Abstract
Cancer prediction models are becoming ubiquitous, yet we generally have no idea whether they do more good than harm. This is because current statistical methods for evaluating prediction models are uninformative as to their clinical value. Prediction models are typically evaluated in terms of discrimination or calibration. However, it is generally unclear how high discrimination needs to be before it is considered “high enough”; similarly, there are no rationale guidelines as to the degree of miscalibration that would discount clinical use of a model. Classification tables do present the results of models in more clinically relevant terms, but it is not always clear which of two models is preferable on the basis of a particular classification table, or even whether either model should be used at all. Recent years have seen the development of straightforward decision analytic techniques that evaluate prediction models in terms of their consequences. This depends on the simple approach of weighting true and false positives differently, to reflect that, for example, delaying the diagnosis of a cancer is more harmful than an unnecessary biopsy. Such decision analytic techniques hold the promise of determining whether clinical implementation of prediction models would do more good than harm.
Introduction
In this paper, I will make a very simple point: cancer prediction models are becoming ubiquitous, yet we generally have no idea whether they do more good than harm. This is because current statistical methods for evaluating prediction models are uninformative as to their clinical value. I conclude by recommending some simple decision analytic tools that can help identify whether or not a prediction model should be used with patients, and if so, which of two or more competing models should be chosen.
Prediction models for cancer are becoming ubiquitous
In August 2008, I searched Medline for “cancer” with either “prediction model” or “prognostic model” or “nomogram”. I retrieved over 8000 hits. Amongst the first few references were several papers showing that molecular markers predict cancer outcomes, such as STARD10 in breast cancer(1), N-terminal pro-brain natriuretic peptide for neuroendocrine tumors(2) and insulin-like growth factor-binding protein 3 and IGF-I plasma in colorectal cancer(3). I also found papers looking at the predictive value of clinical variables, such as age, stage and performance status for lung cancer(4) and prior surgery for renal cancer(5). There were three papers on prediction models in the first 20 papers retrieved: J-CAPRA for prostate cancer(6), five different models (Myriad, Barnetson, Wijnen, MMRpro, and PREMM) for colorectal cancer(7) and a “prognostic index” for follicular lymphoma (8). This rather informal survey is complemented by more systematic reviews. Shariat et al, for example, found over 100 different prediction tools for prostate cancer alone(9).
The glut of prediction models has now expanded outside the scientific literature. Models can be found on numerous sites on the web, including the National Cancer Institute’s web site(www.cancer.gov) and, amusingly, both www.nomogram.org and www.nomograms.org. They can also be found in routine clinical practice, such the “PCPT risk calculator” to determine indication for prostate biopsy(10) and Adjuvant Online(11) or Oncotype DX(12) for decisions about adjuvant therapy for breast cancer. Inclusion criteria for trials now often depend on models: the Gail model for breast cancer has been used to select patients for breast cancer chemoprevention studies and the Kattan nomogram is used to determine eligibility for clinical trials of adjuvant therapy for prostate cancer.
The ubiquity of prediction models in medicine is not restricted to cancer. Perhaps the most well-known medical prediction model is the Framingham model for cardiovascular disease(13), with the APACHE prognostic system for mortality in critical care a close second(14). Repeating my search for prediction but excluding cancer retrieves nearly 25,000 papers. A very cursory look at the first 20 papers identifies a very wide variety of models: mortality after fungal infection(15); aerobic capacity(16); medication compliance(17); heart disease(18–19); Cesarean section(20), low birthweight(21); transfusion requirements after transplant(22); myodysplasia(23) and stroke rehabilitation(24).
No doubt many investigators are driven to generate prediction models by the medical imperative to individualize care, so that decisions are made with respect to the individual patient rather than a group average. But it is tempting to speculate that the reason behind so many models for so many different diseases is that modeling is particularly easy research to do, a low cost and low effort method to produce another paper: one merely takes an existing data set, runs it through some software, and out pops a prediction model.
Do prediction models do more good than harm?
It is widely assumed that a prediction model is in and of itself a good thing. But it is not difficult to show that a prediction model, even if accurate, could have demonstrable harms. For example, there are several prediction models for adjuvant therapy after surgery for breast cancer(11) (12). These models provide patients with estimates of their benefit from adjuvant therapy, by estimating survival with and without chemotherapy. But take a group of 100 women who would have accepted chemotherapy had they been given the group average of a 5% absolute increase in survival, but who instead are estimated by a prediction model to have 2% risk benefit and therefore elect to avoid adjuvant. Two of these women will die as a result of using the prediction model. It might be argued that this does not represent bad medicine, because the harm of these two deaths is outweighed by the benefit of 98 women avoiding the trauma of chemotherapy. However, if the model was even slightly miscalibrated, deaths associated with use of the prediction model would not be offset by reductions in chemotherapy rates. As such, it is clear that prediction models need to be evaluated before they are used to inform clinical decision making.
