Abstract
PML nuclear bodies are matrix-associated domains that recruit an astonishing variety of seemingly unrelated proteins. Since their discovery in the early 1960s, PML bodies have fascinated cell biologists because of their beauty and their tight association with cellular disorders. The identification of PML, a gene involved in an oncogenic chromosomal translocation, as the key organizer of these domains drew instant interest onto them. The multiple levels of PML body regulation by a specific posttranslational modification, sumoylation, have raised several unsolved issues. Functionally, PML bodies may sequester, modify or degrade partner proteins, but in many ways, PML bodies still constitute an enigma.
PML bodies are nuclear structures that seem to sequester, modify, and degrade certain proteins. PML and/or partner sumoylation was proposed to be the glue that holds them together.
PML BODIES, AN INTRODUCTION
PML nuclear bodies (NBs) are spheres of 0.1–1.0 µm in diameter found in most cell-lines and many tissues. They belong to the nuclear matrix, an ill-defined super-structure of the nucleus proposed to anchor and regulate many nuclear functions, including DNA replication, transcription, or epigenetic silencing (Stuurman et al. 1990). The PML protein is the key organizer of these domains that recruits an ever-growing number of proteins, whose only common known feature to date is their ability to be sumoylated (Bernardi and Pandolfi 2007). PML and NBs were proposed to fine-tune a wide variety of processes, through facilitation of partner protein posttranslational modifications (notably sumoylation itself) resulting in partner sequestration, activation, or degradation. Several NBs subtypes have been defined on morphological bases, which all contain an electron-dense shell and an inner core.
PML NBs came to the forefront with the observation that the oncogenic PML/RARA protein disrupts them in a treatment-reversible manner (Daniel et al. 1993; Dyck et al. 1994; Koken et al. 1994; Weis et al. 1994; Zhu et al. 1997). PML NBs are regulated by cellular stress: viral infection (Everett 2006), DNA-damage, transformation (Koken et al. 1995; Terris et al. 1995; Gurrieri et al. 2004), and oxidative stress (Yamada et al. 2001a; Villagra et al. 2006). Moreover, transcription of PML and several genes encoding partner proteins is dramatically enhanced by interferons. Yet, pml-/- mice, which cannot assemble NBs, develop normally and live well, demonstrating that NBs are dispensable for most basic biological functions. Nevertheless, recent data has implicated PML in the control of cellular senescence and stem cell self-renewal, extending the fields of investigation of PML function (Ferbeyre et al. 2000; Pearson et al. 2000; Salomoni and Pandolfi 2002; Ito et al. 2008).
HISTORICAL OVERVIEW
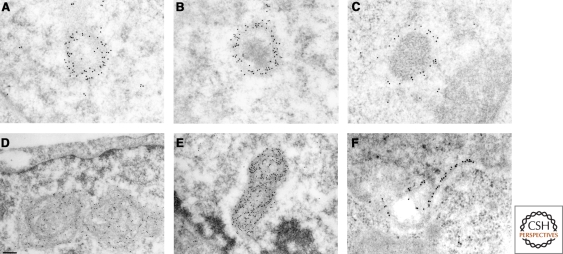
As for several other nuclear structures, electron microscopy and auto-antibodies were the two founding fathers of the field. Work from several pioneers in the early 1960s noted the presence of dense spherical objects by electron microscopy (de Thé et al. 1960). Two classes were morphologically described: Empty ones (fibrillar) and granular ones, with an inner microgranular material proposed to be ribonucleoproteins (Fig. 1). PML NBs were later observed by immunofluorescence using autoimmune sera from primary billiary cirrhosis patients. These allowed the identification of the first NB-associated protein, SP100 (Szostecki et al. 1990), and an initial characterization of these structures (Ascoli and Maul 1991).
Figure 1.
Different types of PML bodies. Immuno-gold electron microscopy of PML NBs in CHO cell line stably overexpressing PML. (A, B, C) “Classical” PML NBs. PML is distributed on a dense electron shell (A, B) or on light halo (C), that can contain a microgranular inner core (B, C) or not (A). (D, E) PML body in arsenic trioxide-treated cell. Note the absence of the inner core and the enrolled fibrillar aspect of the body (D). (F) PML-II targets the inner membrane region of the nuclear envelope in pml-/- MEF cells. Images courtesy of Edmond and Francine Puvion (CNRS, Villejuif, France).
Identification of an anti-nuclear matrix antibody, which labeled the same structures as SP100 (Stuurman et al. 1992), drew the first link between these bodies and the nuclear matrix. The localization of PML, a protein fused to the retinoic acid receptor α(RARA) in the PML/RARA oncoprotein of acute promyelocytic leukemia (APL), to the same nuclear dots as SP100, renewed interest for these domains. The observation that in APL cells PML/RARA disrupted these domains drew instant excitement from the scientific community (Daniel et al. 1993; Dyck et al. 1994; Koken et al. 1994; Weis et al. 1994). Moreover, PML bodies were restored by two different anti-APL therapies, retinoic acid and arsenic trioxide, later shown to trigger PML/RARA degradation (Quignon et al. 1997; Zhu et al. 2001), identifying the first striking parallel between the status of the bodies and that of the cell. Many further studies showed that PML bodies altered in stress conditions, notably viral infections, heat shock, and exposure to heavy metals (Everett 2001; Dellaire and Bazett-Jones 2004; Everett 2006).
PML bodies recruit an ever-increasing number of partner proteins (now in the range of 100), one of the most studied being DAXX, a potent repressor of transcription and modulator of apoptosis. Critically, PML is the actual organizer of the bodies (Ishov et al. 1999; Zhong et al. 2000a; Lallemand-Breitenbach et al. 2001). Among these recruited proteins, one deserves a special mention: an ubiquitin-like protein named SUMO, as PML conjugation by SUMO plays a critical role in recruitment of partners, many of which are sumoylated themselves. Recent studies have focused on more fundamental cell-biology issues: Dynamics of the bodies (Eskiw et al. 2003, Muratani et al., 2002, Chen et al., 2008), relation to other nuclear components (Wang et al. 2004; Batty et al. 2009; Russell et al. 2009), mode of assembly (Shen et al. 2006), and determinants of partner recruitment and actual function in cellulo and in vivo.
THE PML PROTEINS
PML is a member of the TRIM/RBCC family of proteins, many members of which are ubiquitin ligases that generate subcellular structures through autoassembly (Reymond et al. 2001; Meroni and Diez-Roux 2005). Transcription of the PML gene is tightly controlled by interferons α/β or γ, but also by p53 (Stadler et al. 1995; de Stanchina et al. 2004), which both yield a dramatic increase in the number and the size of the bodies. PML harbors an amino-terminal RING finger that directly binds the SUMO E2 ligase UBC9 (Duprez et al. 1999), two RING-like domains, the B boxes (Tao et al. 2008), and a coiled-coil mediating homodimerization (Kastner et al. 1992). For other RBCC/TRIM family members, partner binding-specificity often relies on the carboxyl terminus. For PML, a variety of carboxy-terminal domains generated by alternative splicing yield isoforms (Jensen et al. 2001). When expressing single PML isoform in pml-/- cells, distinct types of PML NBs were observed, implying that isoform-specific sequences contact different nuclear constituents that influence morphogenesis (Beech et al. 2005; Condemine et al. 2006; Weidtkamp-Peters et al. 2008) (Table 1). Yet, because of the coiled-coil, all endogenous isoforms colocalize. The most abundant (but perhaps least studied) isoform, PML-I, harbors an exonuclease-III domain, that targets PML to nucleolar caps in stressed or senescent cells (Condemine et al. 2007). In addition to the nuclear localization signal (NLS) present in all PML isoforms, PML-I harbors a nuclear export signal (NES) that allows shuttling of all isoforms between the two compartments through heterodimer formation (Henderson and Eleftheriou 2000; Beech et al. 2005; Condemine et al. 2006). The most extensively studied isoform, PML-IV, induces senescence in primary human fibroblasts (Bischof et al. 2002) and apoptosis in many other cellular settings, at least in part through p53 activation (Guo et al. 2000).
