITC filter sets.
Coimmunoprecipitation and Homodimerization Inhibition Assay
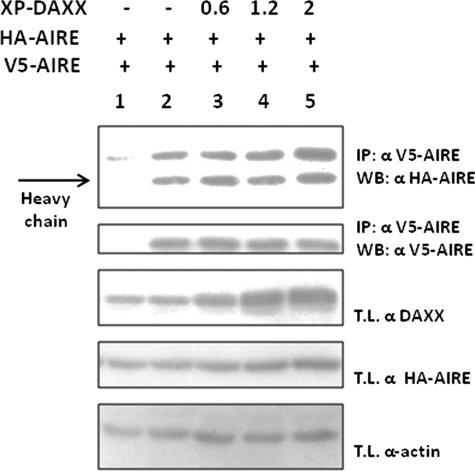
COS-1 cells (∼106) transfected with HA-AIRE constructs described above and pXL5-DAXX were lysed on ice in 0.5 ml of lysis buffer containing 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.2% Nonidet P-40, 5 μm ZnCl2, 30 mm Na4P2O7, 50 mm NaF, 2 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors tablet (Roche Applied Science). Cell lysates were centrifuged at 12,000 × g for 20 min at 4 °C, and supernatants (500–700 μg of protein extracts for each experiment) were preclarified with 50 μl of 50% protein G-Sepharose 4B resin (GE Healthcare) for 4 h at 4 °C. After centrifugation, 3 μl of anti-HA antibody (Covance) were added to the supernatant followed by overnight agitation at 4 °C. To recover protein complexes, 50 μl of 50% protein G-Sepharose 4B resin were added into each sample and incubated by agitation for additional 4 h at 4 °C. Resin was washed four times with PBS containing 0.2% Nonidet P-40, and the immune complexes were released from the resin by boiling in 30 μl of 2× sample buffer. Samples were analyzed by 10% SDS-PAGE and Western blotting using polyclonal antibody against DAXX (diluted 1:3000) using ECL Plus chemiluminescence method (GE Healthcare). In the homodimerization inhibition assay, COS-1 cells were transfected with a constant amount of HA-AIRE and V5-AIRE as well as increasing amounts of Xpress-DAXX (0.6, 1.2, and 2.0 μg). IPs were performed from total lysates of transfected cells using 1 μg of anti-V5 antibody.
GST-Pulldown
Expression of GST fusion proteins were performed following the manufacturer's instructions (GE Healthcare). n brief, Escherichia coli strain BL21SI was transformed with pGEX-2T-GST and pGEX-2T-AIRE (aa 1–160) in 2× YTA broth containing 100 μg/ml ampicillin and induced for protein expression with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 30 °C for 3 h. Bacterial cells were harvested and lysed. Appropriate volumes of MagneGST-glutathione beads (Promega) were added to the supernatants and incubated for 30 min at 4 °C. After washing, the purified proteins were eluted from beads by incubation with 200 μl of Elution Buffer containing the following: 100 mm glutathione, 50 mm Tris-HCl, pH 8.1, 1% Triton X-100, 100 mm NaCl. Then 2 or 5 μg of each purified GST fusion protein were bound with 5 or 10 μl of MagneGST-glutathione beads (Promega) for 1 h in 1× PBS at 4 °C. The proteins-beads complexes were blocked for 1 h with 5% bovine serum albumin in 1× PBS and then incubated with 10 μl of radiolabeled DAXX protein produced with the TnT system (Promega) for 2 h at 4 °C in Binding Buffer containing the following: 10 mm Hepes, pH 8, 0.5 m NaCl, 1 mm MgCl2, 12,5% glycerol, 0.5 mm dithiothreitol. After extensive washes with 1% Nonidet P-40 in 1× PBS, the GST fusion protein-DAXX complex was removed from the beads by competition using 20 μl of Elution Buffer. 10 μl of eluted solution were added with 4 μl of 2× Sample Buffer and analyzed in 12% SDS gel. The gel was than exposed to autoradiography after being fixed and dried.
RESULTS
HSR/CARD Domain of AIRE Interacts with Ser/Pro/Thr-rich Domain of DAXX
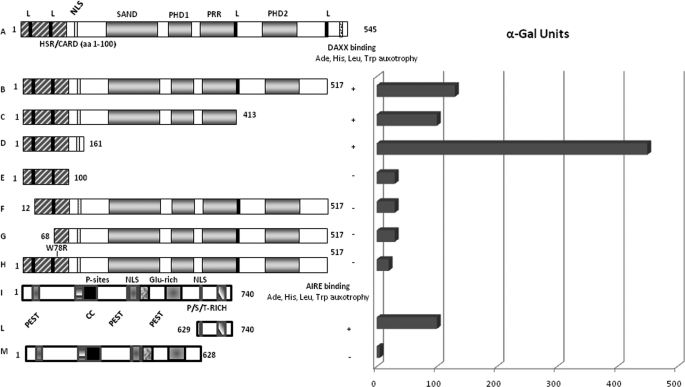
Here, we sought to assess potential AIRE-interacting proteins through the yeast two-hybrid assay. In detail, a GAL4-DNA-BD AIRE fusion protein (pGBKT7-AIRE-(1–517)) was used as a bait to screen a GAL4-AD-tagged cDNA expression library of human thymus where AIRE is highly expressed. We identified seven positive clones that encoded the C-terminal region of DAXX protein (aa 629–740) corresponding to a Ser/Pro/Thr-rich domain. The interaction was then confirmed by retransformation (Fig. 1).
FIGURE 1.
