Abstract
Background
Although fine-needle aspiration (FNA) biopsy of thyroid nodules is very sensitive in detecting thyroid malignancy, it remains ambiguous in 20–30% of cases. Current biomarkers for thyroid cancer lack either the sensitivity or specificity to substantially address this clinical problem. The aim of this study was to investigate the gene expression patterns of human telomerase reverse transcriptase (hTERT) alternative splice variants in benign and malignant thyroid tumors in an attempt to find a more reliable biomarker in the differential diagnosis of thyroid nodules.
Methods
One hundred and thirty-three thyroid tumors from eight histopathological tumor types were collected from patients undergoing thyroid surgery at Johns Hopkins Hospital. Gene expression patterns of hTERT alternative splice variants were investigated in the tumors by nested reverse transcriptase-PCR. Telomerase enzyme activity was evaluated in a subset of 16 samples associated with the different hTERT patterns. Association of c-myc expression and hTERT patterns was also examined.
Results
Malignant thyroid tumors exhibited a greater proportion of the active full-length hTERT transcript (0.57 ± 0.15) than inactive splice variants, α− (0.13 ± 0.02), or β−/α−β− deletion transcripts (0.30 ± 0.11; p < 0.001). The opposite was observed in benign tumors, which exhibited greater proportions of β−/α−β− deletion transcripts (0.64 ± 0.08) than either the full-length (0.19 ± 0.06) or α− deletion transcripts (0.17 ± 0.02; p < 0.001). Similar results were observed among a diagnostically challenging subset of 50 thyroid tumors that were suspicious for malignancy on FNA. Further, increased telomerase enzymatic activity was only associated with expression of the full-length hTERT isoform. In contrast, c-myc expression, which has been implicated in hTERT regulation, correlated with overall hTERT transcription without specificity for expression of the full-length isoform.
Conclusions
These differences in gene expression patterns of hTERT alternative splice variants may provide a useful adjunct to FNA diagnosis of suspicious thyroid tumors.
Introduction
The clinical problem associated with patients who present with a suspicious thyroid nodule continues to place clinicians and patients in situations where decisions about the surgical approach need to be made with inadequate information. Although fine-needle aspiration (FNA) biopsy of a thyroid nodule is very sensitive in the detection of malignancy, it is indeterminate or suspicious in 20–30% of cases (1,2). There are over 100,000 patients each year who present with a suspicious thyroid nodule in the United States (3,4). Terminologies commonly used in suspicious cytopathology reports include the following: follicular or Hürthle cell neoplasm, suspicious for papillary or follicular variant of papillary thyroid cancer, or cellular atypia. Because clinicians often cannot determine malignancy, either pre- or intraoperatively, patients with suspicious thyroid lesions cannot be optimally managed (5–7). This often results in two scenarios: (i) patients who ultimately have a benign lesion on final histopathology may be subjected to unnecessary surgery; (ii) patients with a malignant thyroid nodule may need to undergo a second operation for completion thyroidectomy only after a diagnosis of cancer is rendered on permanent histological section.
There have been several attempts to address this problem through the use of molecular markers. However, an accurate molecular signature of thyroid malignancy has yet to be discovered (8). Some studies have pointed to telomerase as one of the more promising molecular markers for the diagnosis of malignancy. Telomerase is a ribonucleoprotein complex that stabilizes human chromosomes by adding telomere sequence (TTAGGG) repeats to their ends. Telomerase enzyme activity has been detected in 29–100% of thyroid cancers, but also in up to 45% of benign thyroid tumors (9–11). Similarly, expression of its catalytic component human telomerase reverse transcriptase (hTERT) has been documented in up to 90% of thyroid cancers, but also in up to 35% of benign tumors (12,13). This overlap in telomerase enzyme activity and hTERT expression levels between benign and malignant thyroid tumors has therefore precluded the use of telomerase as a useful marker of malignancy.
Alternative splicing of RNA is important in both embryological development and certain disease states. Patterns of alternative splice variants can be both tissue and disease specific (14). The identification of cancer-specific splice variants of certain genes may result in more reliable biomarkers for diagnosis and tumor staging and as potential therapeutic targets (15–17). The hTERT transcript is known to have seven alternative splice sites, including three deletions (Fig. 1A) and four insertions (18,19). There are several possible combinations of these alternative splice sites resulting in a large number of potential variant transcripts, but only a few have been confirmed (20). The four insertions and one deletion (β−, 182 bp) result in premature termination and nonfunctional proteins (20). The α− deletion (36 bp within the reverse transcriptase [RT] motif A) has been shown to be a dominant negative inhibitor of telomerase activity (21). The γ− deletion (189 bp) has been identified in hepatocellular carcinoma cell lines and is also believed to be nonfunctional (19). Telomerase enzyme activity has been shown by several groups to be regulated by posttranscriptional alternative splicing of hTERT (22,23). Further, the patterns of hTERT alternative splice variants are known to vary in ovary, kidney, uterine, and breast cancer, compared to corresponding adjacent normal tissues (22,24–26). However, there has been no study examining the differences in alternative splice variant patterns between benign and malignant tumors that originate from the same tissue type, or the possibility that splice variant patterns might be more specific for malignant disease than overall hTERT transcript levels.
