Bites from apes efficiently transmit the foamy virus to humans in natural settings in central Africa.
Keywords: Interspecies transmission, retrovirus, foamy, hunters, primates, epidemiology, emerging viruses, research
Abstract
Simian virus infections of humans are an increasing public health concern. Simian foamy virus (SFV) infections have been reported in persons occupationally exposed to nonhuman primates and in a few hunters in Cameroon. To better understand this retroviral zoonosis in natural settings, we studied persons who lived in southern Cameroon, near nonhuman primate habitats. First we studied a general population of 1,164 adults; 4 were SFV positive according to serologic and molecular assays. Then we studied 85 persons who reported having been bitten or scratched by nonhuman primates; 7/29 (24.1%) of those who had contact with apes (gorillas or chimpanzees) were SFV positive, compared with only 2/56 (3.6%) of those who had had contact with monkeys. These data demonstrate efficient transmission of SFVs to humans in natural settings in central Africa, specifically following ape bites, and viral persistence in the human host.
A large proportion of viral pathogens that have recently emerged in humans have originated in various animals. After initial interspecies transmission, these viruses have evolved and disseminated into the human population through various distinct mechanisms. However, understanding of the initial steps of the emergence of some viruses and associated diseases remains poor. Microbiologic studies of these high-risk populations are thus necessary to obtain new insights into the early events of this emergence process (1–4).
Nonhuman primates represent a potential source of microbes for humans (1,5–12), e.g., simian immunodeficiency virus and simian T-lymphotropic virus (12–15). Simian foamy viruses (SFVs) are exogenous complex retroviruses, highly prevalent in several animal species in which they cause persistent infections (16–26). Phylogenetic analyses have demonstrated a species-specific distribution of such retroviruses. This species specificity indicates a long-term coevolution of SFVs with their natural hosts (27), which could explain their possible lack of pathogenicity observed in vivo and the persistence of the infection (23,24,28–31). Among nonhuman primate populations, SFV seroprevalence can reach 75%–100% in adults, and SFVs appear to be present at high concentrations in the saliva of infected animals (16–18,22,29,31).
In humans, SFV infection has been reported in 1%–4% of persons occupationally exposed to nonhuman primates in zoos, primate centers, and laboratories, mainly in North America but also in Europe (7–10). More recently, naturally acquired SFV infections were described in a few hunters living in Cameroon (11) and in 1 person who had had contact with Macaca fascicularis in Indonesia (32).
After other studies demonstrated high prevalence and genetic diversity of SFVs in monkeys and apes in Gabon and Cameroon (16,17), we investigated the presence of SFV infection in humans living in these regions. Our goals were to 1) determine, by using specific serologic and molecular methods, the prevalence of SFV infection in the adult population of different ethnic groups (including Pygmies) who lived in rural areas of Cameroon near natural nonhuman primate habitats and who were thus at risk for cross-species transmission; 2) trace the origin of the SFVs infecting these persons by isolation and molecular characterization of the virus; and 3) gain new insights into the epidemiologic determinants and risk factors linked to such naturally acquired retroviral infections, especially the type of nonhuman primates, the circumstances of the contact leading to the infection, and possible intrafamilial transmission of such viruses.
Materials and Methods
Populations
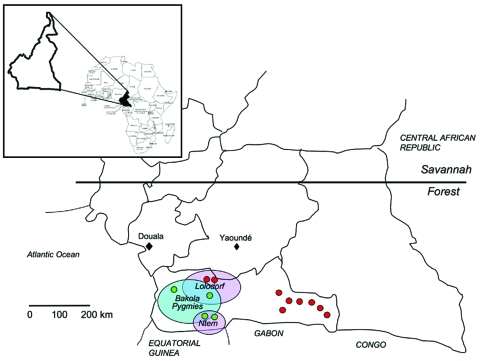
The first study, a retrospective study, was based on a large series of samples collected during 1994–2000 for epidemiologic studies on human T-lymphotropic virus (HTLV)-1 and HTLV-2 as well as human herpesvirus 8 (33,34). The samples originated from adults of 3 ethnic populations: Bakola Pygmies and 2 groups of Bantus, who lived in lowland tropical remote forest areas (Bipindi/Lolodorf and Ntem) in southwestern Cameroon (Figure 1).
Figure 1.
Geographic distribution in Cameroon of the studied populations and the 13 persons infected by simian foamy virus (SFV), according to serologic and molecular results. Red, SFV-positive persons from the hunter study; green, SFV-positive persons from the retrospective study; blue circle, Pygmy area; violet circles, Bantu areas.
The second study, the hunter study, was conducted in 2004–2005 in remote villages near nonhuman primate habitats in the South Province of Cameroon (Figure 1). This study was focused on persons who reported direct and severe contacts (bites, wounds, scratches, other injuries) with animals, especially nonhuman primates, mainly while hunting.
