Abstract
Objective
Obesity and type 2 diabetes are reaching epidemic proportions in Western societies and contribute to substantial morbidity and mortality. The PPARγ and PGC-1α system plays an important role in regulating efficient energy utilization and oxidative phosphorylation both of which are decreased in obesity and insulin resistance.
Design, Methods
We measured metabolic parameters and the expression of PPARγ and PGC-1α mRNA using quantitative real-time PCR in omental and subcutaneous (SC) adipose tissue in an observational study of 153 individuals as well as in SC fat and skeletal muscle in an interventional study of 60 subjects (20 each with normal glucose tolerance, impaired glucose tolerance and type 2 diabetes) before and after intensive physical training for 4 weeks.
Results
PPARγ and PGC-1α mRNA expression in both fat depots as well as in skeletal muscle is associated with markers of insulin resistance and cardiovascular risk. PGC-1α mRNA expression is significantly higher in SC compared to omental fat, whereas PPARγ expression is not significantly different between these fat depots. Skeletal muscle and SC fat PPARγ and PGC-1α mRNA expression increased significantly in response to physical training.
Conclusions
Gene expression of PPARγ and PGC-1α in human adipose tissue is related to markers of insulin resistance and cardiovascular risk. Increased muscle and adipose tissue PPARγ and PGC-1α expression in response to physical training may mediate the beneficial effects of exercise on insulin sensitivity.
Keywords: PPARγ, PGC-1α, adipose tissue, obesity, diabetes, exercise training
Introduction
Obesity and type 2 diabetes, conditions of epidemic proportions in Western societies, are states of insulin resistance associated with increased fatty acid deposits in muscle as well as inefficient energy utilization and oxidative phosphorylation. Peroxisome proliferator activated receptor γ (PPARγ) acts as a transcription factor regulating the expression of several genes involved in metabolic pathways including energy expenditure and oxidative phosphorylation (1). PPARγ is highly expressed in adipose tissue, where its activation with thiazolidinediones may alter fat distribution and adipocyte phenotype and up-regulates genes involved in fatty acid metabolism and triglyceride storage (1). Furthermore, PPARγ activation is associated with potentially beneficial effects on the expression and secretion of several molecules, including adiponectin, IL-6, TNF α, plasminogen activator inhibitor-1 (PAI-1), monocyte chemoattractant protein-1 (MCP-1), and angiotensinogen, as well as a reduction in plasma nonesterified fatty acid supply (1).
PPARγ coactivator 1 α (PGC-1α), identified as the first member of a small family of transcriptional coactivators, is a cofactor for the nuclear hormone receptor PPARγ that is required for the adaptive thermogenic response to lower temperatures and has been linked to tissue-specific metabolic pathways in the adaptive response to environmental and nutritional stimuli. Despite generally low expression in human white adipose tissue (2), PGC-1α mRNA expression has been shown to be downregulated in white adipose tissue of insulin-resistant (3) and morbidly obese (4) subjects. The PGC-1α gene is highly expressed in human skeletal muscle (1, 5) and insulin stimulation increases muscle expression of PGC-1α (6). In addition, expression of PGC-1α is induced by exercise through 5′-AMP-activated protein kinase (AMPK) and calcium, cytokines, and leptin (7). Expression of PGC-1α in skeletal muscle is lower in patients with type 2 diabetes and also in non-diabetic subjects with a positive family history of diabetes (8). Exercise studies using different training duration and intensities revealed heterogeneous results on physical exercise induced changes with both increased (9-11) and unaffected (12-14) PGC-1α expression in skeletal muscle. It still remains unknown, however, whether PGC-1α expression in adipose tissue changes with exercise. Moreover, PGC-1α and PPARγ mRNA expression changes in muscle and fat have not been systematically studied in relation to exercise induced changes of anthropometric and metabolic parameters.
Because exercise improves insulin resistance and the PPARγ pathway is associated with increasing the oxidative phosphorylation pathway, we hypothesized that PGC-1α and PPARγ expression would be upregulated in muscle and downregulated in adipose tissue with intensive exercise. To this end we first explored associations of PPARγ and PGC-1α expression in visceral and subcutaneous adipose tissue with metabolic parameters and insulin sensitivity in paired samples of omental and subcutaneous adipose tissue from 153 individuals with a wide range of obesity, fat distribution, insulin sensitivity and glucose tolerance in the context of a cross-sectional study. We then examined, in the context of an interventional study, the effect of four weeks of intensive exercise on PPARγ and PGC-1α mRNA expression in both skeletal muscle and subcutaneous adipose tissue in 60 subjects with various degrees of glucose tolerance.
Research Design and Methods
Cross-sectional study
Paired samples of visceral and subcutaneous adipose tissue were obtained from 153 consecutively enrolled Caucasian men (n=75) and women (n=78) who underwent open abdominal surgery for gastric banding, cholecystectomy, weight reduction surgery, abdominal injuries, or explorative laparotomy. Inclusion and exclusion criteria have been previously described (15). In brief, patients were included with the following: 1) Absence of any acute or chronic inflammatory disease as determined by a leucocyte count > 7000 Gpt/l, C-reactive protein (CRP) > 5.0 mg/dl or clinical signs of infection, 2) Undetectable antibodies against glutamic acid decarboxylase (GAD), 3) No medical history of hypertension, i.e. systolic blood pressure (SBP) was < 140mmHg and diastolic blood pressure (DBP) was < 85mmHg, 4) No clinical evidence of either cardiovascular or peripheral artery disease, 5) No thyroid dysfunction, 6) No alcohol or drug abuse, 7) No pregnancy. Percent body fat was measured by dual-energy X-ray absorptiometry (DEXA), and abdominal visceral and subcutaneous fat area was calculated using computed tomography scans as described previously (16). Visceral and subcutaneous fat areas were calculated using CT or MRI scans at the level of L4-L5. A ratio of visceral to SC fat area > 0.5 was used as cut-off to define visceral and SC obesity groups. Samples of visceral and subcutaneous adipose tissue were immediately frozen in liquid nitrogen after explantation. Insulin sensitivity was assessed with the euglycemic-hyperinsulinemic clamp method and oral glucose tolerance tests (OGTT) (16). With this, we identified 67 individuals with either type 2 diabetes (T2D, n=36) or impaired glucose tolerance (IGT, n=31). Three days before the OGTT, the patients documented a high-carbohydrate diet. The OGTT was performed after an overnight fast with 75g standardized glucose solution (Glucodex Solution 75g, Merieux, Montreal, Canada). Venous blood samples were taken at 0, 60, and 120min for measurements of plasma glucose concentrations. Insulin sensitivity was assessed with the euglycemic-hyperinsulinemic clamp method as previously described (17). Basal blood samples were taken after an overnight fast and plasma insulin, leptin and adiponectin, free fatty acids, cholesterol were assayed as previously described (17). The study was approved by the ethics committees at the University of Leipzig. All subjects gave written informed consent before taking part in the study.
