Abstract
Tetraploidy can constitute a metastable intermediate between normal diploidy and oncogenic aneuploidy. Here, we show that the absence of p53 is not only permissive for the survival but also for multipolar asymmetric divisions of tetraploid cells, which lead to the generation of aneuploid cells with a near-to-diploid chromosome content. Multipolar mitoses (which reduce the tetraploid genome to a sub-tetraploid state) are more frequent when p53 is downregulated and the product of the Mos oncogene is upregulated. Mos inhibits the coalescence of supernumerary centrosomes that allow for normal bipolar mitoses of tetraploid cells. In the absence of p53, Mos knockdown prevents multipolar mitoses and exerts genome-stabilizing effects. These results elucidate the mechanisms through which asymmetric cell division drives chromosomal instability in tetraploid cells.
Keywords: aneuploidy, apoptosis, centrosome, colon carcinoma, mitotic catastrophe
Introduction
Tetraploid cells are detected in some precancerous lesions such as Barrett's oesophagus and cervical dysplasia, where their presence coexists with the loss of functional p53 (Heselmeyer et al, 1996; Maley et al, 2004). Owing to the increase in the number of chromosomes, perhaps coupled to changes in the geometry of the mitotic machinery (Storchova et al, 2006; Storchova and Kuffer, 2008), tetraploid cells frequently activate the DNA damage response and become genomically unstable. Thus, tetraploidy may be considered as a metastable state that links normal diploidy to cancer-associated aneuploidy (Storchova and Pellman, 2004; Fujiwara et al, 2005; Margolis, 2005).
Numerous tumour suppressor genes including p53 (Margolis, 2005), BRCA1 (Schlegel et al, 2003), LATS2 (Aylon et al, 2006) and APC (Tighe et al, 2004) actively repress tetraploidy, meaning that their removal can either stimulate the spontaneous tetraploidization of cells or facilitate the survival of tetraploid cells generated upon cytokinesis or karyokinesis inhibition. The former has been shown for the knockdown of APC in cultured cells and for the conditional knockout of APC in small intestine epithelia in vivo (Caldwell et al, 2007). The latter has been demonstrated in cultured cancer cell lines that were depleted from p53 by gene knockout or RNA interference (Cross et al, 1995; Andreassen et al, 2001; Castedo et al, 2006a, 2006b), as well as in p53−/− primary mouse mammary epithelial cells (Fujiwara et al, 2005; Senovilla et al, 2009). Moreover, the expression of some oncogenes including Myc (Yin et al, 1999), Aurora-A (Wang et al, 2006) and human papillomavirus (HPV)-encoded E6 (Incassati et al, 2006) can stimulate tetraploidization.
The mechanisms through which tetraploidy favours oncogenesis are complex and have not yet been entirely elucidated. One single tetraploid cell can undergo multipolar mitosis, which often leads to the generation of three or more daughter cells (Storchova and Pellman, 2004). This process causes the near-to-stochastic distribution of chromosomes and hence is lethal for most daughter cells. Nullisomy (the total absence of one particular chromosome) and polysomy (the presence of extra copies of one chromosome), indeed, result in major genetic defects involving the incorrect assembly of multiprotein complexes and fatal linkage disequilibria, which are rarely compatible with cell survival (Zhivotovsky and Kroemer, 2004; Roumier et al, 2005; Ganem et al, 2007). Moreover, during mitosis, the presence of more than two centrosomes in tetraploid cells can also favour merotelic chromosome attachments and hence chromosomal lagging, which may favour chromosome loss or asymmetric distribution among daughter cells, even when the division is bipolar (Ganem et al, 2009).
Multipolar and asymmetric cell division, as they can result from tetraploidy (Levine et al, 1991; Ganem et al, 2007), are commonly observed in malignant lesions and have been suspected to contribute to oncogenesis for over a century (Boveri, 2008). Supernumerary centrosomes, as they are detected in malignant cells (Levine et al, 1991; D'Assoro et al, 2002), can be induced experimentally, and this reportedly suffices to trigger oncogenesis (Basto et al, 2008; Gergely and Basto, 2008). Moreover, ‘anisocytosis' and ‘anisokaryosis' (heterogeneity in cell size and nuclear size, respectively), which presumably result from asymmetric divisions, are well-established histological hallmarks of malignancy (Boveri, 2008; Holland and Cleveland, 2009). It is interesting to note that in some cancer types (e.g., non-small cell lung cancer), these morphological traits of malignancy correlate with the expression of one particular oncogene, Mos (Gorgoulis et al, 2001).
Mos (also called c-Mos) is the first human oncogene cloned, and has been identified as the cellular homologue of the viral oncogene v-Mos, which is encoded by the Moloney murine sarcoma virus (Oskarsson et al, 1980; Watson et al, 1982). The p39Mos protein (hereafter referred to as Mos) has been shown to stimulate the transformation of murine fibroblasts in vitro (Okazaki and Sagata, 1995; Fukasawa and Vande Woude, 1997). Moreover, transfection-enforced overexpression of Mos reportedly inhibits mitotic progression (Wang et al, 1994) and causes the generation of binucleated cells due to the inhibition of cytokinesis (Okazaki et al, 1992; Fukasawa and Vande Woude, 1995). Apparently, Mos can stabilize another oncogene product, c-Fos, (Okazaki and Sagata, 1995) and enhance the expression of cyclins (Rhodes et al, 1997), thereby stimulating cell proliferation. The genetic invalidation of Mos has no obvious phenotypic consequences in mice (Colledge et al, 1994; Hashimoto et al, 1994). However, although Mos−/− male mice exhibit normal reproduction rates, Mos−/− female are nearly infertile (Colledge et al, 1994; Hashimoto et al, 1994), in line with the fact that Mos is strictly necessary for the first meiotic division of oocytes (Sagata et al, 1989a), and then exerts a critical checkpoint function during metaphase II (Sagata et al, 1989b). Both the meiosis-regulatory and the transforming effects of Mos require its serine–threonine kinase activity (Haccard et al, 1993; Okazaki and Sagata, 1995). Known Mos substrates include cyclin B2, tubulin and MEK1 (Roy et al, 1990; Zhou et al, 1991; Sagata, 1997), and the meiotic checkpoint function of Mos depends on its capacity to activate the mitogen-activated protein kinase (MAPK) pathway (Haccard et al, 1993). Thus, very little is known about the role of Mos in somatic cells and on the mechanisms by which Mos can act as an oncogene.
