Abstract
The bacteria cell envelope is a complex multilayered structure that serves to protect these organisms from their unpredictable and often hostile environment. The cell envelopes of most bacteria fall into one of two major groups. Gram-negative bacteria are surrounded by a thin peptidoglycan cell wall, which itself is surrounded by an outer membrane containing lipopolysaccharide. Gram-positive bacteria lack an outer membrane but are surrounded by layers of peptidoglycan many times thicker than is found in the Gram-negatives. Threading through these layers of peptidoglycan are long anionic polymers, called teichoic acids. The composition and organization of these envelope layers and recent insights into the mechanisms of cell envelope assembly are discussed.
Bacteria possess multilayered protective envelopes. Peptidoglycan layers ensure resistance to osmotic pressure; other components, such as lipopolysaccharide, help preserve cell integrity.
It has been well known since the late 1830s that all living organisms are composed of fundamental units called cells. The cell is a finite entity with a definite boundary, the plasma membrane. That means that the essence of the living state must be contained within the biological membrane; it is a defining feature of all living things. Everything that exists outside of the biological membrane is nonliving. Chemists tend to think of membranes as self assembling but biological membranes do not self assemble; they require energy to be established and maintained. In almost all cells this structure is a phospholipid bilayer that surrounds and contains the cytoplasm. In addition to lipid components biological membranes are composed of proteins; the proteins are what make each membrane unique. Despite its obvious importance, membranes and their associated functions remained poorly understood until the 1950s. Before this, many viewed the membrane as a semipermeable bag. This view persisted for a number of years, particularly in the case of bacteria, because it could not be envisioned how such a “simple” organism could have anything but a simple membrane (Rothfield 1971).
The bacterial cell envelope, i.e., the membrane(s) and other structures that surround and protect the cytoplasm, however, is anything but a simple membrane. Unlike cells of higher organisms, the bacterium is faced with an unpredictable, dilute and often hostile environment. To survive, bacteria have evolved a sophisticated and complex cell envelope that protects them, but allows selective passage of nutrients from the outside and waste products from the inside. The following discussion concerns the organization, composition, and the functions of the various layers and compartments that make up this remarkable cellular structure. It is easily appreciated that a living system cannot do what it does without the ability to establish separate compartments in which components are segregated. Specialized functions occur within different compartments because the types of molecules within the compartment can be restricted. However, membranes do not simply serve to segregate different types of molecules. They also function as surfaces on which reactions can occur. Recent advances in microscopy, which are discussed in other articles on this subject, have revealed strikingly nonrandom localization of envelope components. Here, we will highlight recent advances in our understanding of how these extracellular organelles are assembled.
More than 100 years ago Christian Gram (1884) developed a staining procedure that allowed him to classify nearly all bacteria into two large groups, and this eponymous stain is still in widespread use. One group of bacteria retain Christian's stain, Gram-positive, and the other do not, Gram-negative. The basis for the Gram stain lies in fundamental structural differences in the cell envelope of these two groups of bacteria. For our discussion of the Gram-negative bacterial cell envelope we will use Escherichia coli, an extensively-studied organism that has served as a model for understanding a number of fundamental biological processes. In comparing Gram-negative and Gram-positive cell envelopes we will use Staphylococcus aureus as a reference point but will highlight specific differences between it and Bacillus subtilis. B. subtilis is the major Gram-positive model organism and a substantial knowledge base exists for it, but the cell envelope of S. aureus has been studied more extensively because of interest in how surface features mediate interactions with the environment in the course of infection. Care should be taken in generalizing from examples drawn from particular microorganisms. For example, E. coli inhabits the mammalian gut. Accordingly, E. coli and other enteric bacteria must have a cell envelope that is particularly effective at excluding detergents such as bile salts. This need not be a pressing issue for other Gram-negative bacteria, and their envelopes may differ in species- and environmentally specific ways. Nonetheless, the ability to use the Gram stain to categorize bacteria suggests that the basic organizational principles we present are conserved. In addition, many bacteria express an outermost coat, the S-layer, which is composed of a single protein that totally encases the organism. S-layers and capsules, which are coats composed of polysaccharides, are beyond the scope of this review.
THE GRAM-NEGATIVE CELL ENVELOPE
After more than a decade of controversy, techniques of electron microscopy were improved to the point in which they finally revealed a clearly layered structure of the Gram-negative cell envelope (Fig. 1) (Glauert and Thornley, 1969). There are three principal layers in the envelope; the outer membrane (OM), the peptidoglycan cell wall, and the cytoplasmic or inner membrane (IM). The two concentric membrane layers delimit an aqueous cellular compartment that Peter Mitchell (1961) first termed the periplasm. During a similar time frame biochemical methods were developed to isolate and characterize the distinct set of proteins found in the periplasm (Heppel, 1967), and to characterize the composition of both the inner and outer membranes (Miura and Mizushima, 1968; Osborn et al. 1972). Studies since then have only reinforced their basic conclusions.
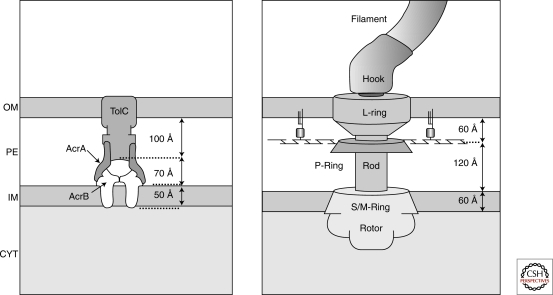
Figure 1.
Transenvelope machines in the Gram-negative cell envelope. The AcrA/B proteins together with TolC form an efflux pump that expels harmful molecules such as antibiotics from the cell directly into the media (Koronakis et al. 2000; Eswaran et al. 2004; Murakami et al. 2006). The flagellar basal body hook structure connects the motor to the flagella (DePamphilis and Adler, 1971). Distances shown provide a reasonable estimate of the size of the cellular compartments shown. PE, periplasm; CYT, cytoplasm.
The Outer Membrane
Starting from the outside and proceeding inward the first layer encountered is the OM. The OM is a distinguishing feature of Gram-negative bacteria; Gram-positive bacteria lack this organelle. Like other biological membranes, the OM is a lipid bilayer, but importantly, it is not a phospholipid bilayer. The OM does contain phospholipids; they are confined to the inner leaflet of this membrane. The outer leaflet of the OM is composed of glycolipids, principally lipopolysaccharide (LPS) (Kamio and Nikaido 1976). LPS is an infamous molecule because it is responsible for the endotoxic shock associated with the septicemia caused by Gram-negative organisms (Raetz and Whitfield 2002). The human innate immune system is sensitized to this molecule because it is a sure indicator of infection.