AIRE interacts with DAXX in yeast two-hybrid assay. Left panel, schematic representation of AIRE and DAXX constructs. A and I, full-length AIRE and DAXX. B, AIRE construct used to screen human thymus library. C–H, AIRE constructs used to map interaction domains. L, DAXX fragment that interacts with AIRE in yeast two-hybrid assay. M, DAXX construct used to map interaction domain. Right panel, quantitative two hybrid assay. α-Galactosidase units detected by the quantitative assay are shown as gray bars. The constructs used in each assay correspond to the aligned left panel. Plus and minus symbols indicate the transformant capabilities to grow on high stringency medium through ADE and HIS reporter gene activation. CC, coiled coil; NLS, nuclear localization signal; PST, Pro/Ser/Thr.
DAXX is a ubiquitously expressed protein with a particularly high expression in the thymus and testes (32). It colocalizes predominantly in PML-nuclear bodies where together with PML the protein influences apoptosis and transcription (33, 34). However, several DAXX-interacting proteins are not located solely in the PML- nuclear bodies indicating that DAXX is also localized in alternative nuclear structures. DAXX was initially identified as a binding protein of Fas death domain and was shown to potentiate Fas-mediated apoptosis (32). Paradoxically, Daxx−/− mice showed embryonic lethality related to widespread apoptosis. These data taken together indicate that DAXX has antiapoptotic function and is essential for embryonic development (35). Biochemically, DAXX may act as a transcriptional regulator that can mainly repress transcription (36–39). Thus, DAXX might regulate cellular processes by modulating transcription of specific genes under different conditions and has been found to take part of a large multiprotein complex that includes core histones and histone deacetylase complexes (HDACs) (40), as well as a chromatin remodeling complex that includes the ATRX protein (41, 42). DAXX can also associate with sumoylated CBP and represses CBP transcriptional activity through HDAC2 recruitment (43). On the other hand, DAXX-mediated transcriptional repression can be mitigated by sumoylated PML that attracts DAXX to the nuclear domain (PODs) (44). However, the mechanism by which DAXX binds to sumoylated PML and sumoylated transcription factors is still largely unclear. As DAXX associates directly with a number of DNA binding transcription factors, including Pax3 and Pax5 (36, 37), ETS1 (38), p53 (39), and its family members p73 and p63 (45), glucocorticoid receptor (46), androgen receptor (47), and Smad4 (48), it is likely that it represents a crucial link between DNA binding factors and transcriptional regulatory complexes. DAXX-mediated transcriptional repression is due to interaction with HDAC (40), and indeed the SPT domain of DAXX recruits histone deacetylases leading to inactive chromatin structure and transcriptional repression.
We carried out deletion studies to map the AIRE domains involved in the interaction with DAXX. PCR fragments encoding deletion derivatives of AIRE were subcloned into pGBKT7-GAL4 DNA-BD and cotransformed in yeast cells with full-length pGAD GAL4-AD-DAXX. We used six truncated constructs lacking both the C- and N-terminal region of the protein and a construct mutated in the HSR/CARD domain (W78R). Only the constructs including the N-terminal portion of the protein are capable to interact with DAXX protein. This region mapped to aa 1–161 contains both the HSR/CARD domain and the bipartite nuclear localization signal motif. Notably the construct carrying the HSR/CARD domain alone failed to interact with DAXX probably because the absence of the nuclear localization signal prevents the proper cellular localization of AIRE needed for the DAXX interaction. Previous studies carried out by the Ilmarinen et al. (11) have clearly shown that specific mutations within the nuclear localization signal prevented the protein from entering the nucleus. Very interestingly, the protein carrying the W78R mutation, known to impair AIRE homodimerization (9), did not interact with DAXX; this fact underlines the importance of HSR/CARD domain. We thus hypothesize that the interaction between AIRE and DAXX might occur as consequence of the correct dimerization of AIRE. These data further strengthen the HSR/CARD importance in the AIRE-DAXX interaction. To establish if the Ser/Pro/Thr-rich region of DAXX protein is the only domain involved in AIRE interaction, we produced by site-specific mutagenesis a pGAD-DAXX construct with a stop codon at position 629. This construct, which lacks the 111 aa of the C-terminal moiety, did not produce transformants when cotransformed with pGBKT7-AIRE (aa 1–517). These results indicated that the Ser/Pro/Thr-rich region is the only domain involved in the interaction with AIRE (Fig. 1). We quantified the strength of interaction by liquid α-galactosidase assay. Unexpectedly, the construct that contains only the DAXX binding domain interacts three times stronger than the full-length AIRE protein, probably because of a better exposure of the domain (Fig. 1).