Here, we developed a cellular model of aneuploidization in which p53−/− cells were driven into tetraploidy, which was followed by multipolar mitosis and re-acquisition of a near-to-diploid chromosome content. We found that Mos was upregulated in p53-deficient tetraploid cells and that it was strictly required for the occurrence of multipolar divisions, presumably because Mos acts as an inhibitor of centrosome coalescence.
Results and discussion
Generation of sub-tetraploid derivatives from p53-deficient tetraploid cells
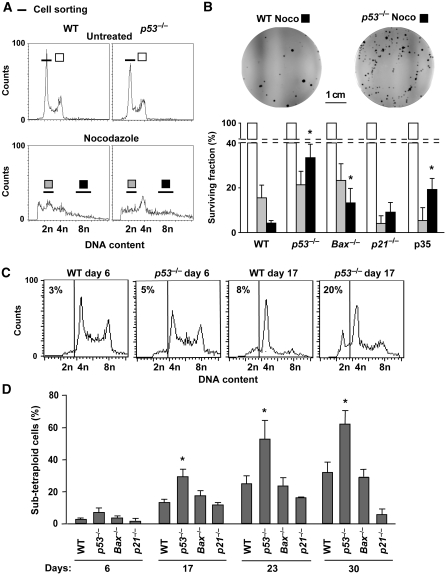
Shortly (2 days) after a 48 h-long treatment with the microtubule poison nocodazole or the cytokinesis inhibitor cytochalasin D, p53−/− human colon carcinoma HCT 116 cell cultures contained a higher fraction of polyploid cells (with a ⩾4n DNA content), yet a lower amount of dying and dead cells (exhibiting the dissipation of the mitochondrial transmembrane potential (ΔΨm) and the breakdown of plasma membranes, respectively) (Kroemer et al, 2007; Galluzzi et al, 2009) than their p53-proficient counterparts (Figure 1A and Supplementary Figure S1). Moreover, fluorescence-activated cell sorter (FACS)-purified cells with an ∼8n DNA content formed colonies more efficiently when they did not express p53 than when they did so (Figure 1B), in line with the notion that p53 deficiency is permissive for the generation and survival of tetraploid cells (Castedo et al, 2006b). Cultures derived from FACS-purified cells with an ∼8n DNA content were characterized by a 4n DNA content in the G1 phase of the cell cycle and by an 8n DNA content in the G2 and M phases, thereby exhibiting bona fide traits of tetraploidy. However, after several passages, p53−/− (but neither p21−/− nor Bax−/−) cultures progressively accumulated a population of cells with a ∼2n DNA content (Figure 1C and D). This was observed in six independent experiments in which the initial contamination with ∼2n cells (measured immediately after FACS purification) was undetectable. To understand the origin of such ∼2n population, we followed the fate of tetraploid cells 1 week after their generation by videomicroscopy, and found that p53−/− cells underwent multipolar (mostly tri- or tetrapolar) divisions—which are associated with Y- and X-shaped metaphases (Figure 2A)—much more frequently than p53+/+ control cells (Figure 2B and Supplementary Videos 1 and 2). These results suggest that cells with a ∼2n DNA content (hereafter referred to as ‘sub-tetraploid') appearing at significant frequencies (⩾10%), as early as 15 days after tetraploidization, might result from a peculiar process of multipolar division. A significant percentage of daughter cells that originated from p53−/− tetraploid cells by multipolar mitosis could enter and terminate normal bipolar divisions (Figure 2C and D, and Supplementary Video 3), suggesting that such sub-tetraploid cells can give rise to a new lineage. The reduction of the chromosomal content was significantly more frequent among p53−/− tetraploid cells than among control ones (Figures 1 and 2), and, in another cell line, was exacerbated by pharmacological inhibition of p53 by cyclic pifithrin-α (Komarov et al, 1999) (Supplementary Figure S2). Thus, the tumour suppressor p53 reduces the probability of sub-tetraploidy.
Figure 1.
