Abstract
Background
Familial hemophagocytic lymphohistiocytosis is a fatal disease characterized by immune dysregulation from defective function of cytotoxic lymphocytes. Three causative genes have been identified for this autosomal recessive disorder (PRF1, UNC13D, and STX11). We investigated the molecular genetics of familial hemophagocytic lymphohistiocytosis in Korea.
Design and Methods
Pediatric patients who fulfilled the HLH-2004 criteria were recruited from the Korean Registry for Histiocytosis. Molecular genetic studies were performed on the patients’ DNA samples by direct sequencing of all coding exons and flanking sequences of PRF1, UNC13D, and STX11.
Results
Forty patients were studied and familial hemophagocytic lymphohistiocytosis mutations were identified in nine; eight patients had UNC13D mutations (89%) and one had a mutation in PRF1. No patient had a STX11 mutation. Notably, four patients had only one UNC13D mutant allele, suggesting that the other mutation was missed by conventional direct sequencing. All UNC13D mutations were deleterious in nature. One known splicing mutation, c.754-1G>C, was recurrent, accounting for 58% of all the mutant alleles (7/12). Five UNC13D mutations were novel (p.Gln98X, p.Glu565SerfsX7, c.1993-2A>G, c.2367+1G>A, and c.2954+5G>A). The one patient with PRF1 mutation was homozygous for a frameshift mutation (p.Leu364GlufsX93), which was previously reported to be the most frequent PRF1 mutation in Japan.
Conclusions
This is the first investigation on the molecular genetics of familial hemophagocytic lymphohistiocytosis in Korea. The data showed that UNC13D is the predominant causative gene in the Korean population. The identification of mutations missed by conventional sequencing would better delineate the mutation spectrum and help to establish the optimal molecular diagnostic strategy for familial hemophagocytic lymphohistiocytosis in Korea, which might need an RNA-based screening strategy.
Keywords: hemophagocytic histiocytosis, familial, FHL, genetics, UNC13D, Korea
Introduction
Hemophagocytic lymphohistiocytosis (HLH) is characterized by fever and hepatosplenomegaly in association with pancytopenia and other laboratory abnormalities including hypertriglyceridemia and hypofibrinogenemia. Histologically, proliferation and infiltration of T-lymphocytes and histiocytes with hemophagocytic activity can be observed in reticuloendothelial systems, bone marrow, and the central nervous system (CNS). HLH can occur as either a primary or a secondary disease. Primary HLH is also known as familial hemophagocytic lymphohistiocytosis (F-HLH or FHL). Secondary HLH often occurs in association with underlying conditions such as infection, malignancy, and autoimmune disease. FHL is a rare and genetically heterogeneous disorder with an autosomal recessive inheritance.1–3 Clinically, FHL usually occurs within the first year of life and typically follows a fatal course, with a median survival of less than 2 months from diagnosis if untreated.2,4–6 A positive family history of the disease and some clinical and laboratory features such as CNS involvement and deficient natural killer (NK) cell activity suggest the diagnosis of FHL, but molecular genetic diagnosis is confirmatory.7–9 Prompt diagnosis of FHL is important because the treatment of choice for this disease is stem cell transplantation, and it has been reported that the time to transplantation is critical for the outcome of the patients.10–12 According to the recent diagnostic criteria for FHL, a molecular genetic diagnosis consistent with FHL is self-sufficient for the diagnosis of the disease.13
Four affected genetic loci have been identified for FHL (FHL1~FHL4). In 1999, the perforin gene (PRF1, MIM170280) on chromosome 10q21 was identified as the disease gene in FHL2.14 More recently, two additional FHL genes, the UNC13D gene (MIM 608898) and the STX11 gene (MIM 603552), have been identified for FHL3 on chromosome 17q25 and FHL4 on 6q24, respectively.15,16 The causative gene for FHL1 (9q21.3-22, MIM267700) is still elusive.17 The three genes, PRF1, UNC13D, and STX11, encode proteins involved in the cytotoxic activity of T lymphocytes.16,18
Data obtained from different populations have shown ethnic differences in the molecular genetics of FHL. There have been no published data, not even a case report, on the genetic background of FHL in Korea. For the first time in this study, the molecular genetics of FHL were investigated in a series of selected candidate pediatric patients in Korea, involving all three FHL genes to identify the spectrum of causative genes and their mutations in this country.
