Abstract
BACKGROUND
Primary biliary cirrhosis is a chronic granulomatous cholangitis, characteristically associated with antimitochondrial antibodies. Twin and family aggregation data suggest that there is a significant genetic predisposition to primary biliary cirrhosis, but the susceptibility loci are unknown.
METHODS
To identify genetic loci conferring a risk for primary biliary cirrhosis, we carried out a genomewide association analysis in which DNA samples from 2072 Canadian and U.S. subjects (536 patients with primary biliary cirrhosis and 1536 controls) were genotyped for more than 300,000 single-nucleotide polymorphisms (SNPs). Sixteen of the SNPs most strongly associated with primary biliary cirrhosis were genotyped in two independent replication sets. We carried out fine-mapping studies across three loci associated with primary biliary cirrhosis.
RESULTS
We found significant associations between primary biliary cirrhosis and 13 loci across the HLA class II region; the HLA-DQB1 locus (encoding the major histocompatibility complex class II, DQ β chain 1) had the strongest association (P = 1.78×10−19; odds ratio for patients vs. controls, 1.75). Primary biliary cirrhosis was also significantly and reproducibly associated with two SNPs at the IL12A locus (encoding interleukin-12α), rs6441286 (P = 2.42×10−14; odds ratio, 1.54) and rs574808 (P = 1.88×10−13; odds ratio, 1.54), and one SNP at the IL12RB2 locus (encoding interleukin-12 receptor β2), rs3790567 (P = 2.76×10−11; odds ratio, 1.51). Fine-mapping analysis showed that a five-allele haplotype in the 3′ flank of IL12A was significantly associated with primary biliary cirrhosis (P = 1.15×10−34). We found a modest genomewide association (P<5.0×10−5) with the risk of disease for SNPs at the STAT4 locus (encoding signal transducer and activator of transcription 4) and the CTLA4 locus (encoding cytotoxic T-lymphocyte–associated protein 4) and 10 other loci.
CONCLUSIONS
Our data show significant associations between primary biliary cirrhosis and common genetic variants at the HLA class II, IL12A, and IL12RB2 loci and suggest that the interleukin-12 immunoregulatory signaling axis is relevant to the pathophysiology of primary biliary cirrhosis. (ClinicalTrials.gov number, NCT00242125.)
Primary biliary cirrhosis is the most common autoimmune liver disease, affecting up to 1 in 1000 women over 40 years of age.1 Treatment for this chronic cholestatic condition remains empirical.2 The granulomatous destruction of interlobular bile ducts that characterizes primary biliary cirrhosis is almost always associated with antimitochondrial antibodies specific for the E2 subunit of the pyruvate dehydrogenase complex.3 The hepatic accumulation of autoreactive T lymphocytes in patients with primary biliary cirrhosis, as well as data from animal models of autoimmune cholangitis, implicate T lymphocytes — CD4+ T helper lymphocytes in particular — in the pathogenesis of primary biliary cirrhosis.4–6
A genetic predisposition for primary biliary cirrhosis has been revealed by analysis of both aggregation data from families and concordance data from twins. The rate of concordance among monozygotic twins is 60%,7 with the sibling relative risk estimated as 10.5.8 The coexistence of other autoimmune diseases in patients and the increased prevalence of such diseases in their families is also consistent with a genetic influence.9,10 Among the genes studied as susceptibility candidates, only HLA has consistently been associated with primary biliary cirrhosis. HLA-DRB1*0801 confers an increased risk of primary biliary cirrhosis in whites, with an odds ratio of 2.4 to 3.3. In addition, the derived population attributable fraction for HLA-DRB1*08 ranges from 2.8 to 8.8%11,12 (for the derivation, see the Supplementary Appendix, available with the full text of this article at NEJM.org). Primary biliary cirrhosis has also been associated — in some, but not all, studies — with variants in CTLA4 (the gene encoding cytotoxic T-lymphocyte-associated protein 4).13–15 To explore the genetic basis of primary biliary cirrhosis, we conducted genomewide association screening of subjects from North America.
METHODS
PATIENTS AND CONTROLS
We used a two-stage design to analyze data from 2072 white subjects (536 patients and 1536 controls) from North America (Fig. 1). Stage 1 consisted of a genomewide association survey of patients with primary biliary cirrhosis and controls from Canada, with additional historical controls from the United States. We carried out replication analyses (stage 2) involving patients and controls from the United States (stage 2a) and additional patients and controls from Canada (stage 2b), as well as fine-mapping studies of all Canadian subjects from the genomewide association screening (stage 1) and replication analysis (stage 2b). We obtained written informed consent from all subjects, and the study was approved by a local institutional ethics committee at each center.
Figure 1. Quality Control and Study Design.
Panel A shows the outcomes of genotyping quality control for single-nucleotide polymorphisms (SNPs) and genomic DNA from individual subjects. The requirements for SNPs to meet quality-control standards were call rates of greater than 95%, P>0.0001 for the test for Hardy–Weinberg equilibrium, and P>0.01 for the test for minor allele frequency. Panel B shows the stages of analysis in the study: from stage 1, the genomewide association screening, through stage 2, the replication analysis, to fine-mapping and haplotype analyses. IL12A denotes the gene encoding interleukin-12α, IL12RB2 the gene encoding interleukin-12 receptor β2, IL23R the gene encoding interleukin-23 receptor, and STAT4 the gene encoding signal transducer and activator of transcription 4.
All patients fulfilled the criteria of the American Association for the Study of Liver Diseases for primary biliary cirrhosis,16 and all patients and controls were whites of European origin (as determined by self-report and, for patients included in stage 1, confirmed by genotyping; see the Supplementary Appendix). Data from a total of 536 patients with primary biliary cirrhosis and 1536 healthy controls (399 healthy volunteers in Toronto who had no history of autoimmune disease, and an additional 1137 historical controls from an M.D. Anderson lung cancer study17) were included in the stage 1 genomewide association analysis. The stage 2a replication analysis included data from 410 patients with primary biliary cirrhosis and 310 controls from the Mayo Clinic Primary Biliary Cirrhosis Registry,14 and stage 2b included data from additional subjects in Canada (116 patients and 896 controls). The genomic data from the Canadian patients and controls in stage 1, which were subject to quality-control standards, and stage 2b (the additional 116 patients and 896 controls in Canada) were included in the fine-mapping analyses. Data from all patients and controls (in stages 1, 2a, and 2b) were included in the combined analysis.
