Abstract
T-cell acute lymphoblastic leukemia (T-ALL) patients frequently display NOTCH1 activating mutations and Notch can transcriptionally down-regulate the tumor suppressor PTEN. However, it is not clear whether NOTCH1 mutations associate with decreased PTEN expression in primary T-ALL. Here, we compared patients with or without NOTCH1 mutations and report that the former presented higher MYC transcript levels and decreased PTEN mRNA expression. We recently showed that T-ALL cells frequently display CK2-mediated PTEN phosphorylation, resulting in PTEN protein stabilization and concomitant functional inactivation. Accordingly, the T-ALL samples analyzed, irrespectively of their NOTCH1 mutational status, expressed significantly higher PTEN protein levels than normal controls. To evaluate the integrated functional impact of Notch transcriptional and CK2 post-translational inactivation of PTEN, we treated T-ALL cells with both the gamma-secretase inhibitor DAPT and the CK2 inhibitors DRB/TBB. Our data suggest that combined use of gamma-secretase and CK2 inhibitors may have therapeutic potential in T-ALL.
Keywords: NOTCH1, protein stabilization, CK2 inhibitors, T-ALL
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) patients frequently display NOTCH1 activating mutations.1 Recently it was demonstrated that Notch1 can negatively regulate PTEN at the transcriptional level.2 It was further suggested that such negative regulation could occur in diagnostic T-ALL cells collected from patients. However, the actual impact of NOTCH1 mutations on PTEN expression and activity in primary T-ALL is still unclear. Although NOTCH1 mutations are expected to occur in approximately 50% of diagnostic T-ALL cases,2, 3 most samples appear to display high PTEN protein levels compared to normal thymocytes.4 The apparently paradoxical increase in PTEN expression results from CK2-mediated phosphorylation of PTEN and consequent PTEN protein stabilization and functional inactivation, which ultimately contributes to hyperactivation of PI3K/Akt oncogenic pathway in T-ALL cells.4 Here, we sought to understand how Notch1-and CK2-mediated regulation of PTEN may be integrated and explored therapeutically in T-ALL.
Design and Methods
Primary samples and T-ALL cell lines
T-ALL cells were obtained at diagnosis from bone marrow or peripheral blood of pediatric patients with high leukemia involvement (85–100%). Samples were enriched by density centrifugation over Ficoll-Paque (GE Healthcare). Normal thymocytes were isolated from thymic tissue obtained from children undergoing cardiac surgery as described.4 Informed consent and institutional review board approval (Gabinete de Investigação Clínica, Instituto Português de Oncologia, and Comitê de Ética em Pesquisa da Faculdade de Ciências Médicas da Universidade Estadual de Campinas) were obtained in accordance with the Declaration of Helsinki. TAIL7, which shares significant similarities with primary leukemia samples,5 TALL-1 and HPB-ALL are PTEN-positive T-ALL cell lines.
NOTCH1 sequencing and mutational analysis
Part of exon 26, the whole exon 27 and the beginning (nt 4677–5255) of exon 28 was amplified from cDNA samples using primers Notch2627F and Notch2627R, and sequenced using the same primers. In some cases, exon 26 and exon 27 were amplified and sequenced from genomic DNA with the intronic primers Notch26F and Notch26R, and Notch27F and Ex27R743, respectively. The region spanning TAD and PEST domains of exon 34 was amplified with primers Notch34TF and Notch34PR, and the resulting 855 bp fragment was sequenced with primers Notch34TF and Ex34PestF. The PEST coding region was also amplified with primers Ex34PestF and Ex34PestR, followed by semi-nested PCR with primers Notch34PF and Ex34PestR, and sequenced with primer Ex34PestR. This strategy covered all mutational hot-spot regions previously reported for NOTCH1.1,6,7 Primer sequences and the PCR protocol are shown in the Online Supplementary Appendix. All mutations were confirmed by sequencing a newly amplified product. When necessary, PCR amplicons were cloned into pGEM-T vector (Promega) and several individual clones were sequenced.
