Abstract
Plants are essential sources of food, fiber and renewable energy. Effective methods for manipulating plant traits have important agricultural and economic consequences. We introduce a rational approach for associating genes with plant traits by combined use of a genome-scale functional network and targeted reverse genetic screening. We present a probabilistic network (AraNet) of functional associations among 19,647 (73%) genes of the reference flowering plant Arabidopsis thaliana. AraNet associations have measured precision greater than literature-based protein interactions (21%) for 55% of genes, and are highly predictive for diverse biological pathways. Using AraNet, we found a 10-fold enrichment in identifying early seedling development genes. By interrogating network neighborhoods, we identify At1g80710 (now Drought sensitive 1; Drs1) and At3g05090 (now Lateral root stimulator 1; Lrs1) as novel regulators of drought sensitivity and lateral root development, respectively. AraNet (http://www.functionalnet.org/aranet/) provides a global resource for plant gene function identification and genetic dissection of plant traits.
What is the best approach for identifying genes for important plant traits? Forward genetics is limited as mutations in many genes may render only moderate or weak phenotypes. Similarly, while reverse genetics allows for directed assay of gene perturbations1, saturated phenotyping for many plant traits is impractical. A pragmatic near-term solution is the computational identification of likely candidate genes for desired traits, allowing for focused, efficient use of reverse genetics. This solution is not unlike rational drug design in which computer-assisted and expert knowledge are combined with targeted screening for the desired drug, or in our case, trait.
One emerging approach for prioritizing candidate genes is network-guided guilt-by-association. In this approach, functional associations are first determined between genes in a genome on the basis of extensive experimental datasets, encompassing millions of individual observations. Such a map of functional associations is often represented as a graph model and referred to as a functional gene network2.
Probabilistic functional gene networks integrate heterogeneous biological data into a single model, enhancing both model accuracy and coverage. Once a suitable network is generated, new candidate genes are proposed for traits based upon network associations with genes previously linked to these traits. Such network-guided screening has been successfully applied to unicellular organisms3, 4 and C. elegans5, 6, and is a proposed strategy for identifying human disease genes4, 5,7-10.
We demonstrate here that this approach successfully identifies genes affecting specified traits for a reference flowering plant, Arabidopsis thaliana, and we introduce a genome-wide, functional gene network for Arabidopsis suitable for prioritizing candidate genes for a wide variety of traits of economic and agricultural importance.
RESULTS
Reconstruction of a genome-wide functional network of Arabidopsis genes (AraNet)
We integrated diverse functional genomics, proteomics and comparative genomics datasets into a genome-wide functional gene network, using data integration and benchmarking methods customized for Arabidopsis genes (see Supplementary Methods). The datasets included mRNA co-expression patterns measured from DNA microarray datasets (Supplementary Table 1), known Arabidopsis protein-protein interactions11-14, protein sequence features including sharing of protein domains, similarity of phylogenetic profiles15-17 or genomic context of bacterial or archaebacterial homologs18-20, and diverse gene-gene associations (mRNA co-expression, physical protein interactions, multiprotein complexes, genetic interactions, literature mining) transferred from yeast21, fly11, 22-24, worm5, and human genes via orthology25 (Supplementary Table 2). In total, 24 distinct types of gene-gene associations, encompassing >50 million individual experimental observations, were scored for their ability to correctly reconstruct shared membership in Arabidopsis biological processes, then incorporated into a single integrated network model, dubbed AraNet. AraNet contains 1,062,222 functional linkages among 19,647 genes (~73% of the total Arabidopsis genes), with each linkage weighted by the log likelihood of the linked genes to participate in the same biological processes.
Integrating data improves network coverage and accuracy, as tested by recovery of known functional associations (Figure 1A, Supplementary Fig. 15). AraNet extends substantially beyond well-characterized Arabidopsis genes (Figure 1B): 23,720 Arabidopsis genes are unannotated with Gene Ontology Biological Process (GO-BP) annotations by reliable experimental evidence13. AraNet includes more than half (7,465) of genes lacking even sequence homology-based annotations (14,847 genes). These genes’ functions can now be hypothesized based upon their network neighborhoods. AraNet implicates specific processes for 4,479 uncharacterized genes using guilt-by-association. 2,986 uncharacterized genes are associated only with other uncharacterized genes in AraNet, suggesting many still uncharacterized cellular processes in plants.
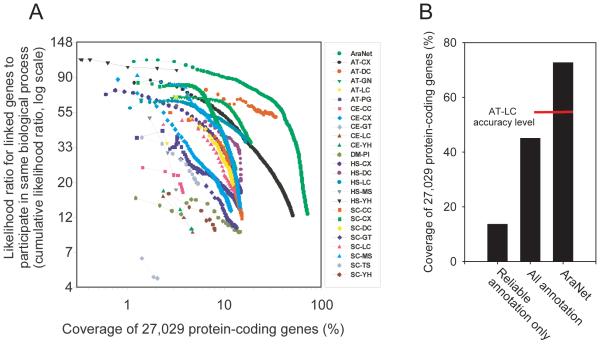
Figure 1.
Construction, accuracy, and coverage of AraNet, a functional gene network for Arabidopsis thaliana. (A) Pairwise gene linkages derived from 24 diverse functional genomics and proteomics data sets, representing >50 million experimental or computational observations, were integrated into a composite network with higher accuracy and genome coverage than any individual data set. The integrated network (AraNet) contains 1,062,222 functional linkages among 19,647 (73%) of the 27,029 protein-coding A. thaliana genes. The plot x axis indicates the log-scale percentage of the 27,029 protein-coding genes13 covered by functional linkages derived from the indicated datasets (curves); the y axis indicates predictive quality of the datasets, measured as the cumulative likelihood ratio of linked genes to share Gene Ontology (GO) ‘biological process’ term annotations, tested using 0.632 bootstrapping and plotted for successive bins of 1,000 linkages each (symbols). Datasets are named as XX-YY, where XX indicates species of data origin (AT, A. thaliana; CE, C. elegans; DM, D. melanogaster; HS, H. sapiens; SC, S. cerevisiae) and YY indicates data type (CC, co-citation; CX, mRNA co-expression; DC, domain co-occurrence; GN, gene neighbor; GT, genetic interaction; LC, literature curated protein interactions; MS, affinity purification/mass spectrometry; PG, phylogenetic profiles; PI, fly protein interactions; TS, tertiary structure; YH, yeast two-hybrid. (B) AraNet spans ~73% of the protein-coding genes, far in excess of current GO biological process annotations for A. thaliana, for which ~12.2 % of genes are annotated by reliable experimental evidence (GO evidence codes IDA, IMP, IGI, IPI) or traceable author statements (GO evidence code TAS), or ~45% annotated by any evidence including computational inferences or sequence homology. The subset of AraNet linkages stronger than the likelihood ratio for literature-curated protein interactions (AT-LC, corresponding to a likelihood ratio of 35:1) covers 55% of the genes.