Evaluation of prediction models
There are a large number of metrics available whereby statisticians can assess prediction models. Here we focus on four of the most common: statistical significance, relative risk, discrimination and calibration. We also discuss a relatively new innovation, the reclassification table.
Statistical significance
Investigators commonly report whether a predictive model, or a predictor within a model, has a statistically significant association with outcome. Over-reliance on p values falls afoul of the traditional distinction between clinical and statistical significance. A model that correctly estimated a minor increase in risk for a small proportion of patients might well have a statistically significant association with outcome, were the sample size large enough, but would have no clinical role.
Relative risk
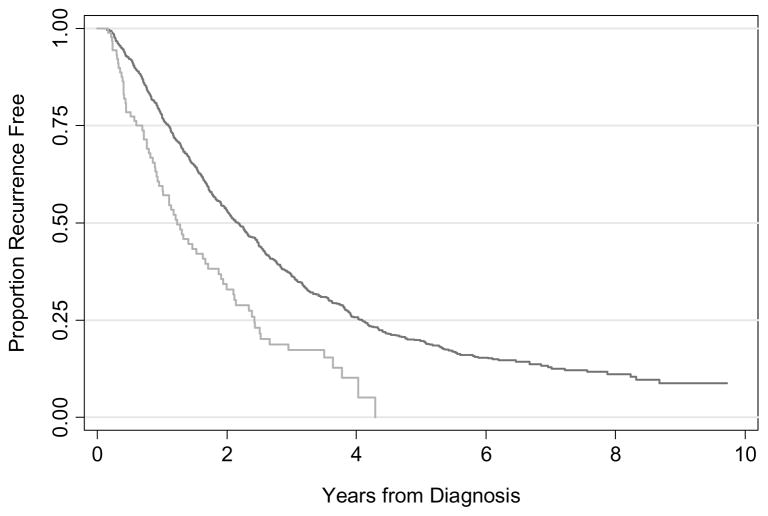
It is very common to see in the literature graphical presentations or numerical estimates of relative risk. For instance, an investigator might report that patients defined as being at high risk from a model had a hazard ratio of 1.85 in comparison to those at low risk, and present a survival curve as in figure 1. This appears to show powerful risk separation, implying that the model should be used in practice. But could a clinician really base treatment on the model? About half of low-risk patients, who constitute 90% of the sample, recur within two years, compared to two-thirds of high-risk patients. For every 100 patients, clinicians who treated on the basis of the model would fail to treat 45 who would otherwise recur and unnecessarily treat 3 patients who would not recur; treating all patients would involve the unnecessary treatment of 50 patients. Use of the model would therefore avoid 47 unnecessary treatments at the expense of failing to treat 45 patients who do require treatment. Given that a cancer recurrence leads to death for most cancers, it is hard to see this as a good trade-off.
Figure 1.
Hypothetical survival curve for a risk prediction model. Grey line: High risk; Black line: low risk.
Discrimination
In place of statistical significance and survival curves, many analysts have advocated measures of discrimination, such as the area-under-the-curve, or its equivalent for time-to-event data, the concordance index. These measures can be interpreted as the probability that, for a randomly selected pair of patients, the patient with the event, or shorter survival, is given a higher risk by the prediction model. Measures of discrimination therefore range from 0.5 (chance) to 1 (perfect discrimination). The concordance index for figure 1 is 0.53, little better than a coin flip, even if statistically significant. In this particular case, the concordance index would correctly identify the model as worthless.
Measures of discrimination have several limitations. One of the most obvious concerns just how high discrimination needs to be in order to be “high enough” to justify use of a model. Clearly a concordance index of 0.53 is too low, but would 0.70 make the model worthwhile? What about 0.60? A similar consideration affects the common use of the concordance index to estimate the value of a new predictor. For example, it has been recommended that investigators should report the concordance index of a model including readily available predictors, such as stage and grade, and then see how much this is improved when the new predictor, such as a molecular marker, is added to the model(25). Again, how much of an increment in predictive accuracy would be enough to justify use of the new marker?