Design and Methods
Patients
Candidate patients for molecular genetic study for FHL were recruited from the Korean Registry for Histiocytosis. All were pediatric patients and fulfilled the HLH-2004 diagnostic criteria.13 With written informed consent, peripheral blood samples were obtained from the patients and family members including parents. The Institutional Review Board of each institution involved approved the study protocol.
Molecular genetic study
DNA samples were extracted from peripheral blood leukocytes using the Wizard Genomic DNA Purification Kit according to the manufacturer’s instructions (Promega, Madison, WI, USA). Nucleotide sequences of all exons and their flanking intronic sequences of PRF1, UNC13D, and STX11 were obtained by polymerase chain reaction (PCR) followed by direct sequencing using primer pairs designed by the authors (available upon request). PCR was performed on a thermal cycler (Applied Biosystems, Foster City, CA, USA), and direct sequencing was performed with the BigDye Terminator Cycle Sequencing Ready Reaction kit on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequence variations, if any, were analyzed with reference to the wild type sequences (GenBank accession n. NM_005041 for PRF1, NM_199242 for UNC13D, and NM_003764 for STX11) using the Sequencher program (Gene Codes Corp., Ann Arbor, MI, USA). We followed the nomenclature guidelines from the Human Genome Variation Society (http://www.hgvs.org/mutnomen/) to describe the mutations identified: for nucleotide numbering, A of the ATG translation initiation codon was numbered +1; at the amino acid level, the first Met was numbered +1. For splicing mutations, the conventional description by intervening sequence (IVS) is also given.
Results
Study patients
Forty patients suspected to have FHL were recruited for the molecular genetic study from the Korean Registry for Histiocytosis. The clinical and laboratory findings of the 40 study patients are shown in Online Supplementary Table S1. The median age at diagnosis was 22 months (range, 1 month – 12 years and 10 months). Twenty-one patients were boys and 19 were girls (male-to-female ratio, 1.1:1). All patients fulfilled the HLH-2004 criteria for the diagnosis of FHL, although NK cell activity was determined in a limited number of patients because the test is not widely available in Korea. No patient was born to a consanguineous couple or had a family history of histiocytosis.
Molecular genetic tests
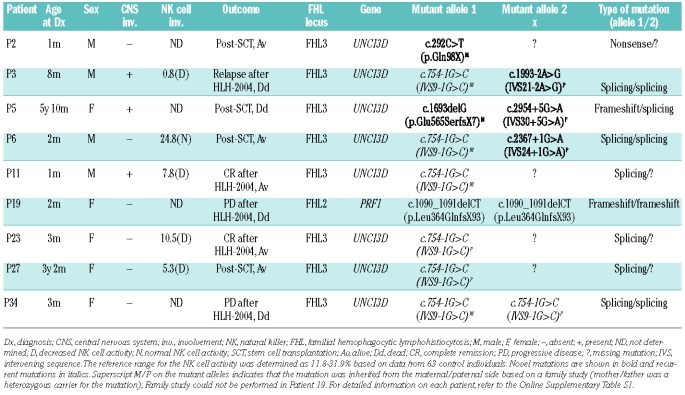
Direct sequencing analyses identified FHL mutations in nine out of the 40 patients (23%; Table 1). Notably, eight patients (89%; 8/9) had mutations in UNC13D (FHL3). Only one patient (11%) had mutations in PRF1 (FHL2). No patient had mutations in STX11. Of note, four patients with UNC13D mutations (50%; 4/8) had only one mutant allele (Table 1; P2, P11, P23, and P27). The 12 mutant alleles observed in the eight patients consisted of six different mutations, all of which were deleterious in nature (4 splicing, 1 nonsense, and 1 frameshift mutations). One known splicing mutation (c.754-1G>C or IVS9-1G>C) was recurrent, accounting for 58% (7/12) of all mutant alleles detected in our series of patients. The other five mutations were novel (c.292C>T [p.Gln98X], c.1693delG [p.Glu565SerfsX7], c.1993-2A>G [IVS21-2A>G], c.2367+1G>A [IVS24+1G>A], and c.2954+5G>A [IVS30+5G>A]). P3 and P6 were compound heterozygotes for two different splicing mutations (IVS9-1G>C and IVS21-2A>G in P3 and IVS9-1G>C and IVS24+1G>A in P6). P5 was a compound heterozygote for a novel frameshift mutation and a novel splicing mutation (Glu565SerfsX7 and c.2954+5G>A). Family studies were performed in all patients, except P19, which demonstrated that all mutations identified were inherited from one or other of the parents (there were no de novo mutations) (Table 1). P19 was the only patient who had mutations in PRF1. This patient was homozygous for a known frameshift mutation, c.1090_1091delCT (p.Leu364GlnfsX93). Her parents refused to participate in the family study.