Evaluating the accuracy of AraNet
We verified the reliability of functional associations in AraNet by testing consistency with known Arabidopsis gene annotations. We applied guilt-by-association in AraNet to identify genes associated with specific biological processes as follows: Each gene in the genome was scored for association with a particular process by summing network edge weights connecting that gene to known genes in that process. A gene’s resulting score corresponds to the naïve Bayes estimate for the gene to belong to that process given network evidence (Figure 2A). Performing cross-validation of this test allows us to assess predictive power with a receiver-operator characteristic (ROC) curve, measuring the true positive prediction rate versus false positive prediction rate as a function of prediction score. We use the area under the ROC curve (AUC) to summarize performance. AUC values of ~0.5 and 1 indicate random and perfect performance, respectively.
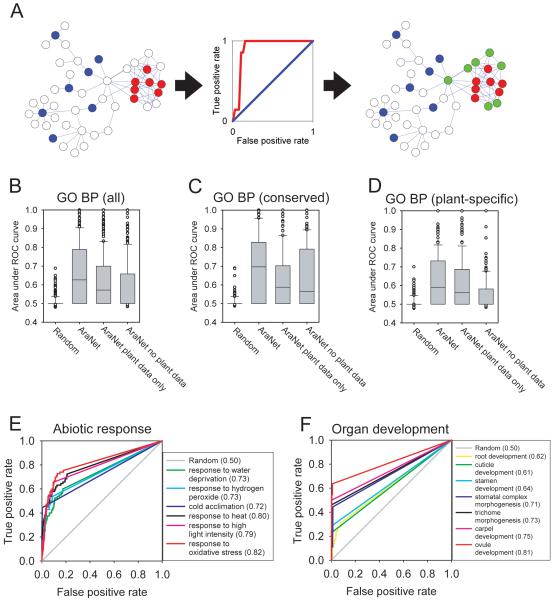
Figure 2.
Predictive power of AraNet for conserved and plant-specific biological processes. AraNet’s predictive capacity was measured using cross-validated receiver operator characteristic (ROC) curve analysis, as illustrated in (A). For a given process, each gene in the A. thaliana genome is rank-ordered by the sum of its network linkage scores to the set of ‘bait’ genes already associated with that process (omitting each bait gene from the bait set for purposes of evaluation). High-scoring genes are most tightly connected to the bait set and are the most likely new candidates to participate in that process. This trend is evident in a ROC plot measuring recovery of bait genes as a function of rank, calculating the true-positive prediction rate (sensitivity; TP/(TP+FN)) versus the false-positive prediction rate (1–specificity; FP/(FP+TN)). If bait genes are highly interconnected (red circles), unlike random genes (blue circles), additional genes connected to the bait genes (green circles) are more likely to be involved in the same process. The area under the cross-validated ROC curve (AUC) provides a measure of predictability, ranging from ~0.5 for random expectation (blue curve) to 1 for perfect predictions (red curve). (B) Distributions of AUC values are plotted for network-based identification of genes for each of the 318 Gene Ontology biological process terms with annotations, (C) for each of the 151 biological process terms with annotations shared between plant and animal or between plant and yeast, and (D) for each of the 167 biological process terms with annotations found in plants but absent from animals and fungi. In all cases, AraNet performs significantly better than for random gene sets of the same sizes. AraNet showed strong predictive power, even when using only Arabidopsis-derived links, although addition of non-plant datasets significantly boosted performance. In bar-and-whiskers plots, the central horizontal line in the box indicates the median AUC and the boundaries of the box indicate the first and third quartiles of the AUC distribution. AraNet specifically identified genes associated with (E) plant abiotic stress response genes and (F) organ developmental processes, as annotated by GO.
Using cross-validation, we tested AraNet’s ability to correctly associate genes with each Gene Ontology biological process, observing significantly better than random predictability for the majority of biological processes (p < 10−53; Wilcoxon signed rank test unless noted otherwise) (Figure 2B). AraNet incorporates data from other organisms; we correspondingly observed higher predictability for evolutionarily conserved processes than for GO processes annotated only with plant genes (p < 10−24, Wilcoxon rank sum test) (Figures 2C,D). Nonetheless, genes were correctly associated with plant-specific processes at significantly higher rates than expected by chance (p < 10−28) (Figure 2D). For example, many important plant traits showed high predictability, including abiotic stress responses (Figure 2E), organ development (Figure 2F), biotic stress responses (Supplementary Fig. 4A), and hormonal signaling (Supplementary Fig. 4B). Tests on two additional independent data sets—the set of reliable GO Cellular Component annotations describing 86 subcellular locations or protein complexes of Arabidopsis proteins13 and the KEGG definitions of 82 Arabidopsis biochemical pathways26—show similarly high predictive power (p ≤ 10−16 and p ≤ 10−14, respectively, compared to chance) (Figure 3A,B). We find that AraNet shows far stronger predictability than previous smaller-scale networks of Arabidopsis genes (Supplementary Table 3, Supplementary Fig. 5)27-30, stemming at least in part from larger coverage, which allows for stronger guilt-by-association due to higher query gene coverage. Thus, AraNet is strongly predictive for many Arabidopsis processes, including those specific to plants.
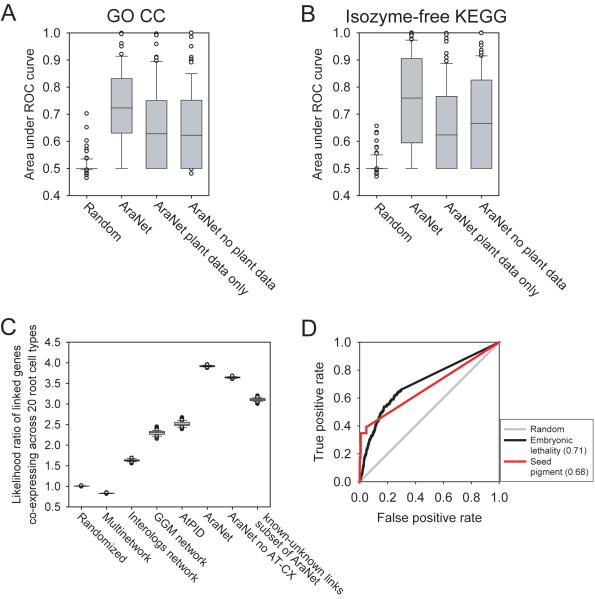
Figure 3.