Table 1.
PML isoforms with specific localizations.
| PML isoform: | Specific localization in pml -/- cells: |
|---|---|
| PML I | Nucleolar upon stress, cytoplasm |
| PML II | Fibrillar/nuclear envelope (lamina?) |
| PML IV | Nucleolar upon stress |
| PML V | Thick shell/nuclear matrix tethering |
| PML VII | Cytoplasmic/early endosomes |
The expression of PML isoforms in pml-/- cells reveals specific localizations, suggesting that the carboxy-terminus drives interactions with specific unidentified partners. From (Beech et al. 2005; Condemine et al. 2006; Weidtkamp-Peters et al. 2008).
PML undergoes several critical posttranslational modifications, notably phosphorylation and sumoylation. PML sumoylation has been implicated in NB-morphogenesis. DNA damage- or stress- activated kinases like ATM, ATR, CHK2, HIPK2, CK2, or ERK phosphorylate PML, possibly regulating PML stability, NB biogenesis and partner association (Engelhardt et al. 2003; Hayakawa and Privalsky 2004; Scaglioni et al. 2006; Gresko et al. 2009) and contributing to DNA repair or apoptosis control.
MORE THAN ONE PML BODY!
It is generally assumed that “PML NBs” designates a single object. However, there is considerable evidence that PML bodies are diverse. Indeed, PML aggregates onto different sites to create an unsuspected repertoire of PML-accumulating domains in response to a variety of stresses (Eskiw et al. 2003; Bernardi and Pandolfi 2007). Before engaging in the catalog of structures that contain PML, one has to realize that contrarily to the common appreciation, the overwhelming majority of the PML protein pool in a cell is actually not NB-bound. In most cell lines, even in vivo, more than 90% of PML has a diffuse nuclear localization, not associated with the nuclear matrix or NBs (Lallemand-Breitenbach et al. 2001) (P. Hemmerich, personal communication). The most extensively studied factor modulating PML distribution has been arsenic trioxide (see later discussion), although several DNA-damage-activated kinases are also important. Stress-induced aggregation may promote aggregation of typical NBs or conversely disperse them into microspeckles (see Table 2). The differences among these bodies can be based on morphology or content, yielding a much more dynamic view of PML than previously thought.
Table 2.
PML NBs are sensitive to cellular stress.
| Stress | PML NB phenotypes | SUMO colocalization | Cell types | Ref | |
| IFNs | Increased number and size | Increased | All | Stadler, 1995 | |
| As2O3 | Large PML shell, decreased number | Increased | All |
Zhu, 1997 Lallemand-Breitenbach, 2001 |
|
| CdCl2/heat shock | Dispersed micro-bodies | No | All | Eskiw, 2003 | |
|
Proteasome Inhibition |
Nucleolus | Yes | Primary |
Condemine, 2007 Mattsson K, 2001 |
|
| Increased number and size | Increased | Transformed |
Lallemand-Breitenbach, 2001 Everett, 1999 |
||
| Actinomycin-D | Large, Peri-nucleolar | ? | Primary + transformed |
Janderova-Rossmeislova, 2007 Condemine, 2007 |
|
| Dispersed micro-bodies | Yes | Transformed | Eskiw, 2004 | ||
| DNA Damage | Doxo, IRgamma, UVc | Large, perinucleolar | Yes | Primary transformed |
Bernardi, 2004 Condemine, 2007 Kurki, 2003 |
| UVc, alkylators, staurosporine DNase | Dispersed micro-bodies | Yes | Primary transformed | Salomoni, 2005, Condemine, 2007 Eskiw, 2003 Eskiw, 2004 |
|
List of exogenous agents that perturb PML NB organization; the status of PML sumoylation followed by NB localization is indicated. The phenotype of PML NBs is dependent on the type of stress and cellular context (transformed cell lines or primary cells).
Structure of the Classical Body
The classical PML body is a spherical object with a diameter of 0.1–1 µm, which may or may not have a micro-granular centre. These bodies, from five to 15 per nucleus in cell-lines, are mostly proteinaceous in nature and do not in general contain RNA or DNA (Boisvert et al. 2000). PML forms the outer shell of the bodies and partners are usually inside, which is easily shown after PML/partner overexpression (Guiochon-Mantel et al. 1995) (Figs. 1, 2). Like several other bodies, PML NBs are present in the interchromosomal space (Bridger et al. 1998), likely explaining why they are often found close or adjacent to other bodies (Wang et al. 2004; Sun et al. 2005; Batty et al. 2009; Russell et al. 2009). Although devoid of DNA, PML NBs may be associated with some specific chromosomal loci, like MHC class I gene cluster region for which PML NBs were proposed to modulate chromatin architecture and transcription (Shiels et al. 2001; Wang et al. 2004; Kumar et al. 2007). One elegant study has found a PML body constantly juxtaposed to a repressor locus, underscoring the links with transcriptional regulation (Tsukamoto et al. 2000). Conversely, chromatin changes occurring during transcription or the cell cycle may modulate PML NBs structure and number (Eskiw et al. 2004; Wang et al. 2004; Ching et al. 2005). PML NBs are profoundly modified during many virus infections (Everett 2006). They may, for example, accumulate viral genomes at their periphery or within their central core during infection of quiescent cells (Everett et al. 2007).
Figure 2.
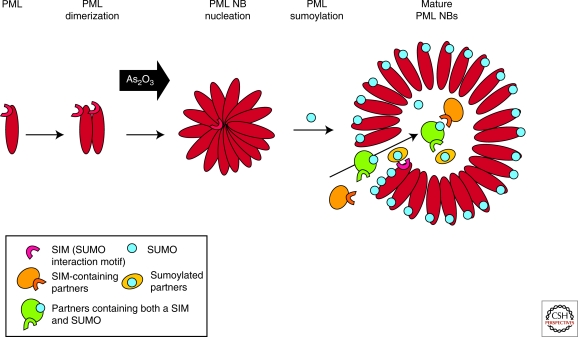
Schematic representation of PML NB biogenesis. PML proteins first dimerize through the RBCC domains and then multimerize to nucleate NBs. PML sumoylation then leads to organization in spherical body. SIM-containing or sumoylated partners (or both) are recruited by the SUMO or SIM of PML into the inner core of the body.
PML NBs, Telomeres and DNA Damage
Alternative lengthening of telomeres (ALT)-associated PML bodies (APBs) are larger structures observed in cell-lines that do not express telomerase and maintain telomere length by homologous recombination. APBs contain two types of DNA double-strand break repair and homologous recombination factors, the Rad50/Mre11/NBS1 complex and Rad51/Rad52, together with the replication factor A (RPA), the helicase BLM and the telomeric repeat-binding factors TRF1 and TRF2 (Wu et al. 2000; Wu et al. 2003). APBs harbor actively replicating telomeres in the S/G2 phase of the cell cycle. Almost all these proteins are sumoylated. Association of the SUMO ligase SMC5/6 complex with APBs is required for TFR1/2 sumoylation and cell survival (Potts and Yu 2007). Recent studies have argued for a role of PML in facilitating these processes (Jiang et al. 2007; Draskovic et al. 2009).