Exercise interventional study
We studied 60 Caucasian men and women with no acute or chronic inflammatory disease, alcohol or drug abuse, or diabetic retinopathy or nephropathy. Based on OGTT results, subjects were categorized into groups of normal glucose tolerance (NGT) (n=20, 9 males, 11 females), impaired glucose tolerance (IGT) (n=20, 9 males, 11 females), and type 2 diabetes (T2D) (n=20, 11 males, 9 females). All subjects were enrolled in 60 minutes of supervised physical training sessions 3 days per week, as previously described (17). In brief, each training session included 20 minutes biking or running, 20 minutes of swimming, and 20 minutes of warming up/cooling down periods. All subjects completed a graded bicycle-ergometer test to volitional exhaustion and had maximal oxygen uptake measured with an automated open circuit gas analysis system at baseline. The highest oxygen uptake/minute reached was defined as the maximal oxygen uptake (VO2max), and subjects subsequently trained at their individual submaximal heart rate using heart rate monitors. At baseline and after 4 weeks of training (48 hours after the last training session), subcutaneous adipose tissue and blood samples were obtained in the fasting state, and DEXA analyses and measurements of anthropometric parameters were performed. All baseline blood samples and tissue samples were collected between 8 and 10 am after an overnight fast. Skeletal muscle biopsies were obtained under local anaesthesia from the right vastus lateralis muscle and immediately frozen in liquid nitrogen (16).
Analysis of PPARγ and PGC1α mRNA expression in adipose tissue and skeletal muscle
Human PPARγ and PGC-1α gene expression was measured by quantitative real-time (RT)-PCR in a fluorescent temperature cycler using the TaqMan assay, and fluorescence was detected on an ABI PRISM 7000 sequence detector (Applied Biosystems, Darmstadt, Germany), after an initial denaturation at 95°C for 10min, followed by 40 PCR cycles, each cycle consisting of 95°C for 15s, 60°C for 1min, and 72°C for 1min. The following primers were used: Adipose tissue (detects PPARγ2): human PPARγ: AGG CGA GGG CGA TCT TGA CAG (sense) and GAT GCG GAT GGC CAC CTC TTT (antisense); Skeletal muscle (detects PPARγ1): human PPARγ: AAA GAA GCC GAC ACT AAA CC (sense) and CTT CCA TTA CGG AGA GAT CC (antisense); human PGC-1α : TGC CCT GGA TTG TTG ACA TGA (sense) and TTT GTC AGG CTG GGG GTA GG (antisense); human 18s rRNA: TGC CAT GTC TAA GTA CGC ACG (sense); TTG ATA GGG CAG ACG TTC GA (antisense). In addition, we measured mRNA expression of the PPARγ target genes lipoprotein lipase (LPL), hormone sensitive lipase (HSL) and fatty acid synthase (FASN) as previously described (18, 19). Expression of PPARγ, PGC-1α, LPL, HSL, FASN and 18s RNA were quantified by using the second derivative maximum method of the Taqman Software (Applied Biosystems, Darmstadt, Germany). Amplification of specific transcripts was confirmed by melting curve profiles at the end of each PCR. The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis.
Statistical analyses
Descriptive characteristics are expressed as mean ± SD comparisons. These were conducted using one-way ANOVA with Bonferroni corrected post hoc tests. Nonparametric Spearman correlation coefficients were calculated to examine the cross-sectional associations of PPARγ and PGC-1α with anthropometric and insulin resistance-related parameters. Analyses were performed first without adjustments and then adjusting for age, gender, and either BMI, percent body fat or WHR. For the interventional study, post hoc comparisons of baseline and after-training measures, expressed as mean ± SD, were conducted using a paired t test within groups of glucose tolerance (NGT, IGT and T2D). Differences in change between groups in measurements were compared by general linear model regression analysis with and without adjustments for BMI. Statistical analyses were performed using SPSS (SPSS, Inc., Chicago, IL) and SAS (SAS Institute, Cary, NC).
Results
PPARγ and PGC-1α mRNA expression in adipose tissue - cross sectional study
We present pair-wise comparisons of descriptive characteristics of subjects participating in both the cross-sectional study (n=153) and exercise intervention study (n=60) in Table 1. We performed additional analyses in subgroups of lean (BMI < 25 kg/m2) and obese (BMI > 30 kg/m2) subjects. Based on CT scans measurement (L4-L5) of abdominal visceral and subcutaneous fat areas, obese subjects were further categorized as predominantly visceral or subcutaneous obese. Predominantly visceral obesity was defined as a ratio of visceral/SC fat area > 0.5, as previously described (16). PPARγ mRNA expression in adipose tissue was not significantly different between omental and subcutaneous fat depots both in lean and in obese individuals (Figure 1A). In obese subjects, omental and SC PPARγ mRNA expression is significantly higher compared to lean (Figure 1A). In addition, individuals with predominantly visceral fat accumulation showed significantly higher PPARγ mRNA expression in both depots compared to SC obese patients (Figure 1A). Despite significantly lower mRNA expression of PGC-1α compared to PPARγ in adipose tissue, subcutaneous is 3-4 fold higher than omental PGC-1α expression in all obesity and fat distribution subclasses (Figure 1B). In addition, SC PGC-1α expression was significantly lower in obese patients compared to lean controls (p<0.05, Figure 1B). Patients with visceral obesity had significantly lower visceral PGC-1α expression than lean subjects (p<0.05, Figure 1B).