Validation of AraNet by independent datasets. AraNet shows strong predictive power for gene annotation sets independent of those used to construct AraNet, plotting predictability for (A) 86 GO cellular component terms, and (B) 82 KEGG metabolic pathways (excluding isozymes). The capacity for making association between genes and cell- and tissue-specific biological processes likely arises from the strong tendency for linked genes to share spatiotemporal expression patterns. This tendency is apparent in (C), in which the co-occurrence of mRNA transcripts across 20 root cell-types31 is significantly greater for network-linked genes than for randomized gene pairs (calculated as in the Supplementary Information). Moreover, this tendency is stronger in AraNet than those in previous, smaller gene networks (Supplementary Table 3)27-30. Genes linked in AraNet were significantly more co-expressed in each root cell-type than gene pairs from random networks (repeating the calculation for 100 randomized networks and plotting the distribution of the 100 resulting odds ratios), with >400% enrichment over random for cell type-specific co-expression across the 20 root cell types in AraNet. This trend cannot be explained simply by the incorporation of Arabidopsis mRNA expression data into AraNet, as a version of the network lacking this data shows similarly high cell-type specificity. (D) AraNet shows predictability for genes affecting embryonic lethality or changes in seed pigmentation, as identified in the SeedGenes database32. AUC values are indicated in parentheses.
Differential contributions of linkages derived from plant versus non-plant data on AraNet’s accuracy
Many AraNet linkages derive from orthology to animals and yeast, organisms evolutionarily distant from Arabidopsis. Therefore, we asked the extent to which non-plant derived linkages contribute to AraNet’s accuracy. A version of AraNet composed only of links from yeast and animal data sharply reduces predictive power (p < 10−4) (Figure 2B). A version of only plant-derived links performs substantially better (p ≤10−11), implying that plant-derived data underlies much of AraNet’s predictive power (Figure 2B). This plant-derived data dependence is stronger when predicting plant-specific than conserved processes (p ≤ 10−19, Wilcoxon rank sum test) (Figure 2C,D).
Any method for linking genes to plant traits must perform well for plant-specific processes, so we examined evidence supporting these cases. Remarkably, even for well-predicted plant-specific pathways (AUC scores ranging from ~0.7 - 1, Figure 4), supporting evidence did not derive entirely from plants. For example, photorespiration genes were identified by combining evidence from Arabidopsis, human and C. elegans. Similarly, trichome differentiation genes were recovered using predominantly yeast-derived evidence. Genes of abscisic acid-mediated signaling drew support from all organisms, including fly protein interactions. Thus, while plant-derived links provide most of AraNet’s predictive power, non-plant-derived linkages help substantially in associating genes with plant-specific processes, as processes unique to plants nonetheless often involve conserved genes with conserved interactions.
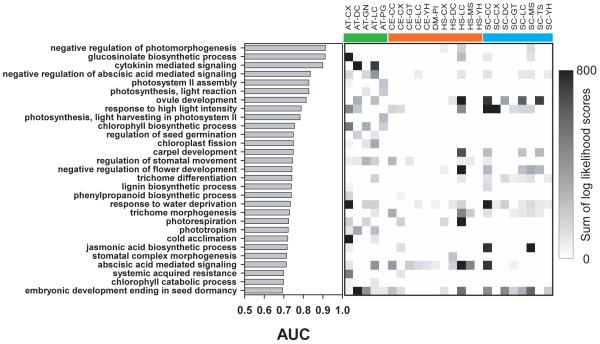
Figure 4.
AraNet correctly associates genes with many processes unique to plants, nonetheless relying at least in part on data from animals and yeast, which contribute evidence for linkages among genes that are broadly conserved but whose roles in Arabidopsis are in plant-specific processes. The performance at associating genes with each of 29 biological processes specific to plants (annotated only with plant genes in GO and are known to occur only in plants) is summarized as the area under a cross-validated ROC curve (AUC). Even though these processes are absent in animals or fungi, the associated genes often have orthologs in these taxa, and AraNet draws upon data from these orthologs in making the associations. Each gray square demarks the support provided by a dataset, measured as a sum of log likelihood scores contributing to that process, with darker gray indicating higher support. Datasets are labeled as in Figure 1.
Linked genes share cell-type specific expression patterns
Many traits in multicellular organisms pertain to specific tissues or cell types. The predictive strength shown by AraNet for such processes raises the question of how a global gene network, incorporating diverse samples and data from orthologs, can correctly identify genes for cell and tissue specific processes. Using measurements of transcript observations in 20 root cell types31 that were not used in building AraNet, we measured the extent to which genes linked in AraNet were spatiotemporally co-expressed in these cells. We find that linked genes show strong cell-specific co-expression in Arabidopsis (Figure 3C)—indeed, far stronger than in previous networks of Arabidopsis genes (Supplementary Table 3)27-30—with linked genes four times more likely to be expressed in the same cell types than random expectation. Thus, although different individual networks were not constructed for each cell-type, such cell- and tissue-specificity is nonetheless at least in part implicitly encoded in AraNet linkages. This correlation between functional association and spatiotemporal co-expression of genes likely enhances prediction strength for many traits, and is evident even for linkages between characterized and uncharacterized genes (Figure 3C), supporting applicability of AraNet to uncharacterized genes.
Associating genes with specific mutant phenotypes
Because linked genes in AraNet tend to operate in the same processes (Figures 1-4), we might expect that they often affect the same phenotypic traits3, 5. This allows association of new candidate genes with traits of interest based on network connections. To test this, we used results from large-scale mutant seed phenotyping32 and analyzed genes whose disruption induced embryonic lethality or changes in seed (embryo) pigmentation. Genes involved in each trait were interlinked significantly better than random expectation (Figure 3D). Unlike AraNet, previous Arabidopsis gene networks27-30 do not significantly predict either phenotype (Supplementary Fig. 6). Thus, AraNet offers a feasible approach for selecting genes likely to be associated with specific plant traits.
Ten-fold enrichment in the discovery rate for genes involved in seed pigmentation
To experimentally test the association of new genes with a trait, we used 23 known seed pigmentation genes (Supplementary Table 4) to search AraNet for new pigmentation genes. Genes in this phenotypic class generally affect chloroplast development or photomorphogenesis, and mutant seedlings show early developmental defects, with albino, pale green, purple or variegated leaves33.
