Abstract
The tumor suppressor p53 is a master sensor of stress that controls many biological functions, including implantation, cell-fate decisions, metabolism, and aging. In response to a defined stress signal such as gamma radiation, the response of p53 is heterogeneous in vivo. Like a complex barcode, the ability of p53 to function as a central hub that integrates defined stress signals into decisive cellular responses, in a time- and cell-type dependent manner, is facilitated by the extraordinary complexity of its regulation. Key components of this barcode are the autoregulation loops, which positively or negatively regulate p53’s activities. Thus, this article focuses on reviewing our current understanding of how autoregulation loops formed between p53 and how its transcriptional targets regulate the activities of p53 at a variety of levels, through mdm2-dependent and -independent pathways. Knowing that a large number of autoregulation loops exist that influence p53’s activity, our future challenge is to elucidate which of these play a central role in regulating p53, under which conditions, in response to what stress, and at which particular stage of our lives. Such knowledge may ultimately lead to the development of more effective anticancer therapeutics.
Autoregulatory loops involving p53 transcription factors and their targets fine-tune stress responses, ensuring that cell-fate decisions are executed appropriately.
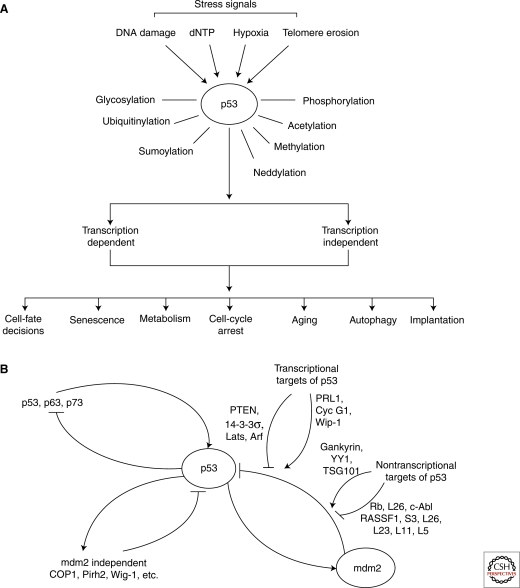
Thirty years after its discovery, we now know that the tumor suppressor p53 is a master sensor of stress. Emerging studies have shown that, in addition to its ability to function as one of the most important tumor suppressors, p53 also controls many other biological functions, including implantation, cell-fate decisions (Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marion et al. 2009; Utikal et al. 2009), metabolism, and aging. Because of its central role in dictating a large number of biological outcomes in response to a variety of stress signals, the activity of p53 is regulated with exquisite precision. The activities of p53, both transcription-dependent and independent, are regulated via its mRNA and protein levels, cellular localization, and ability to bind over 100 cellular proteins and control the expression of thousands of potential target genes. To achieve the level of precision required, p53 is posttranslationally modified by almost all types of protein modification, including phosphorylation, acetylation, glycosylation, ubiquitination, sumoylation, neddylation, and methylation. Importantly, p53’s response to a defined stress signal is heterogeneous in vivo (Fig. 1A). Tissues can be sorted into three groups according to the response of p53 and its downstream effectors to a defined signal: In group 1 tissues, p53 responds and generates a defined cellular response; in group 2 tissues, p53 responds but does not generate a defined cellular response; and in group 3, there is no response by p53. The ability of p53 to function as a central hub that integrates defined stress signals into decisive cellular responses, in a time- and cell-type-dependent manner, is facilitated by the extraordinary complexity of p53’s regulation, like that of a complex barcode (Murray-Zmijewski et al. 2008). Key components of this barcode are the autoregulation loops, which positively or negatively regulate p53’s activities. Thus, this article focuses on reviewing our current understanding of how these loops contribute to the regulation of p53.
Figure 1.
The variety of p53’s responses to differing stress signals is partly achieved through a series of autoregulation loops. The complexity of p53’s regulation enables it to dictate a large number of biological outcomes, in response to a variety of different stress signals (A). Key factors in facilitating this complexity of regulation are the mdm2-dependent, mdm2-independent, and p53 family sibling autoregulation loops (B).