Table 1.
Descriptive and metabolic characteristics of PPARγ and PGC-1α from a cross-sectional study of n=153 subjects categorized as lean, subcutaneous (SC) obese or visceral obese and n=60 subjects categorized in groups of normal glucose tolerance (NGT), impaired glucose tolerance (IGT) and type 2 diabetes (T2D) from separate exercise intervention trial at baseline. Data are expressed as means ± SD and compared using ANOVA with Bonferroni corrections for post-hoc tests. aP<0.05, b P<0.01, c P<0.001 vs. lean or NGT; dP<0.05, e P<0.01, f P<0.001 vs. SC fat or IGT
| Cross-sectional study | Exercise intervention study | |||||
|---|---|---|---|---|---|---|
| Variable | Lean | SC obese | Visceral obese | NGT | IGT | T2D |
| N | 58 | 58 | 37 | 20 | 20 | 20 |
| Age | 50.2 ± 15.7 | 55.3 ± 13.2 | 64.4 ± 11.6 c / e | 32.8 ± 11.0 | 56.0 ± 11.5 c | 53.1 ± 6.7 c |
| Male (n) / Female (n) | 28 / 30 | 28 / 30 | 19 / 18 | 9 / 11 | 9 / 11 | 11 / 9 |
| BMI (kg/m2) | 23.9 ± 1.4 | 35.9 ± 6.8 c | 33.6 ± 6.1 c | 24.3± 1.5 | 29.8 ± 3.9 b | 31.4 ± 3.2 c |
| WHR | 0.85 ± 0.1 | 1.05 ± 0.1 c | 1.13 ± 0.1 c / e | 0.84 ± 0.1 | 1.21 ± 0.2 c | 1.28 ± 0.1 c |
| Body fat % (by DEXA) | 21.6 ± 2.9 | 41.1 ± 19.5 c | 32.9 ± 6.8 c / f | 24.5 ± 3.2 | 35.0 ± 8.3 c | 38.2 ± 8.0 c |
| Fasting plasma glucose (mmol/l) | 5.4 ± 2.9 | 5.9 ± 1.4 b | 5.8 ± 1.0 | 5.2 ± 0.5 | 5.7 ± 0.6a | 6.2 ± 0.6 c / e |
| HbA1C (%) | 5.3 ± 0.2 | 5.8 ± 0.6 c | 5.9 ± 0.7 c | 5.2 ± 0.2 | 5.8 ± 0.2 c | 6.3 ± 0.3 c / f |
| HDL (mg/dl) | 62.0 ± 19.0 | 48.6 ± 14.1 c | 41.6 ± 18.5c | 46.4 ± 8.9 | 63.4 ± 13.1 c | 56.8 ± 12.1a |
| LDL (mg/dl) | 107.1 ± 30.0 | 110.6 ± 28.5 | 136.5 ± 26.8 c / f | 90.5 ± 17.0 | 124.5 ± 20.5 c | 127.6 ± 33.3 c |
| IL-6 (pg/ml) | 0.87 ± 1.3 | 3.67 ± 3.3c | 5.11 ± 2.5c/d | 1.01 ± 0.8 | 3.17 ± 2.7a | 4.7 ± 3.0 c |
| Adiponectin (μg/ml) | 9.7 ± 5.0 | 6.7 ± 3.9 b | 4.8 ± 3.0 c | 8.7 ± 2.5 | 3.4 ± 1.2 c | 3.5 ± 1.9 c |
| VO2 max (ml/kg/min) | 27.7 ± 5.7 | 24.2 ± 4.4 b | 23.0 ± 5.6 c | 33.8 ± 3.4 | 25.2 ± 2.3 c | 21.8 ± 3.2 c / e |
Figure 1. PPARγ and PGC-1α mRNA expression in human adipose tissue.
(A) PPARγ and (B) PGC-1α mRNA expression in visceral (Vis) and subcutaneous (SC) adipose tissue in lean (n=58), predominantly visceral (n=58) or subcutaneously (n=37) obese subjects. Values are means +/- SE. *p<0.05 for differences between the obesity groups and lean subjects for either omental or subcutaneous fat depot. #p<0.05 for visceral versus SC obese for either depot. — p<0.01 visceral versus SC PGC-1α mRNA expression. Correlation between visceral adipose tissue PPARγ mRNA expression and (C) whole body glucose uptake during the steady state of an euglycemic-hyperinsulinemic clamp, adjusted for age, gender and BMI (r=-0.61, p<0.001) and (D) HbA1c (r2=0.64, p<0.001). Correlation between subcutaneous (SC) adipose tissue PPARγ mRNA expression and (E) whole body glucose uptake during the steady state of an euglycemic-hyperinsulinemic clamp, adjusted for age, gender and BMI (r=-0.47, p<0.01) and (F) HbA1c (r=0.49, p<0.01). Data are log transformed to achieve normal distribution.