From AraNet’s top 200 candidate genes, we screened all genes with available homozygous T-DNA insertional mutant lines (Supplementary Table 5). We screened 90 candidate genes (represented by 118 mutant lines), of which 14 genes (represented by 17 lines) exhibited color and morphology defects in young seedlings, reminiscent of seed pigmentation mutants (Supplementary Tables 6, 7). This represents a 10-fold enrichment in the discovery rate of the mutant phenotype (p ≤ 10−12, binomial distribution) over that observed during screens of T-DNA insertional lines33 (see Methods). This discovery rate compares well to animal networks, e.g. discovering 16 tumor suppressor effectors from 170 candidates5.
Of the 14 genes with mutant phenotypes, 3 genes (At5g45620, At4g26430 and At5g50110) exhibited the phenotypes in two alleles, 6 genes in only one of the two alleles, and 5 genes were tested in only one allele (Figure 5A, Supplementary Table 7). The 6 genes in which only one of the two alleles showed phenotype are likely to be untagged and were not characterized further. Expressivity of the phenotypes of the 11 lines representing 8 genes (6 lines for 3 genes and 5 lines for 5 genes) varied among individual plants within the homozygous population, ranging from delayed or failed germination, arrested or delayed development, anthocyanin accumulation, clear or white patches on the shoot to pale green shoot. As expected from known seed pigmentation mutants, survival rate in soil was less than 100% in most lines (Supplementary Table 8).
Figure 5.
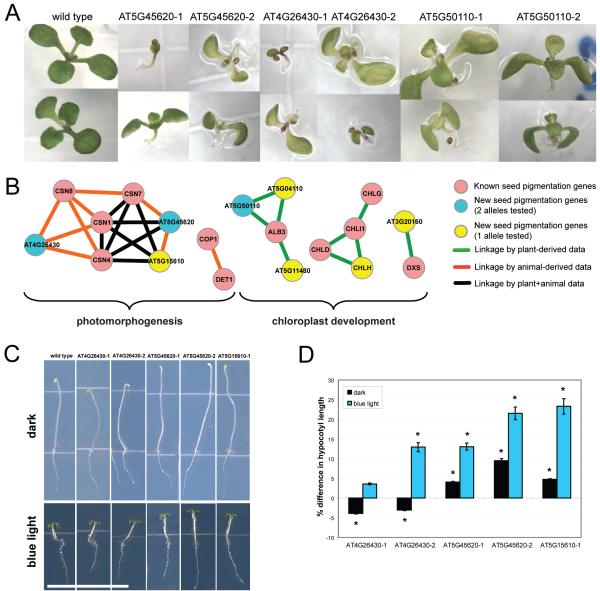
Discovery of new seed (embryo) pigmentation defective genes predicted by AraNet guilt-by-association. (A) Seedling pigmentation defects are apparent in each of two independent alleles for the genes AT5G45620, AT4G26430 (CSN6B), and AT5G50110, all predicted based on network connections to known pigmentation defect genes. (B) Eight new seed pigmentation defective genes are organized into five network components by connections to known seed pigmentation genes, with evidence for the connections coming both from plant- and animal-derived datasets. (C) Mutants linked to known CSN genes show longer hypocotyl length than wild type in dark and under blue light, except CSN6B mutants, which show slightly shorter hypocotyls in dark. Scale bar = 1.3 cm. (D) Most of the differences in hypocotyl length of these mutants are slight (5-25%) but significant. Significant differences from wild type are indicated by asterisks (p-value < 0.01, paired t-test, n = 26 (dark), 32 (blue light)). n indicates the number of plates, each plate containing 7-8 plants of wild type and mutant genotype. Results are from seven (dark) or eight (blue light) independent experiments.
To determine how these genes are associated with seed pigmentation, we examined linkages among the known and newly identified (3 supported by two alleles and 5 by one allele) genes (Figure 5B). These genes form five network components, two belonging to photomorphogenesis and three to chloroplast development (Supplementary Table 9).
The largest component includes members of the COP9 signalosome complex (CSN), an evolutionarily conserved post-translational regulatory complex involved in cell proliferation, response to DNA damage and gene expression34. In plants, the CSN complex is essential for photomorphogenesis35. Two of the 3 genes supported by two independent alleles belong to this component, At4g26430 and At5g45620. At4g26430 (also known as CSN6B) encodes a subunit of CSN, CSN6. Using a single allele, CSN6B has been shown to be genetically redundant to another gene (CSN6A) under white light, though only partially redundant in dark and blue light36. At5g45620 encodes a protein with sequence similarity to a subunit of the lid subcomplex of 26S proteasome but its biological role is unknown13. Supporting evidence for its prediction comes from protein domain co-occurrence with FUS5, FUS6 and COP8 and among their human orthologs (Supplementary Table 5). The CSN and the lid subcomplex of 26S proteasome complexes share structural and functional similarities34, suggesting involvement of other protein degradation machineries in photomorphogenesis and early seedling development. The selfed progeny of mutants that survived to make seeds did not show the seedling defects under standard growth conditions as their progenitors (data not shown). However, when grown in dark or under blue light, the mutants showed slight (5-25%) but significant (p-value <0.01, paired t-test, see Methods) differences in hypocotyl length (Figure 5C-D). CSN6B mutants showed reduced hypocotyl length in dark but slightly increased hypocotyl length when grown under blue light compared to wild type. The other two mutants had longer hypocotyls than wild type under both dark and blue light conditions. All three genes have paralogs in the genome. As double mutants of CSN6A and CSN6B show a constitutive photomorphogenic phenotype, severe seedling dwarfism, and fusca-like defects36, it is possible that At4g26430 and At5g45620 may also show redundancy with their respective paralogs.
The remaining hits from our screen link to each other and known seed pigmentation genes in three components relevant to thylakoid biogenesis and chlorophyll biosynthesis, processes affecting chloroplast development and function (Figure 5B). Supplementary Table 9 details possible roles for the newly discovered genes. These results confirm that AraNet can efficiently associate new genes with a specific phenotypic trait.