Both studies received clearance from national and local authorities. All participants received detailed information about the study and gave consent. Blood samples were collected in 5–10 mL EDTA tubes. Plasma was available from all participants in the retrospective study, whereas for some in the hunter study, only a few drops of blood were taken by fingerstick and conserved on filter paper (Whatman samples) as described (35). See Technical Appendix, for more details.
Serologic Tests, Virus Isolation, and Molecular Studies
We screened by Western blot (WB) all plasma and Whatman samples for the presence of SFV antibodies as described (18,22,26). Plasma was tested at a 1:100 dilution. For each Whatman sample, a 1-cm punch was diluted in 1 mL of phosphate-buffered saline and tested at a 1:8 dilution (Technical Appendix). Virus isolation, electron microscopy, and immunofluorescence (IFA) were performed as described (9,21,26,36; Technical Appendix).
For the molecular studies, genomic DNA was extracted from the peripheral blood buffy coat by using the QIAamp DNA Blood Mini Kit (QIAGEN, Courtaboeuf, France). Two SFV proviral genomic regions (465 bp of the integrase gene and 109 bp of the long terminal repeat [LTR]) were amplified in nested PCR (18,21,37). Integrase PCR products were purified, cloned, and sequenced. The GenBank accession numbers of the 13 new integrase sequences are DQ838495–DQ838507. Phylogenetic analyses were performed as described (18,38,39; Technical Appendix.)
Results
Retrospective Study
The retrospective epidemiologic survey was performed among 1,164 adults (mean age 50.6 years) who lived in the Ocean region of Cameroon (Figure 1; Table 1). The studied populations included 478 Bakola Pygmies (mean age 47.6 years) and 686 Bantus (mean age 52.6 years).
Table 1. Serologic results for simian foamy virus retrospective study, rural Cameroon, 1994–2000.
| Area | Study population |
Test results |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Race | Age range, y | Total no. | Men | Women | Negative | Borderline, no. (%) | Positive, no. (%) | ||
| Bipindi Lolodorf | Bakola Pygmies | 30–82 | 478 | 214 | 264 | 448 | 16 (3.34) | 14 (2.92) | |
| Bantus | 40–83 | 370 | 180 | 190 | 326 | 40 (10.81) | 4 (1.08) | ||
| Ntem | Bantus | 20–78 | 316 | 144 | 172 | 283 | 30 (9.49) | 3 (0.9) | |
| Total | 20–83 | 1,164 | 538 | 626 | 1,057 | 86 (7.38) | 21 (1.8) | ||
Of the 1,164 samples tested by WB assay based on chimpanzee foamy virus antigens, 21 (1.8%) were considered clearly positive (strong reactivity to both p70 and p74 ape proteins, Gag doublet) (Figure 2, panel A), 86 (7.4%) were considered borderline/indeterminate (presence of either a faint gag doublet or of at least a strong band of the right size and 1 or few other bands of often low intensity) (Figure 2, panel C), and the remaining 1,057 samples were considered negative (absence of any band) (Figure 2, panel C; Table 1). The 86 indeterminate samples were then tested by WB assay using antigens from a monkey foamy virus (originating from participant AG16, Figure 2, panel B); all were still indeterminate or negative.
Figure 2.
A) Western blot (WB) results based on chimpanzee (cpz) simian foamy virus (SFV) antigens. B) WB results based on monkey simian foamy virus antigens originating from participant AG16. C) Example of sero-indeterminate samples (lanes 1–7) and negative samples (lanes 8–13), detected by cpzSFV WB. Last lane (POS cpz), serum from an SFV-positive chimpanzee.
DNA was available from 11 of the 21 persons whose WB assay results were positive and from 52 of 86 whose results were borderline/indeterminate. All 63 DNA samples were amplifiable by PCR for β-globin gene. When integrase primers were used, PCR was positive for 4 of 63 samples (Table 2). When LTR primers were used, PCR was positive in 3 of these 4 samples (Table 2).
Table 2. Demographic and epidemiologic features, serologic and PCR results, 13 SFV-infected inhabitants of rural Cameroon*.