GENOTYPING AND QUALITY CONTROL
DNA purification and genotyping methods are described in the Supplementary Appendix. In stage 1, samples from the patients and the Canadian controls were genotyped for 373,400 single-nucleotide polymorphisms (SNPs) with the use of the Illumina HumanHap370 BeadChip. Samples from the M.D. Anderson historical controls were genotyped on the Illumina HumanHap300 BeadChip. Genotyping data were subjected to quality control before the data analysis (see the Supplementary Appendix). Genotypes were called if they exceeded minimum quality-control standards. Individual samples with genotype call rates of less than 95% and SNPs with call rates of less than 95%, minor-allele frequencies of less than 1%, or deviation from the Hardy–Weinberg equilibrium at P<0.0001 were removed. Replication and fine-mapping were performed with the use of the Sequenom MassArray iPLEX genotyping platform.
STATISTICAL ANALYSIS
For the genomewide association analysis, pairwise identity-by-state analysis was performed with PLINK software (version 1.05)18 (http://pngu.mgh. harvard.edu/purcell/plink/) to identify subjects with excess identity-by-descent sharing (PI_HAT >0.25), a measure of the degree of genetic relationship between subjects. One subject from each of the 15 pairs exceeding this threshold was removed from the association analysis; the subject in each pair with the higher average SNP call rate was retained. Hierarchical cluster analysis was performed with the use of PLINK to identify subjects with similar genotypes over the entire genome; the 39 samples that were more than 4 SD from a nearest neighbor were removed from the analysis. Subsequent association analyses were conducted with the use of a conditional analysis adjusting for stratifications among groups of subjects identified by hierarchical clustering. To control alternatively for the potential confounding influences of population stratification, association analysis was also performed on the basis of a principal-components analysis implemented in the EIGENSTRAT method19 with default parameters, in which we adjusted for the 10 eigenvectors having the highest eigenvalues. The lambda values showed minimal inflation (1.085 before and 1.056 after adjustment for eigenvectors); with the use of PLINK, the lambda values were 1.14 without adjustment for clusters and 1.09 with adjustment for clusters.
Allele and genotype associations were assessed by means of PLINK software, and linkage disequilibrium between pairs of SNPs and haplotypes was determined with the use of Haploview software, version 4.1 (www.broad.mit.edu/mpg/haploview). Haplotype block structure was defined according to the criteria established by Gabriel et al.20 and the pairwise estimates of standardized Lewontin's disequilibrium coefficient (D′), whereas the linkage disequilibrium among pairs of SNPs was characterized according to the square of the correlation coefficient (r2).
Combined analyses were carried out with Cochran–Mantel–Haenszel tests and SAS software, with adjustment for potential confounding of the case or control frequency with genotype frequency among groups of subjects according to the stage of analysis or center. Supplementary analyses are described in the Supplementary Appendix.
RESULTS
GENOMEWIDE ASSOCIATION ANALYSIS (STAGE 1)
The most significant results of the stage 1 genomewide association survey are shown in Table 1 and Figures 2 and 3. (The full set is available from the database of Genotypes and Phenotypes [dbGaP]; www.ncbi.nlm.nih.gov/sites/entrez?db=gap, accession number phs000183.v1.p1). The stage 1 results are based on the analysis of the 305,724 SNPs that passed quality-control standards and the 505 patients with primary biliary cirrhosis and 1507 controls who were retained after the pairwise identity-by-state analysis and correction for population stratification.
Table 1.
Results of Genomewide Association (Stage 1) and Replication (Stage 2) Analyses.*
| SNP | Chromosome | Location | Gene | Risk Allele |
Genomewide Association Analysis |
Replication Analysis: U.S. Cohort |
Replication Analysis: Canadian Cohort |
Replication Analysis: Both Cohorts (N = 526 Patients, N = 1206 Controls) |
Combined Analysis (N = 1031 Patients, N = 2713 Controls) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk-Allele Frequency |
EIGENSTRAT P Value |
PLINK P Value |
Odds Ratio (95% Cl) |
Risk-Allele Frequency |
PLINK P Value |
Risk-Allele Frequency |
PLINK P Value |
Bonferroni- Corrected P Value |
P Value | Odds Ratio (95% Cl) |
||||||||
| kb | patients (N = 505) | controls (N=1507) | patients (N = 410) | controls (N = 310) | patients (N = 116) | controls (N = 896) | ||||||||||||
| HLA region | ||||||||||||||||||
| rs2856683 | 6p21.3 | 32763196 | HLA – DQB1 | C | 0.360 | 0.222 | 1.34×10−14 | 8.58×10−17 | 1.99 (1.69–2.34) |
0.334 | 0.228 | 1.44×10−5 | 0.298 | 0.224 | 1.30×10−2 | 3.18×10−5 | 1.78×10−19 | 1.75 (1.55–1.98) |
| rs9275312 | 6p21.3 | 32773706 | HLA – DQB1 | G | 0.227 | 0.128 | 1.99×10−11 | 3.84×10−13 | 2.01 (1.66–2.42) |
|||||||||
| rs9275390 | 6p21.3 | 32777134 | HLA – DQB1 | G | 0.373 | 0.246 | 5.73×10−11 | 1.13×10−13 | 1.81 (1.55–2.11) |