The autoregulation loops described here are limited to those that are formed between p53 and its transcriptional targets. The biological implications of each loop largely depend on the function of the transcriptional target in question and, thus, three major types of autoregulation loops are described: (1) mdm2 dependent; (2) mdm2 independent; and (3) extended autoregulation loops formed among the p53 family siblings (Fig. 1B). Of these, p53/mdm2 is the master autoregulation loop, and it dictates the fate of an organism by controlling the expression level and activity of p53. It is therefore not surprising that this autoregulation loop is itself subject to different types of regulation, which can be divided into two subgroups. The first group uses transcriptional targets of p53 to positively or negatively regulate either the stability of mdm2 and/or its E3 ubiquitin ligase activity, to form a second layer of autoregulation. This multi-layered autoregulation ensures that p53’s activity is controlled with the utmost precision. The second group contains proteins that are not transcriptional targets of p53, and the majority of these act by affecting the stability or enzymatic activity of mdm2. The list of regulators that belong to this group is perhaps the longest, and only a few examples are shown here. Interestingly, some are themselves subject to direct regulation by mdm2 (Rb, L26, and TSG101); hence, they also form mdm2 autoregulation loops. It is important to note that almost all of the regulators shown here are deregulated in human tumors.
Despite the fact that p53 stimulates the production of mdm2 to protect its own interests, the expression level and activity of p53 is also regulated by several mdm2-independent autoregulation loops. These regulate the expression of p53 at the transcriptional, mRNA stability, and protein stability levels. Furthermore, autoregulation loops exist that control the transcriptional activity of p53 by affecting its ability to bind to its targets, and extended autoregulation loops exist among the p53 family siblings. The message emerging from these autoregulation loops is, therefore, simple and clear: p53 is a looped master protein that is both positively and negatively regulated by a series of autoregulation loops, which are able to control its activity with a fine level of precision.
LIFELINE: THE p53/mdm2 NEGATIVE-FEEDBACK LOOP
As a transcription factor, p53 is able to transactivate or transrepress genes that are involved in a large number of biological processes, including cell-cycle arrest, senescence, apoptosis, and autophagy. One of the first and most important transcriptional targets of p53 to be identified, mdm2, binds to p53 and inhibits its transcriptional activity, forming the first identified negative p53 autoregulation loop (Momand et al. 1992; Oliner et al. 1992; Barak et al. 1993). Subsequent studies have demonstrated that mdm2 is also the major E3 ubiquitin ligase of p53, binding and targeting it for proteosome-mediated degradation (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997). Thus, mdm2 is able to negatively regulate p53 in two different ways. The biological significance of the p53/mdm2 autoregulation loop in determining life or death was revealed in an elegant study using mdm2 knockout mice. Mdm2 deficiency is embryonic lethal because of excessive apoptosis, caused by an increase in both p53’s protein level and activity. Remarkably, this lethality can be completely rescued by an absence of p53 (Jones et al. 1995; Montes de Oca Luna et al. 1995). Therefore, p53 plays an essential role in controlling the growth or death of a cell, ultimately deciding the fate of an organism, and “produces” mdm2 to control its own activity.
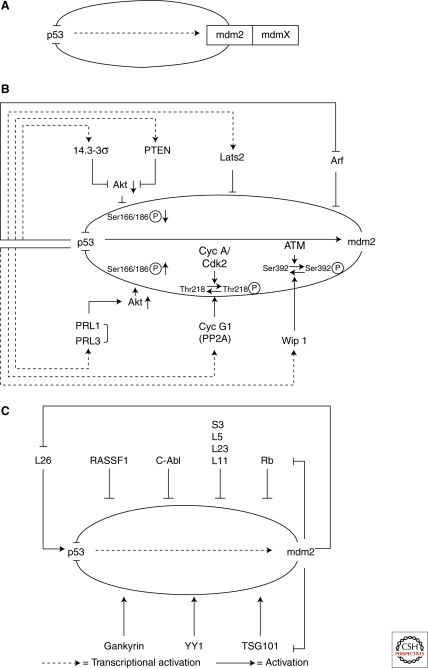
In addition to mdm2, mdmX (a close relative) also plays a key role in controlling the activity of p53. MdmX deficiency is similarly embryonic lethal due to excessive apoptosis and cell-cycle arrest, and, as with mdm2, this lethality is completely rescued by an absence of p53 (Parant et al. 2001; Migliorini et al. 2002). However, despite its structural similarity to mdm2, the ring finger of mdmX does not possess E3 ubiquitin ligase activity. In addition, mdmX alone can not target p53 for degradation, even though it can bind to p53 and inhibit its transcriptional activity. Thus, how mdmX can play such an important role in regulating p53 and why mdm2 fails to compensate for mdmX deficiency to regulate the activity of p53 are important questions. It is possible that mdmX works for mdm2. Supporting evidence for this idea comes from the fact that mdmX binds to, stabilizes, and enhances mdm2’s ability to ubiquitinate and target p53 for degradation. MdmX has not yet been identified to be a direct transcriptional target of p53 and consequently does not form an autoregulation loop with it. It is therefore possible that mdmX acts as a cofactor of mdm2, providing a further level of regulation of p53’s activity (Fig. 2A).