Discovering functions for previously uncharacterized Arabidopsis genes using AraNet
Given that AraNet can successfully associate genes with traits of interest, we wished to test hypothesized roles for uncharacterized A. thaliana genes in planta. AraNet predicts biological roles for 4,479 previously uncharacterized genes. We selected three uncharacterized genes (At1g80710, At2g17900, and At3g05090) based on several criteria: 1) no known biological process assigned; 2) predicted by AraNet to be involved in developmental regulatory processes; and 3) exist as single copy genes. These represent extremely stringent tests of the network-based association method, and are all cases in which sequence homology has failed.
AraNet predicts GO-BP annotations, ordering predictions by the sum of the LLS scores linking a gene to genes already annotated by each term (Supplementary Table 10). For the three genes selected, we tested for physiological phenotypes in the top 10 predicted processes. Two control genes, At1g15772 and At2g34170, were chosen randomly from genes lacking AraNet functional predictions. Mutant plants were confirmed for homozygocity (Supplementary Table 12B) and lack of detectable transcripts (data not shown). Selfed progeny of homozygous plants were subjected to a bank of phenotypic assays based on the top 10 predictions (see Supplementary Information). Of the three mutants, two exhibited phenotypes in the predicted processes.
At1g80710 is a novel regulator of drought sensitivity, now named Drought sensitive 1 (Drs1)
AraNet implicated the gene At1g80710 in the response to water deprivation, among other processes, drawing support from affinity purifications of yeast orthologs (SC-MS)37, 38 (Supplementary Table 10). This gene is expressed in all tissues examined, with highest abundance in flowers Supplementary Fig. 7). We asked whether the ability to retain water differed in the mutants. Under drought, mutant plants retained ~80% of the water of wild type (p ≤ 0.001, unpaired t-test, Figure 6A). Reduced water retention was not observed in control mutants (Supplementary Fig. 8).
Figure 6.
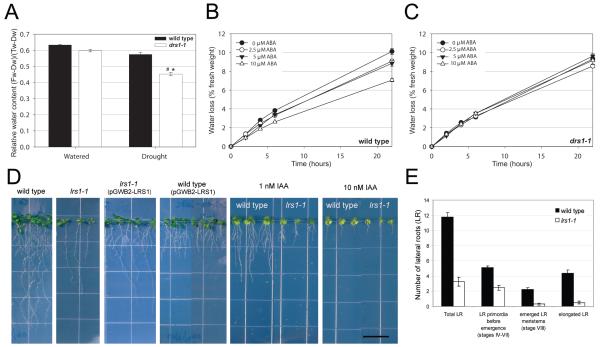
Discovery of new regulators of drought sensitivity and lateral root development from previously uncharacterized genes using AraNet. (A) Plants carrying a T-DNA insertion (drs1-1) in a previously uncharacterized gene, At1g80710, retained significantly less water than wild type under drought. Relative water loss was calculated as (Fw-Dw)/(Tw-Dw) (Fw, fresh weight; Dw, dry weight; Tw, turgor weight). Significant differences between the relative water loss of wild type and mutant plants are indicated by * (p ≤ 0.001, unpaired t-test, n = 15), significant differences between watered and drought conditions of the same genotype by # (p ≤ 0.0001, unpaired t-test, n = 15). (B-C) Transpiration was reduced in wild type plants in the presence of abscisic acid (ABA) in a dosage dependent manner (B) whereas mutant plants were insensitive to ABA (C). (D) The number of lateral roots is strongly reduced in lines carrying a T-DNA insertion (lrs1-1) in another previously uncharacterized gene At3g05090. This phenotype can be complemented by reintroduction of the functional gene. When additional copies of the gene are expressed in a wild type strain, lateral roots increase, while the primary root decreases, in length. 1 nM Auxin (IAA) increases the number and length of lateral roots in both the wild type and mutant seedlings. Contrarily, 10 nM IAA severely reduces the primary root length in both genotypes. Scale bar = 1.4 cm. (E) Different stages of the lateral root (LR) formation are affected in the lrs1-1 mutant. Wild type lateral roots are distributed fairly evenly among LR primordia, emerged LR and elongated LR. The mutant has reduced numbers of the LR in all of these stages, though the reduction is more severe in the emerged and elongated LR than that in the LR primordia. Error bars indicate standard error.
Drought response is mediated by several signaling pathways in Arabidopsis, including the hormone abscisic acid (ABA), transcription factor DREB2A, ERD1, and E3 SUMO ligase SIZ139, 40. To determine whether the reduced water retention upon drought stress is ABA-mediated, we examined the effect of ABA on transpiration of detached leaves. The mutant was insensitive to ABA on water loss whereas the wild type lost significantly less water in the presence of 10 μM ABA (p ≤ 0.0004, unpaired t-test, Figure 6B-C). At this ABA concentration, mutant leaves lost 30% more water than wild type (p ≤ 0.04, unpaired t-test, Figure 6B-C). ABA showed no effect on germination rate in the mutant (data not shown), indicating that not all ABA-mediated processes are affected in the mutant.
Both the water retention and ABA-insensitive water transpiration response segregated as a single recessive Mendelian locus and linked to the T-DNA insertion (Supplementary Table 11, Supplementary Fig. 9). We designate At1g80710 as Drs1 (drought sensitive 1) and the T-DNA insertion allele (Salk_001238C) as drs1-1. An independent T-DNA allele (Salk_149366C) that we designate as drs1-2 exhibited the same phenotypes in relative water content following drought and ABA-insensitive water transpiration (Supplementary Fig. 14) confirming that the phenotypes are linked to mutations in Drs1. Drs1 is a WD-40 repeat family protein containing a DWD (DDB1 binding WD40) motif41. Some DWD-containing proteins are substrate receptors for DDB1-Cul4 ubiquitin ligase machinery in humans, yeast and Arabidopsis41, 42. Combination of AraNet prediction and experimental testing thus demonstrates that Drs1 promotes tolerance to drought stress, possibly mediated by ABA, and suggests involvement of DDB1-Cul4-mediated protein degradation in drought response. Given that the a priori odds of selecting a gene affecting the response to water deprivation are approximately 1 in 318 (currently only 85 of 27,029 Arabidopsis genes are annotated for response to water deprivation), these tests strongly support the network-based approach to rationally associate even entirely uncharacterized genes with plant traits.