| Participant code, sex, ethnicity | Year(s) of sample collection | Age at sample collection, at animal contact, y | Type of animal | Wound location | Serologic results (specimen) | LTR/ Intergase PCR | Integrase sequence | Viral load, copies/500 mg DNA |
|---|---|---|---|---|---|---|---|---|
| 60601, M, Bantu†‡ | 1999 | 67, 30 | Gorilla, monkey | Finger | + (plasma) | +/+ | Gorilla | 10–100 |
| 801001, M, Pygmy†‡§ | 1996, 1998 | 60, 35 | Gorilla | Leg | + (plasma) | +/+ | Gorilla | 100–1,000 |
| 210301, M, Pygmy†‡ | 1996, 2004 | 68, 35 | Gorilla | Leg | + (plasma) | -/+ | Gorilla | 1–10 |
| 51302, F, Bantu†‡§ | 1998 | 40, NK | None? | NK | + (plasma) | +/+ | Chimp. | 100–1,000 |
| CH29, M, Pygmy¶ | 2004, 2005 | 50, 49 | Chimp., gorilla | Finger, foot | + (DBS, plasma) | +/+ | Gorilla | 1–10 |
| CH61, M, Bantu*§¶ | 2004, 2005 | 65, 52 | Gorilla | Hand, arm | + (plasma) | +/+ | Gorilla | 10–100 |
| CH65, M, Pygmy¶ | 2004 | 58, 26 | Gorilla | Head, arm | + (plasma) | +/+ | Gorilla | 1–10 |
| CH66, M, Pygmy¶ | 2004 | 60, 56 | Cerco. nictitans, Chimp. | Hand, foot | + (plasma) | +/+ | Chimp. | ND |
| CH86, M, Bantu†¶ | 2004 | 62, 47 | Gorilla | Hand | + (plasma) | +/+ | Gorilla | 1–10 |
| CH99, M, Bantu¶ | 2004 | 26, 25 | Monkey, species? | Hand | + (plasma) | –/+ | Mandrill | 1–10 |
| CH101, M, Bantu¶ | 2004 | 76, 65 | Gorilla | Hand | + (plasma) | +/+ | Gorilla | 1–10 |
| AG15, M, Bantu¶ | 2004, 2005 | 71, 28 | Chimp. | Hand, foot | + (DBS, plasma) | +/+ | Chimp. | 100–1,000 |
| AG16, M, Bantu¶ | 2004, 2005 | 43, 23 | Monkey Cerco. | Foot | + (DBS, plasma) | –/+ | Cerco. | 1–10 |
*SFV, simian foamy virus; LTR, long terminal repeat; +, positive; –, negative; DBS, dried blood spot; NK, not known; Chimp., chimpanzee; ND, not determined; Cerco., Cercopithecus. The age at contact with nonhuman primate and the type of animal contact concern the results of the field interviews performed for each person found to be infected by SFV. †Wife or husband also tested. ‡Retrospective study participant. §1 (A051302) or 2 (CH61 and 801001) children also tested. ¶Hunter study participant.
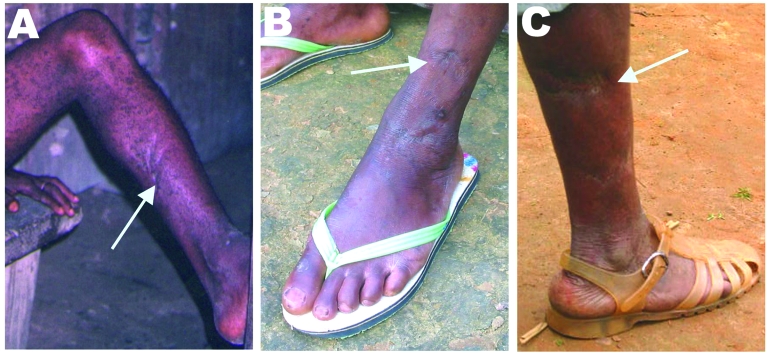
Field interviews indicated that 3 persons (2 Bakola Pygmies [801001 and 210301] and 1 Bantu [60601]) were frequent hunters and had been severely bitten by gorillas 25–35 years ago (Table 2); all 3 had scars on their legs and fingers (Figure 3). The fourth infected person (A051302) was a Bantu woman who did not recall any bites or injuries from monkeys or apes. However, she had had frequent contact with wild game meat from nonhuman primates while butchering and preparing meals, as is common in this area (3,4).
Figure 3.
Wounds resulting from bites or scratches from a nonhuman primate. A) Participant no. 801001. B) Participant no. AG16. C) Participant no. 210301.
Sequence analyses of the 4 integrase gene fragments indicated that the 3 persons bitten by gorillas were infected with a gorilla foamy virus. These 3 sequences were similar to the sequence CAM1083 (96.7%–98.5% identity) reported in a Cameroonian hunter infected by a gorilla foamy virus (11) and to known sequences of foamy virus from gorillas living in Cameroon and closely related to each other (97%–99% identity). The Bantu woman had been infected by a chimpanzee belonging to Pan troglodytes troglodytes, 1 of 2 chimpanzee subspecies endemic to Cameroon (Appendix Figure).
Hunter Study
Our next step was to not only characterize more cases of such interspecies transmission, looking especially for viral acquisition from other nonhuman primates, but also to assess the frequency of such phenomena and to define the parameters that characterize a risk population. Thus, we focused our work on persons who had regular contact with nonhuman primates, hunters in lowland rain forest regions.