With few exceptions, the proteins of the OM can be divided into two classes, lipoproteins and β-barrel proteins. Lipoproteins contain lipid moieties that are attached to an amino-terminal cysteine residue (Sankaran and Wu 1994). It is generally thought that these lipid moieties embed lipoproteins in the inner leaflet of the OM. In other words, these proteins are not thought to be transmembrane proteins. There are about 100 OM lipoproteins in E.coli, and the functions of most of these are not known (Miyadai et al. 2004; but see below). Nearly all of the integral, transmembrane proteins of the outer membrane assume a β-barrel conformation. These proteins are β sheets that are wrapped into cylinders, and we will refer to these outer membrane proteins as OMPs. Not surprisingly, some of these OMPs, such as the porins, OmpF, and OmpC, function to allow the passive diffusion of small molecules such as mono- and disaccharides and amino acids across the OM. These porins have 16 transmembrane β strands, they exist as trimers (Cowan et al. 1992), and they are very abundant; together they are present at approximately 250,000 copies per cell. Other OMPs, such as LamB (18 transmembrane β strands) (Schirmer et al. 1995) or PhoE (16 transmembrane β strainds) (Cowan et al. 1992), exist as trimers as well and they function in the diffusion of specific small molecules, maltose or maltodextrins and anions such as phosphate respectively, across the OM. When induced by the presence of maltose or phosphate starvation, respectively, these proteins are very abundant as well. OmpA is another abundant OMP. It is monomeric, and it is unusual in that it can exist in two different conformations (Arora et al. 2000). A minor form of the protein, with an unknown number of transmembrane strands, can function as a porin, but the major, nonporin form has only eight transmembrane strands, and the periplasmic domain of this form performs a largely structural role (see later discussion). An additional class of OMPs, which are larger β-barrels (20–24 transmembrane β strands), but are present at much lower levels, function as gated channels in the high affinity transport of large ligands such as Fe-chelates or vitamins such as vitamin B-12 (for review see Nikaido 2003).
The OM is essential for the survival of E. coli, but it contains only a few enzymes. For example, there is a phospholipase (PldA) (Snijder et al. 1999), a protease (OmpT) (Vandeputte-Rutten et al. 2001), and an enzyme that modifies LPS (PagP) (Hwang et al. 2002). The active site of all of these enzymes is located in the outer leaflet, or it faces the exterior of the cell (OmpT). Mutants lacking any of these enzymes exhibit no striking phenotypes. The only known function of the OM is to serve as a protective barrier, and it is not immediately obvious why this organelle is essential. But what a barrier it is. Salmonella, another enteric bacterium, can live at the site of bile salt production in the gall bladder (Sinnott and Teall, 1987), and it is generally true that Gram-negative bacteria are more resistant to antibiotics than are their Gram-positive cousins. Indeed, some Gram-negative bacteria, such as Pseudomonas, are notorious in this regard.
LPS plays a critical role in the barrier function of the OM. It is a glucosamine disaccharide with six or seven acyl chains, a polysaccharide core, and an extended polysaccharide chain that is called the O-antigen (Raetz and Whitfield 2002). Traditionally pathogenic E. coli are classified by the antigenic properties of their O-antigen and the major protein (flagellin, termed H) component of the flagella (see later discussion). Hence, E. coli O157:H7. LPS molecules bind each other avidly, especially if cations like Mg++ are present to neutralize the negative charge of phosphate groups present on the molecule. The acyl chains are largely saturated, and this facilitates tight packing. The nonfluid continuum formed by the LPS molecules is a very effective barrier for hydrophobic molecules. This coupled with the fact that the porins limit diffusion of hydrophilic molecules larger than about 700 Daltons, make the OM a very effective yet, selective permeability barrier (Nikaido 2003).
The Peptidoglycan Cell Wall
Bacteria do not lyse when put into distilled water because they have a rigid exoskeleton. Peptidoglycan is made up of repeating units of the disaccharide N-acetyl glucosamine-N-actyl muramic acid, which are cross-linked by pentapeptide side chains (Vollmer et al. 2008). The peptidoglycan sacculus is one very large polymer that can be isolated and viewed in a light microscope. Because of its rigidity, it determines cell shape. The enterics are rod shaped, but cell shapes can vary. For example, vibrios and caulobacters are comma shaped. Recent results suggest that the glycan chains run perpendicular to the long axis of a rod shaped cell, i.e., hoops of glycan chains around the girth of the cell (Gan et al. 2008). Agents such as enzymes or antibiotics that damage the peptidoglycan cause cell lysis owing to the turgor pressure of the cytoplasm. Lysis can be prevented in media of high osmolarity. However, without the peptidoglycan, cells lose their characteristic shape. The resulting cells are called spheroplasts. With E. coli, normal methods of spheroplast production produce nonviable cells, but they can continue metabolism and biosynthesis for hours. However, special methods can be used to produce L forms, which are spherical in shape and can be propagated on high osmolarity media (Joseleau-Petit et al. 2007).
The OM is basically stapled to the underlying peptidoglycan by a lipoprotein called Lpp, murein lipoprotein, or Braun's lipoprotein (Braun, 1975). The lipids attached to the amino terminus of this small protein (58 amino acids) embed it in the OM. Lpp is the most abundant protein in E. coli, more than 500,000 molecules per cell. The ϵ-amino group of the carboxy-terminal lysine residue of one third of these molecules is covalently attached to the diaminopimelate residue in the peptide crossbridge. In addition, proteins such as OmpA bind peptidoglycan noncovalently. Nonetheless, mutants that lack Lpp shed OM vesicles (Yem and Wu 1978).
The Periplasm
The OM and IM delimit an aqueous cellular compartment called the periplasm. The periplasm is densely packed with proteins and it is more viscous than the cytoplasm (Mullineax et al. 2006). Cellular compartmentalization allows Gram-negative bacteria to sequester potentially harmful degradative enzymes such as RNAse or alkaline phosphatase. Because of this, the periplasm has been called an evolutionary precursor of the lysosomes of eukaryotic cells (De Duve and Wattiaux, 1966). Other proteins that inhabit this compartment include the periplasmic binding proteins, which function in sugar and amino acid transport and chemotaxis, and chaperone-like molecules that function in envelope biogenesis (Ehrmann, 2007; see later discussion).
The Inner Membrane
One of the hallmarks of eukaryotic cells is the presence of intracellular organelles. These organelles are defined by limiting membranes, and these organelles perform a number of essential cellular processes. The mitochondria produce energy, the smooth endoplasmic reticulum (ER) synthesize lipids, protein secretion occurs in the rough ER, and the cytoplasmic membrane contains the receptors that sense the environment and the transport systems for nutrients and waste products. Bacteria lack intracellular organelles, and consequently, all of the membrane-associated functions of all of the eukaryotic organelles are performed in the IM. Many of the membrane proteins that function in energy production, lipid biosynthesis, protein secretion, and transport are conserved in bacteria, but their cellular location is different. In bacteria, these proteins are located in the IM.