At3g05090 is a novel regulator of lateral root development, now named Lateral root stimulator 1 (Lrs1)
The second candidate gene, At3g05090, was implicated in cell proliferation and meristem organization, drawing support from phylogenetic profiling of bacterial homologs of Arabidopsis proteins and domain co-occurrence patterns of yeast orthologs (Supplementary Table 10). We examined both shoot and root development in at3g05090-1 seedlings. We did not observe shoot phenotypes, but the number of lateral roots (LR) was significantly reduced (p ≤ 10−37, unpaired t-test, Figure 6D-E, Supplementary Fig. 10). This phenotype segregated as a single recessive Mendelian locus linked to the T-DNA insertion (Supplementary Table 11, Supplementary Fig. 12A). The length of the primary root was shorter than in wild type (Figure 6D) but this phenotype was unlinked to the T-DNA insertion (Supplementary Fig. 12B), showing that the LR phenotype is separable and independent from the primary root phenotype. We designate At3g05090 as Lrs1 (lateral root stimulator 1) and the at3g05090-1 allele as lrs1-1. Homozygous lines transformed with a wild type coding sequence driven under a 35S CaM virus promoter complemented the LR phenotype (Figure 6D, Supplementary Fig. 10). To determine if the LR formation is blocked before the LR meristem emergence, we examined the number of LR primordia and meristems (LR stages IV-VIII43). Wild type lateral roots are distributed fairly evenly among LR primordia, emerged LR and elongated LR (Figure 6E). The mutant has reduced numbers of the LR at all of these stages, though the reduction is more severe in the emerged and elongated LR than in the LR primordia (Figure 6E). Transforming wild type lines with the 35S::LRS1 construct did not increase the number of LR, but we observed a dramatic increase in the length of the LR and decrease in the primary root length (Figure 6D, Supplementary Fig. 10).
Regulation of root architecture and function, modulated by both intrinsic and extrinsic signals, is critical for efficient nutrient and water use for plants. Auxin, a plant hormone, is a key regulator for LR development, including LR initiation, primordium development and emergence44. The reduction in LR number in the mutant and the increase in LR length concomitant with the decrease in the primary root length in the overexpressed lines evoke defects in auxin accumulation or perception44. We thus asked whether exogenous auxin could alleviate the phenotype by growing plants in the presence of native auxin, indole acetic acid (IAA). The LR number was increased by IAA in the mutant (Figure 6D, Supplementary Fig. 11A-B), demonstrating that auxin perception was not altered in the mutant and suggesting that auxin accumulation is compromised in the mutant. Auxin accumulation can be altered by changing synthesis, degradation, sequestration or transport45. To test for auxin transport defects, we examined effects of an auxin transport inhibitor, N-(1-naphthyl)phthalamic acid (NPA) on root growth. NPA decreases both the number and length of LR in both genotypes (Supplementary Fig. 11A-B). Lrs1 encodes another DCAF protein41, suggesting involvement of DDB1-Cul4-mediated protein degradation in lateral root development. These results demonstrate that the lrs1-1 mutant is defective in lateral root development and suggest roles for DDB1-Cul4-mediated protein degradation in regulating auxin accumulation during LR primordium development and LR meristem emergence, consistent with its hypothesized roles in cell proliferation and meristem organization.
DISCUSSION
We demonstrate here that genes can be rationally associated with plant traits through guilt-by-association in a gene network. For this purpose, we created AraNet, a genome-wide gene network for A. thaliana, a reference organism for flowering plants, including many crops. AraNet is the most extensive gene network for any plant thus far; gene annotations derived by network guilt-by-association extend substantially beyond current gene annotations. We validated the network’s predictive power by cross-validation tests, independent pathway and phenotype datasets, cell-specific expression datasets, and by experiments on computationally selected candidate genes.
AraNet generates at least two main types of testable hypotheses. The first type uses a set of genes known to be involved in a specific process as bait to find new genes involved in that process. This test is useful if the bait genes are well-connected (i.e., high AUC). We used the set of genes conferring seed pigmentation defects (AUC = 0.68) as bait and found a 10-fold enrichment in identifying mutants with comparable phenotypes. Of the 318 GO biological processes with ≥5 genes, ~43% have AUCs of at least 0.68 (Supplementary Table 14), suggesting that AraNet will be useful in identifying new genes in nearly half of these biological processes. In practice, this translates into identifying a small set of new genes from a relatively limited scale screen of the top network-predicted candidates (e.g., computer simulations suggest finding an average of 4-7 novel genes from tests of the top 200 candidates for biological processes with AUC >0.6; Supplementary Fig. 12). The second type of hypotheses involves predicting functions for uncharacterized genes. We assayed predicted phenotypes for three uncharacterized genes, two of which showed phenotypes in the predicted processes, response to drought and meristem development. There are 4,479 uncharacterized genes in AraNet (30% of protein-coding genes) with links to characterized genes, suggesting broad utility for AraNet in identifying candidate functions. Both of these modes of operation can be easily performed on the AraNet website.
While AraNet currently shows high accuracy for many processes (Figures 2-4), there are nonetheless specific processes that are poorly represented, with this trend stronger among plant-specific processes (Figure 2D). This trend manifested in our experimental validation of only 2 of 3 tested candidate genes, although these intentionally represented challenging cases lacking any current functional annotation and for which sequence homology approaches had failed. While we observed that non-plant-derived datasets helped identify genes for plant-specific processes, it is clear that more plant datasets will strongly enhance the utility of gene networks for finding trait-relevant genes.
Three major causes underlie such cases of poor predictive performance: First, our current knowledge of genetic factors for a process may be so sparse that AraNet cannot link them efficiently. Second, AraNet may lack linkages or data relevant to the poorly predicted processes. These two trends likely explain the lower performance among plant-specific processes relative to more broadly-studied, evolutionarily conserved processes. Additional plant-specific datasets, e.g. protein interactions, should help here, as should considering both indirect and direct network linkages for ranking candidates. Third, strongly implicated candidate genes that nonetheless test negative for a trait, resulting in apparent false positives, might be masked by epistatic effects, thus actually representing true predictions and false negative assay results. This trend may be reasonably common and has been previously observed in yeast46.
AraNet represents a major step towards the goal of computationally identifying gene-trait associations in plants. This work suggests that gene networks for food and energy crops will be important enablers for enhanced manipulation of traits of economic importance and crop genetic engineering.
METHODS
All analyses are based on the set of 27,029 predicted protein coding loci of Arabidopsis thaliana (genome release version TAIR7)13. Reference and benchmark sets, raw datasets, and the construction and computational validation of AraNet are described in full in the online supplement.