Effect of p53 on the survival and on the genomic stability of tetraploid HCT 116 cells. (A, B) The absence of p53 increases the clonogenic survival of freshly generated polyploid cells. Wild type (WT), p53−/−, Bax−/− and p21−/− diploid human colon carcinoma HCT 116 cells and HCT 116 cells stably transfected with a plasmid encoding the baculoviral inhibitor of caspases p35 (p35) were left untreated or treated with 100 nM nocodazole (Noco) for 48 h. After washing, cells were cultured for additional 48 h in drug-free culture medium, then stained with Hoechst 33342, followed by fluorescence-activated cell sorter (FACS) purification of diploid (2n, white and grey symbols for untreated and nocodazole-treated cells, respectively) or polyploid (>4n, black symbols) cell populations. In A, representative cell cycle distributions of WT and p53−/− cells from one out of three independent experiments are shown. X-axis = Hoechst 33342 fluorescence (DNA content); Y-axis = cell number per channel (counts). In B, the results of clonogenic survival assays carried out on such FACS-purified cells are reported. The upper part shows representative pictures of colonies formed by WT and p53−/− cells as observed upon crystal violet staining 10 days after FACS purification. Columns depict the survival fraction (mean±s.e.m., n=3 parallel wells, normalized for plating efficiency and to control diploid cells) of the diploid and polyploid cell populations with the indicated genotype and corresponding to the FACS-purified populations represented in A. Asterisks indicate statistically significant differences as compared to WT tetraploid cells (Student's t-test, P<0.05). (C, D) Genomic instability of viable polyploid cells. Diploid HCT 116 cells with the indicated genotype were treated for 48 h with nocodazole and tetraploid populations were FACS-purified as in A (>4n, black symbols). Cells then were cultured for the indicated number of days and analyzed for DNA content. Representative cell cycle profiles of WT and p53−/− tetraploid cells are shown in C, whereas quantitative data are reported in D (mean±s.e.m., n=6 independent determinations). In C, the percentages of cells with a sub-tetraploid DNA content (indicating chromosomal loss) are indicated. Statistical significance resulting from the comparison to WT cells is highlighted by asterisks (Student's t-test, P<0.05).
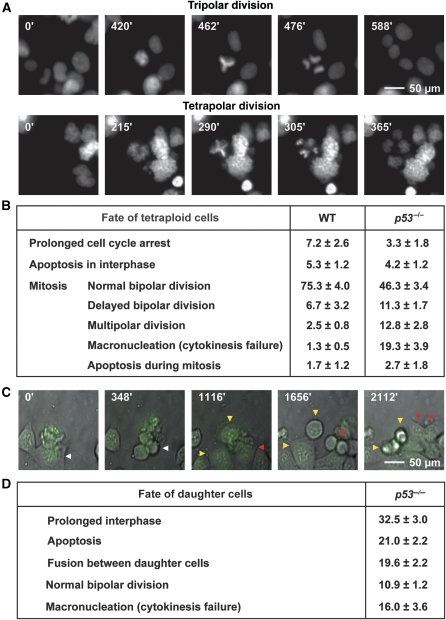
Figure 2.
Multipolar divisions of p53-deficient tetraploid cells. Isogenic wild type (WT) and p53−/− diploid HCT 116 cells expressing a histone 2B–green fluorescent protein (H2B–GFP) chimera were treated for 48 h with nocodazole and then cultured in drug-free medium for additional 48 h. Polyploid cells were fluorescence-activated cell sorter (FACS)-sorted as in Figure 1A (>4n, black symbols) and monitored by fluorescence videomicroscopy for 48 h. In A snapshots taken at the indicated time points exemplify the appearance of tripolar and tetrapolar mitosis, thus exhibiting the representative Y- and X-shaped metaphase, respectively. Full-length movies proving the completion of cell division are available as Supplementary Videos S1 and S2. B reports the percentage of cell cycle arrest, apoptosis induction and various mitotic aberrations as quantified among 150 to 200 mitoses for each genotype (mean ± s.e.m., n=3 independent experiments). In C representative images captured at the indicated time points show that some daughter cells that originated from p53−/− tetraploid HCT 116 cells by multipolar mitosis can enter and terminate normal bipolar divisions (red arrows), whereas others undergo apoptosis (yellow arrows). A full-length movie that demonstrates the completion of a normal cell division by such a daughter cell is available as Supplementary Video S3. D reports the percentage of these cells (arisen from p53−/− tetraploid HCT 116 cells by multipolar mitosis) that underwent the indicated fate by the end of the experiment (as quantified among 150-200 daughter cells, mean±s.e.m., n=3 independent experiments). Note that only cells entering multipolar mitosis within the first 18 h of the assays were tracked for the subsequent 30 h. A full-colour version of this figure is available at The EMBO Journal Online.
Sub-tetraploidy is linked to centrosome defects and aneuploidy
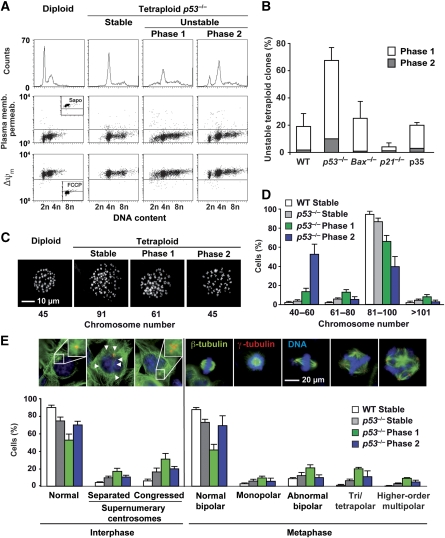
Next, we generated multiple tetraploid clones from isogenic HCT 116 cells. As compared with wild type (WT), Bax−/−, p21−/− or apoptosis-resistant (owing to the expression of the caspase inhibitor p35 from Baculovirus) tetraploid cells, p53−/− tetraploid clones displayed some difficulties in maintaining a stable tetraploid genome. Four weeks after cloning, indeed, only one-third of p53−/− tetraploid clones still exhibited a clean tetraploid DNA content profile, whereas the other two-thirds accumulated viable sub-tetraploid populations (Figure 3A and B), first as rather broad shoulders (‘phase 1') and later as sharper peaks (‘phase 2') of sub-tetraploid cells. Such sharper peaks presumably arise in cultures that are dominated by one or few viable sub-clones with a comparable sub-tetraploid DNA content. Phase 1 and phase 2 unstable sub-tetraploid populations also arose after repeated transfection (approximately once every 5 days) with a small interfering RNA (siRNA) targeting p53 (Supplementary Figure S2). Chromosome counting confirmed that phase 2 unstable p53−/− tetraploid cells frequently contained a roughly diploid number of chromosomes (Figure 3C and D). Phase 1 unstable p53−/− tetraploid cells were characterized by a high frequency of aberrant mitoses that were either monopolar, bipolar characterized by lagging chromosomes or multipolar linked to supernumerary centrosomes. Multipolar mitoses were also more frequent among phase 2 unstable tetraploids as compared to WT or stable p53−/− tetraploids (Figure 3E). As compared to phase 1 cells, phase 2 cells proliferated more quickly, and among them the sub-tetraploid population had shorter duplication times and shorter mitoses than the tetraploid one (Supplementary Figure S3). This explains the outgrowth of sub-tetraploid cells over their tetraploid counterparts. When such sub-tetraploid cells were exposed to nocodazole, they could again tetraploidize and then revert once more to sub-tetraploidy, but this process of reversion was not accelerated (Supplementary Figure S4).