Figure 2.
Control of the p53/mdm2 autoregulation loop. The p53/mdm2 autoregulation loop is central to the control of p53’s varied functions. The loop itself can be regulated by both transcriptional (B) and nontranscriptional (C) targets of p53. MdmX may act as a cofactor of mdm2, providing a further level of regulation (A).
Regardless of the role of mdmX, the need to precisely control p53’s activity was convincingly demonstrated by the phenotypes of a hypomorph mdm2 transgenic mouse, where slightly reduced mdm2 expression (around 30%) resulted in a small increase in p53 protein level and activity. This small change had a profound impact on the ability of p53 to control cell growth in normal development (Mendrysa et al. 2003) and confer tumor resistance at a later age (Mendrysa et al. 2006). In addition, a three- to fourfold increase in mdm2 expression has been shown to augment B-cell proliferation, and reduce their susceptibility to p53-dependent apoptosis, in transgenic mice. These effects were caused by the inhibition of p53 and resultant suppression of p21 expression (Wang et al. 2008). All of these studies argue that whereas the p53/mdm2 autoregulation loop is a lifeline that makes the decision between life and death, a number of factors that affect the loop’s integrity, either by strengthening or weakening it, can have a profound impact on an organism’s quality of life. These factors are likely to be targeted in pathological conditions such as cancer.
CONTROL OF THE p53/mdm2 LIFELINE
As the p53/mdm2 autoregulation loop plays such a pivotal role in controlling p53 activity, it is not surprising that it is itself the center of many other positive and negative regulatory loops. Stress signals, particularly ones that have pathological consequences, result in a large number of break points in the p53/mdm2 circuit. The ultimate consequence of these breakages is a change in p53’s activities, which are either enhanced or reduced. Molecules that cause these breakpoints form additional layers of the p53/mdm2 regulation loop. Interestingly, the existing evidence seems to suggest that the prime target of these layers is mdm2. Many mdm2 regulators are themselves transcriptional targets of p53, or direct targets of mdm2, illustrating once again the extent of the complexity built into our genome to regulate the activity of p53. Mdm2 regulators identified to date can be sorted into two major groups: those that are transcriptional targets of p53 (Fig. 2B) and those that are not (Fig. 2C).
Regulation of the p53/mdm2 Loop by Transcriptional Targets of p53
Apart from being an E3 ubiquitin ligase of p53, mdm2 is itself autoubiquitinated. Mdm2 normally has a protein half life of 20–30 minutes (Olson et al. 1993). Thus, its own expression level is tightly regulated. One of the major regulators of the expression level and stability of mdm2 is phosphorylation. Phosphorylation at Ser166 and Ser186 by protein kinase B/Akt kinase results in the stabilization of mdm2, and is achieved through its increased nuclear localization and reduced autoubiquitination (Zhou et al. 2001; Ogawara et al. 2002; Feng et al. 2004). Interestingly, this regulation of mdm2 turns out to be a focal point of many further regulation loops, formed by a number of recently identified p53 transcriptional targets that positively or negatively regulate p53’s activities. For example, tyrosine phosphatase of regenerating liver 1 and 3 (PRL-1 and PRL-3, respectively), negatively regulate p53 expression by enhancing the ability of Akt to increase the phosphorylation of mdm2 at Ser166 (Min et al. 2009). Two other p53 transcriptional targets, 14-3-3σ and PTEN, also use Akt kinase as the basis for their p53 regulation loop. However, in this case, both 14-3-3σ and PTEN positively regulate p53’s activities. 14-3-3σ binds to and inhibits Akt kinase activity directly, whereas PTEN inhibits Akt kinase activity by inhibiting PI3 kinase, an upstream kinase that activates Akt. Thus, by reducing the phosphorylation of mdm2 at Ser166 and Ser186, 14-3-3σ and PTEN increase p53’s activity by preventing mdm2 from targeting p53 for degradation (Hermeking et al. 1997; Stambolic et al. 2001; Freeman et al. 2003; Yang et al. 2003).
In addition to Akt, a number of other kinases regulate the stability and activity of mdm2. Among them, ATM and cyclin-dependent kinase cyclin A/cdk2 play an important role in regulating its activity. By phosphorylating Thr218 and Ser392, cyclin A/cdk2 and ATM, respectively, reduce the activity of mdm2 (Maya et al. 2001; Zhang and Prives 2001). Like Akt phosphorylation, this regulation of mdm2 is also at the center of a number of autoregulation loops formed by p53 targets, such as cyclin G1 and Wip1. By binding to protein phosphatase 2A, cyclin G1 enhances the ability of PP2A to dephosphorylate mdm2 at T218, a site that is phosphorylated by cyclin A/cdk2 and negatively regulates mdm2’s activity (Okamoto et al. 2002). Similarly, Wip1, another serine/threonine protein phosphatase, dephosphorylates mdm2 at Ser395, a site that is phosphorylated by ATM kinase and inhibits mdm2’s ability to degrade p53. Thus, in both situations, the dephosphorylation of mdm2 stabilizes it and enhances its ability to ubiquitinate and degrade p53.