|||||||||
| rs7775228 | 6p21.3 | 32766057 | HLA – DQB1 | G | 0.207 | 0.122 | 1.90×10−10 | 1.70×10−10 | 1.87 (1.54–2.27) |
|||||||||
| rs2395148 | 6p21.3 | 32429532 | C6orf10 | A | 0.055 | 0.019 | 1.50×10−9 | 5.62×10−10 | 3.24 (2.20–4.77) |
0.065 | 0.026 | 8.19×10−4 | 0.068 | 0.025 | 2.77×10−4 | 8.94×10−5 | 3.62×10−14 | 2.87 (2.16–3.82) |
| rs9277535 | 6p21.3 | 33162839 | HLA – DPB1 | G | 0.326 | 0.236 | 7.26×10−9 | 8.28×10−9 | 1.60 (1.36–1.87) |
0.318 | 0.248 | 3.85×10−3 | 0.331 | 0.250 | 7.83×10−3 | 2.32×10−3 | 3.92×10−11 | 1.50 (1.33–1.70) |
| rs3806156 | 6p21.3 | 32481676 | BTNL2 | A | 0.457 | 0.354 | 1.31×10−8 | 1.27×10−9 | 1.58 (1.37–1.84) |
0.428 | 0.361 | 0.01 | 0.42 | 0.360 | 0.07 | 0.07 | 1.11×10−9 | 1.42 (1.27–1.58) |
| rs9357152 | 6p21.3 | 32772938 | HLA – DQB1 | A | 0.826 | 0.741 | 3.88×10−8 | 7.25×10−8 | 1.65 (1.37–1.97) |
|||||||||
| rs3135363 | 6p21.3 | 32497626 | BTNL2 | A | 0.784 | 0.718 | 4.46× 10−8 | 3.72×10−7 | 1.56 (1.30–1.85) |
|||||||||
| rs9277565 | 6p21.3 | 33164875 | HLA – DPB1 | A | 0.276 | 0.198 | 5.94×10−8 | 8.32×10−8 | 1.58 (1.34–1.87) |
|||||||||
| rs2281389 | 6p21.3 | 33167774 | HLA – DPB1 | G | 0.236 | 0.168 | 1.58×10−7 | 2.38×10−7 | 1.59 (1.33–1.90) |
|||||||||
| rs660895 | 6p21.3 | 32685358 | HLA – DRB1 | G | 0.277 | 0.195 | 3.41×10−7 | 4.68×10−8 | 1.60 (1.35–1.90) |
|||||||||
| rs9501626 | 6p21.3 | 32508322 | HLA – DRA | A | 0.173 | 0.110 | 4.43×10−7 | 6.83×10−7 | 1.68 (1.37–2.07) |
|||||||||
| Non-HLA regions | ||||||||||||||||||
| rs6441286 | 3q25.33–q26 | 161211572 | IL12A | G | 0.497 | 0.390 | 1.20×10−8 | 3.25×10−8 | 1.51 (1.30–1.75) |
0.488 | 0.371 | 1.11×10−5 | 0.471 | 0.395 | 2.55×10−2 | 6.80×10−5 | 2.42×10−14 | 1.54 (1.38–1.72) |
| rs3790567 | 1p31.2 | 67594965 | IL12RB2 | A | 0.341 | 0.240 | 4.51×10−8 | 8.60×10−8 | 1.54 (1.32–1.81) |
0.311 | 0.241 | 5.19×10−3 | 0.340 | 0.246 | 1.72×10−3 | 0.005 | 2.76×10−11 | 1.51 (1.33–1.70) |
| rs574808 | 3q25.33–q26 | 161215677 | IL12A | T | 0.678 | 0.574 | 3.81×10−7 | 5.34×10−7 | 1.47 (1.27–1.72) |
0.668 | 0.564 | 6.97×10−5 | 0.677 | 0.588 | 9.26×10−3 | 2.78×10−5 | 1.88×10−13 | 1.54 (1.37–1.73) |
| rs6838639 | 4q27 | 123118615 | TRPC3 | G | 0.804 | 0.729 | 1.36×10−6 | 3.45×10−7 | 1.59 (1.33–1.92) |
0.764 | 0.780 | 0.48 | 0.727 | 0.735 | 0.80 | 1.00 | 2.35×10−3 | 1.23 (1.08–1.40) |
| rs3790565 | 1p31.2 | 67583944 | IL12RB2 | C | 0.258 | 0.183 | 2.50×10−6 | 1.41×10−6 | 1.53 (1.29–1.82) |
0.232 | 0.195 | 0.10 | 0.269 | 0.184 | 1.91×10−3 | 0.05 | 1.24×10−8 | 1.46 (1.28–1.67) |
| rs9964104 | 18q21 | 50751695 | CCDC68 | A | 0.687 | 0.606 | 8.37×10−6 | 2.77×10−5 | 1.39 (1.19–1.61) |
0.642 | 0.643 | 0.97 | 0.639 | 0.626 | 0.71 | 1.00 | 2.13×10−4 | 1.25 (1.11–1.40) |
| rs3124607 | 9q34.3 | 138534660 | NOTCH1 | G | 0.730 | 0.665 | 9.85×10−6 | 6.45×10−6 | 1.45 (1.24–1.70) |
|||||||||
| rs16833239 | 2q32 | 191648505 | STAT4 | G | 0.962 | 0.921 | 1.55×10−5 | 2.60×10−5 | 2.13 (1.47–3.03) |
0.934 | 0.924 | 0.48 | 0.933 | 0.922 | 0.54 | 1.00 | 4.67×10−5 | 1.65 (1.30–2.10) |
| rs10222962 | 4p15 | 32036553 | PCDH7 | G | 0.111 | 0.070 | 1.55×10−5 | 1.67×10−5 | 1.70 (1.33–2.18) |
|||||||||
| rs2211312 | 13q33.3 | 107203928 | FAM155A | A | 0.925 | 0.878 | 1.78×10−5 | 5.08×10−5 | 1.75 (1.33–2.33) |
0.879 | 0.887 | 0.63 | 0.865 | 0.878 | 0.59 | 1.00 | 0.61 | 1.05 (0.88–1.24) |
| rs907092 | 17q21 | 35175785 | IKZF3 | A | 0.522 | 0.449 | 2.05×10−5 | 7.04×10−6 | 1.40 (1.21–1.62) |
0.536 | 0.487 | 0.07 | 0.492 | 0.456 | 0.18 | 0.24 | 7.61×10−6 | 1.29 (1.15–1.44) |
| rs9303277 | 17q21 | 35229995 | IKZF3 | A | 0.571 | 0.495 | 2.75×10−5 | 3.95×10−5 | 1.41 (1.22–1.66) |
|||||||||
| rs4679904 | 3q26.1 | 161823590 | ARF7 | G | 0.798 | 0.719 | 2.79×10−5 | 5.38×10−6 | 1.52 (1.27–1.79) |
0.767 | 0.710 | 0.02 | 0.748 | 0.730 | 0.55 | 0.51 | 1.13×10−6 | 1.38 (1.21–1.57) |
| rs2305480 | 17q12 | 35315722 | GSDMB | A | 0.509 | 0.438 | 3.66×10−5 | 1.41×10−5 | 1.38 (1.19–1.60) |
|||||||||
| rs6140113 | 20p13 | 691770 | C20orf54 | G | 0.891 | 0.835 | 3.81×10−5 | 3.75×10−5 | 1.61 (1.30–2.04) |
0.849 | 0.831 | 0.36 | 0.765 | 0.813 | 0.05 | 1.00 | 0.34 | 1.08 (0.93–1.25) |
| rs10488631 | 7q32.1 | 128381419 | IRF5 – TNPO3 | G | 0.168 | 0.117 | 4.57×10−5 | 2.14×10−5 | 1.55 (1.26–1.90) |
0.167 | 0.113 | 4.72×10−3 | 0.130 | 0.090 | 0.14 | 0.05 | 1.52×10−7 | 1.52 (1.30–1.78) |
| rs6748358 | 2q33 | 204465150 | CTLA4 | C | 0.574 | 0.497 | 4.91×10−5 | 1.41×10−5 | 1.39 (1.19–1.62) |
|||||||||
The single-nucleotide polymorphisms (SNPs) are listed in order of decreasing significance as indicated by the EIGENSTRAT P values; only those with P values of less than 5.0×10−7 in the HLA region and less than 5.0×10−5 in non-HLA regions are shown. The EIGENSTRAT and PLINK P values are for the comparison of allele frequency between patients and controls. For the joint replication analysis (stages 2a and 2b), P values were calculated with a Cochran–Mantel–Haenszel method of combining allele-frequency counts, and the Bonferroni correction was applied. For the combined analysis (stages 1 and 2), P values and odds ratios were calculated with the use of the Cochran–Mantel–Haenszel method of combining allele frequency counts.