During 2004–2005, field missions were initiated in remote villages of Bantus and Baka Pygmies in different areas of south Cameroon. In each village we specifically asked for persons who had had direct contact and severe bites, scratches, wounds, other injuries from animals, mainly nonhuman primates.
This study included 102 persons, 84 men and 18 women, most of them adults (mean age 40 years, range 2–80 years). Of these 102, 29 (28.4%) had had contact with apes (gorillas, chimpanzees), and 56 (54.9%) with monkeys (Cercopithecus nictitans), mandrills, and a few other small monkeys not precisely identified). Thus, 85 of 102 had been in contact with nonhuman primates. Contact with rats, elephants, warthogs, duikers, squirrels, porcupines, and leopards was reported by 17 (16.6%).
From the 102 persons, we obtained 61 plasma samples and 41 dried blood spots. All samples were tested by WB, and 10 (9.7%) were clearly SFV seropositive (Figure 2). Of 15 specimens that were indeterminate/borderline, WB based on monkey FV antigens (originating from participant AG16) showed them all to be negative or indeterminate.
PCR performed on the available DNA (from the 10 WB-seropositive, 8 sero-indeterminate, and 33 seronegative persons) gave positive results for the integrase gene in 9 of the 10 WB-positive samples (Table 2) and negative results for the others. The LTR PCR was positive for 7 of 9 integrase-positive samples and none of the 42 others.
All 9 SFV-positive persons belonged to the group of 85 persons who had had known contact and bites or scratches from apes or monkeys. Thus, the subsequent epidemiologic analysis was restricted to these 85 (71 men, 14 women; mean age 39 years). According to univariate analysis, foamy virus–positive serologic results were associated with the type of nonhuman primate encountered (monkeys 3.6% vs. apes 24.1%, p = 0.003) and the type of encounter (pets 0% vs. hunting 16.1%, p = 0.022) (Table 3). No other studied risk factor (except age at time of contact) was significantly associated with positive results.
Table 3. Univariate analysis results for risk factors for simian foamy virus, 85 persons, rural Cameroon*.
| Risk factor | Total no. tested | Positive, no. (%) | p value |
|---|---|---|---|
| Age at contact, y | |||
| >45 | 65 | 4 (6.2) | |
| <45 | 20 | 5 (25) | 0.017 |
| Sex | |||
| Male | 71 | 9 (12.7) | |
| Female | 14 | 0 | 0.159 |
| Ethnicity | |||
| Bantu | 72 | 6 (8.3) | |
| Pygmy | 13 | 3 (23.1) | 0.112 |
| Type of animal interaction | |||
| Pet† | 29 | 0 | |
| Hunted | 56 | 9 (16.1) | 0.022 |
| Type of nonhuman primate | |||
| Monkey | 56 | 2 (3.6) | |
| Ape | 29 | 7 (24.1) | 0.003 |
| Wound type | |||
| Scratches | 9 | 0 | |
| Bites | 76 | 9 (11.8) | 0.275 |
| Wound location | |||
| Upper body | 31 | 2 (6.5 ) | |
| Lower body | 54 | 7 (13) | 0.348 |
| Scars | |||
| Absent | 12 | 0 | |
| Present | 73 | 9 (12.3) | 0.198 |
*Only the 85 persons with nown severe bites or scratches from a nonhuman primate. Univariate analyses were performed by using STATA (StataCorp LP, College Station, TX, USA) software with the χ2 and Fisher exact tests with critical p value = 0.05. †Most pets were Cercopithecus nictitans and mandrills; some were small chimpanzees.
Among the 56 persons who had received severe bites or scratches from nonhuman primates while hunting, 7 (36%) of the 19 that had encountered an ape were infected with SFV, in contrast to only 2 (5.4%) of 37 who had had contact with a small monkey (p<0.05) (data not shown). To determine possible intrafamilial transmission of SFVs, we tested 4 wives and 1 husband of 5 of the index case-participants as well as 5 of their children (Table 2). All were seronegative according to WB.
Of the 9 SFV-positive persons, 7 had been severely bitten by a gorilla (4 persons) or chimpanzee (1 person) 1–53 years ago while hunting (Table 2); some displayed large scars on the legs, arms, feet, or fingers (Figure 3). Hunters CH66 and CH29 had been severely bitten by 2 different animals in 2 separate hunting incidents. The 2 other SFV-positive persons were adult men who had been bitten by a small monkey, including a mandrill and a C. nictitans (Table 2).
Phylogenetic analyses of the 9 integrase products indicated that all belonged to the large clade of the African SFVs with 5 strains from gorilla, 2 from chimpanzee, 1 from mandrill, and 1 closely related to Cercopithecus strains (Appendix Figure). The 2 hunters who had been bitten by 2 different animals were infected with chimpanzee (CH66) and gorilla (CH29) foamy viruses, respectively.