Moreover, despite often being described providing information as to “predictive accuracy”, a measure of discrimination such as the area-under-the-curve is entirely unaffected by whether or not a prediction is a good one. As a simple illustration, consider table 1. All three models have an area-under-the-curve of 0.75, which is normally seen as pretty good. Model 1 gives close the true risk for the two groups of patients; the area-under-the-curve for models 2 and 3 are identical even though the actual risks given to patients are wildly inaccurate.
Table 1. Risks from three models.
Discrimination is the same in each case despite very different risk predictions. Only model 1 is well calibrated.
| Observed outcome | Predicted risk of cancer from the model | |||||
|---|---|---|---|---|---|---|
| Cancer | No cancer | Model 1 | Model 2 | Model 3 | ||
| Risk group from model | High risk | 15 (60%) | 10 (40%) | 60% | 99% | 0.1% |
| Low risk | 5 (14%) | 30 (86%) | 14% | 98% | 0.09% | |
Calibration
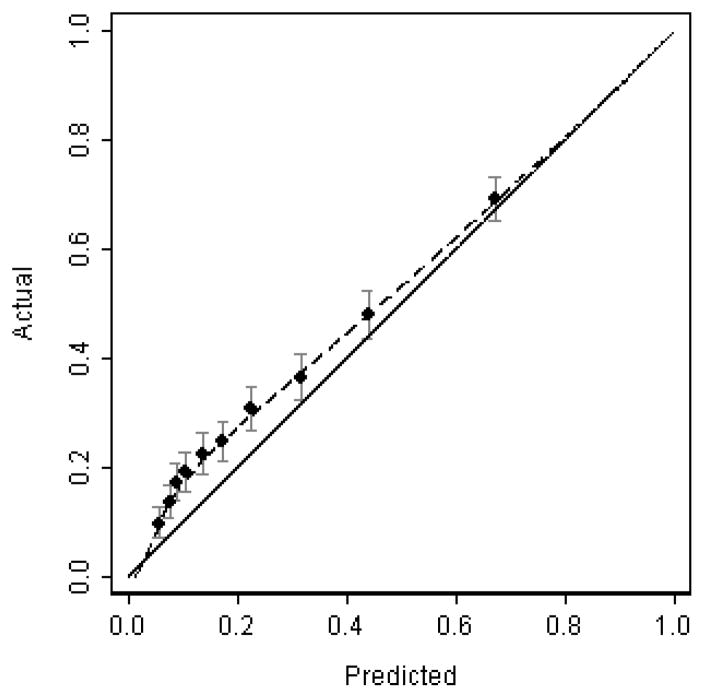
In model 2, the cancer rate for a group of patients given a 98% risk was only 14%. This is known as miscalibration: a model is well calibrated if for every 100 patients given a risk of x%, close to x actually have the event. Calibration is normally illustrated graphically in terms of a calibration plot (see figure 2). The x-axis represents the predicted outcome from the model, and the y-axis represents the actual outcome that was observed. It is clear from figure 2 that the model underestimates risk for patients at low risk, although it is reasonably well calibrated for those at high risk.
Figure 2.
Hypothetical calibration plot
One immediate and obvious drawback to calibration is that it is unclear as to how it should be interpreted. How much miscalibration would be “too much” so as to entail that a model is not of value? Part of the problem is that typical methods for assessing calibration do not provide numbers of clear utility.
A test of statistical significance can be applied to calibration, the Hosmer–Lemeshow test, but its value is highly questionable. The null hypothesis is that “the model is well calibrated”; because we cannot accept the null hypothesis on the basis of a high p value, the test cannot tell us what we really want to know (i.e. good calibration), only that there is insufficient evidence of miscalibration. On a more philosophical level, it is implausible that any non-trivial model would be perfectly calibrated implying that statistical tests of calibration should always give a low p value, given a sufficient sample size.
Another method to gauge calibration is to compare observed and predicted probabilities. This can be done either in terms of an absolute risk difference (e.g. predicted risk was 5% higher than observed risk) or in relative terms (e.g. an odds ratio of 1.25 between observed and predicted risk). Giving a single comparison of observed and predicted probabilities has limited value: a small overall difference in risk may conceal that the model overestimates low risk and underestimates high risk; conversely, an apparently large difference in risk may result from a limited number of clinically irrelevant misclassifications, such as from 90% to 50%. Accordingly, analysts often break up the data into quantiles and compare risk within each quantile. However, it is only by having a large number of quantiles, such as ten, that one can be sure that an average for a quantile is not obscuring important differences for different risk strata within a quantile. Yet larger number of quantiles make results less interpretable – what do 10 separate odds ratios really mean? – and increases problems of sampling variation: even were a study to have as many 1000 patients, 10 quantiles would leave only 100 patients per quantile, giving a confidence interval around a proportion of ± 6 – 10%.