Quantitative RT-PCR
Total RNA (1μg) was reverse transcribed using the ImProm II Reverse Transcriptase enzyme (Promega) and random hexamers. The quantitative assessment of HES1, MYC and PTEN transcripts was made by Q-PCR on a StepOne Real-Time PCR System (Applied Biosystems). PCR products were cloned into the pGEM-T Easy vector (Promega) and standard curves were obtained by serial dilutions of uncut plasmid. HES1, MYC and PTEN transcript values were normalized with respect to the number of ABL transcripts. PCR reactions were performed in 15 μL containing 5 μL of diluted cDNA (~5X dilution), 7.5 pmol of each primer, and 7.5 μL of SYBR Green Master Mix (Roche). Primers and PCR protocols are shown in the Online Supplementary Appendix. Q-PCR expression values were transformed into log values. Experiments were carried out in duplicates.
Western blot
Cells were lysed in 50mM Tris-HCl pH 8.0, 150mM NaCl, 5mM EDTA, 1% (v/v) NP-40, 1mM Na3VO4, 10mM NaF, 10mM NaPyroph, 1mM 4-(2-aminoethyl) benzenesulfonyl (AEBSF), 10μg/ml leupeptin, 10 μg/mL aprotinin, 1 μg/mL Pepstatin, resolved by 10% SDS-PAGE, transferred onto nitrocellulose membranes, and immunoblotted with the following antibodies: PTEN, P-PTEN (S380) and Notch Val1744 (Cell Signaling Technology), ZAP-70 (Upstate), and Actin (Santa Cruz Biotechnology). Densitometry analysis of PTEN and Actin was performed using Image Quant 5.2 software. Each band was analyzed with a constant frame.
Analysis of cell size, cell number, proliferation and cell viability
Cells were cultured in 24-well plates as 2×106 cells/mL at 37°C with 5% CO2 in RPMI-1640 medium supplemented with 10% FBS in the presence or absence of DRB/TBB and/or DAPT, and analyzed after three, four or seven days. Cell size was analyzed by flow cytometry, as described.8 Total cell counts were calculated by trypan blue exclusion using a hemocytometer. Proliferation was assessed as described.9 Briefly, cells were cultured in flat-bottom 96-well plates and incubated with 3H-thymidine (27×10−3 mBq/well) for 16 hours prior to harvest. 3H-thymidine incorporation was assessed using a liquid scintillation counter. Viability was evaluated by flow cytometry analysis of FSC x SSC pattern, as described.8 Minimum tested dose of each inhibitor (1, 5 or 10 μM DAPT; 12.5 or 25 μM DRB/TBB) that originated at least a 10% inhibitory effect was identified for each cell line, for each functional assay, and used in combination for the assessment of cooperative effects. For primary T-ALL cells, the TBB dose had been previously determined4 and DAPT was tested at a single, high concentration (5 μM).
Statistical analysis
Differences between populations were calculated using an unpaired two-tailed Mann-Whitney test, Student’s t test, or One-Way ANOVA, as appropriate. Differences were considered significant for P<0.05.
Results and Discussion
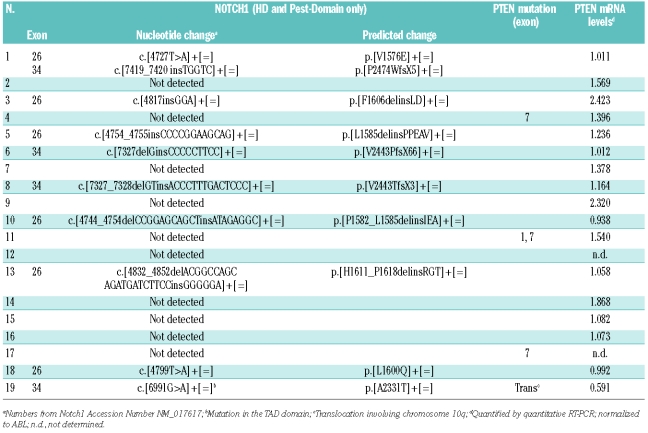
We classified 19 diagnostic T-ALL specimens as NOTCH1 wild-type (NOTCH-WT) or mutated (NOTCH-mut) after sequencing of NOTCH1 mutational hot spots (exons 26, 27, and 34) and also part of exon 28 (nt 5209–5221), recently shown to be altered in T-ALL.7 Our data show that 50% of the patients displayed NOTCH1 mutations (Table 1).