Thus, for each of the 9 case-participants, the match was nearly perfect between the history of contact with a given nonhuman primate species (mainly through severe bites that had occurred decades ago) and the simian virus sequence that was found in the infected person (Table 2).
In Vivo Virus Persistence
Because each of the 6 persons from whom we obtained 2 samples (plasma, dried blood spots, or both) at different times was SFV positive by WB, persistent infection was evident for each person. The duration of this persistent infection was 1–8 years.
Isolation of 2 New Foamy Virus Strains
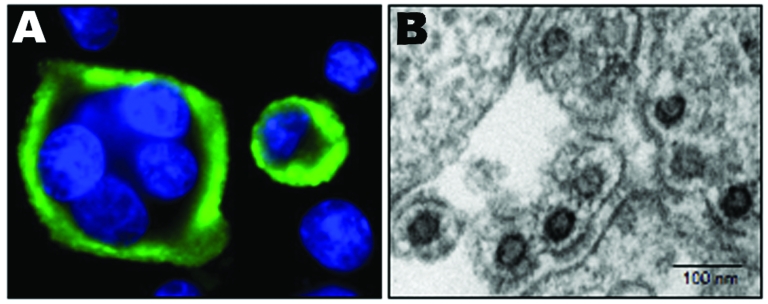
SFV was assayed for 2 persons (AG15 and AG16) from whom blood was available for culture. Giant-cell formation and syncytia were first observed for AG15’s sample after 26 days of coculturing, whereas cytopathic effect (CPE) was detected only after 33 days for AG16’s sample. The destruction of the monolayer of BHK-21 was quite rapid (2–4 days) after the first appearance of the CPE. Syncytia and giant cells showed a strong and clear specific immunofluorescence (Figure 4).
Figure 4.
Immunofluorescence and electron microscopy results. A) Typical multinucleated giant cells with a clear seroreactivity of AG16 antigens, determined by using an immunofluorescence assay with positive anti–foamy virus serum, on BHK-21–infected cells cocultivated with stimulated peripheral blood mononuclear cells. B) Electron microscopy of ultrathin sections from cells infected by AG16 foamy virus.
Electron microscopic analyses of cultured cells with a strong CPE demonstrated the presence of multinucleated giant cells. Typical foamy virus particles (diameter 100–110 nm) were frequently observed with several envelope spikes and a spherical central core (Figure 4). Budding of such virus particles was observed, mainly from the membrane surface of the endoplasmic reticulum.
PCR was performed on DNA extracted from the viral isolates after 2 months of culture. Comparative sequence analyses of the integrase product showed 100% nucleotide identity for AG16 (Cercopithecus strain) and 99.8% for AG15 (chimpanzee strain) between the SFV sequences from the peripheral blood mononuclear cell uncultured DNA and the cultured viral isolate.
Foamy Virus Load in Buffy Coat
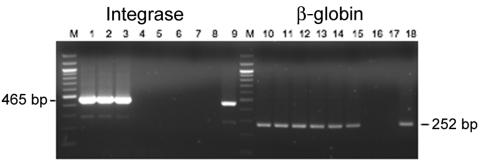
To determine the peripheral blood viral load in persons infected by SFVs and to check whether the discrepancies in the results between the 2 PCR assays (integrase and LTR) could be related to a low viral load (reaching the limits of our PCR sensitivity), we used a semiquantitative PCR assay (18). Of the 13 infected persons, 7 (Table 2) had a very low viral load, 1–10 copies in 500 ng of total DNA. For only 4 (all of them positive for both nested PCRs), the viral load was higher, 100–1,000 copies in 500 ng of total DNA (Figure 5; Table 2).
Figure 5.
Semiquantitative PCR for integrase and β-globin genes using AG15 peripheral blood buffy-coat DNA. Lanes 1–7 and 10–16, serial dilutions of the DNA from 500 ng to 0.5 pg; lanes 8 and 17, negative controls; lanes 9 and 18, positive controls; M, 100-bp ladder.
Discussion
Animal reservoirs are one of the most important sources of emerging infectious diseases that threaten humans. Recent zoonotic transmission of retroviruses has led to the emergence of HIV-1 and HIV-2 in humans (13). Nonhuman primates are natural hosts for other retroviruses. Although SFVs have been recently shown to infect persons occupationally exposed to nonhuman primates in zoos and primate centers, little is known about modes of cross-species transmission of these viruses in their natural habitat.
In the current study of adults living in central African regions with high nonhuman primate diversity, ≈2% of 1,164 persons showed clear seroreactivity to SFVs and at least 4 were persistently infected with SFV, with detectable viral sequences in their peripheral leucocyte DNA. These results confirm and extend to other areas of Cameroon the original findings published by Wolfe et al., who found that 10 (1%) of 1,099 of a comparable population had antibodies to SFV with a positive PCR for only 3 of them (11). These data, combined with the findings of our hunter study, which identified 9 more SFV-infected persons, demonstrate infection by a large diversity of SFVs in persons from geographically isolated areas. Such retroviral zoonosis is thus widespread and occurs in diverse villages where hunters are frequently in contact with nonhuman primates (3,4). In another context, a model has predicted that in Bali, Indonesia, for every 1,000 visitors to a monkey temple, approximately 6 will be infected with SFV (40).