AIRE and DAXX Interact in Mammalian Cells
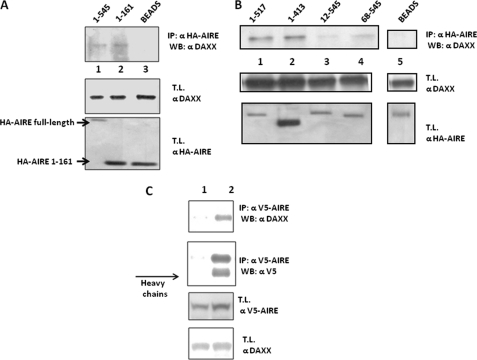
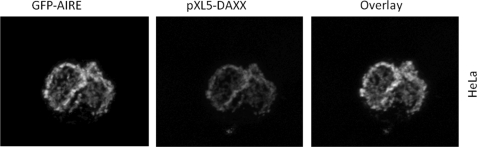
To investigate the complex formation in vivo, we expressed in transiently transfected COS-1 cells pXL5-DAXX and full-length HA-tagged N- and C-terminal deletions of AIRE. HA-AIRE constructs were precipitated from cell lysates with anti-HA antibody. Immunoprecipitates (IPs) were analyzed for the presence of DAXX protein by Western blotting. Only the AIRE constructs containing the intact N-terminal portion interacted with DAXX. Hence, these data indicate that AIRE and DAXX interact “in vivo” (Fig. 2, A and B). Immunoprecipitated fractions were silver-stained to detect fainter bands corresponding to the potential AIRE-DAXX complex partners. Attempts to characterize the additional bands by mass spectrometry technology failed to identify other partners. The AIRE construct that contains only the first 100 amino acids did not express in mammalian cells. Lack of expression could be explained by the proteasome degradation of ubiquitinylated AIRE as it was restored in a proteasome inhibition assay (Fig. 3). It is noteworthy that the same construct, when expressed in yeast and prokaryotic cells as a GAL4 or GST fusion protein, was stabilized and correctly expressed (data not shown). By transient expression of V5-AIRE constructs and immunoprecipitation with anti-V5 from total cell lysates, we found that AIRE interacts also with endogenous DAXX. Indeed, DAXX protein was detected by Western blots in immunoprecipitated fractions (Fig. 2C). To further confirm this finding, we performed a colocalization study in HeLa cells using a fusion GFP-AIRE construct. The AIRE-DAXX complex was detected by anti-rabbit DAXX primary antibody and anti-rabbit secondary antibody conjugated with Texas Red fluorochrome. Confocal microscopy images showed the colocalization of AIRE and DAXX within the nucleus (Fig. 4).
FIGURE 2.
Coprecipitation of DAXX with AIRE in COS-1 cells. A, coprecipitation of transfected DAXX with transfected HA-AIRE and HA-AIRE-(1–161) in COS cells. Cells were transfected with full-length HA-AIRE and HA-AIRE (aa 1–161). IP was performed from lysates using antibodies against HA (lanes 1 and 2) or beads only (lane 3). Upper panel shows the coprecipitated DAXX of the HA-AIRE IPs. 2nd and 3rd panel from the top show the relative amount of transfected DAXX and transfected HA-AIRE. WB, Western blot. T.L., total lysate. B, coprecipitation of transfected DAXX and HA-AIRE constructs in COS cells. Cells were transfected with HA-AIRE and DAXX. Lanes 1 and 2 of the top panel show DAXX amounts of anti-HA immunoprecipitations performed on cell lysates transfected with HA-AIRE constructs lacking C-terminal region; lanes 3 and 4 of the same panel show DAXX amount of anti-HA IPs performed on cell lysates transfected with HA-AIRE constructs lacking N-terminal region. 2nd and 3rd panel from the top show the transfected amount of DAXX and HA-AIRE respectively. C, coprecipitation of endogenous DAXX with transfected V5-AIRE in COS cells. Cells were transfected with V5-AIRE, and IP was performed from lysates using antibodies against V5 (lane 2) or beads only (lane 1). Upper panel shows the coprecipitated endogenous DAXX of the V5-AIRE IP. 2nd panel from the top shows the immunoprecipitated V5-AIR, and lower panels show anti-V5-AIRE expression and endogenous DAXX levels on total lysates.
FIGURE 3.
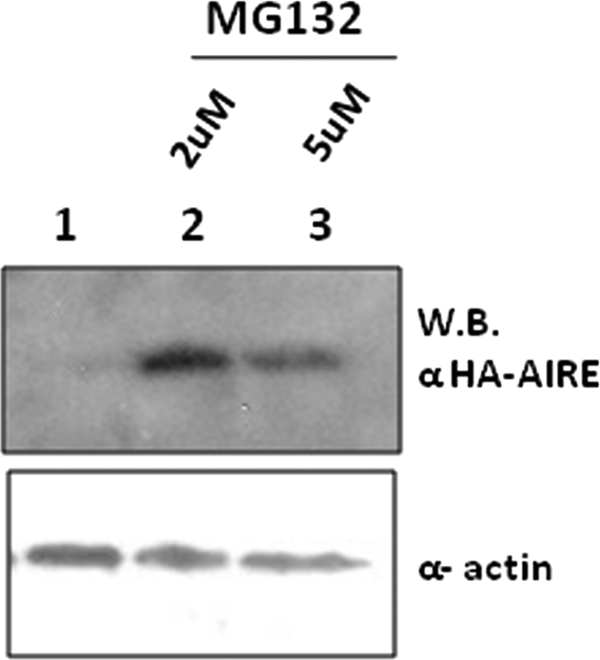
HA-AIRE (aa 1–100) is ubiquitinylated in COS-1 cells. COS-1 cells were transfected with HA-AIRE (aa 1–100). After 24 h from transfection cells were treated with 2 and 5 μm MG132 (lanes 2 and 3) or left untreated (lane 1). Upper panel shows anti-HA-AIRE Western blotting (WB). Bottom panel shows the relative amounts of α-actin.
FIGURE 4.
Colocalization of GFP-AIRE with DAXX. AIRE and DAXX expression constructs were transiently cotransfected in HeLa cells and then immunostained with Texas Red anti-rabbit secondary antibody. Nuclei were visualized by 4′,6-diamidino-2-phenylindole staining (data not shown).
AIRE and DAXX Do Not Interact in Vitro
To identify whether the interaction between AIRE and DAXX proteins was direct or mediated by other proteins, we performed a GST-pulldown assay. DAXX did not copurify with the GST-AIRE fusion protein (aa 1–161) that retained the same fraction of radiolabeled DAXX compared with GST alone (data not shown). This piece of data indicates that the association between DAXX and AIRE is weak and is likely to require other scaffold proteins that increase the stability of the complex. However, the failed interaction could be due to the lack of the post-translational modifications such as phosphorylation, acetylation, or sumoylation that can occur in yeast but not in bacteria.