PML is phosphorylated by several DNA-damage activated kinases, including ATM, ATR, HIPK2, or CHK2. Several studies have implicated PML in DNA-repair, through the recruitment to or release from NBs of many proteins like BLM (Bischof et al. 2001), the Mre11 complex (Carbone et al. 2002), WRN, or TOPBP1 (Dellaire and Bazett-Jones 2004). Some of these proteins are localized in NBs in unstressed conditions, whereas others are only associated with NBs after DNA-damage. Although NBs partially overlap with γH2AX foci, their possible localization on the damage sites is still disputed (Boe et al. 2006). Initial studies localized NBs with BLM on BrdU-positive putative DNA repair sites after irradiation (Bischof et al. 2001). Yet, in later reports, PML-NBs were proposed to retain BLM away from DSB through BLM sumoylation, which could be released by phosphorylation (Eladad et al. 2005; Rao et al. 2005). In response to DNA damage, PML NBs appear to sense the damaged sites, yielding an increased number of microbodies by a fission mechanism. The exact role of PML (bystander or actor) in theses diverse processes is still unclear. Yet, pml-/- cells have a very high rate of sister chromatin exchange (Zhong et al. 1999), implying that PML regulates at least some aspects of homologous recombination.
Nucleolus and RNAs
Electron microscopy studies noted that a subset of bodies contains a granular core, typical for the presence of ribonucleoproteins (Dupuy-Coin et al. 1972). Some of these granular bodies were proposed to bud from the nucleolus (Dupuy-Coin and Bouteille 1972). This most likely reflects the presence of specific RNA-binding proteins and is often observed in hormone-stimulated primary cells (Vagner-Capodano et al. 1982). What determines the presence of the microgranular core inside PML NBs is unknown. We have observed this granular core in a subset of PML-labeled structures (Fig. 1). The nature of this micro-granular core deserves a renewed exploration. Some studies showed the presence of nascent RNA within a subset of the bodies (LaMorte et al. 1998; Fuchsova et al. 2002; Kiesslich et al. 2002), whereas others did not (Boisvert et al. 2000). Finally, a series of papers has linked PML bodies and mRNA export and translation through eukaryotic translation initiation factor 4E (Lai and Borden 2000; Cohen et al. 2001; Culjkovic et al. 2006; Borden and Culjkovic 2009). Heterogeneity of the results may reflect analysis of different cellular systems, but these issues clearly deserve additional studies.
In primary or senescent cells, others and we have observed larger structures that contain nucleolar elements, senescence-associated NBs (SANBs) (Condemine et al. 2007; Janderova-Rossmeislova et al. 2007). Interestingly, in this setting, NB-associated proteins are not present within the inner core, but directly on the PML shell (Condemine et al. 2007). Formation of these domains requires PML-I, which contains a nucleolar targeting domain, possibly the exonuclease-III homology domain (Condemine et al. 2007). The nucleolar constituent recognized by this domain and the consequence of PML delocalization for nucleolar function are unknown. Yet, upon genotoxic stress or inhibition of transcription, ATR-mediated PML phosphorylation relocalizes PML to nucleolar caps, where PML was proposed to sequester the p53 ubiquitin ligase MDM2 (Bernardi et al. 2004; Shav-Tal et al. 2005) (Table 2).
Other Subnuclear Compartments
Studies performed in human embryonic stem cells (HESC) have revealed a further unsuspected heterogeneity in PML NBs (Butler et al. 2009). PML NBs assemble in “rosettes” surrounding DNA centromeres or are distributed in tracks bridging two centromeres. These tracks can cross the nucleus and were frequently observed filing lamina gaps at the nuclear envelope. These PML structures do not contain SUMO, SP100, or DAXX proteins and their role in stem cell fate remains to be explored. The PML-II isoform is associated with the inner membrane of nuclear envelope in somatic cells (Condemine et al. 2006) (Fig. 1). NBs are associated with centromeres in proteasome inhibitor-treated cells in the G2 phase of the cell cycle (Everett et al. 1999a). Finally, NBs containing pericentromeric satellite DNA together with HP1, BRCA1, ATRX, and DAXX were proposed to play a role in the re-establishment of the condensed heterochromatic state of late-replicating satellite DNA in ICF syndrome and possibly in normal cells (Luciani et al. 2006).
Cytoplasmic PML
Among PML isoforms some are devoid of NLS and yield purely cytoplasmic bodies, when expressed in pml-/- cells. Similarly, because PML-I has a nuclear export signal, it forms some cytoplasmic PML-labeled bodies (Condemine et al. 2006). Cytoplasmic PML was proposed to regulate TGFβ signaling by controlling the association of Smad2/3 with SARA and accumulation of SARA and TGFβ-receptor in the early endosomes (Lin et al. 2004) (Table 1).
NB DYNAMICS
Basal Status
NBs have been the focus of several studies using imaging technologies such as FRET or FRAP (Tsukamoto et al. 2000). These have shown that PML is a stable component of the bodies and that partner proteins are more mobile, although they are transiently retained in NBs (Boisvert et al. 2001; Weidtkamp-Peters et al. 2008). The exchange rates of the different PML isoforms between NBs and nucleoplasm showed a clear difference for PML-V, which forms peculiar thick-shelled NBs and might anchor NBs onto the nuclear matrix (Condemine et al. 2006; Weidtkamp-Peters et al. 2008). The bodies themselves are not very mobile, although fusions and fissions can be observed through the cell cycle progression (see later discussion). Some bodies labeled by GFP-SP100 are smaller and more dynamic than typical PML NBs (Muratani et al. 2002; Wiesmeijer et al. 2002). Telomeres quickly get in and out of APBs, whereas their PML shell is quite immobile (Molenaar et al. 2003; Jegou et al. 2009), similar to that of SANBs (Condemine et al. 2007). When interpreting these studies, one has to remember that some were performed in transiently transfected cells where posttranslational modifications, which are essential determinants of partner residence, are unlikely to be complete.
Cell-Cycle Analysis
Analyses of NBs during the cell cycle have provided evidence for duplication by a fission mechanism during S phase (Dellaire et al. 2006c) and have dissected NB reformation during the M/G1 transition (Dellaire et al. 2006b; Chen et al. 2008). During mitosis, PML proteins remain aggregated, but are phosphorylated, become desumoylated and release partners (Everett et al. 1999b). Before nuclear membrane breakdown in prometaphase, PML NBs lose their chromatin-tethering, resulting in increased mobility (Chen et al. 2008). Yet, FRAP performed during mitosis showed that PML proteins did not exchange, demonstrating that mitotic bodies are highly stable (Dellaire et al. 2006c). PML associates with nuclear membranes and nucleoporins during mitosis, facilitating reformation of the nuclear envelope during the telophase/G1 transition (Puvion-Dutilleul et al. 1995; Jul-Larsen et al. 2009). Finally, during telophase to G1 transition, SP100 and DAXX re-enter the nucleus and then bind preformed NBs, SP100 first and later DAXX, consistent with the critical role of PML in NB-nucleation.
Stress Response
Apart from arsenic trioxide that promotes NB formation (see below), heat shock or heavy metals induce reversible NB fragmentation by budding of highly mobile micro-bodies that are devoid of SUMO and most partners (Table 2). In contrast, transcriptional inhibition or DNA damage induce formation of microspeckles or nucleolar caps depending on the type of cells (transformed or primary), with a maintained SUMO-colocalization (Table 2) (Eskiw et al. 2003; Nefkens et al. 2003; Dellaire et al. 2006a). Single cell studies after stress recovery have shown that the initial size, location, and number of NBs was somehow restored, suggesting that NBs may assemble on predetermined sites. A full exploration of the role of PML phosphorylation for the regulation of PML mobility and NB formation would be important to clarify these issues.
IS SUMO THE NB-ASSEMBLY GLUE?