Table 1.
Mutations identified in nine Korean patients with familial hemophagocytic lymphohistiocytosis.
Clinical and laboratory findings of Korean patients with familial hemophagocytic lymphohistiocytosis mutations
The age of the nine patients with FHL mutations ranged from 1 month to 70 months (median, 3 months) at the time of diagnosis of HLH (Table 1). Five patients were boys and four were girls (male-to-female ratio, 1.3:1). All of these patients except for P2 had fever at diagnosis. All patients had splenomegaly and liver dysfunction. CNS involvement was documented in three patients at initial diagnosis. One patient developed CNS involvement during the course of HLH-2004 treatment (P19). All patients except for P34 were found to have hemophagocytic histiocytes in bone marrow. NK cell activity was determined in five of the nine patients; four had decreased activity, while P6 had normal activity. Liver biopsy was performed in P34, which demonstrated increased histiocytes with periportal inflammation. One patient (P27) had laboratory findings indicative of Epstein-Barr virus infection. All patients were treated according to the HLH-2004 protocol. Three patients achieved complete remission. Four patients (P2, P5, P6, and P27) underwent stem cell transplantation. P2 received an allogeneic bone marrow transplant from an unrelated donor, and is currently alive at 10 months post-transplant with routine follow-up visits. P5 underwent double unrelated cord blood transplantation, but she died from cytomegalovirus pneumonia 2 months after the transplant. P6 was grafted with peripheral blood stem cells from his elder sister who was proven to have normal UNC13D alleles in the family study. He survived graft-versus- host disease after transplantation and is currently on routine follow-up 9 months post-transplant. P27 recently received a stem cell transplant from her younger brother who does carry the patient’s mutation. P11 and P23 are waiting for stem cell transplantation. Two patients (P3 and P19) died from CNS involvement during the course of HLH-2004 treatment. P19, who had PRF1 mutations, died from disease relapse with CNS involvement during the course of HLH-2004 treatment. P34 followed a waxing-and-waning course and eventually died from progressive disease.
Discussion
The present study is the first investigation on the molecular genetics of FHL in Korea, involving all three known FHL genes (PRF1, UNC13D, and STX11). According to the data, the majority of Korean patients with FHL had mutation( s) in UNC13D (FHL3). Although the number of FHL patients identified in this study was limited, the data convincingly showed that UNC13D is the major FHL gene among Koreans. Recent studies reported different proportions of FHL genes across populations. In non-Asian populations, PRF1 mutations were identified in 6–58% of FHL patients.22–25 UNC13D mutations were not identified in any Nordic patients, but found in 8% of Turkish and 17% of German and Middle East patients.24,25 STX11 mutations (FHL4) were identified in approximately 20% of FHL patients from Turkish/Kurdish families.15,24 Among Asian countries, data on the molecular genetics of FHL are available only from Japan;18,19,26,27 FHL2 and FHL3 accounted for approximately 19% and between 20% and 25% of cases of FHL in Japan, respectively. This contrasts with our result that FHL3 accounted for 89% of cases of FHL in Korea. This might be explained by the presence of two highly frequent mutations of PRF1 in Japanese patients26 and also by the recurrent splicing mutation (c.754-1G>C) of UNC13D in Korean patients. Taken together, the contribution of FHL2 to all the cases of FHL is lower in Korea than in any other country/population for which data have been reported, including Japan. The frequency of mutation- negative cases after screening PRF1, UNC13D, and STX11 was 78% in the present study, which is comparable to that reported in Japan (75%), but lower than the 93% in Nordic patients and 81% in Germany.19,24,25 The frequency was relatively low in Turkish patients (42%) in whom STX11 mutations are frequent and in the Middle East (18%) where PRF1 mutations are frequent.25