DAXX Represses the Transactivation Property of AIRE
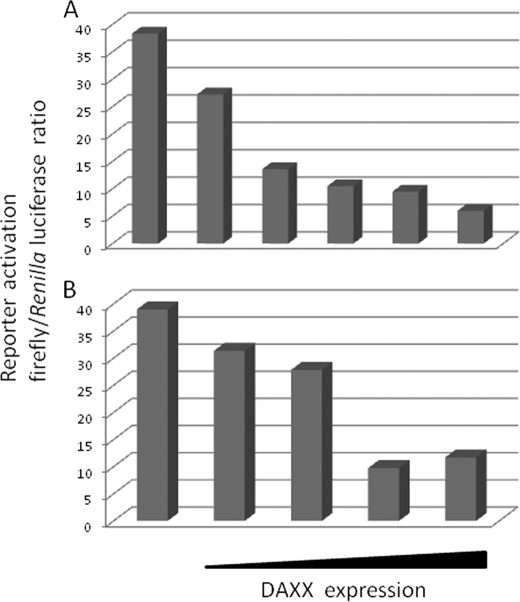
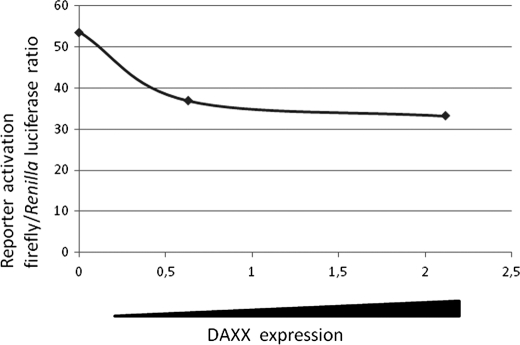
Because DAXX can act as a transcriptional coregulator, the identification of DAXX interaction with AIRE prompted us to examine whether DAXX was involved in regulating AIRE transcriptional activity. To test this hypothesis, we performed transactivation experiments by cotransformation of pEF5HA-AIRE with an increasing amount of pXL5-DAXX in the presence of the pMG3-insulin/Luc promoter reporter plasmid. These experiments were carried out both in COS-1 and in HeLa cells. Consistent with previous reports, AIRE induced the expression of the human insulin promoter reporter activity (28). However the coexpression of DAXX suppressed the transcriptional activity in a dose-dependent manner. As shown in Fig. 5, overexpression of DAXX reduced by 70% the transactivation of the insulin promoter in both HeLa and COS-1 cells.
FIGURE 5.
Transactivation properties of AIRE are negatively regulated by DAXX. A and B, activation of pMG3-insulin/Luc reporter gene in transfected HeLa (A) and COS-7 cells (B). HeLa and COS-7 cells were cotransfected with a pMG3-insulin/Luc firefly luciferase reporter, a control Renilla luciferase reporter, equal amounts of pEF5HA-AIRE, and increasing amounts of DAXX. Cells were cultured for 24 h and then lysed, and luciferase activity was measured on cell lysates. The average in ratio between firefly and Renilla luciferase activities of lysates was plotted versus AIRE and DAXX expression in same lysates as assessed by anti-HA and anti-DAXX blots.
DAXX Does Not Alter the Formation of AIRE Homodimers
Homodimer formation is strongly needed for AIRE to properly transactivate. To determine whether the mechanism by which DAXX represses the transcriptional activity of AIRE requires the formation of the AIRE-AIRE complex, we examined if AIRE homodimers could be coimmunoprecipitated in COS-1 cells lysates. HA-AIRE and V5-AIRE were overexpressed in the presence of increasing amounts of Xpress-DAXX. No difference was noticed in the complex formation even at higher concentrations of DAXX (Fig. 6). These results suggest that the repression induced by DAXX was not caused by the disruption of AIRE homodimers.
FIGURE 6.
Homodimerization inhibition assay. DAXX has no influence on AIRE homodimer formation. COS-1 cells were transfected with V5-AIRE, HA-AIRE, Xpress-DAXX, and IPs were performed from lysates using antibodies against V5 (lanes 2–5) or beads only (lane 1). Upper panel shows anti AIRE-HA blot (homodimer formation). 2nd panel from top shows V5-AIRE. 3rd panel shows endogenous and transfected DAXX levels (lanes 1–5). Lower panels show anti-HA and α-actin blots on total lysates (T.L.). WB, Western blot.
DAXX-mediated Repression of AIRE-dependent Transcription Requires HDACs
To test the hypothesis that HDACs could be involved in the DAXX-mediated transcriptional repression, we performed transactivation assays in the presence of TSA, a HDACs inhibitor. Cells were cotransfected with AIRE, insulin promoter reporter gene, and increasing amounts of DAXX in the presence of TSA. As shown in Fig. 7, down-regulation of insulin promoter reporter gene was relieved by TSA despite the increasing amount of DAXX. To confirm HDAC involvement in AIRE repression, we performed coimmunoprecipitation experiments in mammalian cells. As shown in Fig. 8 AIRE is able to interact with endogenous HDAC1 and HDAC2. In summary, our data suggest that histone deacetylation is involved in DAXX-mediated repression of AIRE transcriptional activity.
FIGURE 7.
Presence of TSA abolishes the negative influence operated by DAXX protein on the transactivation properties of AIRE. Activation of pMG3-insulin/Luc reporter in transfected HeLa. HeLa cells were cotransfected with a pMG3-insulin/Luc firefly luciferase reporter, a control Renilla luciferase reporter, equal amounts of pEF5HA-AIRE, and an increasing amounts of DAXX. Cells were left untreated for 5 h after transfection and then cultured for 24 h in presence of TSA (100 ng/ml). Cells were then lysed, and luciferase activity was measured on cell lysates. The average in ratio between firefly and Renilla luciferase activities of lysates was plotted versus AIRE and DAXX expression in same lysates as assessed by anti-HA and anti-DAXX blots.