We then examined age, gender and BMI-adjusted associations of PPARγ and PGC-1α expression in adipose tissue with anthropometric and metabolic variables among the 153 subjects enrolled in the cross-sectional study (supplementary Table). Both visceral and SC PPARγ mRNA expression significantly correlate with whole body glucose uptake during the steady state of euglycemic-hyperinsulinemic clamp and HbA1c (Figure 1C-F). In addition, PPARγ mRNA expression significantly correlates with parameters of fat distribution (WHR, waist circumference, supplementary Table), glucose metabolism (fasting plasma glucose, 2h OGTT glucose, supplementary Table), lipid metabolism (total and LDL cholesterol, supplementary Table) as well as with adipose tissue mRNA expression of PPARγ target genes lipoprotein lipase (LPL), hormone sensitve lipase (HSL), and fatty acid sythase (FASN) (supplementary Table). There were significant negative correlations between PPARγ mRNA expression in both fat depots and adiponectin serum concentration (supplementary Table).
PGC-1α expression in visceral fat is significantly correlated with adiponectin and percent body fat (negative relationship) (supplementary Table). SC PGC-1α expression correlates with circulating adiponectin, serum FFA, leptin and IL-6 concentrations (supplementary Table). For all of the above parameters the same correlation patterns (at different significance levels) persist when adjusting for age, gender and percent body fat or WHR (data not shown).
Subcutaneous adipose tissue and skeletal muscle PPARγ and PGC-1α mRNA expression in response to 4 weeks of intensive exercise training
60 Caucasian men and women completed a 4 weeks training program and were studied after being divided into subgroups with NGT (n=20), IGT (n=20) glucose tolerance or T2D (n=20) (Table 1). The training effect was confirmed by a significant improvement in VO2 max in all groups. As determined by matched paired t-test (p<0.05), all subjects had a significant increase in VO2 max after the training period (Figure 2A). In addition, we observed significant increased adiponectin serum concentrations after the training program in all glucose tolerance groups (Figure 2B). 4 weeks of physical training resulted in significant decreases in BMI, WHR, and % body fat in all glucose tolerance groups and insulin sensitivity significantly improved in the IGT and T2D groups as previously shown (17).
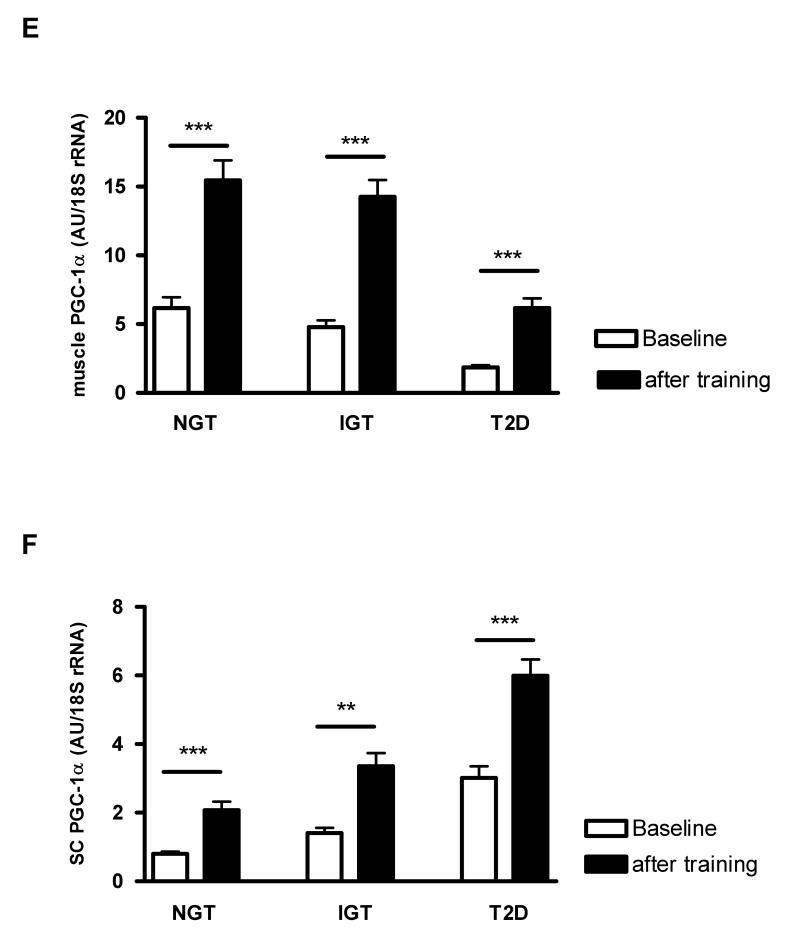
Figure 2. Effect of 4 weeks of intensive exercise program on skeletal muscle and subcutaneous (SC) adipose tissue PPARγ and PGC-1α mRNA expression in normal glucose tolerant (NGT), individuals and patients with impaired glucose tolerance (IGT) or type 2 diabetes (T2D).
(A) VO2max, (B) adiponectin serum concentration, (C) skeletal muscle PPARγ mRNA, (D) SC adipose tissue PPARγ mRNA, (E) skeletal muscle PGC-1α mRNA, (F) SC adipose tissue PGC-1α mRNA in groups of NGT (n=20), IGT (n=20), and T2D (n=20) before and 4 weeks after an intensive exercise program. Data are means +/- SEM. ***, p< 0.001, **, p< 0.01 between baseline and after 4 weeks of intensive physical training.
PPARγ mRNA expression in skeletal muscle was 1.5 fold higher in subjects with NGT as compared to IGT (p=0.15) and 4.2 fold higher in NGT than in patients with T2D (p<0.001). PGC-1α mRNA expression in skeletal muscle was 1.3 fold higher in subjects with NGT as compared to IGT (p<0.05) and 3.3 fold higher in NGT than in patients with T2D (p<0.001). We found age, gender and BMI-adjusted associations of PPARγ and PGC-1α expression in muscle with anthropometric and metabolic variables among the 60 subjects enrolled in the interventional study (supplementary Table). PPARγ mRNA expression in muscle significantly correlates with parameters of glucose metabolism (fasting plasma glucose, 2h OGTT glucose, supplementary Table) and lipid metabolism (total and LDL cholesterol, supplementary Table), but in an opposite direction than in visceral fat tissue. PGC-1α expression in muscle is significantly correlated with WHR and with parameters of insulin sensitivity such as whole body glucose uptake during the steady state of euglycemic-hyperinsulinemic clamp and adiponectin serum concentration (supplementary Table). For all of the above parameters the same correlation patterns (at different significance levels) persist when adjusting for age, gender and percent body fat or WHR (data not shown).