Given that numerical estimates of calibration are questionable, investigators reporting calibration tend to do so in rather vague terms: for example, “because the dots are relatively close to the dashed line, the predictions calculated with use of our nomogram approximate the actual outcomes”(26); “the calibration [plot] demonstrated virtually perfect agreement”(22); “the plotted points were rather close to the 45° line” (27); “calibration …. [was] reasonably accurate”(28). This weakens the value of calibration as a measure to assess the value of prediction models.
Discrimination vs. calibration
A standard approach to evaluating prediction models is to report both discrimination and calibration. The natural problem of having two separate measures is that there is no clear course of action when they conflict. For example, in a paper comparing two prediction models, “Partin” and “Gallina”, Zorn et al. reported that the Gallina model had better discrimination (0.81 vs. 0.78) whereas the Partin model had better calibration(29). This led the authors to the rather equivocal conclusion that: “limitations [of each model] need to be acknowledged and considered before their implementation into clinical practice”.
Classification tables
Hazard ratios, p values, calibration plots and concordance indices have little or no direct relevance to clinical practice. For example, the area-under-the-curve gives the probability that a doctor would correctly identify which of two patients had a disease. But the doctor’s job is not to make guesses between pairs of patients sitting in clinic, it is to make a treatment decision about an individual.
Classification tables are a way to express the results of prediction models in clinical terms. As an example, take the question of whether it would be worth adding information on breast density to a model for whether a woman will be diagnosed with breast cancer. The discrimination and calibration of the “breast density” model have been described(30), but it is not immediately clear what the resulting concordance index of 0.66 means in real terms. The classification table approach is to divide patient into categories of risk, assuming different treatments for each. A simple example is given in table 2, which shows data adapted from Janes et al(31). We assume that women at high risk – defined as having a risk from the statistical model of ≥1.67% - will be treated differently than those at low risk, such as by being advised to use Tamoxifen as a chemopreventive. Table 2 shows that using the standard model will lead to 211,900 women being treated for 4568 potentially preventable cancers; use of the breast density model would involve 193,164 treatments and 4633 cancers. The breast density model is clearly preferable because it involves fewer women being treated but more cancers prevented.
Table 2. Classification table for use of breast density in breast cancer prediction.
Numbers in brackets give frequency and percentage of events. “High risk” is defined as a 5-year risk of breast cancer ≥ 1.67%.
| Risk for model incorporating breast density | ||||
| Low risk | High risk | Total | ||
| Risk for standard prediction model | Low risk | 379,084 (0.90%) | 38,245 (2.06%) | 417,329 (4216, 1.01%) |
| High risk | 56,981 (1.27%) | 154,919 (2.48%) | 211,900 (4568, 2.16%) | |
| Total | 436,065 (4151, 0.95%) | 193,164 (4633, 2.40%) | 629,229 (1.40%) | |
The great advantage here is that the classification model gives results that mean something in the clinic and that can be used as a direct guide for clinical practice. That said, classification tables have two major drawbacks. First, they can only be used to compare two models, they cannot be used to determine whether an individual model should be used in clinical practice. As such, it is plausible that the breast density model is superior to the risk prediction model without breast density and yet neither should be used in the clinic because both are of less value than a strategy of treating all women. Second, classification tables are less easy to interpret when one model reduces both true and false positives. Table 3 shows the breast cancer study when the cut-point for high risk is set at 1% rather than 1.67%. Such a probability threshold might be used for a decision for intensive versus routine screening. The breast density model involves fewer women subjected to intensive screening (379,270 vs. 413,827) but finds fewer cancers early (7023 vs. 7208). On the one hand, the classification table gives numbers than mean something clinically: using the new model will reduce the number of women screened by nearly 35,000 but will delay the diagnosis of 185 cancers; on the other hand, it is not immediately clear whether a reduction in screening of 35,000 is worth those 185 extra cancers.
Table 3. Classification table for use of breast density in breast cancer prediction, using an alternative definition of high risk.