Apart from affecting mdm2 phosphorylation, some p53 transcriptional targets regulate the p53/mdm2 loop by directly influencing mdm2’s E3 ligase activity. One of the best known examples of this is the tumor suppressor protein p19Arf, the expression of which is repressed by p53 (Bates et al. 1998). By binding to mdm2, p19Arf inhibits the E3 ligase activity of mdm2 (Zhang et al. 1998). Recently, another p53 target Lats2 (large tumor suppressor 2) was shown to similarly regulate p53. Lats2 stabilizes p53 by binding to mdm2 and inhibiting its E3 ligase activity (Aylon et al. 2006). Together, these examples support the notion that mdm2 is a prime target for additional regulatory activity.
The number of p53 transcriptional targets identified to date that fulfil the criteria necessary to be described as autoregulation loops is increasing, and many of them seem to predominantly affect the phosphorylation status or E3 ligase activity of mdm2. Significantly, many of these transcriptional targets are connected to other important signaling pathways such as PI3 kinase/Akt, ATM, and cyclin/cdk. The PI3 kinase/Akt signaling pathway is essential in controlling cell growth and metabolism. ATM is the key DNA damage response pathway, whereas cyclin/cdk is essential in controlling cell proliferation or cell-cycle arrest. Thus, these various autoregulation loops connect cell signals involved in cell growth, metabolism, DNA damage and repair, as well as a variety of cell-cycle checkpoints, to the lifeline of p53’s regulation that is the p53/mdm2 autoregulation loop.
Regulation of the p53/mdm2 Loop by Nontranscriptional Targets of p53
A large number of proteins that are not transcriptional targets of p53 have been identified that affect p53’s activity by interfering with the p53/mdm2 circuit (Fig. 2C). Of these, ribosomal proteins including L11, L5, L23, and S7 are among the best studied. By binding to a defined acidic, zinc finger domain of mdm2, these proteins increase p53 stability by inhibiting the E3 ligase activity of mdm2 (Lohrum et al. 2003; Zhang et al. 2003; Dai and Lu 2004; Dai et al. 2004; Zhu et al. 2009). Interestingly, a number of human cancer-associated MDM2 alterations have been reported, a number of which target this region and disrupt the interaction of L5 and L11 with mdm2 (Lindstrom et al. 2007). Gankryin protein, on the other hand, can bind to mdm2 and promote its ability to degrade p53. YY1, another transcription factor, also enhances p53 degradation by increasing mdm2/p53 binding. Interestingly, some of these regulators of mdm2 are themselves targets of mdm2, and form a new type of autoregulation loop that affects the activity of p53. The first known example of such a loop is that formed by the retinoblastoma protein Rb. By binding to a region of mdm2 that is similar to the one that p19Arf recognizes, Rb prevents mdm2 from degrading p53 (Hsieh et al. 1999). Rb is also targeted by mdm2 through the proteasome pathway. Rb, mdm2, and the C8 subunit of the 20S proteasome have been shown to interact both in vivo and in vitro, and mdm2 promotes Rb’s interaction with C8 (Sdek et al. 2005). Hence, by binding to Rb, mdm2 mediates its degradation. C-Abl similarly protects p53 from inhibition by mdm2. Phosphorylation of human mdm2 (Hdm2) at Tyr394 by c-Abl impairs its ability to inhibit p53; thus, c-Abl contributes to the maximum accumulation of p53 in response to DNA damage (Goldberg et al. 2002). Recently, a Ubc-domain-containing protein called TSG101 was also reported to be regulated by mdm2. By interfering with the E2 enzyme activity that ubiquitinates mdm2, TSG101 enhances its stability, resulting in a decrease in p53 expression (Li et al. 2001). In addition, it was recently demonstrated that the tumor suppressor RASSF1A promotes mdm2’s self-ubiquitination and degradation, by disrupting the MDM2-DAXX-HAUSP complex. RASSF1 also partially contributes to p53-dependent checkpoint activation in response to DNA damage, therefore playing an important role in regulating the p53-mdm2 pathway (Song et al. 2008).