FIGURE 8.
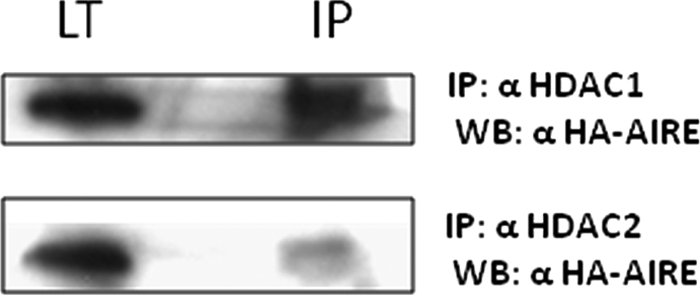
Coprecipitation of endogenous HDAC1 and HDAC2 with transfected HA-AIRE in COS cells. COS-1 cells were transfected with full-length HA-AIRE. IPs were performed from lysates using antibodies against HDAC1 and HDAC2. Upper panel shows the coprecipitated HA-AIRE of anti-HDAC1 IP; bottom panel shows the coprecipitated HA-AIRE of anti-HDAC2 IP. LT, total lysate.
DISCUSSION
Our study aimed to better understand the molecular basis of self-reactive immune processes and consequently to give a contribution into finding an innovative way to prevent and treat autoimmune diseases. The purpose of this study was to identify proteins that interact with AIRE and regulate its activity. The discovery of new potential AIRE interactors might provide more insights on the mechanism of action of AIRE and on its regulation. To test our hypothesis, we performed a yeast two-hybrid screening using AIRE (aa 1–517) as a bait. Upon screening of a human thymus library, we identified seven positive clones coding for the C-terminal region of the DAXX protein, corresponding to the Ser/Pro/Thr-rich domain, which is already known to bind Pax3, Pax5, p53, glucocorticoid receptor, and HDAC2. The AIRE domain involved in this interaction mapped to the HSR/CARD domain, a key motif responsible for the homodimerization, nuclear localization, and transactivation activity of AIRE. This observation was consistent with the fact that the W78R mutation localized in this domain abolishes the interaction between AIRE and DAXX. To confirm the AIRE-DAXX interaction, we performed colocalization experiments in mammalian cells and carried out coimmunoprecipitation assays by transient transfections of full-length DAXX and AIRE and their truncation derivatives. To further validate the AIRE-DAXX interaction, we performed coimmunoprecipitation experiments with endogenously expressed DAXX. Furthermore, GST-pulldown experiments showed that the AIRE-DAXX interaction is weak and requires the presence of protein mediators assembled in multiprotein complexes. As resulted in vitro, the failed interaction could be explained by the lack of post-translational modifications such as phosphorylation, acetylation, or sumoylation that can occur in yeast but not in bacteria. Interestingly, by using transactivation experiments, we detected that DAXX strongly repressed the transcriptional activity of AIRE and that histone deacetylation was involved in DAXX-mediated transcriptional repression. Our experimental data also showed that DAXX repression did not influence the formation of AIRE homodimers. The interaction mapped within the HSR/CARD domain and influenced the transcriptional activity of AIRE. This domain was previously shown by other authors to mediate the interaction between AIRE and PIAS1 protein (28), and here we further demonstrated the significance of the N-terminal HSR/CARD domain in protein-protein interactions. The new functional characterization of the AIRE/CARD domain proposes for AIRE new biological scenarios and unexpected biochemical pathways. Indeed, CARD proteins are involved in two major pathways: regulation of caspase activation both in the context of apoptosis and inflammation and in the regulation of NF-κB activation related to the innate/adaptative immune responses. Furthermore, the CARD protein family may be divided in four subfamilies based on their overall domain structures (50–53) as follows: 1) the NBD-CARDs that contain a nucleotide binding domain in addition to the CARD domain; 2) the coiled-coil CARDs similar to NBD-CARDs with the central NBD replaced by a coiled coil motif; 3) the bipartite CARDs that contain CARD and additional motifs, either a kinase domain, a DD motif, or a PYD motif; 4) the CARD-only proteins, which contain a CARD domain but do not possess any additional recognizable motif.
The AIRE protein may belong to the last category and act as a positive or negative regulator of the multidomain CARDs (NBD and coiled coil CARDs) by competing for binding to their targets. The dimerization process, a very well known process for AIRE previously described in detail by our research group and other authors, is also characteristic of CARD-containing proteins. This domain mediates highly specific protein-protein interactions via homophilic bridges. This dimerization is also crucial for the proper subcellular localization of the proteins. Given that dimerization of CARD proteins is critical for their function, it appears clear how the CARD-mediated interaction between AIRE and DAXX plays an important role in regulating AIRE activity. In fact the interaction resulted in a substantial reduction of AIRE transactivation capability.