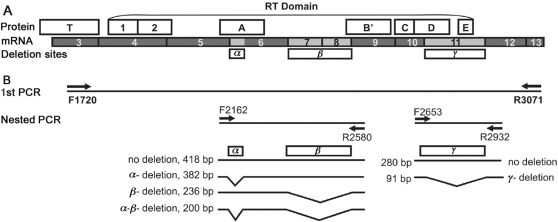
FIG. 1.
Diagram of human telomerase reverse transcriptase (hTERT) alternative splice variants. (A) Structure of hTERT deletion variants. Locations of telomerase-specific T motif, seven conserved reverse transcriptase (RT) motifs (1, 2, A, B′, C, D, and E), exons 3–13, and deletion sites are indicated. (B) The alternative splice sites (α−,β−, and γ−) are depicted with the respective resulting transcripts. Primers F1720 and R3071 were used for the first PCR reaction. Nested primers F2162 and R2580 were used to amplify the region containing the α− and β− deletions, resulting in four possible PCR products. Nested primers F2653 and R2932 were used to amplify the region containing the γ− deletion, resulting in two possible PCR products.
In this study, we examined a cohort of 133 thyroid tumors for hTERT alternative splice variant patterns, focusing on four isoforms—full-length, α− deletion, β− deletion, and α−β− deletion—to determine whether differentiating patterns exist between benign and malignant tumors. Herein, we report a significant difference in these patterns between benign and malignant thyroid tumors, with malignant tumors demonstrating more of the full-length, active isoform. In addition, because c-myc has been reported by others to be associated with hTERT gene expression, but not telomerase enzyme activity (27), we investigated the relationship of hTERT alternative splice patterns with c-myc expression. Finally, we further validated the diagnostic performance of hTERT splice patterns on a clinically relevant subset of thyroid tumors with an associated suspicious FNA diagnosis.
Materials and Methods
Tumor tissues
One hundred and thirty-three thyroid tumors were collected under Johns Hopkins Institutional Review Board approval from patients undergoing thyroid surgery. Samples included 60 malignant (28 papillary thyroid cancers, 24 follicular variant of papillary thyroid cancers, 5 follicular cancers, and 3 Hürthle cell cancers) and 73 benign lesions (31 adenomatoid nodules, 21 follicular adenomas, 12 Hürthle cell adenomas, and 9 Hashimoto's thyroiditis nodules). Follicular and Hürthle cell cancers are relatively infrequent thyroid tumors resulting in the limited sample numbers. Samples were snap frozen in liquid nitrogen and stored at −80°C until use. Among these 133 samples, a subset of 50 tumors had suspicious FNA cytology reports.
RT-PCR and nested PCR
Total RNA was isolated from each tumor with Trizol (Invitrogen, Carlsbad, CA) and purified with RNeasy Mini Kit (Qiagen, Valencia, CA). Reverse transcription was performed with 1 μg of total RNA and oligo(dT) primers by SuperScript II reverse transcriptase (Invitrogen). hTERT alternative splice variants were amplified by nested PCR using primers designed according to GenBank accession no. AF015950 (Fig. 1B). The first round of amplification spanned a region that included all α−, β−, and γ− deletion sites with forward primer F1720, 5′-GCTGCTCAGGTCTTTCTTTTAT-3′, and reverse primer R3071, 5′- GGAGGATCTTGTAGATGTTGGT-3′. PCR was performed in 25 μL of reaction mixture using 1 μL of the cDNA and Platinum Taq DNA polymerase (Invitrogen) by incubation at 94°C for 2 minutes, followed by 25 amplification cycles of 94°C for 30 seconds, 54°C for 30 seconds, and 72°C for 90 seconds, and a final extension at 72°C for 5 minutes. The second round of PCR was carried out with 1 μL of the first-round PCR product, nested primer sets, and the Platinum Taq DNA polymerase. The nested primer set for hTERT α− and β− transcript variants, forward F2162 5′-CCGCCTGAGCTGTACTTTGTC-3′ and reverse R2580 5′-CAGAGCAGCGTGGAGAGGAT-3′, produced four possible products, α+β+ (418 bp), α−β+ (382 bp), α+β− (236 bp), and α−β− (200 bp), respectively, by incubation at 94°C for 2 minutes, followed by 25 amplification cycles of 94°C for 20 seconds, 57.3°C for 20 seconds, and 72°C for 30 seconds, and a final extension at 72°C for 2 minutes. The hTERT γ− transcript was amplified using the nested primer set, forward F2653 5′-GGTGGATGATTTCTTGTTGGT-3′ and reverse R2932 5′-GGTGAGACTGGCTCTGATGG-3′, yielding two possible products, 280 (γ+) and 91 bp (γ−) in length, by incubation in a similar fashion with the exception of a different annealing temperature of 55.5°C. Amplified products were electrophoresed on 2% agarose gels with Nucleic Acid Gel Stain (Cambrex, Rockland, ME) and visualized under ultraviolet light. The densitometric value of each hTERT transcript was quantified using Quantity One image analysis software (version 4.5.2; BioRad, Hercules, CA). The relative gene expression level of each transcript was reported as a relative proportion of all the hTERT transcripts present in the same sample (28). GAPDH served as an internal control.