Figure 3.
Chromosome instability and centrosome amplification in p53−/− tetraploid HCT 116 clones. (A, B) p53 deficiency increases the percentage of unstable tetraploid clones. Tetraploid HCT 116 clones were generated from wild type (WT), p53−/−, Bax−/−, p21−/− and p35-expressing parental cells as described in Supplementary Data. Cell cycle distribution and apoptosis-related parameters were evaluated 4 weeks after cloning by multiparametric cytofluorometry upon staining with Hoechst 33342 (which measures DNA content), the mitochondrial transmembrane potential (ΔΨm)-sensitive dye 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3)) and propidium iodide (PI, an exclusion dye that only stains dead cells). Representative plots are shown in A, the inserts therein showing positive controls for ΔΨm dissipation (as obtained by treating the cells for 30 min with 100 μM protonophore carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, FCCP) and plasma membrane permeabilization (as resulting from a 2 min-long incubation in 0.5% (w/v) saponin (Sapo)). Tetraploid clones were classified according to genomic instability (as evaluated by the quantification of viable sub-tetraploid populations) in stable (sub-tetraploid cells <10%) and unstable (sub-tetraploid cells >10%). Unstable clones were subdivided into ‘phase 1' and ‘phase 2' clones, characterized by broad and sharp sub-tetraploid peaks, respectively. B reports the frequency of unstable clones generated from parental tetraploid cells of the indicated genotype. Frequency was calculated among 50–100 clones for each genotype (mean±s.e.m., n=3 independent series of clones). (C, D) Chromosome count in tetraploid clones. Representative examples of 4′,6 diamidino-2-phenylindole (DAPI)-stained metaphase spreads with the corresponding number of chromosomes are shown in C. For WT and p53−/− clones of the indicated type, D reports the percentage of cells containing 40–60, 61–80, 81–100 or >101 chromosomes, as quantified among 100 metaphases per condition (mean±s.e.m., n=4 different clones in independent assessments) (E) Centrosome amplification correlates with genomic instability. WT and p53−/− clones of the indicated class were cultured on glass coverslips and subjected to immunofluorescence detection of mitotic spindles (β-tubulin staining, green fluorescence) and centrosomes (γ-tubulin staining, red fluorescence). Nuclei were counterstained with Hoechst 33342 (emitting in blue). According to the number of centrosomes, interphase cells were divided in normal (1 or 2 centrosomes) and abnormal (more than 2 centrosomes, either congressed in a single pole or not), whereas metaphases were classified as monopolar (2 congressed centrosomes), normal and abnormal bipolar (2 centrosomes, the latter exhibiting the misalignment of one or more chromosomes), and multipolar (tripolar, tetrapolar or of higher-order polarity, characterized by 3, 4 or more centrosomes, respectively). Representative immunofluorescence microscopy images of each category are shown. The percentage of occurrence of each category is reported, as quantified among 150 to 200 cells for WT and p53−/− clones of the indicated type (mean±s.e.m., n=4 distinct clones). A full-colour version of this figure is available at The EMBO Journal Online.
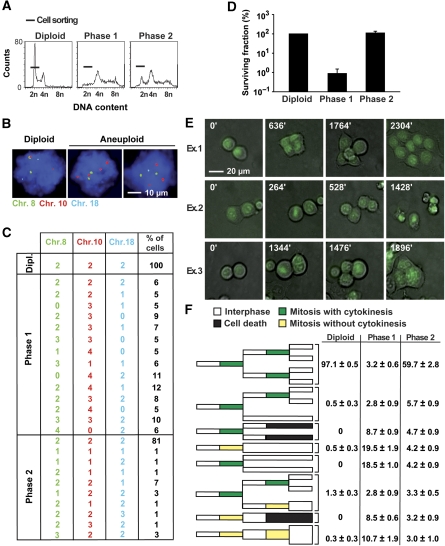
Fluorescence in situ hybridization (FISH) carried out during interphase proved that a large portion of sub-tetraploid cells (which were FACS-purified from unstable p53−/− tetraploid clones) was aneuploid (Figure 4A-C). Thus, especially in phase 1 cultures, nullisomies (which are always lethal) were frequently detected (Figure 4C). Accordingly, most (>99%) of such phase 1 sub-tetraploid cells failed to form stable offspring in clonogenic assays and died (Figure 4D). In phase 2 cultures, the frequency of aneuploid cells was lower, and nullisomies were infrequent (Figure 4C), presumably because viable cells (which efficiently form clones, Figure 4D) had been positively selected. To further explore the behaviour of sub-tetraploid cells, we generated phase 1 and phase 2 clones from p53−/− tetraploid cells expressing histone H2B fused to the N-terminus of green fluorescent protein (H2B–GFP, which labels chromatin in green and hence allows for monitoring chromosome movement), and followed the fate of their sub-tetraploid derivatives after FACS purification (Figure 4E, F and Supplementary Figure S5, and Supplementary Videos 4, 5 and 6). Although phase 1 sub-tetraploid cells rarely (∼3%) engaged in subsequent cycles of division, phase 2 sub-tetraploid cells did so much more frequently (∼60%). Nonetheless, cell cycle blockade, mitosis without cytokinesis and cell death occurred more frequently among sub-tetraploid cells than among their diploid progenitors (Figure 4E, F and Supplementary Figure S5, and Supplementary Videos 4, 5 and 6).