Table 1.
Alterations in NOTCH1 gene, predicted Notch1 protein changes and PTEN mRNA levels in primary T-ALL.
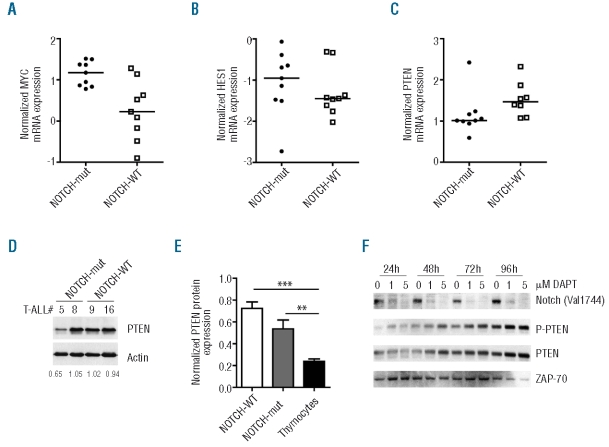
As determined by Q-PCR, NOTCH-mut patient samples displayed higher mRNA levels of the Notch target gene MYC than NOTCH-WT specimens (P=0.004, two-tailed Mann-Whitney test; Figure 1A). These observations are in accordance with previous studies demonstrating the importance of MYC as a downstream effector of Notch-mediated leukemogenesis,10–12 and with recent evidence that MYC mRNA levels are up-regulated in primary T-ALL cells with NOTCH1 activating mutations.13 Although NOTCH-mut T-ALL also presented a tendency for higher HES1 levels (median HES1/ABL =−0.954 vs. −1.449) the difference was not statistically significant (P=0.4363; Figure 1B). Importantly, PTEN mRNA levels were significantly decreased in NOTCH-mut primary T-ALL cells (P=0.021; Figure 1C and Table 1). In addition, NOTCH-mut T-ALLs displayed a tendency to have lower PTEN protein levels than NOTCH-WT samples (median = 0.769 vs. 0.481, P=0.158; Figure 1D and E). Moreover, the gamma-secretase inhibitor (GSI) DAPT up-regulated PTEN protein expression in the primary-like leukemia cell line TAIL7 (Figure 1F), indicating that Notch inhibition results in increased PTEN protein expression. Altogether, our data suggest that activating mutations of NOTCH1 transcriptionally down-regulated PTEN in primary leukemia, in agreement with what was demonstrated in immortalized T-ALL cell lines.2
Figure 1.
NOTCH1 mutations associate with decreased PTEN expression in primary T-ALL. Diagnostic T-ALL specimens classified as NOTCH1 mutated (NOTCH-Mut; n=9) or wild-type (NOTCH-WT; n=9, except for PTEN analysis n=8) were compared by Q-PCR for mRNA expression of (A) HES1 (P=0.436), (B) MYC (P=0.004), and (C) PTEN (P=0.021). Transcript copy numbers were normalized with respect to the number of ABL transcripts and transformed into logarithmic values. (D) NOTCH-WT and NOTCH-Mut samples were analyzed by Western blot for PTEN protein expression. Actin was used as a loading control. Densitometry analysis values of PTEN normalized to Actin are shown for each case. (E) PTEN protein levels in NOTCH-WT T-ALL (n=3), NOTCH-mut T-ALL (n=5) and normal thymocytes (n = 6) were evaluated by densitometry analysis after Western Blot and normalized to the Actin loading control. ** P<0.01; *** P<0.0001. (F) Primary-like leukemia TAIL7 cells were cultured with or without 1 or 5μM DAPT. At the indicated time points, cells were lysed and evaluated by Western blot for the expression of cleaved (activated) Notch (Val1744), PTEN, and P-PTEN(S380). ZAP-70 was used as a loading control. Data are representative of two independent experiments.