Statistical analysis
For analysis of the hTERT alternative splice variant data in thyroid tumors, the following comparisons were performed: (i) between malignant (n = 60) and benign (n = 73) thyroid tumors, and (ii) between malignant (n = 19) and benign (n = 31) thyroid lesions that had corresponding suspicious or indeterminate FNA cytology. These cytologies included suspicious for papillary thyroid cancer or follicular variant of papillary thyroid cancer, thyroid neoplasm, follicular neoplasm, Hürthle cell neoplasm, and neoplasm. Because the data were recorded as the proportion of transcripts in each respective gel lane (full-length, α−, and β−/α−β− deletion), a comparison of equal proportions between tumor types was done. This comparison was based on a standardized difference statistic in multinomial probabilities and tested using a permutation approach. For the purpose of analysis, the α−β− deletion was considered in the same category as the β− deletion because both variants produce nonfunctional proteins.
Receiver operating characteristic analysis
A receiver operating characteristic (ROC) analysis was done to evaluate the use of relative proportions of hTERT splice variants to classify tumors as either benign or malignant. The following three splice variants were quantified: (i) full-length hTERT transcript, (ii) α− deletion transcript, and (iii) β−/α−β− deletion transcript (β−/α−β− deletion was defined as the sum of relative proportions for β− and α−β− deletion transcripts). Since the three ROC curves corresponding to each transcript (full, α− and β−/α−β− deletions) were from the same sample, the method of Delong et al. (29) was implemented for the comparison of estimated areas under each curve. Once a transcript variant was identified as a preferable diagnostic tool, thresholds were reported for (i) simultaneously maximizing sensitivity and specificity (30) and (ii) maximizing specificity while also retaining a sensitivity greater than 50%. This second approach was chosen to minimize the probability of false positives, since FNA already provides a high level of sensitivity.
Quantitative telomerase enzyme activity assay
Telomerase enzyme activity assay was performed on a subset of 16 of the 133 samples using the Quantitative Telomerase Detection Kit (US Biomax, Ijamsville, MD) and according to the manufacturer's instructions. Briefly, for each sample, protein from twelve 10-μm cryosections was extracted in 100 μL CHAPS lysis buffer at 4°C. The protein concentration was determined using Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA). Heat-inactivated controls were performed by preincubating extracts at 85°C for 10 minutes. For each assay, 1 μg protein was added to a 25 μL quantitative telomerase detection (QTD) reaction mix. Reactions were performed in 96-well plates on an ABI prism 770-sequence detector. The extension reactions were run for 20 minutes at 25°C, followed by 40 cycles of PCR amplification, and a melting curve analysis was performed. A standard curve was constructed using a dilution series of the telomerase standard substrate provided by the manufacturer and used to calculate relative amounts of the TRAP assay product. The reaction products were then electrophoresed on a 10% polyacrylamide gel, and the telomerase hexamer ladders visualized by ethidium bromide staining.
Real-time PCR for c-myc
Real-time RT-PCR for c-myc was performed on a subset of 23 of the 133 samples using the synthesized first-strand cDNA from total RNA isolated from thyroid tumors. Assays-on-demand Gene Expression products were used for c-myc (Hs00153408_m1) and GAPDH (Hs99999905_m1) (Applied Biosystems, Foster City, CA). Reactions were performed in a 20 μL reaction volume containing 1× Taq Man universal PCR master mix (Applied Biosystems), 1× Gene expression assay mix (primers and TaqMan minor groove binder [MGB] probe dye-labeled with FAM, 6-carboxyfluorescine), and 1 μL cDNA. Reactions were performed on an ABI7300HT sequence detection system machine (Applied Biosystems). All PCR reactions were performed in triplicate. Fluorescence was quantified with the Sequence detection system software, version 2.0 (Applied Biosystems).
Results
hTERT alternative splice variant patterns in thyroid tumors
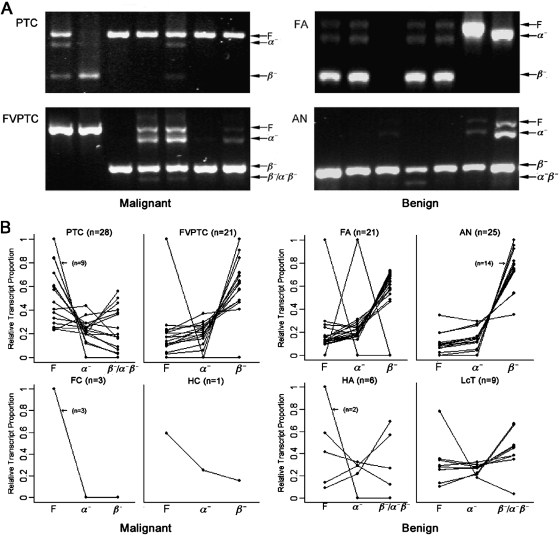
hTERT gene expression was detected in 114 of the 133 (86%) thyroid tumors (Table 1). No tumor exhibited a γ− deletion splice variant, and only 4/133 exhibited an α−β− deletion variant. Representative gels are shown in Figure 2A. The hTERT splice variant patterns present in papillary thyroid cancers, follicular variant of papillary thyroid cancers, follicular adenomas, and adenomatoid nodules (the four most common tumor types that can be suspicious on thyroid FNA) are depicted. The gels demonstrate the prominent presence of full-length hTERT gene expression in papillary thyroid cancer and the progressive loss thereof in follicular variant of papillary thyroid cancer and the benign tumors (follicular adenoma and adenomatoid nodule). There is also a concomitant gain of the inhibitory α− deletion, nonfunctional β−, and α−β− deletion patterns in the benign tumors.