Figure 4.
Characterization and fate of the sub-tetraploid offspring of p53−/− tetraploid cells. (A–C) An important fraction of sub-tetraploid cells are aneuploid. Sub-tetraploid cells derived from phase 1 and 2 unstable p53−/− tetraploid HCT 116 clones or the 2n population of the parental p53−/− diploid cell line were fluorescence-activated cell sorter (FACS)-purified as indicated in A. Thereafter, nuclei from the sorted populations were subjected to fluorescence in situ hybridization (FISH) with probes specific for the centromeric region of chromosome 8, 10 and 18 (green, red and blue fluorescence, respectively). Representative immunofluorescence microphotographs are shown in B. In C the percentage of wild type (WT) diploid or p53−/− sub-tetraploid cells characterized by the indicated chromosomal setup is reported, as scored among ⩾100 metaphases for each cell type (mean±s.e.m., n=3 independent determinations). Alternatively, FACS-purified cells obtained as in A were subjected to clonogenic survival assays (D). Columns depict the survival fraction (mean±s.e.m., n=3 independent experiments carried out in triplicate with at least three distinct clones for each type) of WT diploid or sub-tetraploid cells derived from p53−/− tetraploid clones of the indicated class. (E, F) Videomicroscopy of sub-tetraploid cells. p53−/− diploid cells and tetraploid HCT 116 clones expressing a histone 2B–green fluorescent protein chimera (H2B–GFP) were subjected to FACS-purification of diploid or sub-tetraploid populations as in A. Purified cells with a ∼2n DNA content were then monitored by fluorescence videomicroscopy for 40 h. Representative snapshots taken at the indicated time points illustrate sub-tetraploid cells that undergo two consecutive rounds of symmetric division (example 1), divide into two daughter cells that die by apoptosis (example 2), or undergo mitosis followed by incomplete cytokinesis, resulting in increased ploidy (example 3). Full-length movies can be found in Supplementary Videos S4, S5 and S6. F provides a graphic representation and quantitative data (%, mean±s.e.m., n=3 independent experiment, 100-150 cells analyzed) about the fate of WT diploid or p53−/− sub-tetraploid cells derived from clones of the indicated type. Only cells entering mitosis within the first 10 h were tracked for subsequent 30 h. Please note that cytokinesis failure (leading to an increase in ploidy as well as in cell volume) is depicted with a duplication in bar thickness. The entire catalogue of the observed cell fates is available in Supplementary Figure S5.
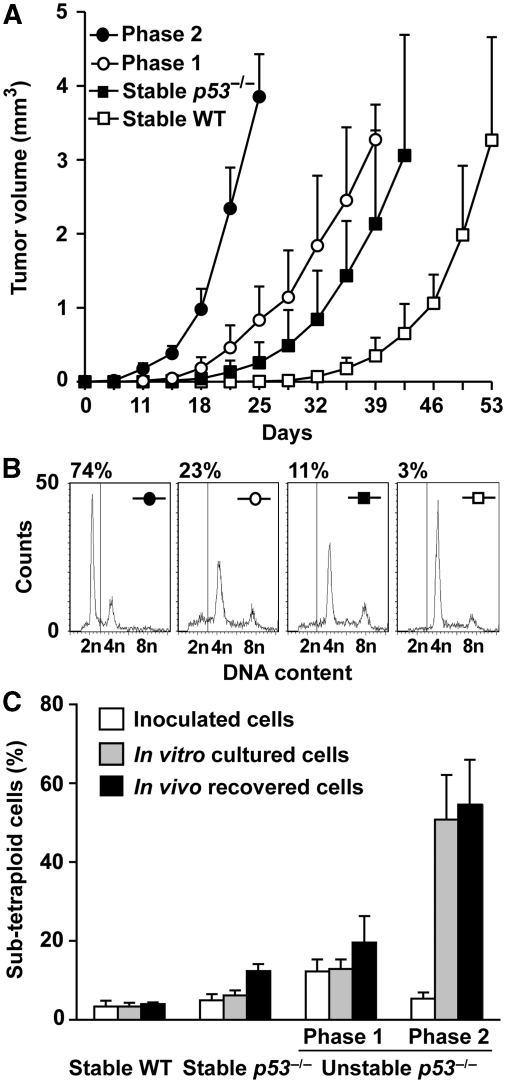
It is important to note that upon inoculation into immunodeficient (nu/nu) mice, tetraploid cells FACS-purified from unstable tetraploid clones (and in particular established phase 2 cultures) formed tumours more rapidly than stable (p53−/− and p53+/+) tetraploid cells (Figure 5A), corroborating the idea that multipolar divisions might favour the selection of aggressive tumour variants. After in vivo selection, sub-tetraploid tumour cells could be recovered at similar frequencies as after an equivalent period of in vitro culture (Figure 5B and C) and were particularly frequent among tumours that arose from unstable phase 2 clones. Thus, it appears that the tetraploidization of p53−/− cells can accelerate tumour progression in the context of the emergence of a sub-tetraploid cancer cell population arising from multipolar mitoses.