Numbers in brackets give frequency and percentage of events. “High risk” is defined as a 5-year risk of breast cancer ≥ 1%.
| Risk for model incorporating breast density | ||||
|---|---|---|---|---|
| Low risk | High risk | Total | ||
| Risk for standard prediction model | Low risk | 176,831 (0.66%) | 38,571 (1.08%) | 215,402 (1576, 0.73%) |
| High risk | 73,128 (0.82%) | 340,699 (1.94%) | 413,827 (7208, 1.74%) | |
| Total | 249,959 (1761, 0.70%) | 379,270 (7023, 1.85%) | 629,229 (1.40%) | |
Simple decision analytic methods for evaluating prediction models
A decision analytic approach to the evaluation of prediction models is based on two principles. First, models may influence medical decisions and second, decisions have consequences that can be directly incorporated into analyses by using weights.
In discussing the value of the breast density model, we asked whether reducing by 35,000 the number of women referred to more intensive screening is worth delaying the diagnosis of 185 breast cancers. This depends on whether a delayed diagnosis is more than 35,000 ÷ 185 = 189 times worse than unnecessary screening. As it turns out we know that it is not. Decision theory states that the harm of unnecessary treatment, relative to a missed treatment, is given by pt ÷ (1 − pt) where pt is the probability threshold. We used a probability threshold of 1% to define high risk, implying that a delayed cancer is 99 times worse than unnecessarily intensive screening. Therefore the large decrease in screening offsets the relatively small number of delayed cancer diagnoses.
This type of analysis can be formalized in the following equation:
where n is total sample size. The net benefit is in the unit of true positives – a patient with disease duly being treated - and thus can be interpreted as “the number of true positives per patient adjusted for the number of false positives”. Because net benefit incorporates both true positives and false positives it can be used as a single metric to compare two models: whichever has the highest net benefit is optimal.
Table 4 gives net benefits from the breast cancer data set for all reasonable strategies, whether using a model (with breast density or otherwise), or just treating or not treating all women. Looking at the probability threshold of 1.67%, the net benefit for the model with breast density is higher than the standard prediction model. This is to be expected given that fewer women are treated with more cancers potentially prevented. Note that the strategy of treating all women is inferior for that of treating no women: this makes sense because the prevalence (1.4%) is less than the threshold probability. Table 4 also shows that use of the model with breast density is the optimal strategy at a threshold probability of 1%, supporting our informal analysis that the number of women avoiding unnecessary intensive screening is more than 99 times greater than the number of delayed cancer diagnoses.
Table 4.
Net benefit for alternative strategies for breast cancer prediction
| Probability threshold 1% | |||||
|---|---|---|---|---|---|
| True positives | False positives | Net benefit calculation | Net benefit per 1000 | ||
| Screen all women intensively | 8784 | 620445 |
|
4.00 | |
| Standard prediction model | 7208 | 406619 |
|
4.93 | |
| Prediction model with breast density | 7023 | 372247 |
|
5.19 | |
| No intensive screening | 0 | 0 |
|
0 | |
| Probability threshold 1.67% | |||||
| True positives | False positives | Net benefit calculation | Net benefit per 1000 | ||
| Screen all women intensively | 8784 | 620445 |
|
−2.79 | |
| Standard prediction model | 4568 | 207332 |
|
1.66 | |
| Prediction model with breast density | 4633 | 188531 |
|
2.27 | |
| No intensive screening | 0 | 0 |
|
0 | |
Table 4 shows only two probability thresholds. It is reasonable to suppose that patients and clinicians might vary as to how they weight the relative harms and benefits of intensive screening or Tamoxifen as against earlier diagnosis or prevention of breast cancer. In addition, the breast cancer prediction models might be used to inform other decisions, such as whether to screen at all or perhaps use a chemopreventive drug more toxic than Tamoxifen, and these decisions would involve different probability thresholds. We have previously proposed varying the threshold probability over a reasonable range and presenting the net benefits graphically, what is termed a decision curve(32). The methodology of decision curve analysis is therefore:
Select a threshold probability pt
Define a positive test as p̂≥ pt where p̂is the predicted probability of disease from the model; for a binary diagnostic test, p̂is 1 for positive and 0 for negative.
Calculate net benefit
Repeat for a range of pt
Repeat steps 1 – 4 for all models and for the strategy of treating all patients (p̂ = 1 for all patients)
Plot net benefit against pt for all models, tests and markers and for the strategy of treating all patients.