We recently demonstrated that most primary T-ALL samples have higher PTEN protein levels than normal human thymocytes.4 This stems from the fact that leukemia cells over-express CK2, which mediates phosphorylation and consequent protein stabilization, but functional inactivation, of PTEN.4 In this context, it is tempting to hypothesize that T-ALL samples with decreased PTEN mRNA expression due to NOTCH1 mutations may have stabilized, although inactivated, PTEN protein, as a consequence of CK2-mediated phosphorylation. In line with this possibility, we found that not only NOTCH-WT but also NOTCH-mut samples had significantly higher PTEN protein levels than normal human thymocytes (Figure 1E). Furthermore, GSI-dependent upregulation of PTEN protein in TAIL7 cells was paralleled by PTEN phosphorylation in the CK2 target residue Ser380 (Figure 1F). Our data suggest that the impact of Notch1-mediated PTEN transcriptional down-regulation may be partially compensated at the protein level by CK2-dependent PTEN phosphorylation and stabilization. This could explain why: 1) NOTCH-mut patients display somewhat lower but similar PTEN protein levels to other T-ALL cases, and 2) both NOTCH-WT and NOTCH-mut samples express higher PTEN protein levels than normal thymocytes.
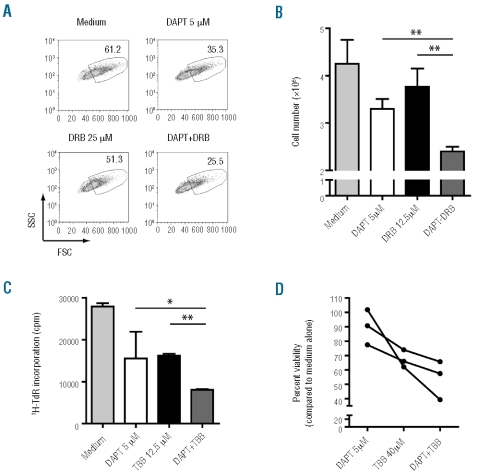
Despite the opposite effects on PTEN protein levels, Notch1 and CK2 ultimately contribute to PTEN inactivation by complementary mechanisms. Therefore, we speculated that the combination of GSI and CK2 inhibitors could have a cooperative effect in restoring PTEN function and thereby targeting leukemia cells. This is particularly relevant given the relative inefficacy of GSI treatment in T-ALL patients14 and recent clinical data in patients with cervical cancer indicating that CK2 antagonists are well tolerated and may have antitumor efficacy.15 Hence, we treated PTEN-positive T-ALL cell lines with GSI DAPT and/or the CK2-specific inhibitors DRB or TBB. Analyses of cell size (Figure 2A and Online Supplementary Figure S1), total cell counts (Figure 2B and Online Supplementary Figure S2) and proliferation (Figure 2C and Online Supplementary Figure S3) revealed that combination of GSI and CK2 inhibitors have a mild but consistent cooperative effect in diminishing leukemia proliferation. Similar results were obtained regarding cell viability using diagnostic primary leukemia cells (Figure 2D).
Figure 2.
GSI and CK2 inhibitors collaborate in decreasing T-ALL cell proliferation. PTEN-positive T-ALL cell lines HPB-ALL (A and C) and TAIL7 (B), and primary T-ALL cells (D), were used to test the cooperative impact of combined treatment with the GSI DAPT and the CK2 inhibitors DRB or TBB, at the indicated concentrations (see Design and Methods section). (A) Cells were cultured for 72h and evaluated for alterations in cell size by forward scatter flow cytometry analysis. (B and C) After seven days of culture, total cell counts (B) were calculated by trypan blue exclusion using a hemocytometer, and proliferation (C) was evaluated by assessing DNA synthesis by 3H-thymidine incorporation. *P<0.05; **P<0.01. (D) Primary T-ALL cells from 3 different patients were assessed for cell viability by flow cytometry at 72h of culture. P=0.020; One-way ANOVA.