Table 1.
Thyroid Tumors Analyzed for hTERT Alternative Splice Variant Patterns by Subtype
| |
Malignant (n = 60) |
Benign (n = 73) |
||||||
|---|---|---|---|---|---|---|---|---|
| Final histology | PTC (n = 28) | FVPTC (n = 24) | FC (n = 5) | HC (n = 3) | FA (n = 21) | AN (n = 31) | HA (n = 12) | LcT (n = 9) |
| hTERT positive (n = 114) | 28 | 21 | 3 | 1 | 21 | 25 | 6 | 9 |
| hTERT full-length >0.33a (n = 41) | 24 (86%) | 4 (19%) | 3 (100%) | 1 (100%) | 1 (5%) | 1 (4%) | 4 (67%) | 3 (33%) |
An hTERT full-length expression cut point of 0.33 corresponded to a specificity of 0.85 and a sensitivity of 0.60.
hTERT, human telomerase reverse transcriptase; AN, adenomatoid nodule; FA, follicular adenoma; FC, follicular carcinoma; FVPTC, follicular variant of papillary thyroid carcinoma; HA, Hürthle cell adenoma; HC, Hürthle cell carcinoma; LcT, lymphocytic thyroiditis nodule; PTC, papillary thyroid carcinoma.
FIG. 2.
Human telomerase reverse transcriptase (hTERT) alternative splice variant patterns in thyroid tumors. (A) Gels demonstrating hTERT alternative splice variant patterns present in representative thyroid tumors. One blank lane in FA represents an hTERT-negative tumor. (B) Plots of relative proportions of full-length (F), α− deletion, and β−/α−β− deletion transcripts for the eight thyroid tumor types (only tumor types that exhibited the combination of α−β− deletion are labeled β−/α−β−). AN, adenomatoid nodule; FA, follicular adenoma; FC, follicular carcinoma; FVPTC, follicular variant of papillary thyroid carcinoma; HA, Hürthle cell adenoma; HC, Hürthle cell carcinoma; LcT, lymphocytic thyroiditis nodule; PTC, papillary thyroid carcinoma.
Statistical analysis of all thyroid nodules
Overall, we found significant differences in the proportions of the various transcripts between malignant and benign thyroid tumors (p < 0.001). On average, the malignant tumors exhibited larger proportions of full-length hTERT transcripts (0.57 ± 0.15) than either the α− (0.13 ± 0.02) or β−/α−β− deletion transcripts (0.30 ± 0.11; Fig. 2B). This was true for all malignant tumor types except follicular variant of papillary thyroid cancer. In contrast, the benign tumors exhibited greater proportions of β−/α−β− deletion transcripts (0.64 ±0.08) than either the full-length (0.19 ± 0.06) or α− deletion transcripts (0.17 ± 0.02, Fig. 2B).
Analysis of suspicious thyroid nodules
In a subset analysis, we repeated our hTERT splice variant assay on 50 thyroid tumors with the preoperative diagnosis of suspicious FNA (Table 2), the cytological category most in need of additional molecular diagnostic tools. Thirty-eight of the 50 (76%) were hTERT positive. The results in this subset were similar to the original cohort, with malignant tumors exhibiting greater proportions of full-length transcripts compared to α− and β−/α−β− deletion transcripts, while among the benign tumors (with the exception of Hürthle cell adenomas), greater proportions of β−/α−β− deletion transcripts were observed compared to full-length or α− deletion transcripts.
Table 2.
hTERT Gene Expression in the Subset of Thyroid Nodules with Preoperative Suspicious FNA Diagnosis
| |
Malignant (n = 19) |
Benign (n = 31) |
||||||
|---|---|---|---|---|---|---|---|---|
| Final histology | PTC (n = 4) | FVPTC (n = 9) | FC (n = 5) | HC (n = 1) | FA (n = 14) | AN (n = 4) | HA (n = 8) | LcT (n = 1) |
| hTERT positive (n = 38) | 4 | 9 | 3 | 1 | 14 | 4 | 2 | 1 |
| hTERT full-length >0.59a (n = 11) | 4 | 1 | 3 | 1 | 1 | 0 | 1 | 0 |
An hTERT full-length expression cut point of 0.59 corresponded to a specificity of 0.90 and a sensitivity of 0.53.
hTERT, human telomerase reverse transcriptase; AN, adenomatoid nodule; FA, follicular adenoma; FC, follicular carcinoma; FNA, fine needle aspiration; FVPTC, follicular variant of papillary thyroid carcinoma; HA, Hürthle cell adenoma; HC, Hürthle cell carcinoma; LcT, lymphocytic thyroiditis nodule; PTC, papillary thyroid carcinoma.