SUMO is conjugated to target proteins on the side chain of lysine residues, creating a branched peptide and significantly changing the binding properties of the protein. SUMO has been implicated in multiple pathways, mostly as a regulator of protein interactions (Hay 2005). Conjugated SUMO may interact with a short motif, known as the SUMO interaction motif (SIM) (Minty et al. 2000; Hecker et al. 2006). SUMO-1 was first identified as a PML partner in a two-hybrid screen (Boddy et al. 1996). PML was shown to be sumoylated on three lysine residues: K65 in the RING finger, K160 in the B1 box, and K490 in the NLS. PML also contains a SIM. Accordingly, intermolecular interactions between the PML SUMO and SIM were proposed to underlie NB biogenesis (Matunis et al. 2006; Shen et al. 2006). Sumoylation-defective Ubc9-/- cells indeed show NB defects (Nacerddine et al. 2005). Moreover, because most partner proteins associated with classical PML NBs are sumoylated and many contain a SIM, SIM/SUMO interactions may also account for partner recruitment as well as sequestration.
Studies on NB-biogenesis have been greatly facilitated by arsenic trioxide. Arsenic trioxide, a very potent therapeutic agent in APL (Zhu et al. 2002; Kogan 2009), was first shown to target PML/RARA for degradation through its PML moiety (Chen et al. 1997; Zhu et al. 1997; Muller et al. 1998; Shao et al. 1998). Arsenic trioxide is the only factor that regulates the partitioning of PML between the nucleoplasm and the nuclear matrix, promoting in a sequential manner NB-formation, PML sumoylation, partner recruitment, and PML degradation (Lallemand-Breitenbach et al. 2001; Lallemand-Breitenbach et al. 2008) (Fig. 2). Critically, PML sumoylation on K160/K65 follows matrix-targeting and is required for partner recruitment (Ishov et al. 1999; Zhong et al. 2000a; Lallemand-Breitenbach et al. 2001). Yet, how arsenic trioxide initiates the SUMO-independent transfer from the soluble diffuse nucleoplasmic form toward the insoluble matrix fraction and why matrix association is followed by sumoylation of these two sites remains unexplained to date.
SUMO/SIM interactions were initially proposed to underlie both formation of the PML mesh and recruitment of partners. Although appealing, this model has been significantly challenged by recent evidence. Specific PML isoforms that do not harbor the SIM yield normal bodies (Weidtkamp-Peters et al. 2008). Conversely, analysis of several partner proteins has shown that their SIM is essential, whereas their sumoylation is dispensable for NB-targeting (Takahashi et al. 2005; Lin et al. 2006; Cho et al. 2009). Taken together, SIM/SUMO are unlikely to play a fundamental role in PML aggregation to create the PML mesh, but may be critical for partner recruitment.
WHAT ARE THE FUNCTIONS OF NBs?
PML influences or regulates key processes such as transcription, apoptosis, senescence, response to DNA-damage or resistance to micro-organisms, which have all been extensively reviewed elsewhere (Salomoni and Pandolfi 2002; Everett 2006; Bernardi and Pandolfi 2007; Bernardi et al. 2008). Two novelties have emerged: (1) the protective role of PML, DAXX, and SP100 against many viral infections (Everett et al. 2006; Tavalai et al. 2006; Everett et al. 2008; Tavalai et al. 2008; Kyratsous and Silverstein 2009), at least in the absence of the viral proteins that disrupt NBs; (2) the role of PML in normal or cancer stem cell fate (Ito et al. 2008; Li et al. 2009; Regad et al. 2009). PML-enforced stem cell self-renewal may rely on its ability to modulate the AKT pathway through regulation of AKT phosphorylation or through the localization of its regulator PTEN (Trotman et al. 2006; Song et al. 2008; Ito et al. 2009). TR2, a modulator of the critical Oct4 stem cell transcriptional regulator, is dependent on PML for its sumoylation (Park et al. 2007; Gupta et al. 2008). The importance of PML in “stemness” highlights the distinctly uncommon morphology of NBs in ES cells (Butler et al. 2009) and questions the nature of the PML isoforms that they actually express.
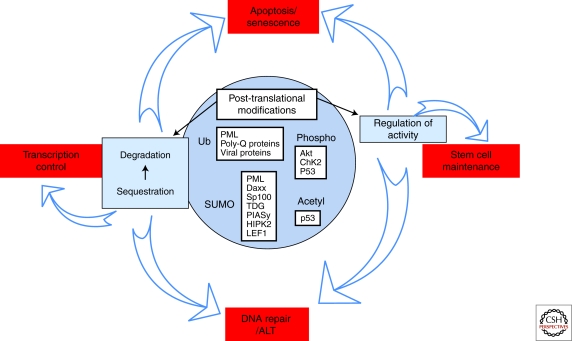
How are these functions actually achieved? The current models envision PML as glue, whose major function is to recruit and concentrate partners within NBs. Recruitment of partners together with many protein-modifying enzymes, could, in principle, enhance posttranslational modifications, yielding activation, sequestration or degradation. There is evidence to support all three of these processes (Fig. 3).
Figure 3.
A general function for PML NBs: Integrated posttranslational protein modifications? PML NBs regulate posttranslational modifications of partner proteins like sumoylation, ubiquitination, but also phosphorylation or acetylation. These modifications regulate a wide variety of partners, leading to modulation of biological processes like transcription, apoptosis/senescence, DNA repair, or ALT and stem cell self-renewal.
Partner Posttranslation Modifications
Perhaps the most studied posttranslational modifications have been those on the tumor suppressor p53. A striking finding has been the concentration of p53-modifying enzymes (CBP, HDM2, HIPK2, and HAUSP) within NBs. PML-enhanced acetylation, sumoylation, and phosphorylation occurring in NBs all appear to enhance p53 function. In that respect pml-/- cells are distinctly impaired in their ability to undergo senescence (Ferbeyre et al. 2000; Pearson et al. 2000; de Stanchina et al. 2004), whereas conversely PML-IV overexpression triggers senescence by a pathway involving both p53 and Rb (Bischof et al. 2002; Mallette et al. 2004; Bischof et al. 2005). The complexity of the multiple mechanisms through which PML enhances p53 function and some apparent contradictions in the published data plea for a renewed exploration of this critical issue in primary cells, using siRNA or gene excision rather than PML/partner overexpression.
Kinase activity may be affected by translocation into NBs: PP2A dephosphorylates p-Akt within PML NBs (Trotman et al. 2006), whereas PP1A was proposed to dephosphorylate RB (Regad et al. 2009). In contrast, recruitment into NBs may favor auto-phosphorylation of some kinases, such as CHK2 (Yang et al. 2002; Yang et al. 2006).
There are also some indications that PML can directly enhance global protein sumoylation in yeast (Quimby et al. 2006) and NBs were proposed to enhance sumoylation of specific partner proteins (Park et al. 2007). Because many NB-associated proteins contain a SIM, this may enhance partner sequestration within NBs.
Partner Sequestration
Sequestration or “depot” was the first proposed function of PML NBs (Negorev and Maul 2001). This sequestration is evident by the relative accumulation of the nucleoplasmic and the NB-associated form of PML-partners, which very significantly varies between individual partners and levels of PML expression, as well as sumoylation. A well-studied sequestered partner is DAXX, a potent repressor that partitions between sumoylated proteins, including PML and many transcription factors. Sequestration of DAXX by NB-associated, sumoylated PML releases transcriptional repression by DNA-bound sumoylated transcription factors (Li et al. 2000; Lehembre et al. 2001; Lin et al. 2003; Lin et al. 2006). Sequestration of DAXX also regulates apoptosis. In particular, DAXX enhances Fas-triggered caspase activation, possibly through release of ASK1 kinase activity (Torii et al. 1999; Zhong et al. 2000b; Salomoni and Khelifi 2006), although opposite results have been observed in other cell-types (Meinecke et al. 2007). Sequestration of DAXX may also modulate other critical regulators, such as HAUSP, ultimately deubiquitinating the PTEN tumor suppressor (Song et al. 2008). In any case, DAXX, which exerts pro- or anti-apoptotic functions, is a critical partner of PML involved in many of it properties (Maul et al. 2000).