Targeted reverse genetics screening for seed pigmentation mutants
We searched AraNet with 23 confirmed seed pigmentation mutants (Supplementary Table 4) from the SeedGenes database32 as bait. We retrieved the homozygous T-DNA insertional lines for the top 200 candidates from the SIGnAL database1 and obtained the stocks from the Arabidopsis Biological Resource Center (Supplementary Tables 5, 6). Seven to 9 seeds for each line were sterilized as described below (Plant Material). Seeds were stratified at 4°C for 2 days in dark and grown in MS media with 1% sucrose under continuous illumination of 50-80 μmol/m2·s at 22°C. Seedlings were observed under a dissecting microscope (Leica MZ125) 6 days after germination and followed up 10-12 days after germination. For each of the lines where sufficient seeds were available, the assay was conducted at least twice. T-DNA insertions and genotypes of the seeds for the 11 lines with the mutant phenotypes described in this study (6 lines representing two alleles of three genes and 5 lines representing single alleles of five genes) were confirmed using PCR as described in the online supplement (Supplementary Table 12B).
To determine the significance of the discovery rate, we used the results of a large-scale screening of T-DNA insertional mutants for embryo defective or seed pigmentation mutants32 as the background rate. This study found ~1260 seed pigmentation mutants from screening 120,000 T-DNA lines. Since this was a forward genetics screening whereas our screen was reverse-genetic (i.e., preselected for intragenic insertions), we adjusted the total number of lines in the background to 84,000 based on the genome-wide distribution of T-DNA insertion sites of 70% insertion events in intragenic regions1.
Candidate selection for uncharacterized genes and experimental validation of mutant phenotypes
To test the predictive power of AraNet in planta, we analyzed mutant phenotypes of genes of unknown function, whose biological roles were inferred by the annotations of the neighbors of these genes in AraNet. Of the 27,029 protein-encoding genes in Arabidopsis, 14,847 have no information about the biological processes in which they are involved. More than half of these uncharacterized genes (7,465 genes) are included in AraNet. Of these, 4,479 genes are inferred to be associated with specific biological processes based upon annotated AraNet neighbors (using only IDA, IMP, IGI, IPI, IEP, and TAS evidence). To test the accuracy of such inferences made by AraNet, we chose three genes to characterize experimentally. These were chosen on the basis of available homozygous knock-out lines, absence of paralogs and AraNet inferences of involvement in specific biological processes. The genes chosen were At3g05090, At1g80710, and At2g17900, whose top 10 AraNet predictions are shown in Supplementary Table 10. From the predictions, we assayed all of the phenotypes that could be measured with available resources at Carnegie. For At1g80710, the following were tested: response to water deprivation (rank 3); trichome differentiation (rank 8); leaf development (rank 9). For At3g05090, the following phenotypes were tested: trichome differentiation (rank 1); leaf development (rank 3); cell proliferation (rank 5); meristem organization (rank 8); and regulation of flower development (rank 10). For At2g17900, the following were tested: meristem organization (rank 2); leaf morphogenesis (rank 3); hyperosmotic salinity response (rank 4); brassinosteroid mediated signaling (rank 5); multidimensional cell growth (rank 6); response to auxin stimulus (rank 7); detection of brassinosteroid stimulus (rank 8). In addition, we selected two genes randomly, At1g15772 and At2g34170, which were included in AraNet but were neighbors of other uncharacterized genes, to test specificity of all observed phenotypes.
Plant Material
Seeds of homozygous T-DNA knock-out mutants were obtained from the Arabidopsis Biological Resource Center. The stock numbers for the 118 seed pigmentation candidate genes are listed in Supplementary Table 6. Seeds for the five uncharacterized genes were SALK_059570C (At3g05090), SALK_001238C (At1g80710), SALK_127952C (At2g17900), Salk_118634C (At1g15772) and Salk_099804C (At2g34170). For experiments conducted in soil, seeds were sown in soil (Premier Pro-mix HP #0439P) supplemented with fertilizer (Osmocote Classic, Hummert International #07-6300). Seeds were stratified at 4°C in the dark for 2 days and grown under 16/8 hours of light/dark (90-100 μmol/m2·s) and 30% humidity at 22°C. For experiments conducted in agar plates, seeds were surface-sterilized with 15% commercial bleach (6.25% sodium hypochlorite) containing a few drops of Tween-20 detergent and rinsed with sterile water 5 times. Seeds were sown on agar plates containing 0.43% Murashige and Skoog (MS) salts, 0.5% MES, 0.5% sucrose, 0.8% agar, pH 5.7. Plants on agar plates were grown under constant illumination of 50-80 μmol/m2·s at 22°C. For root assays, 50 mL of the MS medium was prepared poured into 100 × 100 × 15mm square plates (Fisher Scientific #08-757-11A) one day prior to planting to minimize plate-to-plate variability.
T-DNA insertions were confirmed, and genetic linkage, complementation, and overexpression tests were performed as described in the online supplement.
Visible phenotype assays
The following traits were observed by naked eye and using dissecting (Leica MZ125) and compound (Nikon Eclipse E600 with Nomarski optics) microscopes throughout the lifecycle of the mutant plants: trichome differentiation (observations made on rosette and cauline leaves and sepals). leaf development and morphogenesis, cell proliferation, meristem organization and multidimensional cell growth (observations made on leaf, floral, inflorescence and root organs). To detect phenotypes in the regulation of flower development, floral organs and flowering time were observed under long days (16/8 of light/dark) and short days (8/16 of light/dark).
Hypocotyl length measurements
Seeds were germinated and grown vertically for 4 days in dark or under 4 μM/m2·s continuous blue light. Seven to eight seeds of mutant and wild type Columbia were planted per plate and two plates per genotype were tested in each experiment. Each condition was tested in 7-8 independent experiments. Hypocotyl length was measured using ImageJ on photographs of the plates after 4 days of growth. The average hypocotyl length of each genotype was determined from each plate and the difference in hypocotyl length between wild type and mutant was determined using one-tailed, paired t-test.
Root length and number measurements
Seeds were germinated and grown vertically. Root measurements were taken 10-11 days after germination. The LR number was counted using a dissecting microscope or from digital images of plants using ImageJ. Different stages of the LR were determined using a compound microscope. The root length was measured by tracing the length of the root using ImageJ on digital images of the seedlings.
Hormone response assays
Auxin response
Auxin and auxin transport inhibitor treatments were carried out as described47. Seeds were sown on MS agar medium and grown under continuous light. After 4 days, seedlings were transferred to either MS agar medium (control) or MS agar medium containing 1 nM, 5 nM, 10 nM or 30 nM indole acetic acid (IAA, Sigma #I288G) or 1 nM, 10 nM, 100 nM or 1μM of naphthylphthalamic acid (NPA, Chem Service #PS-343). Both wild type (Col-0) and mutant seedlings were transferred to the same plates. On the tenth or eleventh day of growth, the primary root length, number of lateral roots, and length of lateral roots were measured as described above. Significant differences were determined by unpaired Student’s t-test. Each experiment was conducted with 2-3 plates of 7-10 plants each of wild type and mutant per plate. At least three independent experiments were carried out for each hormone assay.