ROC analysis
Altogether, 114 cases that were hTERT gene expression positive were included in the ROC analysis. Since malignant tumors exhibited a greater proportion of full-length transcripts, we focused on this transcript as a diagnostic tool, and this approach resulted in an area under the curve (AUC) of 0.79. Based on the simultaneous maximization method, a full-length transcript threshold of 0.22 corresponded to a sensitivity and specificity of 0.74. Similar results were observed for the 38 hTERT-positive samples from the subset with suspicious FNAs, with an estimated AUC of 0.69 and, based on a full-length threshold of 0.17, a sensitivity and specificity of 0.67.
In addition to the above approach using equal maximization of sensitivity and specificity, we also examined the full-length threshold associated with the largest observed specificity for a given sensitivity no less than 0.50. By applying these criteria to all samples, a full-length transcript threshold of 0.33 achieved a specificity of 0.85 for a given sensitivity of 0.60 (Table 1). Among the subset of suspicious thyroid nodules, a full-length threshold of 0.59 corresponded to a specificity of 0.90 for a given sensitivity of 0.53 (Table 2), thereby yielding very promising results for a diagnostic strategy that could provide a very high specificity.
Quantitative telomerase enzyme activity analysis
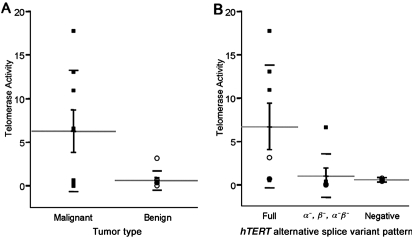
We also tested a subset of 16 thyroid tumors for functional telomerase activity. The malignant tumors (n = 8) showed significantly higher average telomerase enzyme activity (Fig. 3A) than the benign samples (n = 8) (t test; p = 0.03). Several of the thyroid cancers that exhibited minimal capsular invasion or follicular variant of papillary thyroid cancer morphology had telomerase activity values similar to the benign samples. Further, only alternative splice variant patterns showing a preponderance of full-length transcript were significantly associated with high levels of telomerase enzyme activity (χ2 test; p = 0.02; Fig. 3B).
FIG. 3.
Telomerase enzyme activity in a subset of 16 thyroid tumors. (A) Telomerase activity found in benign versus malignant tumors. (B) Telomerase activity found in tumors with different human telomerase reverse transcriptase (hTERT) splice patterns, full-length, α−, β−/α−β−, or hTERT negative. Results are plotted in arbitrary units. The long horizontal bars indicate means; vertical bars, the standard error of the means; and the short horizontal bars, standard deviations. (▪), malignant tumors; (○), benign tumors.
c-myc expression and hTERT alternative splice variant patterns
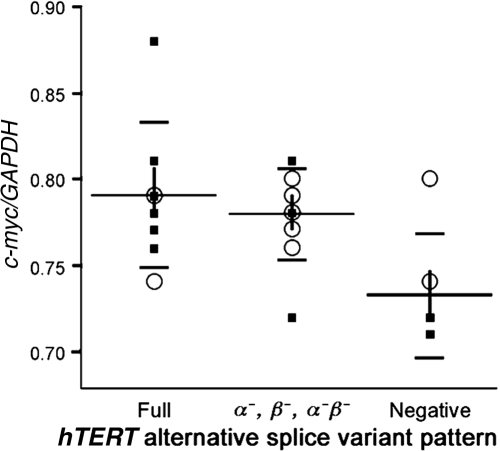
Next, we studied the correlations between c-myc and hTERT gene expression. Similar to others (31), we observed a statistically significant association between c-myc and hTERT gene expression–positive samples. However, we also documented that this correlation did not vary among the different specific splice variant patterns (Fig. 4). Of the 23 samples tested for c-myc gene expression (13 malignant and 10 benign), the following three groups were defined: (i) full-length hTERT (n = 8); (ii) α− and β−/α−β− deletion variants hTERT (n = 9); and (iii) negative hTERT (n = 6). A comparison of mean differences between each pair of groups, with respect to c-myc gene expression, was conducted based on a χ2 test. No difference was seen between full-length and α− and β−/α−β− deletion variants. However, a significant difference in mean c-myc was observed between negative and full-length hTERT groups (p = 0.003) and between negative and α− and β−/α−β− deletion variant groups (p = 0.018). These data suggest that c-myc gene expression correlates with overall hTERT gene expression, regardless of whether or not hTERT is expressed in its active form.
FIG. 4.
c-myc gene expression versus human telomerase reverse transcriptase (hTERT) alternative splice variant patterns. The ratios of c-myc and GAPDH mRNA levels determined by real-time RT-PCR are shown for the three categories of hTERT splice variants. Long horizontal bars represent means; vertical bars, the standard error of the means; and short bars, standard deviations. (▪), malignant tumors; (○), benign tumors.