Although a large number of autoregulation loops have been identified that regulate the activities of mdm2 to ensure its effective regulation of p53, it is puzzling to notice that the majority, if not all, of p53 regulators target mdm2 but not mdmX, even though mdmX is as critical as mdm2 in controlling the life or death decision of p53. It is tempting to make the conclusion in this case that p53 forms an autoregulation loop with mdm2 but not with mdmX. Whether mdmX is a second-class citizen in the p53 regulatory hierarchy or will turn out to be the “X” factor may prove pivotal to our future understanding of p53’s function.
OUT OF THE p53/mdm2 MASTER LOOP
The expression level and activities of p53 are regulated by a large number of molecules that are independent of mdm2. Many of them form autoregulation loops to either directly regulate the transcriptional activity of p53 or regulate its activity by altering its expression level. How a particular autoregulation loop affects p53’s activity depends on the individual biological properties of the p53 transcriptional targets involved.
Regulating p53 Expression Independently of mdm2
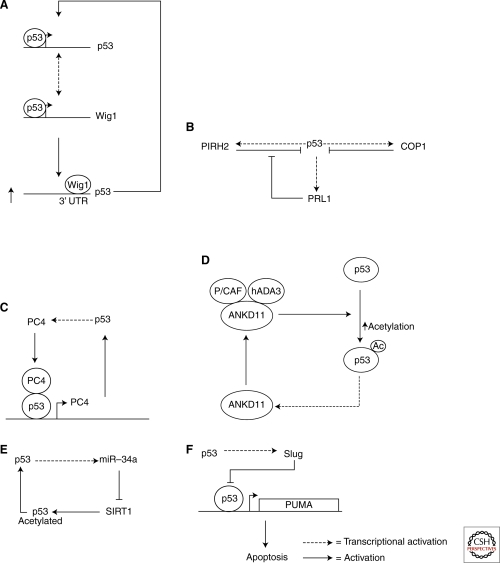
A number of studies have shown that p53 expression is regulated at a variety of levels, and that autoregulation loops exist at all of these. p53 itself forms an autoregulation loop because it is able to induce its own transcription by binding to its promoter (Wang and El-Deiry 2006). The mRNA stability of p53 is also regulated by its own transcriptional target Wig1 (Fig. 3A). By binding to the 3′UTR via an AU-rich element, Wig-1 stabilizes p53 mRNA and forms a positive-feedback loop with p53 (Vilborg 2009). Most autoregulation loops are involved in regulating the stability of p53, however, reflecting the fact that this is how most stress signals exert their effects.
Figure 3.
Mdm2-independent autoregulation of p53. A large number of molecules autoregulate p53 independently of mdm2. These include Wig1 (A), PRL1 (B), PC4 (C), ANKD11 (D), miR-34a (E), and Slug (F).
Apart from mdm2, other E3 ubiquitin ligases have been identified, such as COP1, Pirh2, CARP, and Torpor. Interestingly, some of them also form autoregulation loops with p53. The best studied of these are the p53/COP1 and p53/Pirh2 loops (Fig. 3B). Both COP1 and Pirh2 are transcriptional targets of p53. By binding to p53, they mediate its ubiquitination and degradation independently of mdm2. It is interesting to note that p53/Pirh2 autoregulation is also subject to a further regulation by a newly identified p53 target, PRL-1, which reduces p53’s stability by indirectly enhancing the expression of Pirh2 mRNA.
Direct Regulation of p53’s Activity
As it is a transcriptional coactivator, PC4 plays a critical role in transcription, DNA replication, repair, and cellular transformation. PC4 was also recently identified as a p53 transcriptional target. PC4 binds p53 and recruits it to its own promoter, enhancing its transcription (Fig. 3C). Thus, PC4 forms a positive autoregulation loop to regulate its own expression. This mode of action does not apply to all other p53 coactivators, however. For Ankryin repeat domain 11 (ANKD11), it is the acetylation of p53 that ANKD11 uses to control the activity of p53. ANKD11 binds to p53 acetyltransferases and cofactors P/CAF and hADA3, and stimulates the transcriptional activity of p53 by enhancing its acetylation (Neilsen et al. 2008). As it is a transcriptional target of p53, ANKD11 consequently forms a positive p53 autoregulation loop (Fig. 3D).
Influencing the acetylation of p53 to control its transcriptional activity is also used by the autoregulation loop formed by p53/miR-34a/SIRT1. The function of micro RNA (miRNA) is to silence gene expression by binding and degrading its target RNA, and miR-34a was the first miRNA to be identified as a p53 transcriptional target. Interestingly, p53 is not itself a target of miR-34a. In fact, p53-induced miR-34a forms a positive autoregulation loop with p53 by stimulating p53-induced apoptosis. This is achieved by altering the acetylation status of p53, as miR-34a silences SIRT1 (silent information regulator 1) expression. SIRT1 is an NAD (nicotinamide adenine dinucleotide)-dependent deacetylase that deacetylates proteins including p53. Thus, like that of ANKD11, the positive autoregulation loop formed by miR-34a functions by enhancing p53’s acetylation (Fig. 3E).