Figure 2. Results of the Genomewide Association Analysis.
The y axis represents the level of significance (from the EIGENSTRAT method) for each single-nucleotide polymorphism on each chromosome along the x axis. The dotted line indicates the threshold for genomewide association significance.21 The P values were adjusted in EIGENSTRAT for the 10 eigenvectors having the highest eigenvalues. HLA-DQB1 denotes the gene encoding major histocompatibility complex class II DQ β chain 1, IL12A the gene encoding interleukin-12α, IL12RB2 the gene encoding interleukin-12 receptor β2, and STAT4 the gene encoding signal transducer and activator of transcription 4.
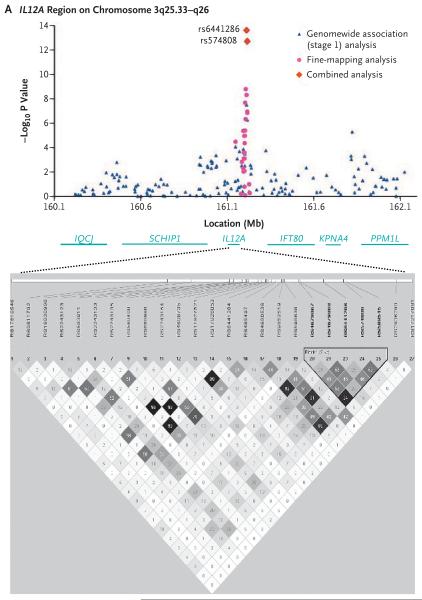
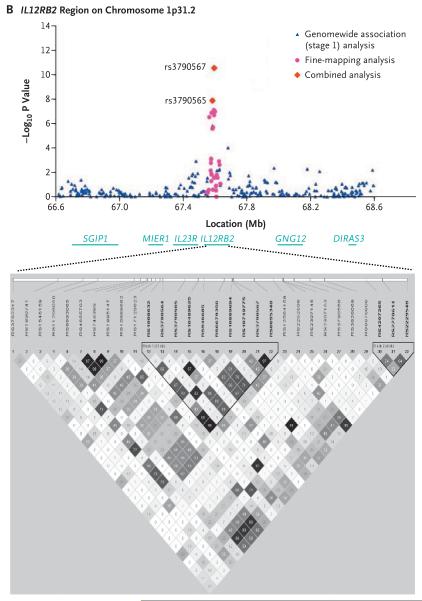
Thirteen SNPs across the HLA region on chromosome 6p21.3 and three SNPs from two distinct non-HLA regions at the IL12A locus, which encodes interleukin-12α (3q25.33–q26), and the IL12RB2 locus, which encodes interleukin-12 receptor β2 (1p31.2), met a significance threshold for genomewide association of P<5.0×10−7 (calculated with the EIGENSTRAT method). The most significantly associated markers according to the EIGENSTRAT method — rs2856683, rs9275312, rs9275390, and rs7775228 — map to the region between the HLA-DQB1 gene, which encodes major histocompatibility complex (MHC) class II, DQ β chain 1, and the HLA-DQA2 gene, which encodes MHC class II, DQ α chain 2. The P values from the PLINK analysis for these associations were between 1.70×10−10 and 8.58×10−17, with odds ratios for patients as compared with controls that ranged from 1.81 to 2.01. Highly significant association signals (according to the PLINK analysis) were also shown for nine other SNPs mapping in or near genes within the HLA region: C6orf10, which encodes chromosome 6 open reading frame 10 (P = 5.62×10−10); HLA-DPB1, which encodes MHC class II, DP β chain 1 (P = 8.28×10−9); BTNL2, which encodes butyrophilin-like 2 (P = 1.27×10−9); and HLA-DRA, which encodes MHC class II, DR α chain (P = 6.83×10−7). The C6orf10 locus had the strongest association with the risk of primary biliary cirrhosis (odds ratio, 3.24). Haplotype analysis, however, revealed that 4 of the 13 SNPs (rs2395148, rs3135363, rs2856683, and rs9357152 [in the C6orf10, BTNL2, HLA-DQB1 and HLA-DQB1 genes, respectively]) accounted for all of the HLA-associated risk for primary biliary cirrhosis (Table 1 in the Supplementary Appendix). Similar levels of association were found in a separate analysis of the genomewide data from Canadian controls only (Table 2 in the Supplementary Appendix).
Among the non-HLA loci, the strongest signals of association with primary biliary cirrhosis were found for two SNPs in the IL12A locus, rs6441286 (P = 3.25×10−8; odds ratio for patients vs. controls, 1.51) and rs574808 (P = 5.34×10−7; odds ratio, 1.47), and one SNP in the IL12RB2 locus, rs3790567 (P = 8.60×10−8; odds ratio, 1.54). A significant association with disease was also seen with a second IL12RB2-region SNP, rs3790565 (P = 1.41×10−6).