Our study demonstrates efficient transmission of SFVs to persons in natural settings in central Africa, specifically after the persons had been bitten while hunting, and a viral persistence in the human host. Indeed, >35% of the hunters bitten severely (often with soft tissue crushing, tearing, and bleeding) by a gorilla or a chimpanzee were SFV infected. This strongly suggests that in a natural situation, contact of human blood with the saliva of an adult ape or monkey is the key factor for SFV transmission to humans. This situation is similar to that of persons occupationally exposed to nonhuman primates in zoos and primate centers, as nearly all of them reported having been bitten by monkeys or apes (5,9,10). Some studies have shown that SFVs are present at high concentration in the saliva (with viral replication) of infected animals (29,31). We recently provided evidence that Macaca tonkeana mostly acquire SFVs through severe bites, mainly young adults when they compete for sex partners (18). In our study, contact with pets was not found to be associated with SFV infection. This might be because pet bites mainly cause superficial tissue damage and rarely cause serious wounds and because some of the animals are probably not SFV infected due to their young age when captured.
In our study, SFV integrase or LTR sequences were not detected in several of the persons who were confirmed seropositive by WB. Although the presence of divergent SFVs could explain such discrepancy, low viral load in the blood samples is more likely, because our PCR primers have been shown to amplify a large variety of African SFVs but also several rather divergent Asian SFVs (16–18,22). This lack of detection of FV sequences by PCR may also indicate nonspecific reactivity with SFV Gag antigens. Lack of SFV sequences has also been recently reported in the peripheral blood mononuclear cell DNA of 7 of 10 African hunters who were SFV seropositive according to WB (11).
We provide the first data, to our knowledge, on the quantification of viral load of SFVs in humans. Our results, based on 13 infected persons, indicate a low viral load in most persons but a large range (1–1,000 copies in 500 ng of total peripheral blood leukocytes DNA). These viral loads are comparable to those in wild-born chimpanzees (16) and captive M. tonkeana (18).
Our work did not demonstrate the presence of SFV in the spouses of 5 index case-participants and in 5 of their children. Combined with the scarce published findings on this topic, these results suggest that SFV transmission among humans does not occur easily by sexual contact or saliva exposure (8–10).
Another concern is the illness and death that might be associated with these retroviral persistent infections after interspecies transmission. The apparent lack of pathogenicity of SFV infection in humans, which is still based on a limited number of cases, contrasts strongly with the massive in vitro lytic properties of these viruses in monkey and human cells (8–10,18,30). The selection bias inherent in the enrollment of healthy persons in our study, as well as in those enrolled by Wolfe et al. (11) and Switzer et al. (10), greatly limits the ability to identify any potential acute or severe associated diseases. A case-control study based on a larger number of SFV-infected persons would help shed light on possible chronic diseases or biological abnormalities associated with human SFV infection. SFV infection in immunocompromised persons, especially those with HIV infection, could also heighten public health concerns because such coinfection is probable in central African areas where HIV-1 is highly endemic.
Supplementary Material
Ethnographic and field survey information
Phylogenetic tree generated on a 425-bp fragment of the integrase simian foamy virus (SFV) gene. The 13 new SFV sequences described in this study are shown in red. Numbers at each node indicate the percentage of bootstrap samples (1,000 replicates); only values >60% are shown. The branch lengths are drawn to scale with the bar indicating 0.1-nt replacement per site. The tree was rooted by using the Asian Macaca mulatta (MmuSFVmac) sequence.
Acknowledgments
We thank Sebastien Chevalier for help with the immunofluorescence assay experiments and Marie-Christine Prevost for electron microscopy studies.
This study was supported financially by the CNRS-URA3015 and the Institut Pasteur de Paris. S.C. was supported by a fellowship from the University of Milan, the Virus Cancer Prevention Association, and the Ligue Nationale Contre le Cancer.
Biography
Dr Calattini is a researcher whose primary interest is the molecular epidemiology of retroviruses, especially spumaviruses and human T-lymphotropic viruses. She is studying the interspecies transmission of such retroviruses from nonhuman primates to humans in central Africa.