A related situation is the concentration of histones and their chaperones in senescent cells. Formation of a specific type of heterochromatin that silences proliferation-associated genes, senescence-associated heterochromatin foci (SAHF), is one of the first known senescence-associated events. The latter is initiated by the concentration of two histone chaperones HIRA and ASF1a, together with the heterochromatin protein HP1, within PML NBs (Zhang et al. 2005; Ye et al. 2007a; Ye et al. 2007b). What initiates their translocation into NBs and how transient concentration of heterochromatin-associated histones and their chaperones into NBs later favor SAHF formation is not understood.
Partner Degradation
Several unstable proteins have been localized to PML bodies (Anton et al. 1999; Smith et al. 2004), whereas proteins that are impaired in their degradation aggregate with PML, SUMO, and ubiquitin (Yamada et al. 2001b). NBs concentrate proteasomes and ubiquitin (Fabunmi et al. 2001; Lallemand-Breitenbach et al. 2001; Lafarga et al. 2002; Lallemand-Breitenbach et al. 2008). Arsenic trioxide-induced PML degradation has linked PML bodies and protein degradation. Arsenic trioxide triggers an initial sumoylation of K160, followed by proteasome-dependent degradation, a very controversial proposal at the time this was discovered (Zhu et al. 1997; Lallemand-Breitenbach et al. 2001). Yet, identification of a yeast SUMO-dependent ubiquitin ligase (Geoffroy and Hay 2009) and realization that its human ortholog RNF4 localizes to NBs (Hakli et al. 2005) led to renewed interest in these findings. The initial arsenic trioxide-induced K160 sumoylation triggers a secondary polyubiquitination by RNF4 and proteasome-mediated degradation in NBs (Lallemand-Breitenbach et al. 2008; Tatham et al. 2008). There is also some evidence for partner degradation within NBs (Kitagawa et al. 2006; Qin et al. 2006; St-Germain et al. 2008), but how general this is and what the specific signals are that promote partner degradation within NBs is currently unknown.
Note that for the same pathway, a combination of those different mechanisms may cooperate. For example, modulation of p53 function may involve both enhanced p53 modifications and HDM2 sequestration. Similarly, regulation of AKT function involves both dephosphorylation in NBs and sequestration of the DAXX/HAUSP regulators of PTEN activity, to control the critical AKT/TOR/FOXO pathway for stem cell fate (Ito et al. 2009).
CONCLUDING REMARKS AND OPEN QUESTIONS
Because PML can aggregate with so many cellular components, forced overexpression can yield many artifacts, notably by titration of partners or aggregation of unstable proteins (Takahashi et al. 2004; Bernardi and Pandolfi 2007). Paradoxically, there are only few studies in which PML functions were investigated in stable transfectants or in pml-/- cells or in which siRNA were used to inactivate the endogenous PML proteins. Even these experimental approaches preclude assessing the role of the bodies per se, compared with that of the diffuse nucleoplasmic fraction of PML. For example, PML-IV-initiated senescence is unaffected by the viral protein CMV IE1, which disrupts NBs (Ahn and Hayward 1997; Bischof et al. 2002). Separating the PML NB functions from those of unassembled PML may be difficult until ad hoc mutants are described.
Another critical issue concerns isoforms. The vast majority of studies have used the PML-IV isoform, which is expressed at very low levels in most cell lines (Stuurman et al. 1992; Condemine et al. 2006), questioning their actual relevance. Finally, future studies will have to address the paradoxical critical role of PML in multiple key processes and the surprisingly modest phenotype of pml-/- mice. Perhaps one clue to this issue is that stress response has not been fully explored in these animals, despite some important contributions (Wang et al. 1998) and that stem cell defects may have relatively few manifestations in adults.
The PML field has been extremely active, yielding over the past years a large number of remarkable results. Yet, some very fundamental issues remain to be explained. What is the biochemical and physiological basis for NB-assembly? What is the micro-granular core? What are the specific functions of different isoforms? Why are so many NB-associated proteins transcriptionally controlled by interferons, including PML itself (Stadler et al. 1995; Grötzinger et al. 1996; Engelhardt et al. 2001; Fabunmi et al. 2001)? These issues will certainly be clarified in the years to come, maintaining a vivid and exciting field.
ACKNOWLEDGMENTS
We apologize to our many colleagues whose work could not be cited because of space restrictions. We warmly thank Edmond and Francine Puvion for their electron microscopy images and many discussions at various stages of this project, Stan Fakan for helpful comments, Morgane Le Bras and Julien Ablain for critical reading of the manuscript. Work in the laboratory is supported by Ligue Nationale contre le Cancer, INSERM, CNRS, University Paris Diderot, Institut Universitaire de France, Institut National du Cancer and Canceropole programs.
Footnotes
Editors: David Spector and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Ahn JH, Hayward GS 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol 71:4599–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton LC, Schubert U, Bacik I, Princiotta MF, Wearsch PA, Gibbs J, Day P, Realini C, Rechsteiner M, Bennink J, et al. 1999. Intracellular localization of proteasomal degradation of a viral antigen. J Cell Biol 146:113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli CA, Maul GG 1991. Identification of a novel nuclear domain. J Cell Biol 112:785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty E, Jensen K, Freemont P 2009. PML nuclear bodies and their spatial relationships in the mammalian cell nucleus. Front Biosci 14:1182–1196 [DOI] [PubMed] [Google Scholar]
- Beech SJ, Lethbridge KJ, Killick N, McGlincy N, Leppard KN 2005. Isoforms of the promyelocytic leukemia protein differ in their effects on ND10 organization. Exp Cell Res 307:109–117 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8:1006–1016 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Papa A, Pandolfi PP 2008. Regulation of apoptosis by PML and the PML-NBs. Oncogene 27:6299–6312 [DOI] [PubMed] [Google Scholar]
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP 2004. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol 6:665–672 [DOI] [PubMed] [Google Scholar]
- Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J 2001. Regulation and localization of the bloom syndrome protein in response to dna damage. J Cell Biol 153:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Kirsh O, Pearson M, Itahana K, Pelicci PG, Dejean A 2002. Deconstructing PML-induced premature senescence. Embo J 21:3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Nacerddine K, Dejean A 2005. Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways. Mol Cell Biol 25:1013–1024 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS 1996. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971–982 [PubMed] [Google Scholar]
- Boe SO, Haave M, Jul-Larsen A, Grudic A, Bjerkvig R, Lonning PE 2006. Promyelocytic leukemia nuclear bodies are predetermined processing sites for damaged DNA. J Cell Sci 119:3284–3295 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, Hendzel MJ, Bazett-Jones DP 2000. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol 148:283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Kruhlak MJ, Box AK, Hendzel MJ, Bazett-Jones DP 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J Cell Biol 152:1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL, Culjkovic B 2009. Perspectives in PML: A unifying framework for PML function. Front Biosci 14:497–509 [DOI] [PubMed] [Google Scholar]
- Bridger J, Herrmann H, Munkel C, Lichter P 1998. Identification of an interchromosomal compartment by polymerization of nuclear-targeted vimentin. J Cell Sci 111:1241–1253 [DOI] [PubMed] [Google Scholar]
- Butler JT, Hall LL, Smith KP, Lawrence JB 2009. Changing nuclear landscape and unique PML structures during early epigenetic transitions of human embryonic stem cells. J Cell Biochem 107:609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone R, Pearson M, Minucci S, Pelicci PG 2002. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21:1633–1640 [DOI] [PubMed] [Google Scholar]