The IM is a phospholipid bilayer. In E. coli the principal phospholipids are phosphatidyl ethanolamine and phosphatidyl glycerol, but there are lesser amounts of phosphatidyl serine and cardiolipin. Other minor lipids include polyisoprenoid carriers (C55), which function in the translocation of activated sugar intermediates that are required for envelope biogenesis (Raetz and Dowhan 1990).
Transenvelope Machines
Certain types of surface appendages such as flagella (DePamphilis and Adler 1971; Macnab 2003), which are required for bacteria motility; Type III secretion systems (Kubori et al. 1998), which inject toxins into the cytoplasm of eukaryotic host cells during the infection process; and efflux pumps (Koronakis et al. 2000; Eswaran et al. 2004; Murakami et al. 2006; Symmons et al. 2009), which pump toxic molecules such as antibiotics from the cell clear across the cell envelope into the surrounding media and are responsible in part for much of the antibiotic resistance in pathogenic bacteria, are molecular machines that are made up of individual protein components that span the peptidoglycan and are located in all cellular compartments. The structures of some of these machines are known at sufficient resolution to provide meaningful insight into the size of the various cellular compartments in E. coli. As shown in Figure 1, these size predictions are in reasonable agreement. However, it should be noted that experimental measurements of the volume of the periplasm, for example, vary widely (Stock et al. 1977).
Envelope Biogenesis
All of the components of the Gram-negative cell envelope are synthesized either in the cytoplasm or at the inner surface of the IM. Accordingly, all of these components must be translocated from the cytoplasm or flipped across the IM. Periplasmic components must be released from the IM, peptidoglycan components must be released and polymerized, and OM components must be transported across the aqueous, viscous periplasm and assembled into an asymmetric lipid bilayer. All of this construction takes place outside of the cell in a potentially hostile environment that lacks an obvious energy source. It seems clear that there is no ATP out there for example. In this section we will summarize what is currently known about the assembly of the major envelope components; proteins, including lipoproteins, LPS, and phospholipids.
All proteins, of course, are synthesized in the cytoplasm. Proteins destined for the periplasm or the OM are made initially in precursor form with a signal sequence at the amino terminus. The signal sequence targets them for translocation from the cytoplasm (Driessen and Nouwen 2008). This translocation reaction is catalyzed by an essential, heterotrimeric IM protein complex called SecYEG (Van den Berg et al. 2004). The signal sequence and this heterotrimeric membrane protein complex are conserved throughout biology (Rapoport 2007). The essential ATPase SecA, together with the proton motive force, drives this translocation reaction (Zimmer et al. 2008). Periplasmic and OM proteins are generally translocated in post-translational fashion, i.e., synthesis and translocation are not coupled. Proteins must be secreted in linear fashion from the amino to the carboxy terminus like spaghetti through a hole; SecYEG cannot handle folded molecules. The cytoplasmic SecB chaperone maintains these secreted proteins in unfolded form until they can be secreted (Randall and Hardy 2002). During the secretion process the signal sequence is proteolytically removed by Signal Peptidase I (Paetzel et al. 2002). Other components of the Sec translocon, such SecD, SecF, and YajC, perform important but nonessential function(s) during translocation, perhaps facilitating release of secreted proteins into the periplasm. Once released, periplasmic proteins are home, but it seems likely that chaperones function to prevent misfolding and aggregation. For example, the periplasmic protein MalS, which contains disulfide bonds, requires the periplasmic disulfide oxidase DsbA for proper folding. In the absence of the DsbA the periplasmic protease/chaperone DegP (HtrA) can substitute (Spiess et al. 1999).
Periplasmic chaperones function to protect OMPs during their transit through the periplasm. Three such proteins have been well characterized and shown have general chaperone activity: SurA, which also functions as a peptidyl-proline isomerase (Behrens and Gross 2001; Bitto and McKay 2003), Skp (Chen and Henning 1996; Walton et al. 2009), and the aforementioned DegP (Krojer et al. 2008; Shen et al. 2009). Genetic analysis indicates that these three proteins function in parallel pathways for OMP assembly; SurA functions in one pathway; DegP/Skp function in the other. Mutants lacking either one of these pathways are viable, but cells cannot tolerate loss of both (Rizzitello et al. 2001). Mutants lacking SurA and Skp, or SurA and DegP are not viable and they show massive defects in OMP assembly. By definition then, these chaperone pathways are redundant. However, this redundancy does not reflect equal roles in OMP assembly. The major OMPs, which account for most of the protein mass of the OM, show preference for the SurA pathway (Sklar et al. 2007) as does the minor OMP LptD (Vertommen et al. 2009; see later). At present no OMP that prefers the DegP/Skp pathway has been identified. It may be that many minor OMPs show no pathway preference. The primary role of the DegP/Skp pathway may be to rescue OMPs that have fallen off the normal assembly pathway, particularly under stressful conditions. It is also possible that other periplasmic proteins have chaperone function that is important for the assembly of a subset of OMPs.
The periplasmic chaperones deliver OMPs to a recently identified assembly site in the OM termed the Bam complex (Fig. 2). This complex is composed of a large β-barrel protein, BamA (aka YaeT or Omp85), and four lipoproteins, BamBCDE (aka YfgL, NlpB, YfiO, and SmpA respectively) (Wu et al. 2005; Sklar et al. 2007). In addition to the β-barrel domain BamA has a large amino-terminal periplasmic domain composed of five POTRA (polypeptide transport associated). The structure of a large fraction of the BamA periplasmic domain has been determined (Kim et al. 2007). Each of the four visible POTRA domains has a nearly identical fold, despite the fact that the amino acid sequence identity between them is very low. In E. coli the first two POTRA domains are not essential for the life of the organism. Nevertheless, BamA is highly conserved in Gram-negative bacteria, and in these organisms there are always five POTRA domains. There are homologs of BamA in both mitochondria and chloroplasts (Moslavac et al. 2005), which are thought to be derived from Gram-negative bacteria. These homologs have one, two, or three POTRA domains, and the proteins function to assemble β-barrel proteins in the OM of these organelles. BamD is the only essential lipoprotein in the Bam complex (Malinverni et al. 2006), and it is highly conserved in Gram-negative bacteria as well. The remaining three lipoproteins are not essential, and they are conserved to varying degrees. We do not yet understand the mechanism of β-barrel folding nor do we understand the functions of the individual proteins in the Bam complex, but there is evidence suggesting that the POTRA domains of BamA may template folding by a process termed β augmentation (Kim et al. 2007).