We identified 12 mutant alleles of UNC13D from eight patients (Table 1). All UNC13D mutations identified were deleterious in nature, with splicing mutations being the most common (83% of all mutant alleles). One known splicing mutation c.754-1G>C was recurrent, and accounted for 58% of the mutations (7/12). It is conceivable that c.754-1G>C is a founder mutation in Korea. The remaining five mutations were novel: three splicing mutations (c.1993-2A>G, c.2367+1G>A, and c.2954+5G>A), one nonsense mutation (p.Gln98X), and one frameshift mutation (p.Glu565SerfsX7). This is in accordance with previous studies on UNC13D mutations,16,18,28,29 and suggests the potential role of RNA-based screening in the molecular diagnosis of FHL3. Our data also showed that there is no mutation hotspot in UNC13D, implying that all coding exons and flanking intronic sequences must be screened.18,30 Of note, half of the patients with UNC13D mutations in our series (4/8) had only one mutant allele (Table 1). Considering the autosomal recessive inheritance, we speculated that there would be a hidden mutation missed by conventional direct sequencing. Splicing mutations involving non-conventional splicing sites (e.g., deep intronic mutations) and large deletion or duplication mutations would be the possibilities. Indeed, a recent study reported that splicing defects accounted for a larger proportion of mutations of UNC13D than had been previously recognized, with deep intronic mutations causing aberrant splicing on reverse-transcriptase PCR.28 It was also demonstrated that point mutations in the coding sequence, c.610A>G and c.1847A>G, resulted in aberrant splicing rather than being missense mutations as Met204Val and Glu616Gly, respectively.28,30 Only one patient in our series (P19) had PRF1 mutations (1/9; 11%). She was homozygous for a frameshift mutation (1090_1091delCT or p.Leu364GlnfsX93). Population-specific deleterious mutations have been reported in PRF1, including 1090_1091delCT in Japanese patients.26,31,32 The patients with 1090_1091delCT in Japan were mostly from the western part of the country. Interestingly, P19 was the only patient from the southernmost part of Korea (Pusan), which is close to the western part of Japan. Thus, the patient might share common haplotypes with Japanese patients with 1090_1091delCT. In addition, we could not rule out the possibility of a concurrent large deletion or uniparental disomy of the genomic region harboring PRF1.33
The clinical characteristics of the FHL patients in this study included early age of onset and poor treatment outcome, particularly in association with CNS involvement. No patient had a family history of FHL. This could be due to the low birth rate and also to the fact that consanguineous marriage is illegal in Korea. Since there was only one patient with PRF1mutations (FHL2), it was difficult to compare those with FHL2 and with FHL3. However, our observation is in line with previous studies that reported deleterious mutations in UNC13D were associated with more severe disease and earlier age of onset.16,18,24,29,30 NK cell activity was determined in five patients with FHL mutations, and four of them had decreased activity (Table 1). P6 had normal NK cell activity; however, the diagnosis of FHL was not equivocal in the patient, since he had two mutant alleles both in the consensus splicing sites. Three patients with FHL3 in our series (38%) had CNS involvement at initial diagnosis, which is lower than previously reported rates (range, 60% – 90%).16,27,30 CNS involvement is the most discouraging clinical manifestation in FHL, and the association of CNS involvement with FHL3 might be related to the role of UNC13D in CNS function.34 Two of these three patients succumbed to the disease, and also the single patient with FHL2 developed CNS involvement and died during the course of HLH-2004 treatment.
In conclusion, this is the first report on the molecular genetics of FHL in Korea. Notably, UNC13D was the predominant causative gene of FHL in Korea. The data also showed that a significant proportion of patients appear to have UNC13D mutations that cannot be detected by conventional sequencing. Further investigations are needed to uncover these missed mutations and thereby establish an optimized molecular diagnostic strategy, potentially involving RNA-based screening.