- Chen YC, Kappel C, Beaudouin J, Eils R, Spector DL 2008. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol Biol Cell 19:3147–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Shi XG, Tang W, Xiong SM, Zhu J, Cai X, Han ZG, Ni JH, Shi GY, Jia PM, et al. 1997. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): I. As2O3 exerts dose-dependent dual effects on APL cells. Blood 89:3345–3353 [PubMed] [Google Scholar]
- Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP 2005. PML bodies: A meeting place for genomic loci? J Cell Sci 118:847–854 [DOI] [PubMed] [Google Scholar]
- Cho G, Lim Y, Golden JA 2009. SUMO interaction motifs in SIZN1 are required for PML-NB localization and for transcriptional activation. J Biol Chem 284:19592–19600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. Embo J 20:4547–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine W, Takahashi Y, Le Bras M, de The H 2007. A nucleolar targeting signal in PML-I addresses PML to nucleolar caps in stressed or senescent cells. J Cell Sci 120:3219–3227 [DOI] [PubMed] [Google Scholar]
- Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, de The H 2006. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res 66:6192–6198 [DOI] [PubMed] [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL 2006. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol 175:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel M-T, Koken M, Romagné O, Barbey S, Bazarbachi A, Stadler M, Guillemin M-C, Degos L, Chomienne C, de Thé H 1993. PML protein expression in hematopoietic and acute promyelocytic leukemia cells. Blood 82:1858–1867 [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW 2004. PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13:523–535 [DOI] [PubMed] [Google Scholar]
- de Thé G, Rivière M, Bernhard W 1960. Examen au microscope électronique de la tumeur VX2 du lapin domestique dérivée du papillome de Shope. Bull Cancer 47:570–584 [PubMed] [Google Scholar]
- Dellaire G, Bazett-Jones DP 2004. PML nuclear bodies: Dynamic sensors of DNA damage and cellular stress. Bioessays 26:963–977 [DOI] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, Bazett-Jones DP 2006a. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol 175:55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP 2006b. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J Cell Sci 119:1026–1033 [DOI] [PubMed] [Google Scholar]
- Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP 2006c. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J Cell Sci 119:1034–1042 [DOI] [PubMed] [Google Scholar]
- Draskovic I, Arnoult N, Steiner V, Bacchetti S, Lomonte P, Londono-Vallejo A 2009. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc Natl Acad Sci 106:15726–15731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de Thé H, Hay RT, Freemont PS 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: Implications for nuclear localisation. J Cell Sci 112:381–393 [DOI] [PubMed] [Google Scholar]
- Dupuy-Coin AM, Bouteille M 1972. Developmental pathway of granular and beaded nuclear bodies from nucleoli. J Ultrastruct Res 40:55–67 [DOI] [PubMed] [Google Scholar]
- Dupuy-Coin AM, Kalifat SR, Bouteille M 1972. Nuclear bodies as proteinaceous structures containing ribonucleoproteins. J Ultrastruct Res 38:174–187 [DOI] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WH, Chen JD, Kakizuka A, Evans RM 1994. A novel macromolecular structure is a target of the promyelocyte- retinoic acid receptor oncoprotein. Cell 76:333–343 [DOI] [PubMed] [Google Scholar]
- Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA 2005. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet 14:1351–1365 [DOI] [PubMed] [Google Scholar]
- Engelhardt OG, Boutell C, Orr A, Ullrich E, Haller O, Everett RD 2003. The homeodomain-interacting kinase PKM (HIPK-2) modifies ND10 through both its kinase domain and a SUMO-1 interaction motif and alters the posttranslational modification of PML. Exp Cell Res 283:36–50 [DOI] [PubMed] [Google Scholar]
- Engelhardt OG, Ullrich E, Kochs G, Haller O 2001. Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp Cell Res 271:286–295 [DOI] [PubMed] [Google Scholar]
- Eskiw CH, Dellaire G, Bazett-Jones DP 2004. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J Biol Chem 279:9577–9585 [DOI] [PubMed] [Google Scholar]
- Eskiw CH, Dellaire G, Mymryk JS, Bazett-Jones DP 2003. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J Cell Sci 116:4455–4466 [DOI] [PubMed] [Google Scholar]
- Everett RD 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266–7273 [DOI] [PubMed] [Google Scholar]
- Everett RD 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol 8:365–374 [DOI] [PubMed] [Google Scholar]
- Everett RD, Earnshaw WC, Pluta AF, Sternsdorf T, Ainsztein AM, Carmena M, Ruchaud S, Hsu WL, Orr A 1999a. A dynamic connection between centromeres and ND10 proteins. J Cell Sci 112:3443–3454 [DOI] [PubMed] [Google Scholar]
- Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A 1999b. Cell cycle regulation of PML modification and ND10 composition. J Cell Sci 112:4581–4588 [DOI] [PubMed] [Google Scholar]
- Everett RD, Murray J, Orr A, Preston CM 2007. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol 81:10991–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Parada C, Gripon P, Sirma H, Orr A 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80:7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN 2001. Interferon γ regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J Cell Sci 114:29–36 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev 14:2015–2027 [PMC free article] [PubMed] [Google Scholar]
- Fuchsova B, Novak P, Kafkova J, Hozak P 2002. Nuclear DNA helicase II is recruited to IFN-α-activated transcription sites at PML nuclear bodies. J Cell Biol 158:463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Hay RT 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10:564–568 [DOI] [PubMed] [Google Scholar]
- Gresko E, Ritterhoff S, Sevilla-Perez J, Roscic A, Frobius K, Kotevic I, Vichalkovski A, Hess D, Hemmings BA, Schmitz ML 2009. PML tumor suppressor is regulated by HIPK2-mediated phosphorylation in response to DNA damage. Oncogene 28:698–708 [DOI] [PubMed] [Google Scholar]
- Grötzinger T, Jensen K, Will H 1996. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-γ activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J Biol Chem 271:25253. [DOI] [PubMed] [Google Scholar]
- Guiochon-Mantel A, Savouret JF, Quignon F, Delabre K, Milgrom E, de Thé H 1995. Effect of PML and PML-RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol Endocrinol 9:1791–1803 [DOI] [PubMed] [Google Scholar]
- Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP 2000. The function of PML in p53-dependent apoptosis. Nat Cell Biol 2:730–736 [DOI] [PubMed] [Google Scholar]
- Gupta P, Ho PC, Huq MM, Ha SG, Park SW, Khan AA, Tsai NP, Wei LN 2008. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc Natl Acad Sci 105:11424–11429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP 2004. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst 96:269–279 [DOI] [PubMed] [Google Scholar]
- Hakli M, Karvonen U, Janne OA, Palvimo JJ 2005. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp Cell Res 304:224–233 [DOI] [PubMed] [Google Scholar]
- Hay RT 2005. SUMO: A history of modification. Mol Cell 18:1–12 [DOI] [PubMed] [Google Scholar]
- Hayakawa F, Privalsky ML 2004. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell 5:389–401 [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I 2006. Specification of SUMO1- and SUMO2-interacting Motifs. J Biol Chem 281:16117–16127 [DOI] [PubMed] [Google Scholar]
- Henderson BR, Eleftheriou A 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 256:213–224 [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh E, Strauss J, Maul G 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Pandolfi PP 2009. A novel signaling network as a critical rheostat for the biology and maintenance of the normal stem cell and the cancer-initiating cell. Curr Opin Genet Dev 19:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP 2008. PML targeting eradicates quiescent leukaemia-initiating cells. Nature 453:1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janderova-Rossmeislova L, Novakova Z, Vlasakova J, Philimonenko V, Hozak P, Hodny Z 2007. PML protein association with specific nucleolar structures differs in normal, tumor and senescent human cells. J Struct Biol 159:56–70 [DOI] [PubMed] [Google Scholar]