Figure 5.
Phase 2 unstable p53−/− tetraploid clones form aggressive tumours. (A) Nude mice were subcutaneously xenografted with similar numbers of wild type (WT) or tetraploid cells that were fluorescence-activated cell sorter (FACS)-purified from p53−/− tetraploid clones of the indicated type (stable, phase 1 or 2 unstable). Tumour volumes (mean±s.e.m., n=10 mice for each clone type) were monitored over time. (B, C) Tumour cells were recovered from mice (in vivo) and the DNA-content was assessed by propidium iodide (PI) staining and cytofluorometry. The same was done on aliquots of the FACS-purified parental cells that were kept frozen or that were cultured in vitro for the same time as tumours proliferated in vivo. B reports representative cell cycle distributions of tumour cells of the indicated type upon recovery from mice. Percentages refer to the sub-tetraploid populations. C depicts the percentage of sub-tetraploid cells (mean±s.e.m., n=3 independent assessments) observed for the indicated cell type.
Obligate contribution of Mos to the generation of sub-tetraploid cells
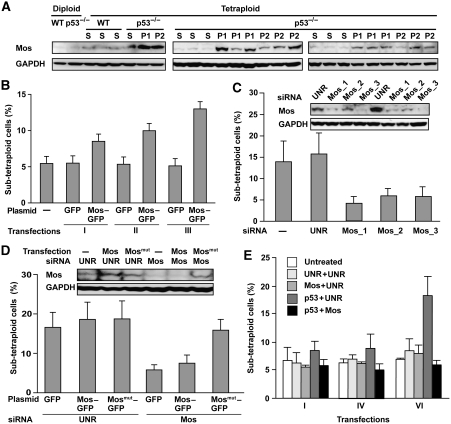
Tetraploidization has been associated with the overexpression of meiosis-specific proteins, in particular Mos (Kalejs et al, 2006). Accordingly, we observed an increase in Mos protein levels that was particularly pronounced in unstable clones (as compared with stable ones) (Figure 6A). We then assessed whether Mos overexpression (as induced by transfection with a polycystronic vector encoding Mos and GFP) might modify the behaviour of stable tetraploid cells. Although Mos-transfected p53+/+ were unable to proliferate (Fukasawa and Vande Woude, 1997; Kalejs et al, 2006), p53−/− tetraploid cells survived Mos overexpression while exhibiting an unstable phenotype, associated with the gradual increase of sub-tetraploid cells in the cultures (Figure 6B and Supplementary Figure S6). Accordingly, depletion of Mos with three different siRNAs had the opposite effect on unstable tetraploid cells and reduced their propensity to generate sub-tetraploid offspring (Figures 6C and Supplementary Figure S6). Such Mos-specific siRNAs had no anti-proliferative effects, ruling out a trivial explanation for their tetraploidy-stabilizing activity (Supplementary Figure S7). Moreover, retransfection of unstable tetraploid cells with a polycystronic construct coding for a non-interferable Mos mutant and GFP annihilated the genome-stabilizing effect of one of the Mos-specific siRNAs, formally excluding off-target effects of this siRNA (Figure 6D). These results could be recapitulated in a completely different experimental system, namely WT HCT 116 cells subjected to repetitive transient transfections with a p53-specific siRNA alone or in combination with a Mos-specific siRNA. In this setting, the emergence of a sub-tetraploid population resulting from recurring depletion of p53 was prevented when Mos was concomitantly downregulated (Figure 6E).
Figure 6.
Correlation between Mos expression and genomic instability in p53−/− tetraploid clones. (A) Mos is upregulated in unstable tetraploid clones. Protein extracts obtained from wild type (WT) or p53−/− clones of the indicated type (S=stable, P1=phase 1, P2=phase 2), were subjected to immunoblotting with antibodies that specifically recognize Mos or glyceraldehyde-3-phosphate dehydrogenase (GAPDH, monitored as a loading control). (B) Mos overexpression triggers genomic instability in stable tetraploid cells. Stable p53−/− HCT 116 cells were transfected for 48 h with a polycystronic plasmid expressing green fluorescent protein (GFP) alone or co-expressing Mos and GFP. The GFP+ populations were fluorescence-activated cell sorter (FACS)-purified, cultured for 48 h, and then subjected to subsequent rounds of transfections with the same constructs at intervals of ∼5 days. Columns depict the percentage of sub-tetraploid cells (mean±s.e.m., n=3 independent experiments) as assessed after one (I) or multiple (II, III) rounds of transfection. Representative cell cycle distributions as observed after three rounds of transfection are illustrated in Supplementary Figure S6. (C) Mos depletion stabilizes unstable p53−/− tetraploid clones. Phase 1 unstable p53−/− tetraploid HCT 116 cells were left untreated (−) or were transfected with a control (UNR) or Mos-silencing siRNAs for 72 h, followed by DNA content analysis. Representative cell cycle distributions are illustrated in Supplementary Figure S6. The percentage of sub-tetraploid cells (mean±s.e.m., n=5 independent experiments) was determined. The efficacy of the small interfering RNA (siRNA)-mediated downregulation of Mos was confirmed by immunoblotting (insert). (D) A non-interferable Mos variant restores genomic instability of originally unstable p53−/− tetraploid cells stabilized by transfection with siRNAs that deplete endogenous Mos. Phase 1 unstable p53−/− tetraploid HCT 116 cells were transfected with a control (UNR) or a Mos-depleting siRNA (Mos_1) for 24 h, followed by further transfection with a control vector encoding green fluorescent protein (GFP) alone or with constructs for the co-expression of GFP and wild type (Mos–GFP) or mutant non-interferable Mos (Mosmut–GFP). After 48 h, Mos expression was controlled by immunoblotting (insert), and 24 h later the percentage of sub-tetraploid cells was quantified (mean±s.e.m., n=3 independent assessments). (E) Mos depletion avoids genomic instability caused by p53 knockdown. Wild type (WT) tetraploid HCT 116 clones were repeatedly (6 times) transfected with a control (UNR), a p53-specific (p53_2) and a Mos-specific (Mos_1) siRNA alone or in combination for 72 h, followed by DNA content analysis and quantification of sub-tetraploid cells. The histogram summarizes data (mean±s.e.m.) from five independent experiments.