Twelve other non-HLA loci with association signals at significance levels of P<5.0×10−5 were identified. These included STAT4, which encodes signal transducer and activator of transcription 4 (rs16833239, P = 2.60×10−5, a locus associated with risks of several other autoimmune diseases and a downstream biologic mediator of interleukin-12 signaling22; CTLA4 (rs6748358, P = 1.41×10−5), a locus previously associated with a risk of primary biliary cirrhosis14,15; and the IRF5–TNPO3 locus (encoding interferon regulatory factor 5 and transportin 3, respectively) (rs10488631) (P = 2.14×10−5which is associated with risk for systemic lupus erythematosus23 and inflammatory bowel disease.24 These 12 loci were more modestly associated with primary biliary cirrhosis than were the IL12A and IL12RB2 loci, but the associations with the disease risk were similar, with odds ratios for patients as compared with controls ranging from 1.38 to 2.13.
INDEPENDENT REPLICATION (STAGE 2) AND COMBINED ANALYSIS
We tested for an association between disease and 16 of the most strongly associated SNPs from the stage 1 analysis, using two independently collected sets of case−control samples from subjects in the United States (stage 2a analysis) and Canada (stage 2b analysis). We found replication of the association (P<0.05 from the PLINK analysis in each replication cohort and P<0.05 with the Bonferroni correction in the joint replication analysis of both cohorts) with the rs2856683, rs2395148, and rs9277535 SNPs at three distinct sites within the HLA region as well as the rs6441286–rs574808 and rs3790567 SNPs at the IL12A and IL12RB2 loci, respectively.
A combined analysis of the genomewide association and replication data sets (from stages 1 and 2) also provided strong evidence (by means of a Cochran–Mantel–Haenszel test) of an association between primary biliary cirrhosis and the loci HLA-DQB1 (P = 1.78×10−19; odds ratio for patients vs. controls, 1.75), C6orf10 (P = 3.62×10−14;odds ratio, 2.87), HLA-DPB1 (P = 3.92×10−11; odds ratio, 1.50), BTNL2 (P = 1.11×10−9; odds ratio, 1.42), IL12A (P = 2.42×10−14; odds ratio, 1.54), and IL12RB2 (P = 2.76×10−11; odds ratio, 1.51). We found no significant effect of antimitochondrial-antibody status on these associations (Table 3 in the Supplementary Appendix). In addition, we found an increasing risk in association with increasing numbers of copies of risk alleles for the IL12A and IL12RB2 loci (Tables 4 and 5 in the Supplementary Appendix).
FINE-MAPPING AND HAPLOTYPE ANALYSES
To refine and further validate the two major non-HLA loci associated with primary biliary cirrhosis (IL12A and IL12RB2), we genotyped tag SNPs across these loci in samples from all Canadian patients and controls (Table 2, and Table 6 in the Supplementary Appendix). To fine-map the IL12A locus, we genotyped 25 SNPs spanning 80 kb and encompassing the IL12A gene and the regions upstream (5′ end, 38 kb) and downstream (3′ end, 32 kb). We observed significant associations for multiple SNPs, with the strongest signal generated by rs4679868, downstream of IL12A (P = 1.58×10−9; odds ratio for patients vs. controls, 1.55). This variant was not genotyped in stage 1. Through haplotype analyses (Table 7 in the Supplementary Appendix), we identified a five-allele haplotype, downstream of IL12A, comprising the rs4679867, rs4679868, rs6441286, rs574808, and rs589545 SNPs (TAGTG), as a major risk haplotype for primary biliary cirrhosis (P = 4.82×10−29). A combined analysis of the Canadian and U.S. samples revealed the presence of this haplotype in 49.1% of all patients (vs. 32.0% of controls) and confirmed the significant association with disease (P = 1.15×10−34; odds ratio, 2.01).
To fine-map the IL12RB2 locus, we genotyped 39 tag SNPs spanning 176.1 kb across the region encompassing IL23R (the gene encoding the interleukin-23 receptor) and IL12RB2. Included in these tag SNPs were IL23R variants previously shown to be associated with Crohn's disease and psoriasis.25,26 SNPs in the intronic regions and regions downstream of IL12RB2 showed the strongest associations with primary biliary cirrhosis; rs6679356 showed the most significant association (P = 7.02×10−8). Moreover, we observed a significant association between primary biliary cirrhosis and a three-SNP haplotype (GTC) downstream of IL12RB2 (present in 35.6% of patients and 25.3% of controls; P = 3.07×10−11; odds ratio for patients vs. controls, 1.53) (Table 8 in the Supplementary Appendix). No significant associations were seen with IL23R SNPs.
Although the association between primary biliary cirrhosis and the rs16833239 SNP in STAT4 was not replicated, in the combined analysis the association was significant (P = 4.67×10−5; odds ratio for patients vs. controls, 1.65), a finding that may be particularly relevant to the associations with markers implicating IL12A and IL12RB2. We therefore carried out additional genotyping of SNPs across the 94.6-kb STAT4 locus, finding modest associations between disease and several intronic SNPs. A SNP in intron 3, rs3024921, showed a significant association (P = 5.76×10−8) in the combined data set (Tables 6 and 9 in the Supplementary Appendix).
Finally, we performed stepwise selection and conditional analysis of the SNPs from the fine-mapping analysis across the IL12A, IL12RB2, and STAT4 loci (Table 10 in the Supplementary Appendix). For each region, we found that multiple alleles underlie the observed associations.
DISCUSSION
We have identified the HLA, IL12A, and IL12RB2 loci as susceptibility loci for primary biliary cirrhosis. Our study provides compelling evidence that all three loci are involved in primary biliary cirrhosis: the successful replication in independent cohorts of the most strongly associated SNP at each of these loci, the high significance levels in the combined analysis, and the identification (through ancillary genotyping) of several other SNPs across the IL12A and IL12RB2 loci that are associated with primary biliary cirrhosis.
Associations between primary biliary cirrhosis and MHC class II alleles have been reported12 but have been only inconsistently replicated in other studies. Our data provide conclusive evidence that this region is involved in primary biliary cirrhosis, with associations shown between disease and variants in the four HLA class II genes (DQB1, DPB1, DRB1, and DRA) and the C6orf10 and BTNL2 genes also at this locus. Among these genes, DQB1, DPB1, and DRB1 have previously been implicated in primary biliary cirrhosis,27 and the association of a BTNL2 truncating mutation identified initially in patients with sarcoidosis28 (another granulomatous disease) has also been seen in patients with other autoimmune diseases. This association may reflect linkage disequilibrium of the BTNL2 mutation with HLA-DQB1–HLA-DRB1 haplotypes that confer a predisposition to disease.29,30 Although strong linkage disequilibrium across this region obscures the identity of the causal alleles under-pinning the associations with primary biliary cirrhosis, haplotype analyses (Table 1 in the Supplementary Appendix) suggest that most of the risk of primary biliary cirrhosis derives from two common haplotypes, AACA (P = 1.09×10−10) and CACA (P = 7.29×10−10), across the C6orf10, BTNL2, and HLA-DQB1 genes. By validating the MHC class II region as a major contributor to the risk of primary biliary cirrhosis, our data support further dissection of this region.