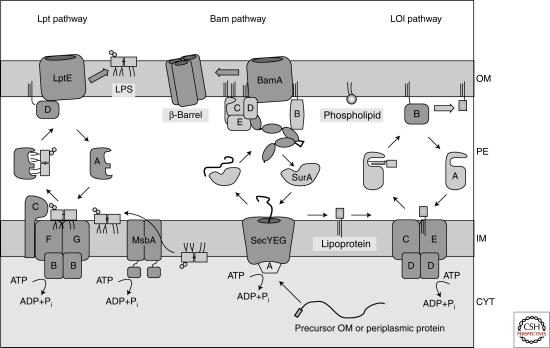
Figure 2.
The cellular machineries required for OM biogenesis. The Lpt pathway, together with MsbA, transports LPS from its site of synthesis to the cell surface. β-barrel proteins and lipoproteins are made initially in the cytoplasm in precursor form with a signal sequence at the amino terminus. The signal sequence directs these precursors to the Sec machinery for translocation from the cytoplasm. Chaperones like SurA deliver beta-barrel proteins to the Bam machinery for assembly in the OM. For OM lipoproteins, after the signal sequence is removed and lipids are attached to the amino-terminal cysteine residue, the Lol machinery delivers them to the OM.
Lipoproteins are made initially with an amino-terminal signal sequence as well, and they too are translocated by the Sec machinery. However, the signal sequence is removed by a different signal peptidase, signal peptidase II (Paetzel et al. 2002). Signal sequence processing of lipoproteins requires the formation of a thioether diglyceride at the cysteine residue, which will become the amino terminus of the mature lipoprotein. Once the signal sequence is removed, an additional fatty acyl chain is added to the cysteine amino group (Sankaran and Wu 1994). These lipid moieties tether the newly formed lipoprotein to the outer leaflet of the IM.
Some lipoproteins remain in the IM, and their biogenesis is complete after signal sequence processing and lipid addition. However, most of the lipoproteins in E. coli are destined for the outer membrane. The Lol system, which transports lipoproteins to the OM has been well characterized (Fig. 2) (Narita and Tokuda, 2006). There is an ABC transporter (LolCDE) in the IM that utilizes ATP hydrolysis to extract the molecule from the IM and pass it to a soluble periplasmic carrier called LolA. LolA delivers the molecule to the OM assembly site, which is the lipoprotein LolB. IM lipoproteins have a “Lol avoidance” signal so that they remain in the IM. The most common Lol avoidance signal is an aspartate residue at position two of the mature lipoprotein.
There is a second protein translocation system in the IM called Tat that translocates folded proteins (Sargent et al. 2006). E. coli uses the Tat system for proteins which have prosthetic groups that must be added in the cytoplasm, and this constitutes a small fraction of the secreted proteins. Other bacteria, such as thermophiles, use the Tat system extensively; presumable because it is easier to fold proteins in the cytoplasm than it is in the hostile environments they live in. In terms of components, the Tat system is remarkable simple; three components. TatB and TatC function to target proteins for translocation by TatA, but how this system recognizes that the substrate is folded, and how it accomplishes the translocation reaction are not yet understood.
Proteins destined for the IM are handled by the Sec machinery as well. However, in general, these proteins are targeted for cotranslational translocation by Signal recognition particle (SRP) and the SRP receptor FtsY (Bernstein 2000). Presumably, posttranslational translocation of these hydrophobic substrates would be inefficient and perhaps dangerous, owing to their great potential for aggregation. Prokaryotic SRP is much simpler than its eukaryotic counterpart. It contains only a single protein, Ffh (fifty four homolog) and an RNA, Ffs (four point five S RNA). Both Ffh and Ffs are GTPases, as are their more complex eukaryotic counterparts.
The transmembrane α-helices, which are characteristic of most biological membrane proteins in both prokaryotes and eukaryotes, serve as secretion signals. The first trans-membrane segment functions as a signal sequence to initiate translocation of the sequences that follow it. These transmembrane sequences tend to be longer and more hydrophobic than typical signal sequences, and this serves as the basis for SRP recognition (Hegde and Bernstein 2006). This signal sequence is not cleaved; it remains attached serving now as a typical transmembrane helix. The second transmembrane helix functions to stop the translocation reaction and this helix exits the SecYEG translocator laterally where it remains in the IM (Van den Berg et al. 2004; Driessen and Nouwen 2008). The third transmembrane helix functions again as an uncleaved signal sequence. These alternating start and stop translocation signals stitch IM proteins into the membrane in stepwise fashion.
Small IM proteins, especially those with small periplasmic domains, can be inserted into the membrane by a second IM translocase called YidC. YidC family members can be found in mitochondria and chloroplasts. Like their mitochondrial homologs, YidC plays an important role in the assembly of energy-transducing membrane proteins such as subunit c of ATPase. YidC may also play a role in the SecYEG-dependent insertion of larger IM proteins during the lateral transfer of the trans-membrane α-helices into the lipid bilayer (Xie and Dalbey 2008).
LPS, including the core polysaccharide, and the O-antigen are both synthesized on the inner leaflet of the IM. LPS is flipped to the outer leaflet of the IM by the ABC transporter MsbA. O-antigen is synthesized on a polyisoprenoid carrier, which then flips it to the outer leaflet. The O-antigen is ligated to the LPS core in the outer leaflet of the IM, a reaction catalyzed by WaaL (Raetz and Whitfield 2002). Note that the common laboratory strain E. coli K-12 does not make the O-antigen. Accordingly, it is termed “rough,” as opposed to the wild-type “smooth” strain. In the last several years a combination of genetics, biochemistry, and bioinformatics was employed to identify seven essential proteins that are required to transport LPS from the outer leaflet of the OM to the cell surface (Fig. 2). These proteins have been termed Lpt (lipopolysaccharide transport); LptA (aka YhbN) (Sperandeo et al. 2007), LptB (aka YhbG) (Sperandeo et al. 2007), LptC (aka YrbK) (Sperandeo et al. 2008), LptD (aka Imp or OstA) (Braun and Silhavy 2002; Bos et al. 2004), LptE (RlpB) (Wu et al. 2006), LptF (aka YjgP) (Ruiz et al. 2008), and LptG (aka YjgQ) (Ruiz et al. 2008). The large β-barrel protein, LptD and the lipoprotein LptE form a complex in the OM. LptA is made with a cleavable signal sequence and resides in the periplasm. LptF and LptG are IM proteins that likely interact with the cytoplasmic protein LptB, a predicted ATPase, to form an ABC transporter that together with the bitopic IM protein LptC, extracts LPS from the IM and passes it to the periplasmic protein LptA for delivery to the OM assembly site, LptD and LptE. An alternative model proposes that all seven proteins together form a transenvelope machine that transports LPS directly from the IM to the cell surface in analogy with efflux pumps. What is clear is that if any of the seven proteins are removed, LPS accumulates in the outer leaflet of the IM (Sperandeo et al. 2008, Ruiz et al. 2008).