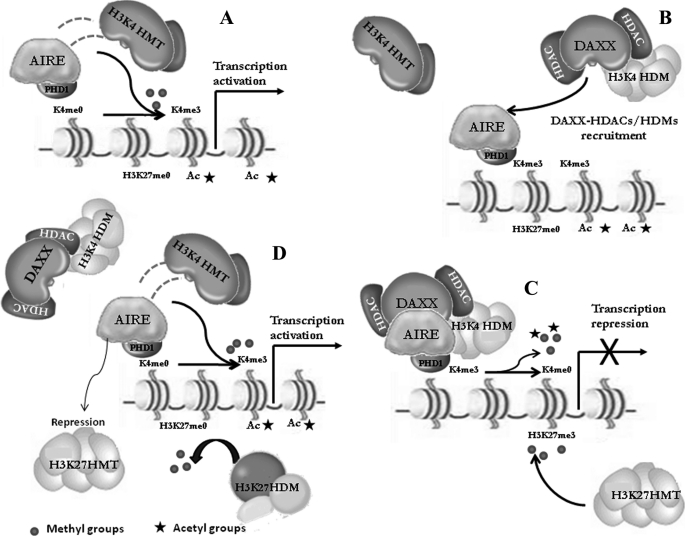
Because both AIRE and DAXX have been described as negative regulators of NF-κB (54, 56),4 it is likely that the interaction might be required for the inhibition of the caspase and NF-κB pathways. Further investigation will be necessary to establish their precise effect on these biological processes. Another level of regulation of the AIRE-DAXX complex could be in epigenetics. Permissive and repressive epigenetic changes are finely harmonized to coordinate the cell type-specific patterns of gene expression that are responsible for cellular maturation and differentiation. In agreement with this, it has been observed that the status of histone methylation can be dynamically regulated by different recently discovered histone demethylases, including the Jumonji (JMJD) and LSD1 family (57–59). To further underline the key role of epigenetic regulators, recent biochemical studies have identified protein complexes having both H3K27 demethylase and H3K4 methyltransferase activities. The finding suggests that, at least in some circumstances, there may be a simultaneous reorganization of both repressive and permissive chromatin states at target promoters (60). DAXX is a protein already known to bind histones and recruit histone deacetylases to repress transcription from target genes. Recent data indicate that AIRE derepresses controlled genes by inducing epigenetic modifications of the chromatin, especially in those tissues where normally restricted genes are ectopically expressed. Recent ChIP on Chip and expression array studies have shown that AIRE works on target gene promoters by inducing H3K4me3 and AcH3 histone modifications typical of active chromatin states. Moreover, AIRE decreases the levels of H3K27me3, an epigenetic marker of transcriptional repression (24). Based on these data, we propose a model to explain the regulatory consequences of the interaction between AIRE and DAXX (Fig. 9). In the first step, AIRE binds histone H3 in its unmethylated K4me0 form, where it recruits a specific H3K4 methyltransferase that, by modifying Lys-4 from the unmethylated to the trimethylated form H3K4me3, leads to an active chromatin state. Histone methylation is a dynamic and reversible process that needs to be promptly counterbalanced in accordance with changes in the phase of the cell cycle. It is also widely accepted that the chromatin repression mechanisms are implemented by the simultaneous occurrence of deacetylation and demethylation. This task requires tight cooperation between histone demethylases and HDACs as highlighted by the RBP2-HDAC complex (30). We suppose that the DAXX-HDAC complex is part of a larger demethylase complex that might mediate AIRE repression through both histone deacetylation and H3K4 demethylation. Very interestingly, a recent work has shown that DAXX colocalizes with the lysine-specific demethylase (LSD)-HDAC1,2-COREST-REST complex further supporting the role of DAXX in complexes with dual demethylase/deacetylase activity (49). Moreover, Lys-4 demethylation is linked to the H3K27 trimethylation, which associates with inactive chromatin states (55). The inactivity of the H3K27 histone methyltransferase complex and the consequent recovery of the active chromatin state might occur through the interaction between AIRE and one or more proteins of the H3K27 complex as supported by our unpublished data,5 which prove the physical interaction of AIRE with a member of the polycomb protein belonging to the H3K27 histone methyltransferase complex. We have mapped the interaction within the repression domain of the polycomb protein that is expected to hinder the repressive capability of the complex. This point represents a topic of considerable interest, and it needs to be studied in depth. In conclusion, we report here the identification of a novel interacting partner for AIRE. Our data further extend the knowledge of the molecular pathways involved in the establishment and maintenance of the immunological tolerance and might give a contribution to the development of therapeutic strategies to prevent and treat autoimmune diseases.
FIGURE 9.
Hypothetical model of action of AIRE-DAXX interaction. The figure shows the proposed working model, which is summarized as follows. A, AIRE-PHD1 binds H3K4me0 and recruits H3K4 histone methyltransferase, which establishes the H3K4me3 mark, characteristic of a transcriptional active chromatin state. B and C, complex DAXX-HDAC-H3K4HDM represses AIRE activity by the simultaneous event of histone deacetylation and Lys-4 demethylation. Subsequently epigenetic repression markers as H3K4me0, H3K27me3, and deacetylation occur. D, complex DAXX-HDAC-H3K4HDM is removed, and AIRE inhibits H3K27 histone methyltransferase. The positive epigenetic markers such as H3K27me0, H3K4me3, and acetylation take place.
Acknowledgments
We thank Alessandro Puddu (Consiglio Nazionale delle Ricerche, Cagliari, Italy) for the help given in confocal microscopy analysis, Asunis Isadora (Consiglio Nazionale delle Ricerche, Cagliari, Italy) for providing pMG3-insulin/Luc reporter plasmid, and Nunzio Bottini (La Jolla Institute for Allergy and Immunology, La Jolla, CA) for pEF5HA vector. We are also grateful to Paolo Moi (Università degli Studi di Cagliari) for sharing plasmids and critically reviewing the manuscript.
This work was supported in part by grants from the Community Sixth Framework Programme, Project “EURAPS”-Autoimmune Polyendocrine Syndrome Type I, 2006–2008.
Supported by “Master and Back” fellowship from the Sardinian Regional Government.
Y. Tao, W. Wheat, C. Williams-Skipp, D. Gutches, D. Wegmann, and R. Scheinman, unpublished data.
A. Meloni, E. Fiorillo, D. Corda, F. Incani, M. L. Serra, A. Contini, A. Cao, and M. C. Rosatelli, unpublished data.
- PHD
- plant homeodomain
- CBP
- CREB-binding protein
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- TRITC
- tetramethylrhodamine isothiocyanate
- NBD
- nucleotide binding domain
- CARD
- caspase recruitment domain
- aa
- amino acid
- GST
- glutathione S-transferase
- PML
- promyelocytic leukemia
- HDAC
- histone deacetylation
- IP
- immunoprecipitation
- TSA
- trichostatin A.