Skeletal muscle PPARγ mRNA expression significantly increased between 2.3 fold in the T2D and 1.5 fold in the IGT group in response the 4 weeks training program (Figure 2C). Expression of PPARγ in SC adipose tissue increased in a parallel pattern in all three groups (Figure 2D). Changes in SC adipose tissue PPARγ mRNA expression significantly correlate with changes in the mRNA expression of PPARγ target genes LPL (r=0.22, p<0.05), HSL (r=0.19, p<0.05), and FASN (r=0.3, p<0.01).
PGC-1α expression significantly increased in all three glucose tolerance groups by ∼ 3 fold in muscle and by ∼ 2 fold in adipose tissue upon physical exercise (Figure 2E, F).
Discussion
Alterations in fatty acid metabolism and mitochondrial dysfunction are critical in the pathogenesis of insulin resistance and the development of type 2 diabetes. PPARγ and the transcriptional coactivator PGC-1α are pivotal in these pathways because they control genes involved in fatty acid oxidation and mitochondrial respiratory chain (1, 5). Mootha et al. (20) identified a set of genes involved in oxidative phosphorylation (OXPHOS), the expression of which was coordinately decreased in human diabetic muscle. Similarly, Patti et al. (8) found the down regulation of OXPHOS not only in individuals with type 2 diabetes but also in their first-degree relatives. In both of these studies, decreased PPARγ and PGC-1α expression was responsible for the down regulation of OXPHOS genes. In addition, the expression of PGC-1α has been shown to be down regulated in adipose tissue of insulin-resistant (3) and morbidly obese (4) subjects.
Here, we extend previous findings by systematically analyzing the anthropometric and metabolic correlates of adipose tissue and skeletal muscle PPARγ and PGC-1α mRNA expression in individuals with a wide range of BMI, fat mass, glucose tolerance and insulin sensitivity both in a cross-sectional study including paired visceral and subcutaneous gene expression and an exercise intervention study. Our study extends a previous pilot study (3) and looks at metabolic parameters and inflammatory markers in association with simultaneous measurement of muscle and adipose tissue expression of PGC-1α and PPARγ in a large study of, collectively, more than 200 individuals. First, we confirm recent findings that the expression of PPARγ is not different between omental and SC fat depots (21, 22). It was previously shown that the expression of PPARγ 1 is fat depot specific, whereas PPARγ 2 was indistinguishable between omental and SC fat (23). Since, our adipose tissue mRNA analysis measured PPARγ 2, our results are in accordance with this previous report (23). Noteworthy, the major PPARγ isoform in non-adipose tissue is PPARγ 1. We therefore measured PPARγ 1 mRNA expression in skeletal muscle biopsies. We find opposite gene expression patterns for adipose tissue PPARγ and PGC-1α mRNA expression. The gene expression of PPARγ in both omental and SC adipose tissue significantly correlates with increasing body weight, fat mass, parameters of impaired glucose metabolism and insulin sensitivity. Noteworthy, strong negative correlations have been found between visceral and SC PPARγ expression and the whole body glucose uptake during the steady state of euglycemic-hyperinsulinemic clamps. Increasing adipose tissue PPARγ expression may represent a compensatory mechanism for the organism in response to deterioration of insulin sensitivity and glucose metabolism. Our data are in accordance with a recent report, demonstrating significantly up regulated PPARγ mRNA expression in the mesenteric adipose tissue of diabetic patients (24). In addition, we confirm the first reported positive association between PPARγ expression and parameters of obesity and impaired glucose metabolism (25). Although, Dubois et al. (26), showed significantly lower adipose tissue PPARγ expression in obese patients with type 2 diabetes compared to patients without diabetes, that study (26) did not include lean healthy controls. We find a significant positive association between PPARγ expression in adipose tissue and BMI, waist circumference, and WHR, whereas a previous report showed the opposite direction of these correlations in obese women (23).
In contrast to observations in adipose tissue, our data demonstrate opposite relationships between PPARγ expression in skeletal muscle and metabolic parameters. In muscle, expression of PPARγ decrease with impairment of glucose tolerance and insulin sensitivity. For example, there were negative correlations between muscle PPARγ expression, fasting plasma glucose, total and LDL cholesterol. Our data support the notion that efficient mitochondrial metabolism and energy utilization which are both directly linked to expression of PPARγ is compromised in insulin resistant states and in patients with type 2 diabetes (20).
Importantly, we also show here that 4 weeks of intensive exercise training significantly increases PPARγ mRNA expression in skeletal muscle independently of different glucose tolerance subgroups. We show here for the first time that increased PPARγ mRNA in muscle is paralleled by significantly increased PPARγ mRNA expression in subcutaneous adipose tissue. Our data are in accordance with recently reported general up regulation of PPARγ mRNA in several tissues of rats (27).
However, PPARγ mRNA expression does only in part determine its activity and post-transcriptional modulation and binding of nuclear cofactors certainly play an important role in PPARγ activation (28, 29). To address this limitation of our study, we measured adipose tissue mRNA of confirmed PPARγ target genes, LPL, HSL, and FASN both in the cross-sectional and in the interventional study. This gene selection was based on results demonstrating that activation of PPARγ by thiazolidinedione treatment causes significant increase in adipose tissue expression of FASN, LPL (30) and HSL (31).