HLA contributions to the risk of primary biliary cirrhosis are proportionately less significant than the contributions to risks of other autoimmune diseases.31 The association of the HLA-DQB1 locus with the risk of primary biliary cirrhosis (odds ratio, 1.75) is similar to the associations of IL12A (odds ratio, 1.54) and IL12RB2 (odds ratio, 1.51). In addition, the population attributable fraction for the IL12A rs6441286 GT and GG risk genotypes (21.5%) is slightly higher than that for HLA-DQB1 rs2856683 CC and CA risk genotypes (21.2%) (Table 11 in the Supplementary Appendix). The IL12RB2 locus also provides a high population attributable fraction, 18.4%. These estimates suggest substantive contributions of all three loci to the risk of primary biliary cirrhosis.
Our data suggest important contributions of the IL12A and IL12RB2 loci to susceptibility to primary biliary cirrhosis. This possibility is consistent with the major immunoregulatory roles of the protein products, interleukin-12 p35 and interleukin-12 receptor β2, which associate with the interleukin-12 p40 and interleukin-12 receptor β1 chains, respectively, to generate interleukin-12 and its receptor. The binding of interleukin-12 to its receptor is thought to modulate autoimmune responses by evoking interferon-γ production, which may in turn inhibit interleukin-23–driven induction of interleukin-17–producing helper T lymphocytes.32,33 The relevance of this pathway to primary biliary cirrhosis is suggested by our finding of associations between primary biliary cirrhosis and several SNPs across the STAT4 gene, which encodes an effector that is integral to interleukin-12 signaling. The SNP rs7574865 in STAT4 intron 3 is known to be associated with risks of rheumatoid arthritis, systemic lupus erythematosus,22 and type 1 diabetes34; our findings suggest that it is associated with primary biliary cirrhosis as well (P = 1.21×10−3; odds ratio, 1.31) (Table 6 in the Supplementary Appendix), although the strongest signals of association at this locus came from intronic SNPs outside the linkage-disequilibrium block tagged by rs7574865.
Similarly, another SNP associated with primary biliary cirrhosis, rs178105416 in the IL12A gene, has previously been shown to be associated with the risk of celiac disease,35 whereas SNPs in the genes encoding interleukin-12β and the interleukin-23 receptor (key components of the signaling pathway of interleukin-12 and its receptor and the related interleukin-23 immunomodulatory axis) are associated with risks for psoriasis and inflammatory bowel disease, although not primary biliary cirrhosis.25,26,36
Our genomewide association study had a statistical power of 82% to detect associations with an odds ratio of 1.5 for patients as compared with controls (see the Supplementary Appendix) and thus provides strong evidence for associations of primary biliary cirrhosis with HLA, IL12A, and IL12RB2 variants. However, the lower statistical power of the study to detect associations with more modest effects, even in the combined analysis (e.g., estimated power of 60% to detect associations with an odds ratio of 1.4) may have impeded the discovery of some risk alleles for primary biliary cirrhosis. Analyses of data from larger and prospectively followed groups of subjects will be required to identify other non-HLA loci influencing risk and to elucidate the relevance of the loci associated with primary biliary cirrhosis to clinically important subphenotypes such as disease progression. Population stratification appeared to have a minimal effect in our analysis, given the minimal inflation of the genomewide chi-square statistics and the similar levels of association in the analysis that was restricted to Canadian subjects.
The causal alleles at the identified risk loci remain unknown. The strongest associations at the IL12A and IL12RB2 loci are with SNPs in the downstream and intronic regions, suggesting that these variants may influence IL12A–IL12RB2 expression. Although it requires further investigation, this possibility is consistent with the development of autoimmune and lymphoproliferative disease in IL12RB2 knockout mice37 and a recent report describing the development of biliary cirrhosis in a child with interleukin-12 deficiency.38 These observations, as well as the cumulative data linking inherited deficiencies of interleukin-12, interleukin-12 receptor, and interferon-γ to increased susceptibility to and severity of mycobacterial and other infectious diseases,39 raise the intriguing possibility that IL12A and IL12RB2 variants associated with primary biliary cirrhosis engender both an impaired response to infection and a potentiated risk of autoimmunity, the former possibly driving the latter.
Precise characterization of the nature and functional sequelae of the HLA and IL12A–IL12RB2 variants that confer a risk of primary biliary cirrhosis remains to be achieved. The association of primary biliary cirrhosis with variants at these loci confirms the critical role of immunogenetic factors in the genesis of this disease. These data point to the possibility that modulation of signaling by interleukin-12 and its receptor may be beneficial in the treatment of patients with primary biliary cirrhosis.
Supplementary Material
Figure 3. Results of Association Analyses and Linkage-Disequilibrium Blocks for the IL12A and IL12RB2 Loci.
Data are shown for the IL12A (encoding interleukin-12α) locus (Panel A) and the IL12RB2 (encoding interleukin-12 receptor β2) locus (Panel B). The results of chi-square analyses are presented in the top plots. Underneath the plots, the organization of the target genes and surrounding loci in humans is shown (not to exact scale). At the bottom, the haplotype block structure is depicted for 25 genotyped SNPs in the IL12A locus (Panel A) and 30 genotyped SNPs in the IL12RB2 locus (Panel B). The block structure is based on criteria established by Gabriel et al.,20 with the use of pairwise estimates of standardized Lewontin's disequilibrium coefficient (D′), whereas the linkage disequilibrium among pairs of SNPs was characterized on the basis of the square of the correlation coefficient (r2). Regions with high r2 values are dark gray, and regions with lower r2 values are lighter gray (i.e., the shade lightens with decreasing r2 values).
Table 2.