Another type of autoregulation loop that directly affect the transcriptional activity of p53 is that formed by p53/Slug/PUMA (Fig. 3F). Being a transcriptional target of p53 and a transcription factor itself, Slug binds directly to the promoter of PUMA and prevents p53 from inducing PUMA’s expression. PUMA is one of the most important p53 targets in mediating p53-induced apoptosis, and this negative autoregulation loop plays an important role, particularly in hematopoietic progenitors (Chipuk et al. 2005; Wu et al. 2005).
In addition to the loops described previously, it is worth noting that a common denominator of p53-inducing stresses is nucleolar disruption. Micropore irradiation has demonstrated that large amounts of nuclear DNA damage fails to stabilize p53 unless the nucleolus is also disrupted. This suggests that that the nucleolus itself acts as a stress sensor that maintains low levels of p53, that are automatically elevated when nucleolar function is impaired in response to stress. As ribosomal biosynthesis predominantly occurs in the nucleolus, this theory also helps to explain cell-cycle-related variations in p53 levels that correlate with phases of nucleolar assembly and disassembly as the cycle progresses (Rubbi and Milner 2003).
CONTROLS AMONG SIBLINGS
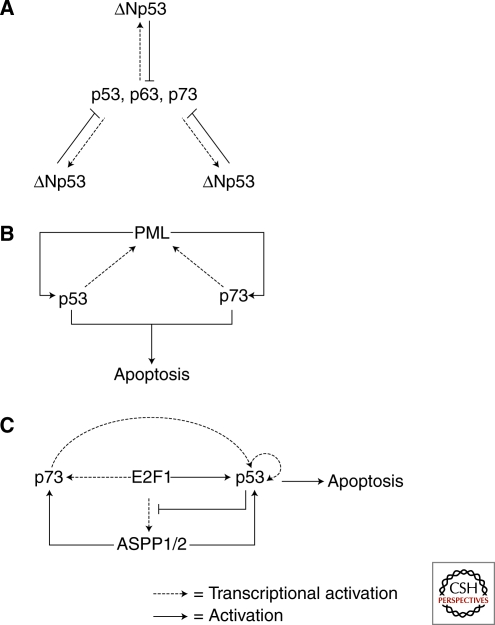
Autoregulation loops are not only restricted to p53. The activities of the other p53 family members, p63 and p73, are also regulated by a number of autoregulation loops. Most interesting of all is the cross regulation achieved among the p53 siblings by the various loops that they form. In vertebrates, there are three family members: p53, p63, and p73. The most homologous regions among these three are their DNA binding domains. Therefore, they are all able to bind to p53-responsive sites and transactivate or transrepress genes. Interestingly, there is a p53 binding site in intron 3 of the p63 and p73 (Yang et al. 1998; Yang et al. 2000) genes and intron 4 of the p53 gene (Bourdon et al. 2005). As a result, p53 and its siblings use these p53 binding sites to transactivate the expression of their amino-terminal truncated and transcriptionally inactivated splice variants: ΔNp53, ΔNp63, and ΔNp73. Because ΔNp53, ΔNp63, and ΔNp73 have the same DNA binding domains as p53, p63, and p73, they can all compete with transcriptionally active p53, p63, and p73 to bind their DNA targets. Therefore, ΔNp53, ΔNp63, and ΔNp73 act as dominant–negative inhibitors of p53, p63, and p73, and form negative autoregulation loops (Fig. 4A). Importantly, these autoregulation loops are highly conserved, and the autoregulation of p53 and ΔNp53 has been reported in zebrafish (Chen et al. 2009).
Figure 4.
Autoregulation loops formed among the p53 family siblings. The three members of the p53 family form highly conserved negative autoregulation loops (A). In addition, p53 and p73 form a positive autoregulation loop with PML (B), and another with E2F1/ASPP1,2 (C).