At baseline, we find significant correlations between PPARγ mRNA expression and mRNA expression of FASN, HSL and LPL in both fat depots. Moreover, increased expression of PPARγ in subcutaneous adipose tissue 4 weeks after the exercise training program significantly correlates with changes in LPL, HSL and FASN adipose tissue mRNA expression, suggesting that increased PPARγ mRNA expression in response to training may be of functional relevance for the expression of its target genes.
Analysis of PGC-1α mRNA expression in human adipose tissue revealed significantly higher PGC-1α expression in SC compared to omental adipose tissue independently of obesity and fat distribution. This is in contrast to a previous study, which did not find significant fat depot specific expression of PGC-1α (4). In addition, we found significantly decreased PGC-1α mRNA expression with increasing body fat mass, visceral fat distribution and impaired glucose metabolism. Our data confirm and extend previous results demonstrating reduced PGC-1α mRNA expression in adipose tissue in patients with obesity and insulin resistance (3, 4). Both omental and SC adipose tissue PGC-1α mRNA expression positively correlates with circulating adiponectin, suggesting that PGC-1α may play a role in adiponectin secretion. This hypothesis is further supported by recent data showing an association of the PGC-1α Gly482Ser polymorphism with circulating adiponectin in patients with diabetes (32).
In parallel to the observed increase in skeletal muscle PPARγ mRNA expression upon physical training, we found that 4 weeks of intensive exercise training significantly increases PGC-1α mRNA expression in skeletal muscle and simultaneously in SC adipose tissue independently of different glucose tolerance subgroups. Our study is the first to demonstrate increased PGC-1α mRNA expression in adipose tissue upon physical training despite relatively low baseline PGC-1α mRNA expression in adipose tissue. Previous work has suggested that intensive exercise leads to increased levels of PGC-1α expression in muscle of mice (33) and healthy male human subjects (9, 10). However, two recent studies looked specifically at PGC-1α expression in human muscle after exercise and found somehow conflicting results (13, 14). Heilbronn et al. (14) found unchanged PGC-1α expression in muscle after a 6 weeks training program. Differences in the study population, training intensity and timing of muscle biopsies (36 versus 48 hours post exercise in our study) may contribute to the divergent results. Our data of increased PGC-1α expression in muscle are in accordance with another recent report that acute exercise increases PGC-1α expression despite a delayed response in obese patients (13). It has been shown that PGC-1α is preferentially expressed in muscle enriched for slow oxidative type I fibers (5). In this context, we could previously show that reduced oxidative enzyme activity in muscle of type 2 diabetic patients is most likely due to a reduction in slow oxidative fibers (34). Therefore, reduced PGC-1α expression in muscle of patients with IGT or T2D may be due to altered muscle fiber distribution with impaired glucose metabolism. Noteworthy, up regulation of PGC-1α in muscle of transgenic mice in vivo was shown to be sufficient to improve exercise performance under different exercise paradigms as well as increase peak oxygen uptake (33). This suggests that increased muscle PGC-1α expression in response to physical training may mediate the beneficial effects of exercise on insulin sensitivity. How does exercise regulate the expression of transcription factors? In relation to exercise, potential stimuli include stretch and muscle tension, the pattern of motor nerve activity and the resultant calcium transients, the energy charge of the cell and substrate availability, oxygen tension and circulating hormones (35). Exercise training leads to skeletal muscle fiber type transformation, mitochondrial biogenesis, and increased GLUT4 protein expression (36). Muscle contractions lead to rapid and robust increases in the activation of signaling factors within skeletal muscle cells representing the initial step in the exercise-induced regulation of gene expression (37). Recently, Röckl et al. proposed a model how different signaling pathways may be involved in these training related adaptations (36): Changes in the cellular energy status stimulate AMP-activated protein kinase in the presence of the AMPK kinase, LKB1. AMPK may be involved in exercise related muscle adaption through increasing PGC-1α expression, which is potentiated by a positive feedback loop through myocyte-enhancing factor 2 (MEF2). Alternative pathways, how exercise may regulate the expression of PGC-1α are contraction-induced activation of p38 mitogen activated protein kinase (p38 MAPK) and increased intracellular Ca2+ levels leading to activation of the Ca2+/calmodulin-dependent phosphatase, calcineurin, as well as Ca2+/calmodulin-dependent protein kinases (CaMKs) (36).
Some limitations of our study need to be discussed. We recognize the potential for uncontrolled confounding of the large cross sectional study and that observational studies do not address causality or mechanisms. Nevertheless, they are useful for developing hypotheses for further research using interventions such as the one used in the second part of our study. Although we examined 4 weeks of intensive exercise in the largest study to date, future studies are needed to examine other interventions including dietary and/or pharmacological interventions. Information regarding medication use by our subjects is not known. Finally, we recognize that our interventional study group had a relatively small sample size; nevertheless, our results correlate with other published data and support our hypothesis that gene expression of PPARγ and PGC-1α in muscle increases with intensive physical exercise. Taken together, our data suggest that PPARγ and PGC-1α expression in muscle is directly associated with changes in metabolic parameters implying a direct and possibly causal role of these molecules in the regulation of these parameters. In contrast, expression of PPARγ and PGC-1α in fat is apparently compensatory.
In conclusion, gene expression of PPARγ and PGC-1α in human adipose tissue is related to fat mass, fat distribution and markers of insulin resistance, glucose tolerance, adipose tissue function and inflammation. Increased muscle and adipose tissue PPARγ and PGC-1α expression in response to physical training may mediate the beneficial effects of exercise on insulin sensitivity.
Supplementary Material
Acknowledgments
Funding: This work was supported by grants from Deutsche Forschungsgemeinschaft (DFG): Clinical Research group “Atherobesity” KFO 152 (project BL 833/1-1 to MB and project FA 476/4-1 to MF), NIH grants DK58785, DK79929, DK 081913, DK58845 and a discretionary grant from BIDMC.