REFERENCES
- 1.Ahonen P., Myllärniemi S., Sipilä I., Perheentupa J. (1990) N. Engl. J. Med. 322, 1829–1836 [DOI] [PubMed] [Google Scholar]
- 2.Perheentupa J., Miettinen A. (1999) 150, 313–325 [PubMed] [Google Scholar]
- 3.Perheentupa J. (2002) Endocrinol. Metab. Clin. North Am. 31, 295–320 [DOI] [PubMed] [Google Scholar]
- 4.Anderson M. S., Venanzi E. S., Klein L., Chen Z., Berzins S. P., Turley S. J., von Boehmer H., Bronson R., Dierich A., Benoist C., Mathis D. (2002) Science 298, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 5.Derbinski J., Gäbler J., Brors B., Tierling S., Jonnakuty S., Hergenhahn M., Peltonen L., Walter J., Kyewski B. (2005) J. Exp. Med. 202, 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liston A., Lesage S., Wilson J., Peltonen L., Goodnow C. C. (2003) Nat. Immunol. 4, 350–354 [DOI] [PubMed] [Google Scholar]
- 7.Gavanescu I., Kessler B., Ploegh H., Benoist C., Mathis D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4583–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittaz L., Rossier C., Heino M., Peterson P., Krohn K. J., Gos A., Morris M. A., Kudoh J., Shimizu N., Antonarakis S. E., Scott H. S. (1999) Biochem. Biophys. Res. Commun. 255, 483–490 [DOI] [PubMed] [Google Scholar]
- 9.Halonen M., Kangas H., Rüppell T., Ilmarinen T., Ollila J., Kolmer M., Vihinen M., Palvimo J., Saarela J., Ulmanen I., Eskelin P. (2004) Hum. Mut. 23, 245–257 [DOI] [PubMed] [Google Scholar]
- 10.Tao Y., Kupfer R., Stewart B. J., Williams-Skipp C., Crowell C. K., Patel D. D., Sain S., Scheinman R. I. (2006) Mol. Immunol. 43, 335–345 [DOI] [PubMed] [Google Scholar]
- 11.Ilmarinen T., Melén K., Kangas H., Julkunen I., Ulmanen I., Eskelin P. (2006) FEBS J. 273, 315–324 [DOI] [PubMed] [Google Scholar]
- 12.Ramsey C., Bukrinsky A., Peltonen L. (2002) Hum. Mol. Genet. 11, 3299–3308 [DOI] [PubMed] [Google Scholar]
- 13.Ferguson B. J., Alexander C., Rossi S. W., Liiv I., Rebane A., Worth C. L., Wong J., Laan M., Peterson P., Jenkinson E. J., Anderson G., Scott H. S., Cooke A., Rich T. (2008) J. Biol. Chem. 283, 1723–1731 [DOI] [PubMed] [Google Scholar]
- 14.Gibson T. J., Ramu C., Gemünd C., Aasland R. (1998) Trends Biochem. Sci. 23, 242–244 [DOI] [PubMed] [Google Scholar]
- 15.Aasland R., Gibson T. J., Stewart A. F. (1995) Trends Biochem. Sci. 20, 56–59 [DOI] [PubMed] [Google Scholar]
- 16.Björses P., Halonen M., Palvimo J. J., Kolmer M., Aaltonen J., Ellonen P., Perheentupa J., Ulmanen I., Peltonen L. (2000) Am. J. Hum. Genet. 66, 378–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitkänen J., Doucas V., Sternsdorf T., Nakajima T., Aratani S., Jensen K., Will H., Vähämurto P., Ollila J., Vihinen M., Scott H. S., Antonarakis S. E., Kudoh J., Shimizu N., Krohn K., Peterson P. (2000) J. Biol. Chem. 275, 16802–16809 [DOI] [PubMed] [Google Scholar]
- 18.Meloni A., Incani F., Corda D., Cao A., Rosatelli M. C. (2008) Mol. Immunol. 45, 805–809 [DOI] [PubMed] [Google Scholar]
- 19.Uchida D., Hatakeyama S., Matsushima A., Han H., Ishido S., Hotta H., Kudoh J., Shimizu N., Doucas V., Nakayama K. I., Kuroda N., Matsumoto M. (2004) J. Exp. Med. 199, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bottomley M. J., Stier G., Pennacchini D., Legube G., Simon B., Akhtar A., Sattler M., Musco G. (2005) J. Biol. Chem. 280, 11505–11512 [DOI] [PubMed] [Google Scholar]
- 21.Org T., Chignola F., Hetényi C., Gaetani M., Rebane A., Liiv I., Maran U., Mollica L., Bottomley M. J., Musco G., Peterson P. (2008) EMBO Rep. 9, 370–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koh A. S., Kuo A. J., Park S. Y., Cheung P., Abramson J., Bua D., Carney D., Shoelson S. E., Gozani O., Kingston R. E., Benoist C., Mathis D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musco G., Peterson P. (2008) Epigenetics 3, 310–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Org T., Rebane A., Kisand K., Laan M., Haljasorg U., Andreson R., Peterson P. (2009) Hum. Mol. Genet. 18, 4699–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meloni A., Perniola R., Faà V., Corvaglia E., Cao A., Rosatelli M. C. (2002) J. Clin. Endocrinol. Metab. 87, 841–846 [DOI] [PubMed] [Google Scholar]
- 26.Abramson J., Giraud M., Benoist C., Mathis D. (2010) Cell 140, 123–135 [DOI] [PubMed] [Google Scholar]
- 27.Liiv I., Rebane A., Org T., Saare M., Maslovskaja J., Kisand K., Juronen E., Valmu L., Bottomley M. J., Kalkkinen N., Peterson P. (2008) Biochim. Biophys. Acta 1783, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilmarinen T., Kangas H., Kytömaa T., Eskelin P., Saharinen J., Seeler J. S., Tanhuanpää K., Chan F. Y., Slattery R. M., Alakurtti K., Palvimo J. J., Ulmanen I. (2008) Mol. Immunol. 45, 1847–1862 [DOI] [PubMed] [Google Scholar]