Like LPS, phospholipids are synthesized in the inner leaflet of the IM. MsbA can flip these molecules to the outer leaflet of the IM (Doerrler et al. 2004), but it is likely that other mechanisms to flip these molecules also exist. How phospholipids reach the OM is not known. What is known is that phospholipids added into the OM reach the IM very quickly (Jones and Osborn 1977). This is true even for lipids like cholesterol which are not naturally found in bacteria. This could suggest sites of IM-OM fusion, or hemi-fusion, that allow intermembrane phospholipid trafficking by diffusion, a hypothesis made by Manfred Bayer (Bayer 1968) long ago that has remained highly controversial.
THE GRAM-POSITIVE CELL ENVELOPE
The Gram-positive cell envelope differs in several key ways from its Gram-negative counterpart (Fig. 3). First and foremost, the outer membrane is absent. The outer membrane plays a major role in protecting Gram-negative organisms from the environment by excluding toxic molecules and providing an additional stabilizing layer around the cell. Because the outer membrane indirectly helps stabilize the inner membrane, the peptidoglycan mesh surrounding Gram-negative cells is relatively thin. Gram-positive bacteria often live in harsh environments just as E. coli does—in fact, some live in the gut along with E. coli—but they lack a protective outer membrane. To withstand the turgor pressure exerted on the plasma membrane, Gram-positive microorganisms are surrounded by layers of peptidoglycan many times thicker than is found in E. coli. Threading through these layers of peptidoglycan are long anionic polymers, called teichoic acids, which are composed largely of glycerol phosphate, glucosyl phosphate, or ribitol phosphate repeats. One class of these polymers, the wall teichoic acids, are covalently attached to peptidoglycan; another class, the lipoteichoic acids, are anchored to the head groups of membrane lipids (Neuhaus 2003). Collectively, these polymers can account for over 60% of the mass of the Gram-positive cell wall, making them major contributors to envelope structure and function. In addition to the TAs, the surfaces of Gram-positive microorganisms are decorated with a variety of proteins, some of which are analogous to proteins found in the periplasm of Gram-negative organisms (Dramsi et al. 2008). Because there is no outer membrane in Gram-positive organisms to contain extracellular proteins, all these proteins feature elements that retain them in or near the membrane. Some contain membrane-spanning helices and some are attached to lipid anchors inserted in the membrane. Others are covalently attached to or associated tightly with peptidoglycan (Scott and Barnett 2006). Still others bind to teichoic acids. Studies on S. aureus have shown that the composition of surface-expressed proteins can change dramatically depending on environmental cues or growth conditions, reflecting the important role of the cell envelope in adapting to the local environment (Pollack and Neuhaus 1994). The major structural elements of Gram-positive cell walls, excluding capsules, will be described below.
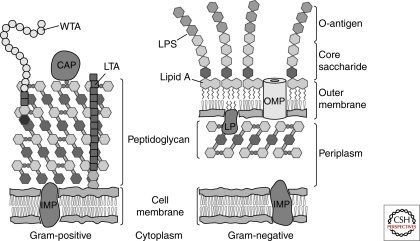
Figure 3.
Depiction of Gram-positive and Gram-negative cell envelopes: CAP = covalently attached protein; IMP, integral membrane protein; LP, lipoprotein; LPS, lipopolysaccharide; LTA, lipoteichoic acid; OMP, outer membrane protein; WTA, wall teichoic acid.
Gram-positive Peptidoglycan
The chemical structure of peptidoglycan in Gram-positive organisms is similar to that in Gram-negatives in that it is composed of a disaccharide-peptide repeat coupled through glycosidic bonds to form linear glycan strands, which are crosslinked into a meshlike framework through the peptide stems attached to the disaccharide repeat. The major difference between Gram-positive and Gram-negative peptidoglycan involves the thickness of the layers surrounding the plasma membrane. Whereas Gram-negative peptidoglycan is only a few nanometers thick, representing one to a few layers, Gram-positive peptidoglycan is 30–100 nm thick and contains many layers.
There are many differences among Gram-positive organisms with respect to the details of peptidoglycan structure, but perhaps the most notable difference relates to the peptide crosslinks between glycan strands (Vollmer 2008; Vollmer et al. 2008). S. aureus contains crosslinks in which the peptides are connected through a pentaglycine branch extending from the third amino acid of one of the stem peptides. This pentaglycine branch is assembled by a set of nonribosomal peptidyl transferases known as FemA, B, and X (Ton-That et al. 1998; Rohrer and Berger-Bachi 2003). Staphylococci can tolerate, albeit with difficulty, the loss of FemA or B, but not of FemX, which attaches the first glycine unit to the stem peptide (Hegde and Shrader 2001; Hubscher et al. 2007). Many Gram-positive organisms contain branched stem peptides, but B. subtilis does not; the stem peptides and crosslinks in this organism are identical in structure to those found in E. coli.
Branched stem peptides in S. aureus and other Gram-positive organisms play a variety of roles. Chief among these roles, they serve as attachment sites for covalently-associated proteins (discussed in more detail later). They have also been implicated in resistance to beta lactam antibiotics (Chambers 2003). Beta lactams inactivate transpeptidases that catalyze the peptide crosslinking step of peptidoglycan synthesis by reacting with the active site nucleophile of transpeptidases. Transpeptidases that couple branched stem peptides are mechanistically similar to those that couple unbranched stem peptides; however, their substrate specificity is sufficiently different that they only recognize unbranched stem peptides, and some of them are resistant to beta lactams (Rohrer and Berger-Bachi 2003; Pratt 2008; Sauvage et al. 2008). For example, methicillin-resistant S. aureus strains express a transpeptidase, PBP2A, that couples only pentaglycine-branched substrates. Many other Gram-positive organisms are also thought to harbor low affinity PBPs that preferentially recognize and couple branched stem peptides. It is thus speculated that the evolution of branched peptides in the peptidoglycan biosynthetic pathway may be an adaptation that enables escape from beta lactams. As described in the following section, however, branched stem peptides also play other important roles.