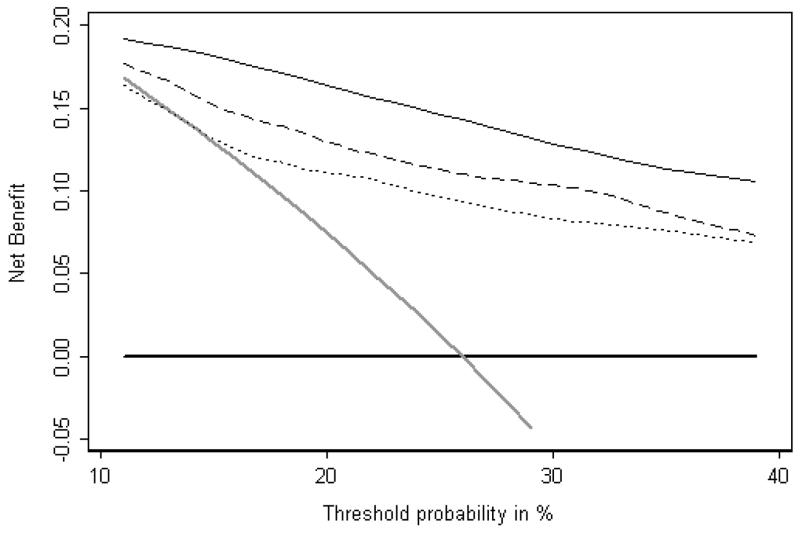
Figure 3 gives a published example of a decision curve. The data set consisted of men undergoing biopsy for prostate cancer; the purpose of the study was to determine whether molecular markers other than PSA were of value for predicting biopsy outcome. The x axis shows threshold probabilities in the range 10 – 40%. This range was chosen on the grounds that it would be unusual for a patient to accept a biopsy if his risk of cancer was less than 10% or conversely, refuse biopsy until his risk was close to 50%. Net benefit, as shown on the y axis is highest for the “four kallikrein model” across the entire range of threshold probabilities. As a result, we can conclude that, irrespective of patient preference, the optimal clinical results will be obtained by determining indication for biopsy on the basis of the four kallikrein model.
Figure 3.
Decision curve analysis: models for the prediction of prostate biopsy. Thin black line: statistical model involving four kallikrein markers. Dashed line: statistical model for free and total PSA. Dotted line: standard clinical model. Grey line: biopsy all men. Thick black line: biopsy no man.
Decision curve analysis is a very flexible technique and can be extended to case control studies and time-to-event data, such as in studies of cancer survival(33). It can also incorporate the harms of testing, for example, if a model requiring data from a test that was invasive or expensive. Papers, tutorials, software code and data sets for decision curve analysis can be found at www.decisioncurveanalysis.org.
Conclusion
We have shown that traditional statistical methods for evaluating prediction models are uninformative as to clinical value. Measures such as p values, relative risks and the concordance index cannot generally tell us whether a model is worth using at all, which of two models is preferable, or whether it is worth obtaining data on an additional predictor. Calibration is a necessary if insufficient condition for use of a model but lacks a clear summary statistic. The recently developed classification tables do produce results in clinically relevant terms but can only be used to compare models – not to determine whether a model is worth using at all – and even then may not give an unambiguous indication of which model is preferable.
Prediction models are sometimes used only for the purposes of informing patients about their likely outcome (“what are my chances, doc?”). In such cases, where no decision is at stake, it may not be unreasonable to depend on discrimination, calibration or classification as a general measure of accuracy. However, when medical decisions depend on the results of prediction model – such as whether to use Tamoxifen as a chemopreventive, or whether to biopsy a man for prostate cancer – then a decision analytic methodology must be used to evaluate models in terms of their consequences. There exist very simple decision analytic tools, requiring only basic math, which incorporate considerations such as it being more harmful to delay a diagnosis of cancer than to biopsy a man unnecessarily. These tools provide a clear indication to which of two models is preferable, whether a model is worth using at all and whether an additional predictor is worth measuring. Wider adoption of decision analytic methods will provide better insights as to whether any of the plethora of new prediction tools do more good than harm.
Acknowledgments
Supported in part by funds from David H. Koch provided through the Prostate Cancer Foundation, the Sidney Kimmel Center for Prostate and Urologic Cancers and P50-CA92629 SPORE grant from the National Cancer Institute to Dr. P. T. Scardino.