- 29.Tonooka A., Kubo T., Ichimiya S., Tamura Y., Ilmarinen T., Ulmanen I., Kimura S., Yokoyama S., Takano Y., Kikuchi T., Sato N. (2009) Biochem. Biophys. Res. Commun. 379, 765–770 [DOI] [PubMed] [Google Scholar]
- 30.Hayakawa T., Ohtani Y., Hayakawa N., Shinmyozu K., Saito M., Ishikawa F., Nakayama J. (2007) Genes Cells 12, 811–826 [DOI] [PubMed] [Google Scholar]
- 31.Pitkänen J., Rebane A., Rowell J., Murumägi A., Ströbel P., Möll K., Saare M., Heikkilä J., Doucas V., Marx A., Peterson P. (2005) Biochem. Biophys. Res. Commun. 333, 944–953 [DOI] [PubMed] [Google Scholar]
- 32.Yang X., Khosravi-Far R., Chang H. Y., Baltimore D. (1997) Cell 89, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong S., Salomoni P., Ronchetti S., Guo A., Ruggero D., Pandolfi P. P. (2000) J. Exp. Med. 191, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. (1999) J. Cell Biol. 147, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaelson J. S., Bader D., Kuo F., Kozak C., Leder P. (1999) Genes Dev. 13, 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollenbach A. D., Sublett J. E., McPherson C. J., Grosveld G. (1999) EMBO J. 18, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E. J., Chen J. D. (2000) Mol. Cell. Biol. 20, 1784–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li R., Pei H., Watson D. K., Papas T. S. (2000) Oncogene 19, 745–753 [DOI] [PubMed] [Google Scholar]
- 39.Zhao L. Y., Liu J., Sidhu G. S., Niu Y., Liu Y., Wang R., Liao D. (2004) J. Biol. Chem. 279, 50566–50579 [DOI] [PubMed] [Google Scholar]
- 40.Hollenbach A. D., McPherson C. J., Mientjes E. J., Iyengar R., Grosveld G. (2002) J. Cell Sci. 115, 3319–3330 [DOI] [PubMed] [Google Scholar]
- 41.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T. L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10635–10640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J., Wu S., Liu H., Stratt R., Barak O. G., Shiekhattar R., Picketts D. J., Yang X. (2004) J. Biol. Chem. 279, 20369–20377 [DOI] [PubMed] [Google Scholar]
- 43.Kuo H. Y., Chang C. C., Jeng J. C., Hu H. M., Lin D. Y., Maul G. G., Kwok R. P., Shih H. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16973–16978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin D. Y., Lai M. Z., Ann D. K., Shih H. M. (2003) J. Biol. Chem. 278, 15958–15965 [DOI] [PubMed] [Google Scholar]
- 45.Zhao L. Y., Colosimo A. L., Liu Y., Wan Y., Liao D. (2003) J. Virol. 77, 11809–11821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muromoto R., Sugiyama K., Yamamoto T., Oritani K., Shimoda K., Matsuda T. (2004) Biochem. Biophys. Res. Commun. 316, 827–833 [DOI] [PubMed] [Google Scholar]
- 47.Lin D. Y., Fang H. I., Ma A. H., Huang Y. S., Pu Y. S., Jenster G., Kung H. J., Shih H. M. (2004) Mol. Cell. Biol. 24, 10529–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang C. C., Lin D. Y., Fang H. I., Chen R. H., Shih H. M. (2005) J. Biol. Chem. 280, 10164–10173 [DOI] [PubMed] [Google Scholar]
- 49.Gu H., Roizman B. (2009) J. Virol. 83, 4376–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lalioti V. S., Vergarajauregui S., Pulido D., Sandoval I. V. (2002) J. Biol. Chem. 277, 19783–19791 [DOI] [PubMed] [Google Scholar]
- 51.Bouchier-Hayes L., Martin S. J. (2002) EMBO Rep. 3, 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong G. S., Jung Y. K. (2002) J. Biochem. Mol. Biol. 35, 19–23 [DOI] [PubMed] [Google Scholar]
- 53.Hiscott J., Lin R., Nakhaei P., Paz S. (2006) Trends Mol. Med. 12, 53–56 [DOI] [PubMed] [Google Scholar]
- 54.Martinon F., Tschopp J. (2004) Cell 117, 561–574 [DOI] [PubMed] [Google Scholar]
- 55.Pasini D., Hansen K. H., Christensen J., Agger K., Cloos P. A., Helin K. (2008) Genes Dev. 22, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park J., Lee J. H., La M., Jang M. J., Chae G. W., Kim S. B., Tak H., Jung Y., Byun B., Ahn J. K., Joe C. O. (2007) J. Mol. Biol. 368, 388–397 [DOI] [PubMed] [Google Scholar]
- 57.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 58.Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 59.De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., Natoli G. (2007) Cell 130, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 60.Agger K., Christensen J., Cloos P. A., Helin K. (2008) Curr. Opin. Genet. Dev. 18, 159–168 [DOI] [PubMed] [Google Scholar]