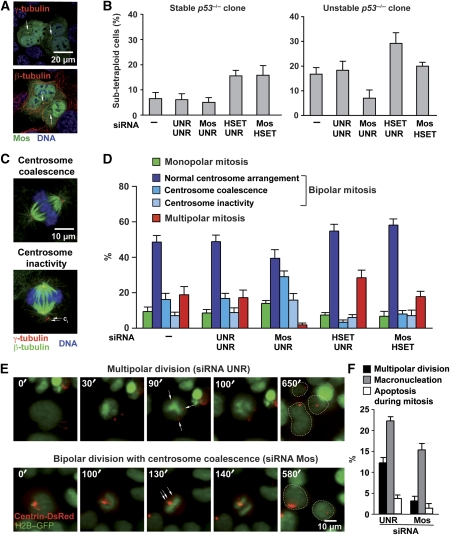
In line with reports indicating that Mos can associate with centrosomes (Wang et al, 1994; Doxsey et al, 2005; Nogales-Cadenas et al, 2009), we found that a Mos–GFP fusion protein localized to centrosomes in HCT 116 cells (Figure 7A and Supplementary Figure S8). To control specificity, we demonstrated that the expression of Mos–GFP (but not of GFP alone) was abolished by a Mos-specific (but not by a control) siRNA (Supplementary Figure S9). Therefore, we investigated whether Mos might influence centrosome dynamics, and in particular whether Mos might affect centrosome clustering, one mechanism relying on the molecular motor HSET that allows for the maintenance of tetraploid genomes (Kwon et al, 2008). Upon siRNA-mediated knockdown of HSET (Supplementary Figure S10), stable p53−/− tetraploid cell cultures progressively accumulated a sub-tetraploid population and this effect was not counteracted by the simultaneous knockdown of Mos (Figure 7B). In phase 1 unstable p53−/− tetraploid cells, Mos depletion limited the generation of a sub-tetraploid offspring (Figure 7B) while resulting in the conversion of multipolar to bipolar mitoses, in which supernumerary centrosomes either congressed to two centrosome pairs or lost their connection to the β-tubulin network and were inactivated (Figure 7C, D and Supplementary Figure S8). In this setting, HSET depletion lead to an increase in multipolar divisions paralleled by a decrease in bipolar mitoses characterized by centrosome coalescence (Figure 7D), correlating with a further increase in the generation of sub-tetraploid cells (Figure 7B). These effects were reduced by the simultaneous depletion of Mos (Figure 7B and D), in line with the hypothesis that Mos modulates centrosome dynamics, thereby affecting the frequency of multipolar mitoses in tetraploid cells. To directly evaluate the putative effects of Mos on centrosome clustering, we transiently transfected a (phase 2) unstable p53−/− tetraploid clone that had previously been engineered for the expression of GFP–H2B with a centrin–DsRed fusion-encoding construct, allowing for the simultaneous monitoring of chromosomal and centrosomal dynamics by videomicroscopy (Figure 7E). Cells that were left untreated or were transfected with a control siRNA frequently underwent multipolar divisions, which were characterized by the presence of more than two centrin–DsRed-positive dots. However, upon transfection with a Mos-specific siRNA the frequency of multipolar (or failed) divisions dropped (Figure 7E and F). In line with immunofluorescence microscopy results (Figure 7C, D and Supplementary Figure S8), we observed an increase in the incidence of bipolar divisions that exhibited centrosome coalescence. Taken together, these data strongly suggest that the knockdown of Mos favours centrosome clustering, correlating with the suppression of multipolar mitosis.
Figure 7.