Acknowledgments
the authors thank the physicians who enrolled their patients in the Korean Registry for Histiocytosis and sent samples for molecular genetic studies. We also thank the patients and family members who participated in this study.
Footnotes
Funding: this work was supported by the grant #2008-138 from Asan Institute of Life Science, Seoul, Korea, a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea #A080588-18, and by grant #CRS-108-02-1 from the Clinical Research Development Program at Samsung Medical Center, Seoul, Korea.
Authorship and Disclosures
HJK and JJS were the principal investigators who coordinated the research and take primary responsibility for the paper. HSY and HJK wrote the paper. HSY, KHY, KWS, HHK, HJK, HYS, HSA, JYK, YTL, KWB, HJI, and JJS recruited patients and their family members and provided clinical data and blood samples. KHJ, KOL, STL, and HSC (Hae-Sun Chung) took part in the molecular genetic studies and interpretation of the results. HJK, SHK, CJP, and HSC (Hyun-Sook Chi) provided hematologic data and related laboratory work-ups including natural killer cell activity.
The authors report no potential conflicts of interest.
The online version of this article has a Supplementary Appendix.
References
- 1.Henter JI, Elinder G, Ost A. Diagnostic guidelines for hemophagocytic lymphohistiocytosis. The FHL Study Group of the Histiocyte Society. Semin Oncol. 1991;18(1):29–33. [PubMed] [Google Scholar]
- 2.Janka GE. Familial hemophagocytic lymphohistiocytosis. Eur J Pediatr. 1983;140(3):221–30. doi: 10.1007/BF00443367. [DOI] [PubMed] [Google Scholar]
- 3.Loy TS, Diaz-Arias AA, Perry MC. Familial erythrophagocytic lymphohistiocytosis. Semin Oncol. 1991;18(1):34–8. [PubMed] [Google Scholar]
- 4.Arico M, Janka G, Fischer A, Henter JI, Blanche S, Elinder G, et al. Hemophagocytic lymphohistiocytosis. Report of 122 children from the International Registry. FHL Study Group of the Histiocyte Society. Leukemia. 1996;10(2):197–203. [PubMed] [Google Scholar]
- 5.Henter JI, Arico M, Elinder G, Imashuku S, Janka G. Familial hemophagocytic lymphohistiocytosis. Primary hemophagocytic lymphohistiocytosis. Hematol Oncol Clin North Am. 1998;12(2):417–33. doi: 10.1016/s0889-8588(05)70520-7. [DOI] [PubMed] [Google Scholar]
- 6.Henter JI, Samuelsson-Horne A, Arico M, Egeler RM, Elinder G, Filipovich AH, et al. Treatment of hemophagocytic lymphohistiocytosis with HLH-94 immunochemotherapy and bone marrow transplantation. Blood. 2002;100(7):2367–73. doi: 10.1182/blood-2002-01-0172. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan KE, Delaat CA, Douglas SD, Filipovich AH. Defective natural killer cell function in patients with hemophagocytic lymphohistiocytosis and in first degree relatives. Pediatr Res. 1998;44(4):465–8. doi: 10.1203/00006450-199810000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Imashuku S, Hyakuna N, Funabiki T, Ikuta K, Sako M, Iwai A, et al. Low natural killer activity and central nervous system disease as a high-risk prognostic indicator in young patients with hemophagocytic lymphohistiocytosis. Cancer. 2002;94(11):3023–31. doi: 10.1002/cncr.10515. [DOI] [PubMed] [Google Scholar]
- 9.Haddad E, Sulis ML, Jabado N, Blanche S, Fischer A, Tardieu M. Frequency and severity of central nervous system lesions in hemophagocytic lymphohistiocytosis. Blood. 1997;89(3):794–800. [PubMed] [Google Scholar]