Overall, our studies support the notion that NOTCH1 mutations can lead to decreased PTEN mRNA expression in primary T-ALL.2 However, this negative effect on PTEN expression appears counterbalanced, at least in part, by CK2-mediated phosphorylation and stabilization of PTEN protein, which are associated with PTEN functional inactivation and consequent hyperactivation of PI3K/Akt pathway.4 Evidently, these observations do not exclude the possibility that other Notch-dependent mechanisms, in addition to PTEN transcriptional downregulation, may promote PI3K/Akt pathway activation in T-ALL cells. For example, it was recently shown that NOTCH1 up-regulates IL-7Rα expression in T-ALL cells.16 This is particularly interesting given that the PI3K/Akt pathway is essential for IL-7/IL-7R-mediated survival, growth and proliferation of primary T-ALL cells in vitro.8
There is evidence from both mouse models and human cancers that PTEN is happloinsufficient for tumor suppression and that the exact level of PTEN activity in a cell is a key determinant of cancer progression.17 In primary T-ALL cells, PTEN activity appears to be negatively regulated at the transcriptional level by Notch, and posttranslationally by CK2. Our data indicate that targeting simultaneously these two molecular mechanisms can lead to more effective inhibition of leukemia cell viability and proliferation in vitro. These observations may have important therapeutic implications for the treatment of diagnostic T-ALL.
Acknowledgments
The authors would like to thank the contribution of patients and the clinical teams involved in providing primary leukemia samples, Dr. Miguel Abecasis for providing thymic specimens, and Dr. Adolfo Ferrando for providing some of the NOTCH sequencing primers.
Footnotes
Funding: this work was supported by grants from Fundação para a Ciência e a Tecnologia (FCT; POCI/SAU-OBS/58913 and PTDC/SAU-OBD/69974), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; 08/10034-1), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; 401122/2005-0). AS and PYJ have FCT SFRH and FAPESP PhD scholarships, respectively.
The Online version of this article has a Supplementary Appendix.
Authorship and Disclosures
AS, PYJ, ABS and DR performed research and analyzed data. SRB coordinated the recruitment and treatment of patients. JAY designed research, analyzed data, supervised the work, and contributed to the writing of the manuscript. JTB designed research, analyzed data, supervised the work and wrote the paper.
The authors reported no potential conflicts of interest.
References
- 1.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 2.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447(7147):966–71. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118(11):3762–74. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barata JT, Boussiotis VA, Yunes JA, Ferrando AA, Moreau LA, Veiga JP, et al. IL-7-dependent human leukemia T-cell line as a valuable tool for drug discovery in T-ALL. Blood. 2004;103(5):1891–900. doi: 10.1182/blood-2002-12-3861. [DOI] [PubMed] [Google Scholar]
- 6.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108(4):1151–7. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 7.Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112(3):733–40. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barata JT, Silva A, Brandao JG, Nadler LM, Cardoso AA, Boussiotis VA. Activation of PI3K Is Indispensable for Interleukin 7-mediated Viability, Proliferation, Glucose Use, and Growth of T Cell Acute Lymphoblastic Leukemia Cells. J Exp Med. 2004;200(5):659–69. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barata JT, Cardoso AA, Nadler LM, Boussiotis VA. Interleukin-7 promotes survival and cell cycle progression of T-cell acute lymphoblastic leukemia cells by down-regulating the cyclin-dependent kinase inhibitor p27(kip1) Blood. 2001;98(5):1524–31. doi: 10.1182/blood.v98.5.1524. [DOI] [PubMed] [Google Scholar]
- 10.Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26(21):8022–31. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward- loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103(48):18261–6. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson Gedman A, Chen Q, Kugel Desmoulin S, Ge Y, Lafiura K, Haska CL, et al. The impact of NOTCH1, FBW7 and PTEN mutations on prognosis and down-stream signaling in pediatric T-cell acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Leukemia. 2009;23(8):1417–25. doi: 10.1038/leu.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15(1):50–8. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solares AM, Santana A, Baladron I, Valenzuela C, Gonzalez CA, Diaz A, et al. Safety and preliminary efficacy data of a novel casein kinase 2 (CK2) peptide inhibitor administered intralesionally at four dose levels in patients with cervical malignancies. BMC Cancer. 2009;9:146. doi: 10.1186/1471-2407-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Garcia S, Garcia-Peydro M, Martin-Gayo E, Ballestar E, Esteller M, Bornstein R, et al. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7R{alpha} gene expression in early human thymopoiesis and leukemia. J Exp Med. 2009;206(4):779–91. doi: 10.1084/jem.20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133(3):403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]