- Jegou T, Chung I, Heuvelman G, Wachsmuth M, Gorisch SM, Greulich-Bode KM, Boukamp P, Lichter P, Rippe K 2009. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol Biol Cell 20:2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K, Shiels C, Freemont PS 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223–7233 [DOI] [PubMed] [Google Scholar]
- Jiang WQ, Zhong ZH, Henson JD, Reddel RR 2007. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene 26:4635–4647 [DOI] [PubMed] [Google Scholar]
- Jul-Larsen A, Grudic A, Bjerkvig R, Boe SO 2009. Cell-cycle regulation and dynamics of cytoplasmic compartments containing the promyelocytic leukemia protein and nucleoporins. J Cell Sci 122:1201–1210 [DOI] [PubMed] [Google Scholar]
- Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M-P, Durand B, Lanotte M, Berger R, Chambon P 1992. Structure, localization and transcriptional properties of two classes of retinoic acid receptor α fusion proteins in acute promyelocytic leukemia (APL): Structural similarities with a new family of oncoproteins. EMBO J 11:629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesslich A, von Mikecz A, Hemmerich P 2002. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J Struct Biol 140:167–179 [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Kajiho H, Negishi T, Ura S, Watanabe T, Wada T, Ichijo H, Katada T, Nishina H 2006. Release of RASSF1C from the nucleus by Daxx degradation links DNA damage and SAPK/JNK activation. EMBO J 25:3286–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan SC 2009. Curing APL: Differentiation or destruction? Cancer Cell 15:7–8 [DOI] [PubMed] [Google Scholar]
- Koken MHM, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thépot J, Juhlin L, Degos L, Calvo F, de Thé H 1995. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene 10:1315–1324 [PubMed] [Google Scholar]
- Koken MHM, Puvion-Dutilleul F, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. 1994. The t(15;17) translocation alters a nuclear body in a RA-reversible fashion. EMBO J 13:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S 2007. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol 9:45–56 [DOI] [PubMed] [Google Scholar]
- Kyratsous CA, Silverstein SJ 2009. Components of nuclear domain 10 bodies regulate varicella-zoster virus replication. J Virol 83:4262–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M, Berciano MT, Pena E, Mayo I, Castano JG, Bohmann D, Rodrigues JP, Tavanez JP, Carmo-Fonseca M 2002. Clastosome: A subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol Biol Cell 13:2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HK, Borden KL 2000. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene 19:1623–1634 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H 2008. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10:547–555 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, et al. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As(2)O(3)-induced PML or PML/retinoic acid receptor α degradation. J Exp Med 193:1361–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMorte VJ, Dyck JA, Ochs RL, Evans RM 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci 95:4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehembre F, Muller S, Pandolfi PP, Dejean A 2001. Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene 20:1–9 [DOI] [PubMed] [Google Scholar]
- Li W, Ferguson BJ, Khaled WT, Tevendale M, Stingl J, Poli V, Rich T, Salomoni P, Watson CJ 2009. PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proc Natl Acad Sci 106:4725–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Leo C, Zhu J, Wu X, O’Neil J, Park EJ, Chen JD 2000. Sequestration and inhibition of daxx-mediated transcriptional repression by PML. Mol Cell Biol 20:1784–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Bergmann S, Pandolfi PP 2004. Cytoplasmic PML function in TGF-β signalling. Nature 431:205–211 [DOI] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, et al. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24:341–354 [DOI] [PubMed] [Google Scholar]
- Lin DY, Lai MZ, Ann DK, Shih HM 2003. Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. J Biol Chem 278:15958–15965 [DOI] [PubMed] [Google Scholar]
- Luciani JJ, Depetris D, Usson Y, Metzler-Guillemain C, Mignon-Ravix C, Mitchell MJ, Megarbane A, Sarda P, Sirma H, Moncla A, et al. 2006. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J Cell Sci 119:2518–2531 [DOI] [PubMed] [Google Scholar]
- Mallette FA, Goumard S, Gaumont-Leclerc MF, Moiseeva O, Ferbeyre G 2004. Human fibroblasts require the Rb family of tumor suppressors, but not p53, for PML-induced senescence. Oncogene 23:91–99 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Zhang XD, Ellis NA 2006. SUMO: The glue that binds. Dev Cell 11:596–597 [DOI] [PubMed] [Google Scholar]
- Maul GG, Negorev D, Bell P, Ishov AM 2000. Review: Properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol 129:278–287 [DOI] [PubMed] [Google Scholar]
- Meinecke I, Cinski A, Baier A, Peters MA, Dankbar B, Wille A, Drynda A, Mendoza H, Gay RE, Hay RT, et al. 2007. Modification of nuclear PML protein by SUMO-1 regulates Fas-induced apoptosis in rheumatoid arthritis synovial fibroblasts. Proc Natl Acad Sci 104:5073–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G 2005. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27:1147–1157 [DOI] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D 2000. Covalent modification of p73α by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem 275:36316–36323 [DOI] [PubMed] [Google Scholar]
- Molenaar C, Wiesmeijer K, Verwoerd NP, Khazen S, Eils R, Tanke HJ, Dirks RW 2003. Visualizing telomere dynamics in living mammalian cells using PNA probes. EMBO J 22:6631–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, Dejean A 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J 17:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Gerlich D, Janicki SM, Gebhard M, Eils R, Spector DL 2002. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat Cell Biol 4:106–110 [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9:769–779 [DOI] [PubMed] [Google Scholar]
- Nefkens I, Negorev DG, Ishov AM, Michaelson JS, Yeh ET, Tanguay RM, Muller WE, Maul GG 2003. Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J Cell Sci 116:513–524 [DOI] [PubMed] [Google Scholar]
- Negorev D, Maul GG 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234–7242 [DOI] [PubMed] [Google Scholar]
- Park SW, Hu X, Gupta P, Lin YP, Ha SG, Wei LN 2007. SUMOylation of Tr2 orphan receptor involves Pml and fine-tunes Oct4 expression in stem cells. Nat Struct Mol Biol 14:68–75 [DOI] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, et al. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207–210 [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H 2007. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol 14:581–590 [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F, Chelbi-Alix MK, Koken M, Quignon F, Puvion E, de Thé H 1995. Adenovirus infection induces rearrangements in the intranuclear distribution of the nuclear body-associated PML protein. Exp Cell Res 218:9–16 [DOI] [PubMed] [Google Scholar]
- Qin Q, Inatome R, Hotta A, Kojima M, Yamamura H, Hirai H, Yoshizawa T, Tanaka H, Fukami K, Yanagi S 2006. A novel GTPase, CRAG, mediates promyelocytic leukemia protein-associated nuclear body formation and degradation of expanded polyglutamine protein. J Cell Biol 172:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon F, Chen Z, de Thé H 1997. Retinoic acid and arsenic: towards oncogene targeted treatments of acute promyelocytic leukaemia. Biochim Biophys Acta 1333:M53–M61 [DOI] [PubMed] [Google Scholar]
- Quimby BB, Yong-Gonzalez V, Anan T, Strunnikov AV, Dasso M 2006. The promyelocytic leukemia protein stimulates SUMO conjugation in yeast. Oncogene 25:2999–3005 [DOI] [PubMed] [Google Scholar]
- Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y 2005. Phosphorylation of BLM, dissociation from topoisomerase IIIα, and colocalization with γ-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol 25:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T, Bellodi C, Nicotera P, Salomoni P 2009. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat Neurosci 12:132–140 [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. 2001. The tripartite motif family identifies cell compartments. Embo J 20:2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RA, Adams NM, Stephens DA, Batty E, Jensen K, Freemont PS 2009. Segmentation of fluorescence microscopy images for quantitative analysis of cell nuclear architecture. Biophys J 96:3379–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Khelifi AF 2006. Daxx: Death or survival protein? Trends Cell Biol 16:97–104 [DOI] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP 2002. The role of PML in tumor suppression. Cell 108:165–170 [DOI] [PubMed] [Google Scholar]
- Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B, Teruya-Feldstein J, Tempst P, Pandolfi PP 2006. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 126:269–283 [DOI] [PubMed] [Google Scholar]
- Shao W, Fanelli M, Ferrara FF, Riccioni R, Rosenauer A, Davison K, Lamph WW, Waxman S, Pelicci PG, Lo Coco F, et al. 1998. Arsenic trioxide as an inducer of apoptosis and loss of PML/RARα protein in acute promyelocytic leukemia cells. J Natl Cancer Inst 90:124–133 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D 2005. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 16:2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP 2006. The mechanisms of PML-nuclear body formation. Mol Cell 24:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels C, Islam SA, Vatcheva R, Sasieni P, Sternberg MJ, Freemont PS, Sheer D 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci 114:3705–3716 [DOI] [PubMed] [Google Scholar]
- Smith KP, Byron M, O’Connell BC, Tam R, Schorl C, Guney I, Hall LL, Agrawal P, Sedivy JM, Lawrence JB 2004. c-Myc localization within the nucleus: evidence for association with the PML nuclear body. J Cell Biochem 93:1282–1296 [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP 2008. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Germain JR, Chen J, Li Q 2008. Involvement of PML nuclear bodies in CBP degradation through the ubiquitin-proteasome pathway. Epigenetics 3:342–349 [DOI] [PubMed] [Google Scholar]
- Stadler M, Chelbi-Alix MK, Koken MHM, Venturini L, Lee C, Saïb A, Quignon F, Pelicano L, Guillemin M-C, Schindler C, et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565–2573 [PubMed] [Google Scholar]
- Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci 101:773–784 [DOI] [PubMed] [Google Scholar]
- Stuurman N, Meijne AML, van der Pol AJ, de Jong L, van Driel R, van Renswoude J 1990. The nuclear matrix from cells of different origin. Evidence for a common set of matrix proteins. J Biol Chem 265:5460–5465 [PubMed] [Google Scholar]
- Sun J, Xu H, Subramony SH, Hebert MD 2005. Interactions between coilin and PIASy partially link Cajal bodies to PML bodies. J Cell Sci 118:4995–5003 [DOI] [PubMed] [Google Scholar]
- Szostecki C, Guldner H, Netter H, Will H 1990. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly rrecognized by autoantibodies of patients with primary biliary cirrhosis. J Immunol 145:4338–4347 [PubMed] [Google Scholar]
- Takahashi H, Hatakeyama S, Saitoh H, Nakayama KI 2005. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J Biol Chem 280:5611–5621 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Lallemand-Breitenbach V, Zhu J, de Thé H 2004. PML nuclear bodies and apoptosis. Oncogene 23:2819–2824 [DOI] [PubMed] [Google Scholar]
- Tao H, Simmons BN, Singireddy S, Jakkidi M, Short KM, Cox TC, Massiah MA 2008. Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry 47:2450–2457 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10:538–546 [DOI] [PubMed] [Google Scholar]
- Tavalai N, Papior P, Rechter S, Leis M, Stamminger T 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80:8006–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalai N, Papior P, Rechter S, Stamminger T 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J Virol 82:126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A 1995. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res 55:1590–1597 [PubMed] [Google Scholar]
- Torii S, Egan DA, Evans RA, Reed JC 1999. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J 18:6037–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP 2006. Identification of a tumour suppressor network opposing nuclear Akt function. Nature 441:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL 2000. Visualization of gene activity in living cells. Nat Cell Biol 2:871–878 [DOI] [PubMed] [Google Scholar]
- Vagner-Capodano AM, Bouteille M, Stahl A 1982. Nucleolar ribonucleoprotein release into the nucleoplasm as nuclear bodies in cultured thyrotropin-stimulated thyroid cells: Autoradiographic kinetics. J Ultrastruct Res 78:13–25 [DOI] [PubMed] [Google Scholar]
- Villagra NT, Navascues J, Casafont I, Val-Bernal JF, Lafarga M, Berciano MT 2006. The PML-nuclear inclusion of human supraoptic neurons: A new compartment with SUMO-1- and ubiquitin-proteasome-associated domains. Neurobiol Dis 21:181–193 [DOI] [PubMed] [Google Scholar]
- Wang Z-G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP 1998. PML is essential for multiple apoptotic pathways. Nature Genet 20:266–272 [DOI] [PubMed] [Google Scholar]
- Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol 164:515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P 2008. Dynamics of component exchange at PML nuclear bodies. J Cell Sci 121:2731–2743 [DOI] [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A 1994. Retinoic acid regulates aberrant nuclear localization of PML/RARa in acute promyelocytic leukemia cells. Cell 76:345–356 [DOI] [PubMed] [Google Scholar]
- Wiesmeijer K, Molenaar C, Bekeer IM, Tanke HJ, Dirks RW 2002. Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J Struct Biol 140:180–188 [DOI] [PubMed] [Google Scholar]
- Wu G, Lee WH, Chen PL 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J Biol Chem 275:30618–30622 [DOI] [PubMed] [Google Scholar]
- Wu G, Jiang X, Lee WH, Chen PL 2003. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res 63:2589–2595 [PubMed] [Google Scholar]
- Yamada M, Sato T, Shimohata T, Hayashi S, Igarashi S, Tsuji S, Takahashi H 2001a. Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases. Am J Pathol 159:1785–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Wood JD, Shimohata T, Hayashi S, Tsuji S, Ross CA, Takahashi H 2001b. Widespread occurrence of intranuclear atrophin-1 accumulation in the central nervous system neurons of patients with dentatorubral-pallidoluysian atrophy. Ann Neurol 49:14–23 [DOI] [PubMed] [Google Scholar]
- Yang S, Jeong JH, Brown AL, Lee CH, Pandolfi PP, Chung JH, Kim MK 2006. Promyelocytic leukemia activates Chk2 by mediating Chk2 autophosphorylation. J Biol Chem 281:26645–26654 [DOI] [PubMed] [Google Scholar]
- Yang S, Kuo C, Bisi JE, Kim MK 2002. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat Cell Biol 4:865–870 [DOI] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Kennedy A, Banumathy G, Zhang R, Adams PD 2007a. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol Cell 27:183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD 2007b. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol 27:2452–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8:19–30 [DOI] [PubMed] [Google Scholar]
- Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP 1999. A role for PML and the nuclear body in genomic stability. Oncogene 18:7941–7947 [DOI] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP 2000a. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748–2752 [PubMed] [Google Scholar]
- Zhong S, Salomoni P, Ronchetti S, Guo A, Ruggero D, Pandolfi PP 2000b. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J Exp Med 191:631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lallemand-Breitenbach V, de Thé H 2001. Pathways of retinoic acid- or arsenic trioxide-induced PML/RARα catabolism, role of oncogene degradation in disease remission. Oncogene 20:7257–7265 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen Z, Lallemand-Breitenbach V, de Thé H 2002. How acute promyelocytic leukemia revived arsenic. Nature Reviews on Cancer 2:705–713 [DOI] [PubMed] [Google Scholar]
- Zhu J, Koken MHM, Quignon F, Chelbi-Alix MK, Degos L, Wang ZY, Chen Z, de Thé H 1997. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci 94:3978–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]