- 10.Jabado N, de Graeff-Meeder ER, Cavazzana-Calvo M, Haddad E, Le Deist F, Benkerrou M, et al. Treatment of familial hemophagocytic lymphohistiocytosis with bone marrow transplantation from HLA genetically nonidentical donors. Blood. 1997;90(12):4743–8. [PubMed] [Google Scholar]
- 11.Durken M, Horstmann M, Bieling P, Erttmann R, Kabisch H, Loliger C, et al. Improved outcome in haemophagocytic lymphohistiocytosis after bone marrow transplantation from related and unrelated donors: a single-centre experience of 12 patients. Br J Haematol. 1999;106(4):1052–8. doi: 10.1046/j.1365-2141.1999.01625.x. [DOI] [PubMed] [Google Scholar]
- 12.Horne A, Janka G, Maarten Egeler R, Gadner H, Imashuku S, Ladisch S, et al. Haematopoietic stem cell transplantation in haemophagocytic lymphohistiocytosis. Br J Haematol. 2005;129(5):622–30. doi: 10.1111/j.1365-2141.2005.05501.x. [DOI] [PubMed] [Google Scholar]
- 13.Henter JI, Horne A, Arico M, Egeler RM, Filipovich AH, Imashuku S, et al. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48(2):124–31. doi: 10.1002/pbc.21039. [DOI] [PubMed] [Google Scholar]
- 14.Dufourcq-Lagelouse R, Jabado N, Le Deist F, Stephan JL, Souillet G, Bruin M, et al. Linkage of familial hemophagocytic lymphohistiocytosis to 10q21-22 and evidence for heterogeneity. Am J Hum Genet. 1999;64(1):172–9. doi: 10.1086/302194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.zur Stadt U, Schmidt S, Kasper B, Beutel K, Diler AS, Henter JI, et al. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet. 2005;14(6):827–34. doi: 10.1093/hmg/ddi076. [DOI] [PubMed] [Google Scholar]
- 16.Feldmann J, Callebaut I, Raposo G, Certain S, Bacq D, Dumont C, et al. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3) Cell. 2003;115(4):461–73. doi: 10.1016/s0092-8674(03)00855-9. [DOI] [PubMed] [Google Scholar]
- 17.Ohadi M, Lalloz MR, Sham P, Zhao J, Dearlove AM, Shiach C, et al. Localization of a gene for familial hemophagocytic lymphohistiocytosis at chromosome 9q21.3-22 by homozygosity mapping. Am J Hum Genet. 1999;64(1):165–71. doi: 10.1086/302187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto K, Ishii E, Sako M, Ohga S, Furuno K, Suzuki N, et al. Identification of novel MUNC13-4 mutations in familial haemophagocytic lymphohistiocytosis and functional analysis of MUNC13-4-deficient cytotoxic T lymphocytes. J Med Genet. 2004;41(10):763–7. doi: 10.1136/jmg.2004.021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueda I, Ishii E, Morimoto A, Ohga S, Sako M, Imashuku S. Correlation between phenotypic heterogeneity and gene mutational characteristics in familial hemophagocytic lymphohistiocytosis (FHL) Pediatr Blood Cancer. 2006;46(4):482–8. doi: 10.1002/pbc.20511. [DOI] [PubMed] [Google Scholar]
- 20.Arico M, Danesino C, Pende D, Moretta L. Pathogenesis of haemophagocytic lymphohistiocytosis. Br J Haematol. 2001;114(4):761–9. doi: 10.1046/j.1365-2141.2001.02936.x. [DOI] [PubMed] [Google Scholar]
- 21.Marcenaro S, Gallo F, Martini S, Santoro A, Griffiths GM, Arico M, et al. Analysis of natural killer-cell function in familial hemophagocytic lymphohistiocytosis (FHL): defective CD107a surface expression heralds Munc13-4 defect and discriminates between genetic subtypes of the disease. Blood. 2006;108(7):2316–23. doi: 10.1182/blood-2006-04-015693. [DOI] [PubMed] [Google Scholar]