Because of the similarity in their DNA binding domains, p53 and its siblings often share the same transcriptional targets and are subject to the same autoregulation. Thus, in theory, the roles of inducer and inhibitor within these loops are interchangeable. This unique feature forms the basis of their cross-regulation. One such example is the ability of p73 to bind to the promoter of p53 via a p53 binding site, and induce the expression of p53 mRNA in response to DNA damage caused by therapeutic agents such as etoposide or adriamycin. Thus, the expression of p53 is positively regulated by both p63 and p73 (Wang and El-Deiry 2006). Similarly, p53 and p73 form a positive autoregulation loop with PML (Fig. 4B). PML is the promyelocytic leukemia (PML) tumor suppressor and its expression is induced by both p53 and p73 because of the presence of a p53 binding site in its promoter. Interestingly, PML uses two different routes to activate its inducers. For p53, PML binds the DNA binding domain of p53 and specifically enhances its transcriptional activity on proapoptotic promoters. This is perhaps partly achieved through an increase in the sumo-modification of p53 (Gostissa et al. 1999; Fogal et al. 2000; de Stanchina et al. 2004). For p73, PML uses an intermediate to activate p73. PML binds to and stabilizes YAP (transcriptional coactivator Yes-associated protein), by mediating its sumo-modification. YAP then binds p73 and enhances its transcriptional activity, thus inducing apoptosis in response to DNA damaging signals (Lapi et al. 2008).
Finally, p53 and its siblings can work together to form extended and intertwined autoregulation loops that control specific cellular responses. P73/E2F1/ASPP1,2/p53 is one such example. The transcription factor E2F1 induces the expression of p73, ASPP1, and ASPP2 (Irwin et al. 2000; Lissy et al. 2000; Fogal et al. 2005b). In turn, induced ASPP1 and ASPP2 bind to and stimulate the transcriptional activities of both p53 and p73 (Samuels-Lev et al. 2001; Bergamaschi et al. 2004). E2F1 also binds to and enhances the transcriptional activity of p53 (Fogal et al. 2005a). Thus, in this autoregulation loop, E2F1 activates both p53 and p73 (Fig. 4C). This type of regulation undoubtedly achieves additive or synergistic effects in controlling cellular responses to stress signals.
Although a number of the autoregulation loops listed here are formed between p53 and p73, in theory there is no reason to suggest that p53 and p63, or p63 and p73, cannot form similar autoregulation loops to achieve cross-regulation. However, spatial and temporal regulation of the expression and biological functions of p53 and its siblings dictates which combination is used to form a particular autoregulation loop, in response to a defined stress signal in a cell- and time-dependent manner. The countless possible combinations that result form the basis of the complexity of p53’s regulation and facilitate the exquisite precision of its execution. The possibility of cross-regulation between the p53 family siblings similarly makes it impossible to discount the possibility that any effect on mdm2’s activity, and subsequent effect on p53, would not also affect the autoregulation of p63 and p73.
CONCLUSIONS AND FUTURE PERSPECTIVES
Autoregulation is perhaps one of the most common, yet fundamental, types of regulation that exists in biology, and there is no doubt that autoregulation loops are at the center of p53’s regulation. In this article, a number of autoregulation loops have been identified that are known to regulate the activities of p53 at a variety of levels, through mdm2-dependent and -independent pathways. After 30 years of extensive study, it is clear that p53 is a master sensor of stress. It is also clear that it acts as a central hub that can integrate different defined stress signals into precise and diverse cellular responses. These responses influence a wide variety of important biological processes, including implantation, the determination of cell fate, metabolism, tumor suppression, and aging. In response to a defined stress signal such as γ radiation, the response of p53 is heterogenous in vivo. The complexity of p53’s regulation underlies this heterogeneity. Understanding how, when, and where p53 integrates a particular stress signal into a precise cellular response is the ultimate goal of p53 biology. Knowing that p53’s activity is heavily regulated by a large number of autoregulation loops, our future challenge is to elucidate which of these play a central role in regulating p53’s activities, under which conditions, in response to what stress signal, and at which particular stage of our lives. Such knowledge may ultimately lead to more effective anticancer therapeutics. Several drugs that disrupt the p53/mdm2 autoregulation loop, such as nutlin and RITA, are already under development. Nutlin, in particular, is currently undergoing clinical trials and may prove to be an important development in cancer treatment in the future.
ACKNOWLEDGMENTS
This work is supported by Ludwig Institute for Cancer Research, AICR, and EU active p53 consortium. I would like to give my special thanks to Dr. Claire Beveridge for her critical reading of the manuscript.