Surface Proteins
S. aureus colonizes human skin and mucosal surfaces. Breaches in the epithelium occasionally result in invasive S. aureus infections that are extremely serious. The ability to adhere to host tissue is a crucial first step in effective colonization by S. aureus, and a variety of surface factors are involved in this process. These factors include teichoic acids, which are discussed in the following section, as well as surface proteins that recognize components of host extracellular matrix such as fibronectin, fibrinogen, and elastin (Clarke and Foster 2006).
Some of these surface proteins, called adhesins, are attached via noncovalent ionic interactions to peptidoglycan or teichoic acids, but many are attached covalently to stem peptides within the peptidoglycan layers (Dramsi et al. 2008; Sjoquist et al. 1972; Fischetti et al. 1990). Proteins destined for covalent surface display contain an amino-terminal signal sequence that enables secretion through the cytoplasmic membrane and a carboxy-terminal pentapeptide cell wall sorting motif, which is commonly LPXTG (DeDent 2008). Enzymes called sortases catalyze a transpeptidation reaction between these sorting motifs and the glycine branch of the stem peptide of peptidoglycan precursors. The transpeptidation reaction is thought to occur in two steps: in the first, a nucleophile in the sortase active site attacks the amide bond between the threonine and glycine of the sorting motif to produce a covalent intermediate in which the amino-terminal portion of the protein substrate is attached to the enzyme; in the second, the glycine branch of the stem peptide enters the active site and the nucleophilic amino terminus of the glycine branch attacks the acyl- enzyme intermediate, regenerating the enzyme and forming a new TG amide bond that anchors the protein to the peptidoglycan precursor. The protein-modified peptidoglycan precursor is then incorporated into peptidoglycan (Dramsi et al. 2008; Marraffini et al. 2006).
In S. aureus, more than twenty protein substrates for the major sortase, sortase A, have been identified. In addition to adhesins, these protein substrates include proteins involved in immune system evasion, internalization, and phage binding. A minor sortase, sortase B, is responsible for surface display of proteins involved in iron acquisition, which is necessary for pathogenesis because iron is required for the function of many bacterial enzymes. In other Gram-positive organisms, sorting enzymes similar to SrtA and SrtB covalently couple proteins that comprise pili.
In addition to covalent attachment, Gram-positive organisms have other ways of retaining cell surface proteins. Many proteins involved in peptidoglycan biosynthesis are anchored to the cytoplasmic membrane by membrane spanning helices. Some proteins required for cell-wall degradation are associated noncovalently with peptidoglycan; others appear to be scaffolded and/or activated by teichoic acids or other types of cell surface polymers.
Teichoic Acids
Teichoic acids are anionic cell surface polymers found in a wide range of Gram-positive organisms, including S. aureus and B. subtilis. There are two major types of teichoic acids: wall teichoic acids (WTAs), which are coupled to peptidoglycan, and lipoteichoic acids (LTAs), which are anchored to the cell membrane. Wall teichoic acids are attached via a phosphodiester linkage to the C6 hydroxyl of occasional MurNAc residues in peptidoglycan. Although the structural variations are considerable, the most common WTAs are composed of a disaccharide linkage unit to which is appended a polyribitol phosphate (polyRboP) or polyglycerol phosphate (polyGroP) chain containing as many as 60 repeats. WTAs extend perpendicularly through the peptidoglycan mesh into what has been characterized as a “fluffy” layer beyond. S. aureus produces polyRboP WTAs; B. subtilis produces either polyRboP and polyGroP depending on the strain. The hydroxyls on the RboP or GroP repeats are tailored with other groups, typically d-alanyl esters or glycosyl moieties, and the nature and extent of the tailoring modifications significantly affect the properties and functions of WTAs (see later) (Neuhaus, 2003).
LTAs are similar to WTAs in that they are composed of polyGroP polymers that are often functionalized with d-alanine or a sugar moiety; however, they also differ in a number of ways. For example, they contain glycerol-phosphate repeats of opposite chirality to those found in WTAs. Furthermore, rather than being attached to peptidoglycan, they are anchored to membrane-embedded glycolipids and typically contain fewer GroP repeats. Thus, they extend from the cell surface into the peptidoglycan layers rather than through and beyond. Together, the LTAs and WTAs comprise what has been concisely described as a “continuum of anionic charge” that originates at the Gram-positive cell surface but extends well beyond the peptidoglycan barrier. The importance of this continuum of negative charge is underscored by the fact that Gram-positive organisms lacking WTAs (either because they do not contain the gene clusters or because they are grown under phosphate-limiting conditions) produce other types of polyanionic polymers in which the negative charges are supplied by carboxylate or sulfate groups. Furthermore, although neither LTAs nor WTAs are essential, deleting the pathways for the biosynthesis of either of these polymers produces organisms that have cell division and morphological defects as well as other, less serious growth defects. Moreover, it is not possible to delete both pathways because the genes involved are synthetic lethals (Oku et al. 2009; Morath et al. 2005).
Teichoic acids account for a significant fraction of the cell wall mass in producing organisms and their functions are many, varied, and species-dependent. Because they are anionic they bind cations and thus play a role in cation homeostasis (Marquis et al. 1976). Networks of metal cations between WTAs also influence the rigidity and porosity of the cell wall. The negative charge density on WTAs can be modulated by tailoring modifications that introduce positive charges along the polymer backbone, and these modifications can have a profound effect on the interactions of bacteria with other cells or molecules. For example, in S. aureus, a d-alanine transferase couples d-alanine moieties to free hydroxyls on the polyribitol phosphate backbone. S. aureus strains lacking d-alanine esters are more susceptible to antimicrobial cationic peptides and to lytic enzymes produced by host neutrophils (Collins et al. 2002; Peschel et al.1999; Peschel et al. 2000). They also display reduced autolysin activity, suggesting a role for functionalized WTAs in scaffolding or activating hydrolytic enzymes involved in cell wall synthesis and degradation.
THE CELL ENVELOPE OF CORYNEBACTERINEAE
The Corynebacterineae are a group of bacteria that includes the very important pathogens Mycobacterium tuberculosis and Mycobacterium leprae. These bacteria are generally classified as high G+C Gram positives, however their cell envelope has characteristics of both Gram-positive and Gram-negative bacteria. Indeed, a genome-based phylogeny places them in between Gram positives and Gram negatives (Fu and Fu-Liu, 2002).
The cell envelope of the Corynebacterineae is very complex and this complexity contributes substantially to their virulence. The peptidoglycan layer that surrounds a standard IM contains covalently attached arabinogalactan and this is covalently attached to mycolic acids (Minnikin, 1982). These mycolic acids have very long alkyl side chains (up to C90) that give the bacteria a waxy appearance and account for their resistance to acid decolorization during staining procedures (acid-fast).