- 22.Goransdotter Ericson K, Fadeel B, Nilsson-Ardnor S, Soderhall C, Samuelsson A, Janka G, et al. Spectrum of perforin gene mutations in familial hemophagocytic lymphohistiocytosis. Am J Hum Genet. 2001;68(3):590–7. doi: 10.1086/318796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molleran Lee S, Villanueva J, Sumegi J, Zhang K, Kogawa K, Davis J, et al. Characterisation of diverse PRF1 mutations leading to decreased natural killer cell activity in North American families with haemophagocytic lymphohistiocytosis. J Med Genet. 2004;41(2):137–44. doi: 10.1136/jmg.2003.011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zur Stadt U, Beutel K, Kolberg S, Schneppenheim R, Kabisch H, Janka G, et al. Mutation spectrum in children with primary hemophagocytic lymphohistiocytosis: molecular and functional analyses of PRF1, UNC13D, STX11, and RAB27A. Hum Mutat. 2006;27(1):62–8. doi: 10.1002/humu.20274. [DOI] [PubMed] [Google Scholar]
- 25.Horne A, Ramme KG, Rudd E, Zheng C, Wali Y, al-Lamki Z, et al. Characterization of PRF1, STX11 and UNC13D genotype-phenotype correlations in familial hemophagocytic lymphohistiocytosis. Br J Haematol. 2008;143(1):75–83. doi: 10.1111/j.1365-2141.2008.07315.x. [DOI] [PubMed] [Google Scholar]
- 26.Ueda I, Morimoto A, Inaba T, Yagi T, Hibi S, Sugimoto T, et al. Characteristic perforin gene mutations of haemophagocytic lymphohistiocytosis patients in Japan. Br J Haematol. 2003;121(3):503–10. doi: 10.1046/j.1365-2141.2003.04298.x. [DOI] [PubMed] [Google Scholar]
- 27.Ishii E, Ueda I, Shirakawa R, Yamamoto K, Horiuchi H, Ohga S, et al. Genetic subtypes of familial hemophagocytic lymphohistiocytosis: correlations with clinical features and cytotoxic T lymphocyte/natural killer cell functions. Blood. 2005;105(9):3442–8. doi: 10.1182/blood-2004-08-3296. [DOI] [PubMed] [Google Scholar]
- 28.Santoro A, Cannella S, Trizzino A, Bruno G, De Fusco C, Notarangelo LD, et al. Mutations affecting mRNA splicing are the most common molecular defect in patients with familial hemophagocytic lymphohistiocytosis type 3. Haematologica. 2008;93(7):1086–90. doi: 10.3324/haematol.12622. [DOI] [PubMed] [Google Scholar]
- 29.Rudd E, Bryceson YT, Zheng C, Edner J, Wood SM, Ramme K, et al. Spectrum, and clinical and functional implications of UNC13D mutations in familial haemophagocytic lymphohistiocytosis. J Med Genet. 2008;45(3):134–41. doi: 10.1136/jmg.2007.054288. [DOI] [PubMed] [Google Scholar]
- 30.Santoro A, Cannella S, Bossi G, Gallo F, Trizzino A, Pende D, et al. Novel Munc13-4 mutations in children and young adult patients with haemophagocytic lymphohistiocytosis. J Med Genet. 2006;43(12):953–60. doi: 10.1136/jmg.2006.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clementi R, zur Stadt U, Savoldi G, Varoitto S, Conter V, De Fusco C, et al. Six novel mutations in the PRF1 gene in children with haemophagocytic lymphohistiocytosis. J Med Genet. 2001;38(9):643–6. doi: 10.1136/jmg.38.9.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SM, Sumegi J, Villanueva J, Tabata Y, Zhang K, Chakraborty R, et al. Patients of African ancestry with hemophagocytic lymphohistiocytosis share a common haplotype of PRF1 with a 50delT mutation. J Pediatr. 2006;149(1):134–7. doi: 10.1016/j.jpeds.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Al-Jasmi F, Abdelhaleem M, Stockley T, Lee KS, Clarke JT. Novel mutation of the perforin gene and maternal uniparental disomy 10 in a patient with familial hemophagocytic lymphohistiocytosis. J Pediatr Hematol Oncol. 2008;30(8):621–4. doi: 10.1097/MPH.0b013e31817580fd. [DOI] [PubMed] [Google Scholar]
- 34.Betz A, Telemenakis I, Hofmann K, Brose N. Mammalian Unc-13 homologues as possible regulators of neurotransmitter release. Biochem Soc Trans. 1996;24(3):661–6. doi: 10.1042/bst0240661. [DOI] [PubMed] [Google Scholar]