Footnotes
Editors: Arnold J. Levine and David P. Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M 2006. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev 20:2687–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M 1993. mdm2 expression is induced by wild type p53 activity. EMBO J 12:461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124–125 [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X 2004. ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol 24:1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 19:2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, Lane DP, Peng J 2009. p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev 23:278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR 2005. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309:1732–1735 [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H 2004. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 279:44475–44482 [DOI] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H 2004. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 24:7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW 2004. PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13:523–535 [DOI] [PubMed] [Google Scholar]
- Feng J, Tamaskovic R, Yang Z, Brazil DP, Merlo A, Hess D, Hemmings BA 2004. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem 279:35510–35517 [DOI] [PubMed] [Google Scholar]
- Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J 19:6185–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogal V, Hsieh JK, Royer C, Zhong S, Lu X 2005a. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J 24:2768–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogal V, Kartasheva NN, Trigiante G, Llanos S, Yap D, Vousden KH, Lu X 2005b. ASPP1 and ASPP2 are new transcriptional targets of E2F. Cell Death Differ 12:369–376 [DOI] [PubMed] [Google Scholar]
- Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, Whale AD, Martinez-Diaz H, Rozengurt N, Cardiff RD, et al. 2003. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 3:117–130 [DOI] [PubMed] [Google Scholar]
- Goldberg Z, Vogt Sionov R, Berger M, Zwang Y, Perets R, Van Etten RA, Oren M, Taya Y, Haupt Y 2002. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J 21:3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J 18:6462–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299 [DOI] [PubMed] [Google Scholar]
- Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B 1997. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell 1:3–11 [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420:25–27 [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S 2009. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460:1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JK, Chan FS, O’Connor DJ, Mittnacht S, Zhong S, Lu X 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell 3:181–193 [DOI] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, et al. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407:645–648 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206–208 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460:1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH 1997. Regulation of p53 stability by Mdm2. Nature 387:299–303 [DOI] [PubMed] [Google Scholar]
- Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, Pandolfi PP, Givol D, Strano S, Lu X, Blandino G 2008. PML, YAP, p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell 32:803–814 [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460:1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liao J, Ruland J, Mak TW, Cohen SN 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc Natl Acad Sci 98:1619–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y 2007. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol 27:1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF 2000. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 407:642–645 [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577–587 [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460:1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, et al. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 15:1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME 2003. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol 23:462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, O’Leary KA, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME 2006. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev 20:16–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 22:5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SH, Kim DM, Heo YS, Kim YI, Kim HM, Kim J, Han YM, Kim IC, Yoo OJ 2009. New p53 target, phosphatase of regenerating liver 1 (PRL-1) downregulates p53. Oncogene 28:545–554 [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237–1245 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203–206 [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Slee EA, Lu X 2008. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol 9:702–712 [DOI] [PubMed] [Google Scholar]
- Neilsen PM, Cheney KM, Li CW, Chen JD, Cawrse JE, Schulz RB, Powell JA, Kumar R, Callen DF 2008. Identification of ANKRD11 as a p53 coactivator. J Cell Sci 121:3541–3552 [DOI] [PubMed] [Google Scholar]
- Ogawara Y, Kishishita S, Obata T, Isazawa Y, Suzuki T, Tanaka K, Masuyama N, Gotoh Y 2002. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem 277:21843–21850 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Li H, Jensen MR, Zhang T, Taya Y, Thorgeirsson SS, Prives C 2002. Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol Cell 9:761–771 [DOI] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80–83 [DOI] [PubMed] [Google Scholar]
- Olson DC, Marechal V, Momand J, Chen J, Romocki C, Levine AJ 1993. Identification and characterization of multiple mdm-2 proteins and mdm-2-p53 protein complexes. Oncogene 8:2353–2360 [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29:92–95 [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22:6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8:781–794 [DOI] [PubMed] [Google Scholar]
- Sdek P, Ying H, Chang DL, Qiu W, Zheng H, Touitou R, Allday MJ, Xiao ZX 2005. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell 20:699–708 [DOI] [PubMed] [Google Scholar]
- Song MS, Song SJ, Kim SY, Oh HJ, Lim DS 2008. The tumour suppressor RASSF1A promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex. EMBO J 27:1863–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW 2001. Regulation of PTEN transcription by p53. Mol Cell 8:317–325 [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460:1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Glahder JA, Wilhelm MT, Bersani C, Corcoran M, Mahmoudi S, Rosenstierne M, Grander D, Farnebo M, Norrild B, et al. 2009. The p53 target Wig-1 regulates p53 mRNA stability through and AU-rich element. PNAS 106:15756–15761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM 2008. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene 27:1590–1598 [DOI] [PubMed] [Google Scholar]
- Wang S, El-Deiry WS 2006. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res 66:6982–6989 [DOI] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT 2005. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123:641–653 [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, dominant-negative activities. Mol Cell 2:305–316 [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99–103 [DOI] [PubMed] [Google Scholar]
- Yang HY, Wen YY, Chen CH, Lozano G, Lee MH 2003. 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol 23:7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Prives C 2001. Cyclin a-CDK phosphorylation regulates MDM2 protein interactions. J Biol Chem 276:29702–29710 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23:8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725–734 [DOI] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 3:973–982 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C 2009. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35:316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]