Results of Fine-Mapping and Haplotype Analyses for the IL12A and IL12RB2 Loci.*
| SNP | Chromosomal Position | Location | Minor/Major Alleles | Risk-Allele Frequency | P Value | Odds Ratio (95% Cl) | |
|---|---|---|---|---|---|---|---|
| kb | Patients (N = 621) | Controls (N = 1279) | |||||
| IL12A (chromosome 3q25.33–q26) | |||||||
| rs17810546 | 5′-Flanking region | 161147744 | G/A | 0.93 | 0.88 | 9.77×10−6 | 1.78 (1.38–2.31) |
| rs583911 | Intron 2 | 161193084 | G/A | 0.50 | 0.42 | 4.34×10−6 | 1.39 (1.21–1.60) |
| rs668998 | 3′-Flanking region | 161198245 | G/A | 0.50 | 0.42 | 1.12×10−5 | 1.37 (1.19–1.58) |
| rs6441284 | 3′-Flanking region | 161200962 | G/A | 0.66 | 0.59 | 4.47×10−5 | 1.36 (1.17–1.58) |
| rs485497 | 3′-Flanking region | 161201826 | G/A | 0.56 | 0.48 | 1.09×10−5 | 1.37 (1.19–1.58) |
| rs4680536 | 3′-Flanking region | 161202965 | G/A | 0.66 | 0.58 | 4.08×10−6 | 1.41 (1.22–1.63) |
| rs9852519 | 3′-Flanking region | 161203322 | T/C | 0.47 | 0.37 | 2.11×10−8 | 1.50 (1.30–1.73) |
| rs4679867 | 3′-Flanking region | 161206597 | A/T | 0.69 | 0.60 | 4.52×10−7 | 1.46 (1.26–1.70) |
| rs4679868 | 3'-Flanking region | 161206848 | A/G | 0.49 | 0.39 | 1.58×10 −9 | 1.55 (1.34–1.78) |
| rs6441286 | 3′-Flanking region | 161211572 | G/T | 0.49 | 0.39 | 4.83×10−9 | 1.52 (1.32–1.76) |
| rs574808 | 3′-Flanking region | 161215677 | C/T | 0.68 | 0.59 | 1.15×10−7 | 1.49 (1.29–1.73) |
| rs589545 | 3′-Flanking region | 161216294 | A/G | 0.68 | 0.59 | 1.34×10−7 | 1.49 (1.28–1.72) |
| IL12RB2 (chromosome 1p31.2) | |||||||
| rs11209050 | Intron 3 | 67564324 | A/C | 0.28 | 0.20 | 2.76×10−7 | 1.53 (1.30–1.79) |
| rs1908632 | Intron 8 | 67578394 | G/T | 0.33 | 0.25 | 1.22×10−7 | 1.52 (1.30–1.77) |
| rs3 790565 | Intron 8 | 67583944 | C/T | 0.25 | 0.18 | 2.44×10−6 | 1.50 (1.27–1.79) |
| rs946685 | Intron 8 | 67588303 | A/G | 0.25 | 0.18 | 1.93×10−6 | 1.49 (1.26–1.76) |
| rs6679356 | Intron 9 | 67592782 | C/T | 0.27 | 0.19 | 7.02×10 −8 | 1.57 (1.33–1.86) |
| rs10749775 | Intron 9 | 67594675 | C/A | 0.19 | 0.13 | 1.88×10−7 | 1.66 (1.37–2.03) |
| rs3790567 | Intron 9 | 67594965 | A/G | 0.34 | 0.25 | 8.32×10−8 | 1.52 (1.30–1.77) |
| rs6695348 | Intron 9 | 67599604 | T/C | 0.33 | 0.25 | 9.87×10−8 | 1.52 (1.30–1.77) |
Single-nucleotide polymorphisms (SNPs) across the IL12A locus (encoding interleukin-12α) and the IL12RB2 locus (encoding interleukin-12 receptor β2) were investigated in the Canadian patients and controls (from the stage 1 and stage 2b analyses). The risk allele for each SNP is underlined. SNPs with significant associations (P<5.0×10−5) with primary biliary cirrhosis are shown. The complete data set is provided in Table 8 in the Supplementary Appendix. P values and odds ratios are for the comparison of allele frequency between patients and controls, as calculated with the PLINK method. The most significant association for each of the two loci is shown in bold.
Acknowledgments
Supported by grants from the Canadian Institutes for Health Research (MOP 74621), the Ontario Research Fund (RE01-061), the Canadian Primary Biliary Cirrhosis Society, the U.S. National Institutes of Health (K23 DK68290, RO3 DK78527, and RO1 DK80670), the American Gastroenterological Association, and the A.J. and Sigismunda Palumbo Charitable Trust.
Dr. Hirschfield reports receiving fellowship support from the University of Cambridge Parke Davis Fund, the Canadian Commonwealth Association, and the Canadian Association for the Study of the Liver; and Dr. Mason, grant support from Novartis and Schering-Plough. Dr. Peltekian reports being chair of the Canadian Liver Foundation. Dr. Heathcote reports receiving lecture fees and grant support from Axcan Pharma; and Dr. Siminovitch, research support from the Immunogenomics Program of the McLaughlin Centre for Molecular Medicine, from a Canada Research Chair award, and from the Sherman Family Foundation. No other potential conflict of interest relevant to this article was reported.
We thank all the study subjects and the technical staff of the University Health Network Analytical Genetics Technology Centre for their assistance in this research.