Footnotes
Dr. Vickers and Ms Cronin have no primary financial relationships with any companies directly interested in the subject matter of this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murphy NC, Biankin AV, Millar EK, McNeil CM, O’Toole SA, Segara D, et al. Loss of STARD10 expression identifies a group of poor prognosis breast cancers independent of HER2/Neu and triple negative status. Int J Cancer. 2009 doi: 10.1002/ijc.24826. [DOI] [PubMed] [Google Scholar]
- 2.Korse CM, Taal BG, de Groot CA, Bakker RH, Bonfrer JM. Chromogranin-A and N-Terminal Pro-Brain Natriuretic Peptide: An Excellent Pair of Biomarkers for Diagnostics in Patients With Neuroendocrine Tumor. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.7047. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Albeniz X, Gallego R. Prognostic role of plasma insulin-like growth factor (IGF) and IGF-binding protein 3 in metastatic colorectal cancer. Clin Cancer Res. 2009;15(16):5288. doi: 10.1158/1078-0432.CCR-09-0635. author reply. [DOI] [PubMed] [Google Scholar]
- 4.Stinchcombe TE, Hodgson L, Herndon JE, 2nd, Kelley MJ, Cicchetti MG, Ramnath N, et al. Treatment outcomes of different prognostic groups of patients on cancer and leukemia group B trial 39801: induction chemotherapy followed by chemoradiotherapy compared with chemoradiotherapy alone for unresectable stage III non-small cell lung cancer. J Thorac Oncol. 2009;4(9):1117–25. doi: 10.1097/JTO.0b013e3181b27b33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warren M, Venner PM, North S, Cheng T, Venner C, Ghosh S, et al. A population-based study examining the effect of tyrosine kinase inhibitors on survival in metastatic renal cell carcinoma in Alberta and the role of nephrectomy prior to treatment. Can Urol Assoc J. 2009;3(4):281–9. doi: 10.5489/cuaj.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooperberg MR, Hinotsu S, Namiki M, Ito K, Broering J, Carroll PR, et al. Risk Assessment Among Prostate Cancer Patients Receiving Primary Androgen Deprivation Therapy. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.21.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monzon JG, Cremin C, Armstrong L, Nuk J, Young S, Horsman DE, et al. Validation of predictive models for germline mutations in DNA mismatch repair genes in colorectal cancer. Int J Cancer. 2009 doi: 10.1002/ijc.24808. [DOI] [PubMed] [Google Scholar]
- 8.Federico M, Bellei M, Marcheselli L, Luminari S, Lopez-Guillermo A, Vitolo U, et al. Follicular Lymphoma International Prognostic Index 2: A New Prognostic Index for Follicular Lymphoma Developed by the International Follicular Lymphoma Prognostic Factor Project. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.21.3991. [DOI] [PubMed] [Google Scholar]
- 9.Shariat SF, Karakiewicz PI, Roehrborn CG, Kattan MW. An updated catalog of prostate cancer predictive tools. Cancer. 2008;113(11):3075–99. doi: 10.1002/cncr.23908. [DOI] [PubMed] [Google Scholar]
- 10.Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, Lucia MS, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98(8):529–34. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 11.Olivotto IA, Bajdik CD, Ravdin PM, Speers CH, Coldman AJ, Norris BD, et al. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol. 2005;23(12):2716–25. doi: 10.1200/JCO.2005.06.178. [DOI] [PubMed] [Google Scholar]
- 12.Marchionni L, Wilson RF, Wolff AC, Marinopoulos S, Parmigiani G, Bass EB, et al. Systematic review: gene expression profiling assays in early-stage breast cancer. Ann Intern Med. 2008;148(5):358–69. doi: 10.7326/0003-4819-148-5-200803040-00208. [DOI] [PubMed] [Google Scholar]
- 13.Sheridan S, Pignone M, Mulrow C. Framingham-based tools to calculate the global risk of coronary heart disease: a systematic review of tools for clinicians. J Gen Intern Med. 2003;18(12):1039–52. doi: 10.1111/j.1525-1497.2003.30107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13(10):818–29. [PubMed] [Google Scholar]