Footnotes
Suggested citation for this article: Calattini S, Betsem EBA, Froment A, Mauclère P, Tortevoye P, Schmitt C, et al. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg Infect Dis [serial on the Internet]. 2007 Sep [date cited]. Available from http://www.cdc.gov/eid/content/13/9/1314.htm
References
- 1.Apetrei C, Marx PA. Simian retroviral infections in human beings. Lancet. 2004;364:137–8, author reply 139–40. 10.1016/S0140-6736(04)16620-8 [DOI] [PubMed] [Google Scholar]
- 2.Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nat Med. 2004;10:S70–6. 10.1038/nm1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfe ND. Bushmeat hunting, deforestation, and prediction of zoonotic disease emergence. Emerg Infect Dis. 2005;11:1822–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfe ND, Prosser TA, Carr JK, Tamoufe U, Mpoudi-Ngole E, Torimiro JN, et al. Exposure to nonhuman primates in rural Cameroon. Emerg Infect Dis. 2004;10:2094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks JI, Rud EW, Pilon RG, Smith JM, Switzer WM, Sandstrom PA. Cross-species retroviral transmission from macaques to human beings. Lancet. 2002;360:387–8. 10.1016/S0140-6736(02)09597-1 [DOI] [PubMed] [Google Scholar]
- 6.Heneine W, Schweizer M, Sandstrom P, Folks T. Human infection with foamy viruses. Curr Top Microbiol Immunol. 2003;277:181–96. [DOI] [PubMed] [Google Scholar]
- 7.Sandstrom PA, Phan KO, Switzer WM, Fredeking T, Chapman L, Heneine W, et al. Simian foamy virus infection among zoo keepers. Lancet. 2000;355:551–2. 10.1016/S0140-6736(99)05292-7 [DOI] [PubMed] [Google Scholar]
- 8.Heneine W, Switzer WM, Sandstrom P, Brown J, Vedapuri S, Schable CA, et al. Identification of a human population infected with simian foamy viruses. Nat Med. 1998;4:403–7. 10.1038/nm0498-403 [DOI] [PubMed] [Google Scholar]
- 9.Schweizer M, Falcone V, Gange J, Turek R, Neumann-Haefelin D. Simian foamy virus isolated from an accidentally infected human individual. J Virol. 1997;71:4821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Switzer WM, Bhullar V, Shanmugam V, Cong ME, Parekh B, Lerche NW, et al. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J Virol. 2004;78:2780–9. 10.1128/JVI.78.6.2780-2789.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, et al. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–7. 10.1016/S0140-6736(04)15787-5 [DOI] [PubMed] [Google Scholar]
- 12.Nerrienet E, Meertens L, Kfutwah A, Foupouapouognigni Y, Gessain A. Molecular epidemiology of simian T-lymphotropic virus (STLV) in wild-caught monkeys and apes from Cameroon: a new STLV-1, related to human T-lymphotropic virus subtype F, in a Cercocebus agilis. J Gen Virol. 2001;82:2973–7. [DOI] [PubMed] [Google Scholar]
- 13.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–41. 10.1038/17130 [DOI] [PubMed] [Google Scholar]
- 14.Corbet S, Muller-Trutwin MC, Versmisse P, Delarue S, Ayouba A, Lewis J, et al. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–34. 10.1128/JVI.74.1.529-534.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slattery JP, Franchini G, Gessain A. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 1999;9:525–40. [PubMed] [Google Scholar]
- 16.Calattini S, Nerrienet E, Mauclere P, Georges-Courbot MC, Saib A, Gessain A. Detection and molecular characterization of foamy viruses in Central African chimpanzees of the Pan troglodytes troglodytes and Pan troglodytes vellerosus subspecies. J Med Primatol. 2006;35:59–66. 10.1111/j.1600-0684.2006.00149.x [DOI] [PubMed] [Google Scholar]
- 17.Calattini S, Nerrienet E, Mauclere PMCG-C, Saib A, Gessain A. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from Cameroon and Gabon. J Gen Virol. 2004;85:3313–7. 10.1099/vir.0.80241-0 [DOI] [PubMed] [Google Scholar]
- 18.Calattini S, Wanert F, Thierry B, Schmitt C, Bassot S, Saib A, et al. Modes of transmission and genetic diversity of foamy viruses in a Macaca tonkeana colony. Retrovirology. 2006;3:23. 10.1186/1742-4690-3-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broussard SR, Comuzzie AG, Leighton KL, Leland MM, Whitehead EM, Allan JS. Characterization of new simian foamy viruses from African nonhuman primates. Virology. 1997;237:349–59. 10.1006/viro.1997.8797 [DOI] [PubMed] [Google Scholar]
- 20.Herchenroder O, Renne R, Loncar D, Cobb EK, Murthy KK, Schneider J, et al. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV). Virology. 1994;201:187–99. 10.1006/viro.1994.1285 [DOI] [PubMed] [Google Scholar]