Footnotes
Declaration of interest: The authors have no relevant conflict of interest to disclose.
Publisher's Disclaimer: Disclaimer. This is not the definitive version of record of this article. This manuscript has been accepted for publication in European Journal of Endocrinology, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the journal accepts no responsibility for any errors or omissions it may contain. The definitive version is now freely available at http://dx.doi.org/10.1530/EJE-09-0767.
References
- 1.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocrine Reviews. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 2.Pirinen E, Kuulasmaa T, Pietilä M, Heikkinen S, Tusa M, Itkonen P, Boman S, Skommer J, Virkamäki A, Hohtola E, Kettunen M, Fatrai S, Kansanen E, Koota S, Niiranen K, Parkkinen J, Levonen AL, Ylä-Herttuala S, Hiltunen JK, Alhonen L, Smith U, Jänne J, Laakso M. Enhanced polyamine catabolism alters homeostatic control of white adipose tissue mass, energy expenditure, and glucose metabolism. Molecular and Cellular Biology. 2007;27:4953–4967. doi: 10.1128/MCB.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammarstedt A, Jansson PA, Wesslau C, Yang X, Smith U. Reduced expression of PGC-1 and insulin-signaling molecules in adipose tissue is associated with insulin resistance. Biochemical and Biophysical Research Communications. 2003;301:578–582. doi: 10.1016/s0006-291x(03)00014-7. [DOI] [PubMed] [Google Scholar]
- 4.Semple RK, Crowley VC, Sewter CP, Laudes M, Christodoulides C, Considine RV, Vidal-Puig A, O'Rahilly S. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1alpha is reduced in the adipose tissue of morbidly obese subjects. International Journal of Obesity Related Metabolic Disorders. 2004;28:176–179. doi: 10.1038/sj.ijo.0802482. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 6.Ling C, Poulsen P, Carlsson E, Ridderstråle M, Almgren P, Wojtaszewski J, Beck-Nielsen H, Groop L, Vaag A. Multiple environmental and genetic factors influence skeletal muscle PGC-1α and PGC-1β gene expression in twins. Journal of Clinical Investigation. 2004;114:1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attie AD, Kendziorski CM. PGC-1alpha at the crossroads of type 2 diabetes. Nature Genetics. 2003;34:244–245. doi: 10.1038/ng0703-244. [DOI] [PubMed] [Google Scholar]
- 8.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proceedings of the National Academy of Science USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Dériaz O. Endurance training in humans leads to fiber type specific increases in levels of peroxisome proliferator activated receptor gamma coactivator 1 and peroxisome proliferator activated receptor alpha in skeletal muscle. Diabetes. 2003;52:2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 10.Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. Journal of Physiology. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibala MJ, McGee SL, Garnham AP, Howlett KF, Snow RJ, Hargreaves M. Brief intense interval exercise activates AMPK and p38 MAPK signaling and increases the expression of PGC-1{alpha} in human skeletal muscle. Journal of Applied Physiology. 2009;106:929–934. doi: 10.1152/japplphysiol.90880.2008. [DOI] [PubMed] [Google Scholar]
- 12.Mortensen OH, Plomgaard P, Fischer CP, Hansen AK, Pilegaard H, Pedersen BK. PGC-1beta is downregulated by training in human skeletal muscle: no effect of training twice every second day vs. once daily on expression of the PGC-1 family. Journal of Applied Physiology. 2007;103:1536–1542. doi: 10.1152/japplphysiol.00575.2007. [DOI] [PubMed] [Google Scholar]
- 13.De Filippis E, Alvarez G, Berria R, Cusi K, Everman S, Meyer C, Mandarino LJ. Insulin-resistant muscle is exercise resistant: evidence for reduced response of nuclear-encoded mitochrondrial genes to exercise. American Journal of Physiology, Endocrinology and Metabolism. 2008;294:607–614. doi: 10.1152/ajpendo.00729.2007. [DOI] [PubMed] [Google Scholar]
- 14.Heilbronn LK, Gan SK, Turner N, Campbell LV, Chisholm DJ. Markers of mitochondrial biogenesis and metabolism are lower in overweight and obese insulin resistant subjects. Journal of Clinical Endocrinolgy and Metabolism. 2007;92:1467–1473. doi: 10.1210/jc.2006-2210. [DOI] [PubMed] [Google Scholar]
- 15.Youn BS, Bang SI, Klöting N, Park JW, Lee N, Oh JE, Pi KB, Lee TH, Ruschke K, Fasshauer M, Stumvoll M, Blüher M. Serum progranulin concentrations may be associated with macrophage infiltration into omental adipose tissue. Diabetes. 2009;58:627–636. doi: 10.2337/db08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blüher M, Williams CJ, Klöting N, Hsi A, Ruschke K, Oberbach A, Fasshauer M, Berndt J, Schön MR, Wolk A, Stumvoll M, Mantzoros CS. Gene expression of adiponectin receptors in human visceral and subcutaneous adipose tissue is related to insulin resistance and metabolic parameters and is altered in response to physical training. Diabetes Care. 2007;30:3110–3115. doi: 10.2337/dc07-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blüher M, Bullen JW, Jr, Lee JH, Kralisch S, Fasshauer M, Klöting N, Niebauer J, Schön MR, Williams CJ, Mantzoros CS. Circulating adiponectin and expression of adiponectin receptors in human skeletal muscle: associations with metabolic parameters and insulin resistance and regulation by physical training. Journal of Clinical Endocrinolgy and Metabolism. 2006;91:2310–2316. doi: 10.1210/jc.2005-2556. [DOI] [PubMed] [Google Scholar]