Unlike other Gram-positive bacteria, Corynebacterineae have an OM. This OM appears to be symmetrical unlike the Gram-negative OM. Mycolic acids appear essential for this OM, but how they are organized remains unclear (Hoffmann et al. 2008; Zuber et al. 2008). Mycobacteria have porin proteins in their OM, but the structure of the main porin, MspA, from Mycobacterium smegmatis is quite different from the typical porin proteins of Gram-negative bacteria (Faller et al. 2004). Indeed, mycobacteria have no obvious homologs of the Bam complex members. This might suggest a novel mechanism for the assembly of proteins in the OM of Corynebacterineae.
CONCLUSION
The cell envelopes of bacteria are complex, dynamic structures that play a variety of protective and adaptive roles. The major conserved component of all bacterial cell envelopes is peptidoglycan, which is essential for stabilizing cell membranes against high internal osmotic pressures. But peptidoglycan alone is not enough to enable bacteria to survive in their different environments. In addition to peptidoglycan, the outer membrane of Gram-negative bacteria the, dense array of negatively charged polymers embedded in Gram-positive peptidoglycan and the complex outer layers of the Corynebacterineae play important roles in cell envelope integrity. One of the major challenges in the next decade will be to define the mechanisms by which these complex structures are assembled and regulated in response to changing environmental conditions.
ACKNOWLEDGMENTS
We thank Juliana Malinverni for help with figures and Susan DiRenzo for help preparing the manuscript. The authors were each supported by grants from the National Institute of General Medical Sciences.
Footnotes
Editors: Lucy Shapiro and Richard Losick
Additional Perspectives on Cell Biology of Bacteria available at www.cshperspectives.org
REFERENCES
- Arora A, Rinehart D, Szabo G, Tamm LK 2000. Refolded outer membrane protein A of Escherichia coli formsion channels with two conductance states in planar lipid bilayers. J Biol Chem 275:1594–1600 [DOI] [PubMed] [Google Scholar]
- Bayer ME 1968. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol 53:395–404 [DOI] [PubMed] [Google Scholar]
- Behrens S, Maier R, de Cock H, Schmid FX, Gross CA 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J 20:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HD 2000. The biogenesis and assembly of bacterial membrane proteins. Curr Opin Microbiol 3:203–209 [DOI] [PubMed] [Google Scholar]
- Bitto E, McKay DB 2003. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J Biol Chem 278:49316–49322 [DOI] [PubMed] [Google Scholar]
- Bos MP, Tefsen B, Geurtsen J, Tommassen J 2004. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci 101:9417–9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta 415:335–377 [DOI] [PubMed] [Google Scholar]
- Braun M, Silhavy TJ 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol 45:1289–1302 [DOI] [PubMed] [Google Scholar]
- Chambers HF 2003. Solving staphylococcal resistance to beta-lactams. Trends in Microbiol 11:145–148 [DOI] [PubMed] [Google Scholar]
- Chen R, Henning U 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol 19:1287–1294 [DOI] [PubMed] [Google Scholar]
- Clarke SR, Foster SJ 2006. In Advances in Microbial Physiology, Vol51, pp 187–225 [DOI] [PubMed] [Google Scholar]
- Collins LV, Kristian S, Weidenmaier C, Faigle M, Van Kessel KP, Van Strijp JA, Götz F, Neumeister B, Peschel A 2002. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis 186:214–219 [DOI] [PubMed] [Google Scholar]
- Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP 1992. Crystal structures explain functional properties of two E. coliporins. Nature 358:727–733 [DOI] [PubMed] [Google Scholar]
- DeDent A, Bae T, Missiakas DM, Schneewind O 2008. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EUBO J 27:2656–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R 1966. Functions of lysosomes. Annu Rev Physiol 28:435–492 [DOI] [PubMed] [Google Scholar]
- DePamphilis ML, Adler J 1971. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol 105:384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerrler WT, Gibbons HS, Raetz CR 2004. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem 279:45102–45109 [DOI] [PubMed] [Google Scholar]
- Dramsi S, Magnet S, Davison S, Arthur M 2008. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol Rev 32:307–320 [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77:643–667 [DOI] [PubMed] [Google Scholar]
- Erhmann M 2007. The periplasm ASM Press, Washington, D.C [Google Scholar]
- Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V 2004. Three's company: Component structures bring a closer view of tripartite drug efflux pumps. Current Opinion in Structural Biol 14:741–747 [DOI] [PubMed] [Google Scholar]
- Faller M, Niederweis M, Schulz GE 2004. The structure of a mycobacterial outer-membrane channel. Science 303:1189–1192 [DOI] [PubMed] [Google Scholar]
- Fischetti VA, Pancholi V, Schneewind O 1990. Conservation of a hexapeptide sequence in the anchor region of surface-proteins from Gram-postive cocci. Mol Micro 4:1603–1605 [DOI] [PubMed] [Google Scholar]
- Fu LM, Fu-Liu CS 2002. Is mycobacterium tuberculosis a closer relative to Gram-positive or Gram-negative bacterial pathogens? Tuberculosis 82:85–90 [DOI] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ 2008. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci 105:18953–18957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauert AM, Thornley MJ 1969. The topography of the bacterial cell wall. Annu Rev Microbiol 23:159–198 [DOI] [PubMed] [Google Scholar]
- Gram HCJ 1884. Uber die isolirte Farbung der Schizomyceten in Schnitt- and Trockenpraparaten. Fortschritte der Medizin 2:185–189 [Google Scholar]
- Hegde RS, Bernstein HD 2006. The surprising complexity of signal sequences. Trends Biochem Sci 31:563–571 [DOI] [PubMed] [Google Scholar]
- Hegde SS, Shrader TE 2001. FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J Boil Chem 276:6998–7003 [DOI] [PubMed] [Google Scholar]
- Heppel LA 1967. Selective release of enzymes from bacteria. Science 156:1451–1455 [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H 2008. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci 105:3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher J, Jansen A, Kotte O, Schafer J, Majcherczyk PA, Harris LG, Bierbaum G, Heinemann M, Berger-Bachi B 2007. Living with an imperfect cell wall: Compensation of femAB inactivation in Staphylococcus aureus. BMC Genomics 8:307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE 2002. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci 99:13560–13565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Osborn MJ 1977. Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. J Biol Chem 252:7405–7412 [PubMed] [Google Scholar]
- Joseleau-Petit D, Liebart JC, Ayala JA, D'Ari R 2007. Unstable Escherichia coli L forms revisited: Growth requires peptidoglycan synthesis. J Bacteriol 189:6512–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamio Y, Nikaido H 1976. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15:2561–2570 [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140 [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317:961–964 [DOI] [PubMed] [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914–919 [DOI] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453:885–890 [DOI] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605 [DOI] [PubMed] [Google Scholar]
- Macnab RM 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100 [DOI] [PubMed] [Google Scholar]
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coil. Mol Microbiol 61:151–164 [DOI] [PubMed] [Google Scholar]