REFERENCES
- 1.Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N Engl J Med. 2005;353:1261–73. doi: 10.1056/NEJMra043898. Erratum, N Engl J Med 2006;354:313. [DOI] [PubMed] [Google Scholar]
- 2.Poupon RE, Poupon R, Balkau B. Ursodiol for the long-term treatment of primary biliary cirrhosis. N Engl J Med. 1994;330:1342–7. doi: 10.1056/NEJM199405123301903. [DOI] [PubMed] [Google Scholar]
- 3.Gershwin ME, Mackay IR. The causes of primary biliary cirrhosis: convenient and inconvenient truths. Hepatology. 2008;47:737–45. doi: 10.1002/hep.22042. [DOI] [PubMed] [Google Scholar]
- 4.Irie J, Wu Y, Wicker LS, et al. NOD. c3c4 congenic mice develop autoimmune biliary disease that serologically and pathogenetically models human primary biliary cirrhosis. J Exp Med. 2006;203:1209–19. doi: 10.1084/jem.20051911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oertelt S, Lian ZX, Cheng CM, et al. Anti-mitochondrial antibodies and primary biliary cirrhosis in TGF-beta receptor II dominant-negative mice. J Immunol. 2006;177:1655–60. doi: 10.4049/jimmunol.177.3.1655. [DOI] [PubMed] [Google Scholar]
- 6.Wakabayashi K, Lian ZX, Moritoki Y, et al. IL-2 receptor alpha(−/−) mice and the development of primary biliary cirrhosis. Hepatology. 2006;44:1240–9. doi: 10.1002/hep.21385. [DOI] [PubMed] [Google Scholar]
- 7.Selmi C, Mayo MJ, Bach N, et al. Primary biliary cirrhosis in monozygotic and dizygotic twins: genetics, epigenetics, and environment. Gastroenterology. 2004;127:485–92. doi: 10.1053/j.gastro.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Jones DE, Watt FE, Metcalf JV, Bassendine MF, James OF. Familial primary biliary cirrhosis reassessed: a geographically-based population study. J Hepatol. 1999;30:402–7. doi: 10.1016/s0168-8278(99)80097-x. [DOI] [PubMed] [Google Scholar]
- 9.Watt FE, James OF, Jones DE. Patterns of autoimmunity in primary biliary cirrhosis patients and their families: a population-based cohort study. QJM. 2004;97:397–406. doi: 10.1093/qjmed/hch078. [DOI] [PubMed] [Google Scholar]
- 10.Lazaridis KN, Juran BD, Boe GM, et al. Increased prevalence of antimitochondrial antibodies in first-degree relatives of patients with primary biliary cirrhosis. Hepatology. 2007;46:785–92. doi: 10.1002/hep.21749. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson PT, Baragiotta A, Heneghan MA, et al. HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: a large-scale study. Hepatology. 2006;44:667–74. doi: 10.1002/hep.21316. [DOI] [PubMed] [Google Scholar]
- 12.Invernizzi P, Selmi C, Poli F, et al. Human leukocyte antigen polymorphisms in Italian primary biliary cirrhosis: a multi-center study of 664 patients and 1992 healthy controls. Hepatology. 2008;48:1906–12. doi: 10.1002/hep.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson P, Veeramani S, Baragiotta A, et al. Cytotoxic T-lymphocyte-associated antigen-4 single nucleotide polymorphisms and haplotypes in primary biliary cirrhosis. Clin Gastroenterol Hepatol. 2007;5:755–60. doi: 10.1016/j.cgh.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Juran BD, Atkinson EJ, Schlicht EM, Fridley BL, Lazaridis KN. Primary biliary cirrhosis is associated with a genetic variant in the 3′ flanking region of the CTLA4 gene. Gastroenterology. 2008;135:1200–6. doi: 10.1053/j.gastro.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker EJ, Hirschfield GM, Xu C, et al. CTLA-4/ICOS gene variants and haplotypes are associated with rheumatoid arthritis and primary biliary cirrhosis in the Canadian population. Arthritis Rheum. 2009;60:931–7. doi: 10.1002/art.24412. [DOI] [PubMed] [Google Scholar]
- 16.Heathcote EJ. Management of primary biliary cirrhosis: the American Association for the Study of Liver Diseases practice guidelines. Hepatology. 2000;31:1005–13. doi: 10.1053/he.2000.5984. [DOI] [PubMed] [Google Scholar]
- 17.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 21.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sigurdsson S, Nordmark G, Göring HH, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76:528–37. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dideberg V, Kristjansdottir G, Milani L, et al. An insertion-deletion polymorphism in the interferon regulatory factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum Mol Genet. 2007;16:3008–16. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 25.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cargill M, Schrodi SJ, Chang M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–90. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Invernizzi P, Selmi C, Mackay IR, Podda M, Gershwin ME. From bases to basis: linking genetics to causation in primary biliary cirrhosis. Clin Gastroenterol Hepatol. 2005;3:401–10. doi: 10.1016/s1542-3565(04)00678-0. [DOI] [PubMed] [Google Scholar]
- 28.Valentonyte R, Hampe J, Huse K, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–64. doi: 10.1038/ng1519. Erratum, Nat Genet 2005;37:652. [DOI] [PubMed] [Google Scholar]
- 29.Traherne JA, Barcellos LF, Sawcer SJ, et al. Association of the truncating splice site mutation in BTNL2 with multiple sclerosis is secondary to HLA-DRB1*15. Hum Mol Genet. 2006;15:155–61. doi: 10.1093/hmg/ddi436. [DOI] [PubMed] [Google Scholar]
- 30.Mochida A, Kinouchi Y, Negoro K, et al. Butyrophilin-like 2 gene is associated with ulcerative colitis in the Japanese under strong linkage disequilibrium with HLA-DRB1*1502. Tissue Antigens. 2007;70:128–35. doi: 10.1111/j.1399-0039.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- 31.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goriely S, Neurath MF, Goldman M. How microorganisms tip the balance between interleukin-12 family members. Nat Rev Immunol. 2008;8:81–6. doi: 10.1038/nri2225. [DOI] [PubMed] [Google Scholar]
- 33.Hoeve MA, Savage ND, de Boer T, et al. Divergent effects of IL-12 and IL-23 on the production of IL-17 by human T cells. Eur J Immunol. 2006;36:661–70. doi: 10.1002/eji.200535239. [DOI] [PubMed] [Google Scholar]
- 34.Fung EY, Smyth DJ, Howson JM, et al. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10:188–91. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 35.Hunt KA, Zhernakova A, Turner G, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher SA, Tremelling M, Anderson CA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710–2. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Airoldi I, Di Carlo E, Cocco C, et al. Lack of Il12rb2 signaling predisposes to spontaneous autoimmunity and malignancy. Blood. 2005;106:3846–53. doi: 10.1182/blood-2005-05-2034. [DOI] [PubMed] [Google Scholar]
- 38.Pulickal AS, Hambleton S, Callaghan MJ, et al. Biliary cirrhosis in a child with inherited interleukin-12 deficiency. J Trop Pediatr. 2008;54:269–71. doi: 10.1093/tropej/fmm119. [DOI] [PubMed] [Google Scholar]
- 39.Filipe-Santos O, Bustamante J, Chapgier A, et al. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: molecular, cellular, and clinical features. Semin Immunol. 2006;18:347–61. doi: 10.1016/j.smim.2006.07.010. Erratum, Semin Immunol 2007;19:136-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.