Abscisic acid (ABA) response
The effect of ABA on detached leaf transpiration was determined as described48 with some modifications: The largest, fully-open rosette leaves of 4 week old plants were excised at the bottom of the petioles and were placed into a Parafilm-sealed 1.5 ml centrifuge tubes containing 1.4 ml of 0, 2.5 μM, 5 μM and 10 μM of ABA in an artificial xylem sap solution (15 mM KNO3, 1 mM CaCI2, 0.7 mM MgSO4, and 1mM (NH4)2HPO4, with pH adjusted to 5.0 with 1M phosphoric acid49). Transpiration was measured by weighing total weight of the tubes at times 0, 2, 4, 6, and 22 hours. All of the excisions took place between 10 am and noon (4-6 hours after the onset of illumination). For the F2 linkage test of drs1-1, four leaves were excised from each plant and two were treated with the sap solution and the other two were treated with 10 μM of ABA in the sap solution. For wild type and mutant comparisons, each experiment used 2-4 leaves from 3-4 plants per genotype at each time point and was conducted in triplicate. Four independent experiments were conducted.
Drought response assay
Response to water deprivation was determined by measuring relative water content as described50. Plants were grown in soil under long day conditions (16/8 hr light/dark) under white light of 90-100 μM m−2 s−1 at 22 °C for 4-5 weeks. Watering was stopped for the drought treatment and relative water content was measured on 0, 4th, 7th and 10th day of droughting. Control plants were watered every 2-3 days. To measure relative water content, plants were excised at the shoot/root junction and any bolts were removed, and rosettes were weighed to determine the fresh weight (Fw). The rosettes were then completely submerged in water for 4 hours and weighed to determine the turgid weight (Tw). Rosettes were then dried overnight at 80 °C and weighed to obtain the dry weight (Dw). Three plants from each genotype for each condition were measured. Relative water content was calculated as (Fw – Dw)/(Tw – Dw) and the significance of differences was determined by Student’s t-test. Three plants of each genotype were used for each time point per experiment. Four independent experiments were conducted.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to M. Ahn, A. Noorani and V. Bakshi for technical assistance, J. Shin for assistance on AraNet web designing, T. Nakagawa (Shimane University, Japan) for providing pGWB2, K. Barton for providing lab space and D. Meinke, M. Running, W. Briggs, Z. Wang and K. Dreher for helpful discussions. This work was supported by Carnegie Institution for Science (BA, SYR) and by the National Research Foundation of Korea (NRF) grant funded by the Korean government (No. 2009-0063342, 2009-0070968) and Yonsei University (No. 2008-7-0284, 2008-1-0018) to I.L and from the N.S.F., N.I.H., and Welch (F1515) and Packard Foundations to E.M.M.
References
- 1.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (New York, N.Y. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 2.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 3.McGary KL, Lee I, Marcotte EM. Broad network-based predictability of Saccharomyces cerevisiae gene loss-of-function phenotypes. Genome Biol. 2007;8:R258. doi: 10.1186/gb-2007-8-12-r258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser HB, Plotkin JB. Using protein complexes to predict phenotypic effects of gene mutation. Genome Biol. 2007;8:R252. doi: 10.1186/gb-2007-8-11-r252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee I, et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nature genetics. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 6.Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 7.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 8.Franke L, et al. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. American journal of human genetics. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linghu B, Snitkin ES, Hu Z, Xia Y, Delisi C. Genome-wide prioritization of disease genes and identification of disease-disease associations from an integrated human functional linkage network. Genome Biol. 2009;10:R91. doi: 10.1186/gb-2009-10-9-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huttenhower C, et al. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–1106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermjakob H, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfarano C, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Folter S, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huynen M, Snel B, Lathe W, 3rd, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 18.Bowers PM, et al. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 20.Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee I, Li Z, Marcotte EM. An Improved, Bias-Reduced Probabilistic Functional Gene Network of Baker’s Yeast, Saccharomyces cerevisiae. PLoS ONE. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatr-aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 25.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 26.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui J, et al. AtPID: Arabidopsis thaliana protein interactome database an integrative platform for plant systems biology. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geisler-Lee J, et al. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145:317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez RA, et al. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8:R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma S, Gong Q, Bohnert HJ. An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 2007;17:1614–1625. doi: 10.1101/gr.6911207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 32.Meinke D, Muralla R, Sweeney C, Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends in Plant Science. 2008;13:483–491. doi: 10.1016/j.tplants.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 33.McElver J, et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends in biochemical sciences. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Peng Z, Serino G, Deng XW. Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell. 2001;13:2393–2407. doi: 10.1105/tpc.010248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusmaroli G, Figueroa P, Serino G, Deng XW. Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. The Plant cell. 2007;19:564–581. doi: 10.1105/tpc.106.047571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 38.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 39.Catala R, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Casimiro I, et al. Dissecting Arabidopsis lateral root development. Trends Plant Sci. 2003;8:165–171. doi: 10.1016/S1360-1385(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 44.Fukaki H, Okushima Y, Tasaka M. Auxin-mediated lateral root formation in higher plants. Int Rev Cytol. 2007;256:111–137. doi: 10.1016/S0074-7696(07)56004-3. [DOI] [PubMed] [Google Scholar]
- 45.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, et al. Rational Extension of the Ribosome Biogenesis Pathway Using Network-Guided Genetics. PLoS Biology. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho HT, Cosgrove DJ. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell. 2002;14:3237–3253. doi: 10.1105/tpc.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munns R, King RW. Abscisic Acid Is Not the Only Stomatal Inhibitor in the Transpiration Stream of Wheat Plants. Plant Physiol. 1988;88:703–708. doi: 10.1104/pp.88.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodger JQ, Sharp RE, Marsh EL, Schachtman DP. Relationships between xylem sap constituents and leaf conductance of well-watered and water-stressed maize across three xylem sap sampling techniques. J Exp Bot. 2005;56:2389–2400. doi: 10.1093/jxb/eri231. [DOI] [PubMed] [Google Scholar]
- 50.Giraud E, et al. The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 2008;147:595–610. doi: 10.1104/pp.107.115121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.