- Marquis RE, Mayzel K, Carstensen EL 1976. Cation exchange in cell walls of Gram-positive bacteria. Can J Microbiol 22:975–982 [DOI] [PubMed] [Google Scholar]
- Marraffini LA, DeDent AC, Schneewind O 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Molec Biol Rev 70:192–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P 1961. Approaches to the analysis of specific membrane transport. In Biological structure and function (ed. Goodwin T.W., Lindberg O.), pp. 581–603 Academic Press, New York [Google Scholar]
- Minnikin DE 1982. Lipids: Complex lipids, their chemistry, biosynthesis and roles, p. 95–184. InRatledge C., Stanford J. (ed.). The biology of the mycobacteria, vol 1 Physiology, identification and classification Academic Press, Inc., New York, NY [Google Scholar]
- Miura T, Mizushima S 1968. Separation by density gradient centrifugation of two types of membranes from spheroplast membrane of Escherichia coli K12. Biochim Biophys Acta 150:159–161 [DOI] [PubMed] [Google Scholar]
- Miyadai H, Tanaka-Masuda K, Matsuyama S, Tokuda H 2004. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem 279:39807–39813 [DOI] [PubMed] [Google Scholar]
- Morath S, Von Aulock S, Hartung T 2005. Structure/function relationships of lipoteichoic acids. J Endotoxin Res 11:348–356 [DOI] [PubMed] [Google Scholar]
- Moslavac S, Mirus O, Bredemeier R, Soll J, von Haeseler A, Schleiff E 2005. Conserved pore-forming regions in polypeptide-transporting proteins. FEBS J 272:1367–1378 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW, Nenninger A, Ray N, Robinson C 2006. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J Bacteriol 188:3442–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179 [DOI] [PubMed] [Google Scholar]
- Narita S, Tokuda H 2006. An ABC transporter mediating the membrane detachment of bacterial lipoproteins depending on their sorting signals. FEBS Lett 580:1164–1170 [DOI] [PubMed] [Google Scholar]
- Neuhaus F 2003. A continuum of anionic charge: Structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Micro Molec Biol Rev 67:686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee BL, Sekimizu K 2009. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol 191:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E, Carson J 1972. Mechinism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem 247:3962–3972 [PubMed] [Google Scholar]
- Paetzel M, Karla A, Strynadka NC, Dalbey RE 2002. Signal peptidases. Chem Rev 102:4549–4580 [DOI] [PubMed] [Google Scholar]
- Pautsch A, Schulz GE 2000. High-resolution structure of the OmpA membrane domain. J Mol Biol 298:273–282 [DOI] [PubMed] [Google Scholar]
- Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274:8405–8410 [DOI] [PubMed] [Google Scholar]
- Peschel A, Vuong C, Otto M, Götz F 2000. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrobial Agents and Chemotherapy 44:2845–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JH, Neuhaus FC 1994. Changes in wall teichoic acid during the rod- sphere transition of Bacillus subtilis 168. J Bacteriol 176:7252–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt RF 2008. Substrate specificity of bacterial DD-peptidases (penicillin-binding proteins). Cellular and Molecular Life Sciences 65:2138–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Dowhan W 1990. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem 265:1235–1238 [PubMed] [Google Scholar]
- Raetz CR, Whitfield C 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LL, Hardy SJ 2002. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci 59:1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450:663–669 [DOI] [PubMed] [Google Scholar]
- Rizzitello AE, Harper JR, Silhavy TJ 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol 183:6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer S, Berger-Bachi B 2003. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in Gram- positive cocci. Antimicrobial Agents and Chemotherapy 47:837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield LI 1971. Biological membranes: An overview at the molecular level. In Structure and function of biological membranes (ed. Rothfield L.I.), pp. 3–9 Academic Press, New York [Google Scholar]
- Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci 105:5537–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran K, Wu HC 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem 269:19701–19706 [PubMed] [Google Scholar]
- Sargent F, Berks BC, Palmer T 2006. Pathfinders and trailblazers: A prokaryotic targeting system for transport of folded proteins. FEMS Microbiol Lett 254:198–207 [DOI] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258 [DOI] [PubMed] [Google Scholar]
- Schirmer T, Keller TA, Wang YF, Rosenbusch JP 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science 267:512–514 [DOI] [PubMed] [Google Scholar]
- Scott JR, Barnett TC 2006. Surface proteins of Gram-positive bacteria and how they get there. Ann Rev Microbiol 60:397–423 [DOI] [PubMed] [Google Scholar]
- Shen QT, Bai XC, Chang LF, Wu Y, Wang HW, Sui SF 2009. Bowl-shaped oligomeric structures on membranes as DegP's new functional forms in protein quality control. Proc Natl Acad Sci 106:4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott CR, Teall AJ 1987. Persistent gallbladder carriage of Salmonella typhi. Lancet 1:976. [DOI] [PubMed] [Google Scholar]
- Sjoquist J, Hjelm H, Johansso IB, Movitz J 1972. Localization of Protein-A in bacteria. Eur J Biochem 30:190–194 [DOI] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci 104:6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Kahne D, Silhavy TJ 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21:2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW 1999. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401:717–721 [DOI] [PubMed] [Google Scholar]
- Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Deho G, Polissi A 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol 189:244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Beil A, Ehrmann M 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339–347 [DOI] [PubMed] [Google Scholar]
- Stock JB, Rauch B, Roseman S 1977. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem 252:7850–7861 [PubMed] [Google Scholar]
- Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V 2009. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci 106:7173–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Labischinski H, Berger-Bachi B, Scbneewind O 1998. Anchor structure of staphylococcal surface proteins III. Role of the femA, femB, and femX factors in anchoring surface proteins to the bacterial cell wall. J Biol Chem 273:29143–29149 [DOI] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44 [DOI] [PubMed] [Google Scholar]
- Vandeputte-Rutten L, Kramer RA, Kroon J, Dekker N, Egmond MR, Gros P 2001. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J 20:5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF 2009. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9:2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbial Rev 32:287–306 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167 [DOI] [PubMed] [Google Scholar]
- Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC 2009. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc Natl Acad Sci 106:1772–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245 [DOI] [PubMed] [Google Scholar]
- Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci 103:11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Dalbey RE 2008. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol 6:234–244 [DOI] [PubMed] [Google Scholar]
- Yem DW, Wu HC 1978. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J Bacteriol 133:1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M 2008. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol 190:5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]