- 18.Berndt J, Kralisch S, Klöting N, Ruschke K, Kern M, Fasshauer M, Schön MR, Stumvoll M, Blüher M. Adipose triglyceride lipase gene expression in human visceral obesity. Experimental and Clinical Endocrinology and Diabetes. 2008;116:203–210. doi: 10.1055/s-2007-993148. [DOI] [PubMed] [Google Scholar]
- 19.Berndt J, Kovacs P, Ruschke K, Klöting N, Fasshauer M, Schön MR, Körner A, Stumvoll M, Blüher M. Fatty acid synthase gene expression in human adipose tissue: association with obesity and type 2 diabetes. Diabetologia. 2007;50:1472–1480. doi: 10.1007/s00125-007-0689-x. [DOI] [PubMed] [Google Scholar]
- 20.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1α responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 21.Montague CT, Prins JB, Sanders L, Zhang J, Sewter CP, Digby J, Byrne CD, O'Rahilly S. Depot-related gene expression in human subcutaneous and omental adipocytes. Diabetes. 1998;47:1384–1391. doi: 10.2337/diabetes.47.9.1384. [DOI] [PubMed] [Google Scholar]
- 22.Bortolotto JW, Margis R, Ferreira AC, Padoin AV, Mottin CC, Guaragna RM. Adipose tissue distribution and quantification of PPARbeta/delta and PPARgamma1-3 mRNAs: discordant gene expression in subcutaneous, retroperitoneal and visceral adipose tissue of morbidly obese patients. Obesity Surgery. 2007;17:934–940. doi: 10.1007/s11695-007-9172-5. [DOI] [PubMed] [Google Scholar]
- 23.Giusti V, Verdumo C, Suter M, Gaillard RC, Burckhardt P, Pralong F. Expression of peroxisome proliferator-activated receptor-gamma1 and peroxisome proliferator-activated receptor-gamma2 in visceral and subcutaneous adipose tissue of obese women. Diabetes. 2003;52:1673–1676. doi: 10.2337/diabetes.52.7.1673. [DOI] [PubMed] [Google Scholar]
- 24.Yang YK, Chen M, Clements RH, Abrams GA, Aprahamian CJ, Harmon CM. Human mesenteric adipose tissue plays unique role versus subcutaneous and omental fat in obesity related diabetes. Cellular Physiology and Biochemistry. 2008;22:531–538. doi: 10.1159/000185527. [DOI] [PubMed] [Google Scholar]
- 25.Vidal-Puig AJ, Considine RV, Jimenez-Liñan M, Werman A, Pories WJ, Caro JF, Flier JS. Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. Journal of Clinical Investigation. 1997;99:2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubois SG, Heilbronn LK, Smith SR, Albu JB, Kelley DE, Ravussin E. Look AHEAD Adipose Research Group. Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. Obesity (Silver Spring) 2006;14:1543–1552. doi: 10.1038/oby.2006.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamura T, Yoshida K, Sugawara A, Nagasaka M, Mori N, Takeuchi K, Kohzuki M. Regulation of skeletal muscle peroxisome proliferator-activated receptor gamma expression by exercise and angiotensin-converting enzyme inhibition in fructose-fed hypertensive rats. Hypertension Research. 2004;27:61–70. doi: 10.1291/hypres.27.61. [DOI] [PubMed] [Google Scholar]
- 28.Auwerx J. PPARgamma, the ultimate thrifty gene. Diabetologia. 1999;42:1033–1049. doi: 10.1007/s001250051268. [DOI] [PubMed] [Google Scholar]
- 29.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, Lazar MA. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes and Development. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bogacka I, Xie H, Bray GA, Smith SR. The effect of pioglitazone on peroxisome proliferator-activated receptor-gamma target genes related to lipid storage in vivo. Diabetes Care. 2004;27:1660–1667. doi: 10.2337/diacare.27.7.1660. [DOI] [PubMed] [Google Scholar]
- 31.Deng T, Shan S, Li PP, Shen ZF, Lu XP, Cheng J, Ning ZQ. Peroxisome proliferator-activated receptor-gamma transcriptionally up-regulates hormone-sensitive lipase via the involvement of specificity protein-1. Endocrinology. 2006;147:875–884. doi: 10.1210/en.2005-0623. [DOI] [PubMed] [Google Scholar]
- 32.Okauchi Y, Iwahashi H, Okita K, Yuan M, Matsuda M, Tanaka T, Miyagawa J, Funahashi T, Horikawa Y, Shimomura I, Yamagata K. PGC-1alpha Gly482Ser polymorphism is associated with the plasma adiponectin level in type 2 diabetic men. Endocrine Journal. 2008;55:991–997. doi: 10.1507/endocrj.k08e-070. [DOI] [PubMed] [Google Scholar]
- 33.Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM. Muscle specific expression of PPARγ coactivator-1α improves exercise performance and increases peak oxygen uptake. Journal of Applied Physiology. 2008;104:1304–1324. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- 34.Oberbach A, Bossenz Y, Lehmann S, Niebauer J, Adams V, Paschke R, Schön MR, Blüher M, Punkt K. Altered fiber distribution and fiber-specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type 2 diabetes. Diabetes Care. 2006;29:895–900. doi: 10.2337/diacare.29.04.06.dc05-1854. [DOI] [PubMed] [Google Scholar]
- 35.Hargreaves M, Cameron-Smith D. Exercise, diet, and skeletal muscle gene expression. Medicine & Science in Sports & Exercise. 2002;34:1505–1508. doi: 10.1097/00005768-200209000-00017. [DOI] [PubMed] [Google Scholar]
- 36.Röckl KS, Witczak CA, Goodyear LJ. Signaling mechanisms in skeletal muscle: acute responses and chronic adaptations to exercise. International Union of Biochemistry and Molecular Biology Life. 2008;60:145–153. doi: 10.1002/iub.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benton CR, Wright DC, Bonen A. PGC-1alpha-mediated regulation of gene expression and metabolism: implications for nutrition and exercise prescriptions. Applied Physiology, Nutrition, and Metabolism. 2008;33:843–862. doi: 10.1139/H08-074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.