Discussion
Despite the extensive efforts to identify accurate biomarkers of thyroid malignancy, the clinical need for reliable markers still exists. Alternative splicing of messenger RNA has been shown to play a critical role in tumor development, yet there has been little or no effort made to exploit this phenomenon for better cancer diagnosis (14–16). As demonstrated in our study, the presence of specific tumorigenesis-related splice variants may serve as potential biomarkers for clinical diagnosis. Others and we have examined telomerase enzyme activity and hTERT gene expression as a marker of thyroid malignancy (11,32–34). However, none have documented that telomerase on its own is a useful diagnostic marker in the differential diagnosis of benign from malignant thyroid nodules. Notably, the majority of prior studies tested only telomerase enzyme activity or overall hTERT transcripts, and did not distinguish among the different hTERT splice isoforms. In our study, however, we examined the patterns of hTERT alternative splice variants in an effort to discern differences between benign and malignant thyroid tumors. Because hTERT expression was low in most of the samples, the target concentration produced from a single conventional PCR within 30 cycles was often too low to be detected. Nonspecific products are frequently generated by increasing the amplification cycles with a single set of primers, even with a hot start. Further, quantitative real-time PCR is not applicable for the evaluation of four different hTERT isoforms. We therefore chose to use nested PCR (i) to increase the sensitivity of the assay to be able to detect each splice variant and (ii) as an effective solution to PCR nonspecificity and gene copy limitation. One major concern about nested PCR is that it does not maintain a linear relationship between the amount of final amplified product and the amount of target sequence. Studies indicate, however, that nested PCR will retain its utility for quantitation if the first-round PCR is maintained in the exponential phase (35). Indeed, quantitative nested real-time PCR assay has been developed and used in some studies without apparent distortion in the amplified product ratio (36). Further, we also optimized our nested PCR reaction using thyroid cell lines to ensure accurate product ratios (data not shown). In our study, primers specific for each of the hTERT isoforms were used in the nested PCR. Our results clearly demonstrate significant differences in the patterns of functional and nonfunctional hTERT transcripts in benign versus malignant tumors.
With the exception of follicular variant of papillary thyroid cancer, the malignant tumors exhibited a greater proportion of the hTERT full-length transcript compared to either the α−, or β−/α−β− deletions, whereas the benign tumors exhibited a greater proportion of the β−/α−β− deletion transcripts compared to the full or α− deletion (Fig. 2B). The fact that follicular variant of papillary thyroid cancer showed comparably less full-length transcript than the other thyroid malignancies is in keeping with the notion that the histological evaluation of these tumors is often problematic, with interobserver variation present up to 60% of the time (37–39). Hopefully, the application of molecular markers, such as hTERT, will allow further refinement of the classification of these thyroid tumors in the future (40).
Our primary objective for testing thyroid tumors for differences in hTERT patterns was to improve the specificity of the clinically ambiguous FNA diagnosis of suspicious thyroid lesions. In the 50 tumors that had corresponding suspicious FNA cytology, the same patterns seen in the 133 tumors were observed with the exception of Hürthle cell adenomas. Indeed, ROC analysis revealed that a full-length transcript proportion over 0.33 yielded a specificity of 85% in the diagnosis of thyroid malignancy. Further, setting the cut point of the full-length transcript proportion at 0.59 in the subset with suspicious FNA reports yielded 90% specificity. Since FNA cytology of the thyroid already provides a very high level of sensitivity, a diagnostic tool that maximizes specificity would complement the already high sensitivity of FNA cytology. Our finding of 90% specificity for the suspicious subset of thyroid nodules is indeed very promising.
Dissociation between hTERT expression and telomerase enzymatic activity was previously observed in various normal tissues and tumors (41,42). Because only overall hTERT expression was determined in those studies, the contribution of a particular hTERT transcript to the enzymatic activity was unknown. In this study, the size of most of the clinical samples available precluded assaying telomerase activity on the entire sample set, because only early stage tumors were included. In those samples we were able to assay enzymatically; however, we clearly demonstrated that telomerase activity was associated with the full-length active hTERT transcript, but not with the negative or any of the other transcript variants (Fig. 3A, B). We further clarified the correlation between c-myc expression and telomerase activity (27). Consistent with previous publications that showed c-myc regulating hTERT expression through the hTERT promoter, we demonstrated that c-myc was associated with the overall hTERT expression, but further clarified that c-myc regulation is not specific for any of the hTERT isoforms (Fig. 4). Because telomerase enzyme activity only correlates with full-length hTERT expression, our data further explain the dissociation reported in the literature between c-myc expression and telomerase enzyme activity.
In summary, our splice variant profiling results suggest that patterns of alternative splice variant patterns of hTERT may be a more powerful diagnostic tool in the distinction of benign and malignant thyroid nodules than total hTERT gene expression alone. Our findings likely represent only the beginning of the unraveling of a more complex regulation of telomerase gene expression. Further investigation on preoperative FNA biopsy samples will be required to evaluate the potential role of hTERT alternative splice variant patterns in the differential diagnosis of thyroid nodules.
Acknowledgments
We would like to thank the following individuals for their assistance in thyroid tumor collection: Kimberly Burckhardt, Filomeno D. Apor, Sidney L. Craddock, Jr., Alan Silverio, Terry Emerson, and Dante Trusty.
This work has been partially supported by NIH Grant R21CA81162.