- 15.Parody R, Martino R, Sanchez F, Subira M, Hidalgo A, Sierra J. Predicting survival in adults with invasive aspergillosis during therapy for hematological malignancies or after hematopoietic stem cell transplantation: Single-center analysis and validation of the Seattle, French, and Strasbourg prognostic indexes. Am J Hematol. 2009;84(9):571–8. doi: 10.1002/ajh.21488. [DOI] [PubMed] [Google Scholar]
- 16.Nunes RA, Vale RG, Simao R, Salles BF, Reis VM, Silva Novaes JD, et al. Prediction of VO2max During Cycle Ergometry Based on Submaximal Ventilatory Indicators. J Strength Cond Res. 2009 doi: 10.1519/JSC.0b013e3181b45c49. [DOI] [PubMed] [Google Scholar]
- 17.Schmid-Mohler G, Thut MP, Wuthrich RP, Denhaerynck K, De Geest S. Non-adherence to immunosuppressive medication in renal transplant recipients within the scope of the integrative model of behavioral prediction: a cross-sectional study. Clin Transplant. 2009 doi: 10.1111/j.1399-0012.2009.01056.x. [DOI] [PubMed] [Google Scholar]
- 18.Goldraich L, Beck-da-Silva L, Clausell N. Are scores useful in advanced heart failure? Expert Rev Cardiovasc Ther. 2009;7(8):985–97. doi: 10.1586/erc.09.68. [DOI] [PubMed] [Google Scholar]
- 19.Koenig W, Vossen CY, Mallat Z, Brenner H, Benessiano J, Rothenbacher D. Association between type II secretory phospholipase A2 plasma concentrations and activity and cardiovascular events in patients with coronary heart disease. Eur Heart J. 2009 doi: 10.1093/eurheartj/ehp302. [DOI] [PubMed] [Google Scholar]
- 20.Verhoeven CJ, Oudenaarden A, Hermus MA, Porath MM, Oei SG, Mol BW. Validation of models that predict Cesarean section after induction of labor. Ultrasound Obstet Gynecol. 2009;34(3):316–21. doi: 10.1002/uog.7315. [DOI] [PubMed] [Google Scholar]
- 21.Law LW, Leung TY, Sahota DS, Chan LW, Fung TY, Lau TK. Which ultrasound or biochemical markers are independent predictors of small-for-gestational age? Ultrasound Obstet Gynecol. 2009;34(3):283–7. doi: 10.1002/uog.6455. [DOI] [PubMed] [Google Scholar]
- 22.Massicotte L, Capitanio U, Beaulieu D, Roy JD, Roy A, Karakiewicz PI. Independent validation of a model predicting the need for packed red blood cell transfusion at liver transplantation. Transplantation. 2009;88(3):386–91. doi: 10.1097/TP.0b013e3181aed477. [DOI] [PubMed] [Google Scholar]
- 23.Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDS) Blood. 2009 doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 24.Lin CL, Lin PH, Chou LW, Lan SJ, Meng NH, Lo SF, et al. Model-based Prediction of Length of Stay for Rehabilitating Stroke Patients. J Formos Med Assoc. 2009;108(8):653–62. doi: 10.1016/s0929-6646(09)60386-7. [DOI] [PubMed] [Google Scholar]
- 25.Kattan MW. Judging new markers by their ability to improve predictive accuracy. J Natl Cancer Inst. 2003;95(9):634–5. doi: 10.1093/jnci/95.9.634. [DOI] [PubMed] [Google Scholar]
- 26.Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90(10):766–71. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 27.Kuijpers T, van der Heijden GJ, Vergouwe Y, Twisk JW, Boeke AJ, Bouter LM, et al. Good generalizability of a prediction rule for prediction of persistent shoulder pain in the short term. J Clin Epidemiol. 2007;60(9):947–53. doi: 10.1016/j.jclinepi.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Grover SA, Hemmelgarn B, Joseph L, Milot A, Tremblay G. The role of global risk assessment in hypertension therapy. Can J Cardiol. 2006;22(7):606–13. doi: 10.1016/s0828-282x(06)70283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zorn KC, Capitanio U, Jeldres C, Arjane P, Perrotte P, Shariat SF, et al. Multi-institutional external validation of seminal vesicle invasion nomograms: head-to-head comparison of Gallina nomogram versus 2007 Partin tables. Int J Radiat Oncol Biol Phys. 2009;73(5):1461–7. doi: 10.1016/j.ijrobp.2008.06.1913. [DOI] [PubMed] [Google Scholar]
- 30.Tice JA, Cummings SR, Smith-Bindman R, Ichikawa L, Barlow WE, Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Ann Intern Med. 2008;148(5):337–47. doi: 10.7326/0003-4819-148-5-200803040-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janes H, Pepe MS, Gu W. Assessing the value of risk predictions by using risk stratification tables. Ann Intern Med. 2008;149(10):751–60. doi: 10.7326/0003-4819-149-10-200811180-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]