- 21.McClure MO, Bieniasz PD, Schulz TF, Chrystie IL, Simpson G, Aguzzi A, et al. Isolation of a new foamy retrovirus from orangutans. J Virol. 1994;68:7124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain AI, Shanmugam V, Bhullar VB, Beer BE, Vallet D, Gautier-Hion A, et al. Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses. Virology. 2003;309:248–57. 10.1016/S0042-6822(03)00070-9 [DOI] [PubMed] [Google Scholar]
- 23.Meiering CD, Linial ML. Historical perspective of foamy virus epidemiology and infection. Clin Microbiol Rev. 2001;14:165–76. 10.1128/CMR.14.1.165-176.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saib A. Non-primate foamy viruses. Curr Top Microbiol Immunol. 2003;277:197–211. [DOI] [PubMed] [Google Scholar]
- 25.Schweizer M, Schleer H, Pietrek M, Liegibel J, Falcone V, Neumann-Haefelin D. Genetic stability of foamy viruses: long-term study in an African green monkey population. J Virol. 1999;73:9256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobaly-Tapiero J, Bittoun P, Neves M, Guillemin MC, Lecellier CH, Puvion-Dutilleul F, et al. Isolation and characterization of an equine foamy virus. J Virol. 2000;74:4064–73. 10.1128/JVI.74.9.4064-4073.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Switzer WM, Salemi M, Shanmugam V, Gao F, Cong ME, Kuiken C, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–80. 10.1038/nature03341 [DOI] [PubMed] [Google Scholar]
- 28.Delebecque F, Suspene R, Calattini S, Casartelli N, Saib A, Froment A, et al. Restriction of foamy viruses by APOBEC cytidine deaminases. J Virol. 2006;80:605–14. 10.1128/JVI.80.2.605-614.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenroder O, Spatz W, et al. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology. 1999;257:7–14. 10.1006/viro.1999.9634 [DOI] [PubMed] [Google Scholar]
- 30.Linial M. Why aren't foamy viruses pathogenic? Trends Microbiol. 2000;8:284–9. 10.1016/S0966-842X(00)01763-7 [DOI] [PubMed] [Google Scholar]
- 31.Murray SM, Picker LJ, Axthelm MK, Linial ML. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J Virol. 2006;80:663–70. 10.1128/JVI.80.2.663-670.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones-Engel L, Engel GA, Schillaci MA, Rompis A, Putra A, Suaryana KG, et al. Primate-to-human retroviral transmission in Asia. Emerg Infect Dis. 2005;11:1028–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plancoulaine S, Abel L, Tregouet D, Duprez R, van Beveren M, Tortevoye P, et al. Respective roles of serological status and blood specific antihuman herpesvirus 8 antibody levels in human herpesvirus 8 intrafamilial transmission in a highly endemic area. Cancer Res. 2004;64:8782–7. 10.1158/0008-5472.CAN-04-2000 [DOI] [PubMed] [Google Scholar]
- 34.Gessain A, Mauclere P, Froment A, Biglione M, Le Hesran JY, Tekaia F, et al. Isolation and molecular characterization of a human T-cell lymphotropic virus type II (HTLV-II), subtype B, from a healthy Pygmy living in a remote area of Cameroon: an ancient origin for HTLV-II in Africa. Proc Natl Acad Sci U S A. 1995;92:4041–5. 10.1073/pnas.92.9.4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeannel D, Kourouma K, Fretz C, Zheng YM, Ureta VA, Drame L, et al. Regional differences in human retroviral infections HIV-1, HIV-2, and HTLV-I/II in rural Guinea (west Africa). J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:315–8. 10.1097/00042560-199503010-00016 [DOI] [PubMed] [Google Scholar]
- 36.Tobaly-Tapiero J, Bittoun P, Saib A. Isolation of foamy viruses from peripheral blood lymphocytes. Methods Mol Biol. 2005;304:125–37. [DOI] [PubMed] [Google Scholar]
- 37.Schweizer M, Neumann-Haefelin D. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology. 1995;207:577–82. 10.1006/viro.1995.1120 [DOI] [PubMed] [Google Scholar]
- 38.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- 39.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–3. 10.1093/jhered/92.4.371 [DOI] [PubMed] [Google Scholar]
- 40.Engel G, Hungerford LL, Jones-Engel L, Travis D, Eberle R, Fuentes A, et al. Risk assessment: a model for predicting cross-species transmission of simian foamy virus from macaques (M. fascicularis) to humans at a monkey temple in Bali, Indonesia. Am J Primatol. 2006;68:934–48. 10.1002/ajp.20299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ethnographic and field survey information
Phylogenetic tree generated on a 425-bp fragment of the integrase simian foamy virus (SFV) gene. The 13 new SFV sequences described in this study are shown in red. Numbers at each node indicate the percentage of bootstrap samples (1,000 replicates); only values >60% are shown. The branch lengths are drawn to scale with the bar indicating 0.1-nt replacement per site. The tree was rooted by using the Asian Macaca mulatta (MmuSFVmac) sequence.