Disclosure Statement
Dr. Martha Zeiger works as a consultant for Veracyte, Inc. The other authors have no competing financial interests.
References
- 1.Baloch ZW. Fleisher S. LiVolsi VA. Gupta PK. Diagnosis of “follicular neoplasm”: a gray zone in thyroid fine-needle aspiration cytology. Diagn Cytopathol. 2002;26:41–44. doi: 10.1002/dc.10043. [DOI] [PubMed] [Google Scholar]
- 2.Yoder BJ. Redman R. Massoll NA. Validation of a five-tier cytodiagnostic system for thyroid fine needle aspiration biopsies using cytohistologic correlation. Thyroid. 2006;16:781–786. doi: 10.1089/thy.2006.16.781. [DOI] [PubMed] [Google Scholar]
- 3.Hayat MJ. Howlader N. Reichman ME. Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12:20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 4.Gharib H. Papini E. Thyroid nodules: clinical importance, assessment, and treatment. Endocrinol Metab Clin North Am. 2007;36:707–735. doi: 10.1016/j.ecl.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Chen H. Zeiger MA. Clark DP. Westra WH. Udelsman R. Papillary thyroid cancer: can operative management be solely based on fine needle aspiration? J Am Coll Surg. 1997;184:605–610. [PubMed] [Google Scholar]
- 6.Carling T. Udelsman R. Follicular neoplasms of the thyroid: what to recommend. Thyroid. 2005;15:583–587. doi: 10.1089/thy.2005.15.583. [DOI] [PubMed] [Google Scholar]
- 7.Udelsman R. Westra WH. Donovan PI. Sohn TA. Cameron JL. Randomized prospective evaluation of frozen-section analysis for follicular neoplasms of the thyroid. Ann Surg. 2001;233:716–722. doi: 10.1097/00000658-200105000-00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mechanick JI. Carpi A. Progress in the preoperative diagnosis of thyroid nodules: managing uncertainties and the ultimate role for molecular investigation. Biomed Pharmacother. 2006;60:396–404. doi: 10.1016/j.biopha.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Cheng AJ. Lin JD. Chang T. Wang TC. Telomerase activity in benign and malignant human thyroid tissues. Br J Cancer. 1998;77:2177–2180. doi: 10.1038/bjc.1998.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umbricht CB. Saji M. Westra WH. Udelsman R. Zeiger MA. Sukumar S. Telomerase activity: a marker to distinguish follicular thyroid adenoma from carcinoma. Cancer Res. 1997;57:2144–2147. [PubMed] [Google Scholar]
- 11.Saji M. Westra WH. Chen H. Umbricht CB. Tuttle RM. Box MF. Udelsman R. Sukumar S. Zeiger MA. Telomerase activity in the differential diagnosis of papillary carcinoma of the thyroid. Surgery. 1997;122:1137–1140. doi: 10.1016/s0039-6060(97)90219-1. [DOI] [PubMed] [Google Scholar]
- 12.Saji M. Xydas S. Westra WH. Liang CK. Clark DP. Udelsman R. Umbricht CB. Sukumar S. Zeiger MA. Human telomerase reverse transcriptase (hTERT) gene expression in thyroid neoplasms. Clin Cancer Res. 1999;5:1483–1489. [PubMed] [Google Scholar]
- 13.Ito Y. Yoshida H. Tomoda C. Uruno T. Takamura Y. Miya A. Kobayashi K. Matsuzuka F. Kuma K. Miyauchi A. Telomerase activity in thyroid neoplasms evaluated by the expression of human telomerase reverse transcriptase (hTERT) Anticancer Res. 2005;25:509–514. [PubMed] [Google Scholar]
- 14.Kalnina Z. Zayakin P. Silina K. Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 15.Skotheim RI. Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int J Biochem Cell Biol. 2007;39:1432–1449. doi: 10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 18.Kilian A. Bowtell DD. Abud HE. Hime GR. Venter DJ. Keese PK. Duncan EL. Reddel RR. Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 19.Hisatomi H. Ohyashiki K. Ohyashiki JH. Nagao K. Kanamaru T. Hirata H. Hibi N. Tsukada Y. Expression profile of a gamma-deletion variant of the human telomerase reverse transcriptase gene. Neoplasia. 2003;5:193–197. doi: 10.1016/S1476-5586(03)80051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wick M. Zubov D. Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Gene. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- 21.Colgin LM. Wilkinson C. Englezou A. Kilian A. Robinson MO. Reddel RR. The hTERT alpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia. 2000;2:426–432. doi: 10.1038/sj.neo.7900112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Y. Liu Z. Fang X. Ge Z. Ge N. Jia Y. Sun P. Lou F. Bjorkholm M. Gruber A. Ekman P. Xu D. Differential expression of full-length telomerase reverse transcriptase mRNA and telomerase activity between normal and malignant renal tissues. Clin Cancer Res. 2005;11:4331–4337. doi: 10.1158/1078-0432.CCR-05-0099. [DOI] [PubMed] [Google Scholar]
- 23.Ulaner GA. Hu J-F. Vu TH. Ciudice LC. Hoffman AR. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]
- 24.Ulaner GA. Hu JF. Vu TH. Oruganti H. Giudice LC. Hoffman AR. Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int J Cancer. 2000;85:330–335. [PubMed] [Google Scholar]
- 25.Yokoyama Y. Wan X. Takahashi Y. Shinohara A. Tamaya T. Alternatively spliced variant deleting exons 7 and 8 of the human telomerase reverse transcriptase gene is dominantly expressed in the uterus. Mol Hum Reprod. 2001;7:853–857. doi: 10.1093/molehr/7.9.853. [DOI] [PubMed] [Google Scholar]
- 26.Zaffaroni N. Della Porta C. Villa R. Botti C. Buglioni S. Mottolese M. Grazia Daidone M. Transcription and alternative splicing of telomerase reverse transcriptase in benign and malignant breast tumours and in adjacent mammary glandular tissues: implications for telomerase activity. J Pathol. 2002;198:37–46. doi: 10.1002/path.1178. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg RA. O'Hagan RC. Deng H. Xiao Q. Hann SR. Adams RR. Lichtsteiner S. Chin L. Morin GB. DePinho RA. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- 28.Brambilla C. Folini M. Gandellini P. Daprai L. Daidone MG. Zaffaroni N. Oligomer-mediated modulation of hTERT alternative splicing induces telomerase inhibition and cell growth decline in human prostate cancer cells. Cell Mol Life Sci. 2004;61:1764–1774. doi: 10.1007/s00018-004-4062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLong ER. DeLong DM. Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 30.Gallop RJ. Crits-Christoph P. Muenz LR. Tu XM. Determination and interpretation of the optimal operating point for ROC curves derived through generalized linear models. Understanding Stat. 2003;2:219–242. [Google Scholar]
- 31.Wu KJ. Grandori C. Amacker M. Simon-Vermot N. Polack A. Lingner J. Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 32.Umbricht CB. Conrad GT. Clark DP. Westra WH. Smith DC. Zahurak M. Saji M. Smallridge RC. Goodman S. Zeiger MA. Human telomerase reverse transcriptase gene expression and the surgical management of suspicious thyroid tumors. Clin Cancer Res. 2004;10:5762–5768. doi: 10.1158/1078-0432.CCR-03-0389. [DOI] [PubMed] [Google Scholar]
- 33.Mora J. Lerma E. Telomerase activity in thyroid fine needle aspirates. Acta Cytol. 2004;48:818–824. doi: 10.1159/000326452. [DOI] [PubMed] [Google Scholar]
- 34.Liou MJ. Chan EC. Lin JD. Liu FH. Chao TC. Human telomerase reverse transcriptase (hTERT) gene expression in FNA samples from thyroid neoplasms. Cancer Lett. 2003;191:223–227. doi: 10.1016/s0304-3835(02)00678-x. [DOI] [PubMed] [Google Scholar]
- 35.Haff LA. Improved quantitative PCR using nested primers. PCR Methods Appl. 1994;3:332–337. doi: 10.1101/gr.3.6.332. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi T. Tamura M. Takahashi SN. Matsumoto K. Sawada S. Yokoyama E. Nakayama T. Mizutani T. Takasu T. Nagase H. Quantitative nested real-time PCR assay for assessing the clinical course of tuberculous meningitis. J Neurol Sci. 2007;255:69–76. doi: 10.1016/j.jns.2007.01.071. [DOI] [PubMed] [Google Scholar]
- 37.Zeiger MA. Dackiw AP. Follicular thyroid lesions, elements that affect both diagnosis and prognosis. J Surg Oncol. 2005;89:108–113. doi: 10.1002/jso.20186. [DOI] [PubMed] [Google Scholar]
- 38.Renshaw AA. Gould EW. Why there is the tendency to “overdiagnose” the follicular variant of papillary thyroid carcinoma. Am J Clin Pathol. 2002;117:19–21. doi: 10.1309/CJEU-XLQ7-UPVE-NWFV. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd RV. Erickson LA. Casey MB. Lam KY. Lohse CM. Asa SL. Chan JK. DeLellis RA. Harach HR. Kakudo K. LiVolsi VA. Rosai J. Sebo TJ. Sobrinho-Simoes M. Wenig BM. Lae ME. Observer variation in the diagnosis of follicular variant of papillary thyroid carcinoma. Am J Surg Pathol. 2004;28:1336–1340. doi: 10.1097/01.pas.0000135519.34847.f6. [DOI] [PubMed] [Google Scholar]
- 40.Golub TR. Slonim DK. Tamayo P. Huard C. Gaasenbeek M. Mesirov JP. Coller H. Loh ML. Downing JR. Caligiuri MA. Bloomfield CD. Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 41.Tahara H. Yasui W. Tahara E. Fujimoto J. Ito K. Tamai K. Nakayama J. Ishikawa F. Ide T. Immuno-histochemical detection of human telomerase catalytic component, hTERT, in human colorectal tumor and non-tumor tissue sections. Oncogene. 1999;18:1561–1567. doi: 10.1038/sj.onc.1202458. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura Y. Tahara E. Tahara H. Yasui W. Ide T. Quantitative reevaluation of telomerase activity in cancerous and noncancerous gastrointestinal tissues. Mol Carcinog. 1999;26:312–320. [PubMed] [Google Scholar]