Effects of Mos on centrosome coalescence in p53−/− tetraploid clones. (A) Mos localizes to centrosomes. Immunofluorescence staining for the detection of β- or γ-tubulin (both in red) in p53−/− tetraploid cells transfected with a plasmid for the expression of a Mos–green fluorescent protein (Mos–GFP) chimera. Nuclei were counterstained with Hoechst 33342 (emitting in blue). Yellow dots that result from the overlap of green and red fluorescence signals suggest that Mos and γ-tubulin colocalizes at centrosomes (arrows). Representative immunofluorescence microphotographs are shown for cells in interphase. Single-channel microphotographs are provided in Supplementary Figure S8. (B) Mos depletion partially reverts genomic instability induced by HSET downregulation. Stable and unstable p53−/− tetraploid HCT 116 clones were transfected with a control (UNR), a Mos-specific (Mos_1) or a HSET-specific (HSET_1) small interfering RNA (siRNA) alone or in combination for 72 h, followed by cytofluorometric analysis of DNA content and quantification of sub-tetraploid cells (mean±s.e.m., n=3 independent experiments). (C, D) Increased coalescence and inactivation of supernumerary centrosomes upon Mos depletion. Unstable p53−/− tetraploid HCT 116 cells were co-transfected with the indicated siRNA combinations for 72 h, and then subjected to immunofluorescence staining for the detection of β- and γ-tubulin (green and red fluorescence, respectively). Hoechst 33342 was employed for nuclear counterstaining. C shows representative immunofluorescence microphotographs of supernumerary centrosomes coalescing or being inactivated (ci), which allows for the bipolar organization of spindles. Single-channel microphotographs are provided in Supplementary Figure S8. D depicts the percentage of metaphases (mean±s.e.m., n=3 independent experiments) characterized by the indicated mitotic spindle arrangements, as scored among at least 100 cells per for each condition. All experiments have been performed on at least three distinct clones of each type. (E, F) Live-cell imaging of the Mos effects on centrosomal and chromosomal dynamics. A (phase 2) unstable tetraploid p53−/− GFP–H2B-expressing clone was transiently transfected with a Mos-specific or a control siRNA (UNR) together with a construct coding for centrin–DsRed (E). E shows representative snapshots. Please note that bipolar divisions are characterized by the coalescence of DsRed-positive centrosomes in two clusters. Full-length movies can be found in Supplementary Videos S7 and S8. Hatched lines indicate the position of daughter cells arising from the observed mitoses. The frequency of multipolar divisions and failed cytokineses observed by live-cell imaging is reported in F (mean±s.e.m., n=3 independent experiments).
Concluding remarks
On theoretical grounds, tetraploid cells may generate an aneuploid offspring by two non-exclusive mechanisms. First, as tetraploid cells contain supernumerary centrosomes, they may enter multipolar mitosis, thereby dividing into more than two (usually three or four) daughter cells provided with randomly distributed chromosomes. Most of these cells would be non-viable and would die during the subsequent interphase or after one additional round of (failing) mitosis. However, in exceptional circumstances, the overall genome composition of one daughter cell may be compatible with survival, and rare, particularly fit cells would then out compete their tetraploid parent, as it has been observed in cell cultures and in tumours. Alternatively, tetraploid cells might lose single (or few) chromosomes because of their merotelic attachment, which results from a ‘multipolar spindle intermediate'. In this case, the presence of supernumerary centrosomes enforces an initial multipolar metaphase followed by the clustering of all centrosomes in two poles, which results in a bipolar division, whereby the chromosome that is merotelically attached lags during anaphase and eventually is either lost or distributed into the wrong daughter cell. This mechanism might give rise to a gradual, progressive loss of chromosomes until the achievement of a sub-tetraploid stage.
Irrespective of the molecular details underlying the aneuploidization of tetraploid cells, it appears that centrosome clustering constitutes a prime mechanism for the maintenance of tetraploid genomes. Supernumerary centrosomes found in tetraploid cells would indeed cause multipolar division or merotelic chromosome attachment (and hence chromosome loss), unless they aggregate in two discrete pools and form two microtubule-organizing centres. This aggregation is under the control of molecular motors including HSET, and HSET depletion indeed compromises the stability of tetraploid cells (Kwon et al, 2008). As shown here, overexpression of Mos that occurs in p53−/− tetraploid cells also inhibits centrosome clustering, thereby favouring multipolar divisions and resulting in the reduction of the tetraploid genome to a sub-tetraploid one.
Little information is available on the cross talk between p53 and Mos. In samples from non-small cell lung carcinoma patients, p53 alterations were shown to correlate with high Mos expression, aneuploidy and tumour aggressiveness (Gorgoulis et al, 2001). As a possible scenario, the inactivation of p53 would favour the presence of supernumerary centrosomes (Fukasawa et al, 1996; Carroll et al, 1999) while augmenting the probability of cells to survive the tetraploidization process (Castedo et al, 2006a; Senovilla et al, 2009). Tetraploid cells that lack an intact p53 system would then upregulate Mos, in turn favouring aneuploidization. Beyond such an oncogenic cooperation at the functional level, additional interactions between p53 and Mos may be envisioned. Reportedly, p53-deficient cells tolerate higher levels of Mos than their p53-proficient counterparts (Fukasawa and Vande Woude, 1997; Kalejs et al, 2006), suggesting that the ‘oncogenic stress' mediated by ectopic expression of this meiosis-restricted protein may kill cells or arrest their cycle if the p53 system is intact. Thus, it is conceivable that the absence of p53 may be permissive for the upregulation of Mos (Fukasawa and Vande Woude, 1997; Gorgoulis et al, 2001; Kalejs et al, 2006), which must occur at the post-transcriptional level, as Mos mRNA levels were not increased in tetraploid cells (data not shown). However, the exact molecular mechanisms explaining the unscheduled expression of Mos in tumour cells remain elusive.
The precise oncogenic mode of action of Mos is an ongoing conundrum. Enforced Mos expression in fibroblasts reportedly causes centrosome amplification (Saavedra et al, 1999). However, although siRNA-mediated knockdown of Mos in tetraploid cells apparently did not decrease the number of centrosomes, Mos expression inversely correlated with the coalescence of supernumerary centrosomes, as if Mos acted as an inhibitor of centrosome clustering. Accordingly, the depletion of Mos favoured centrosome coalescence while switching multipolar to bipolar mitoses, and this effect was inhibited by the simultaneous knockdown of the kinesin HSET, which is indispensable for centrosome clustering. Based on these results, we conclude that Mos may exert its oncogenic activity, at least in part, through the induction of multipolar, asymmetric cell division. It remains to be determined whether the oncogenic activity of Mos is mediated by its direct interaction with centrosomes or by hitherto unknown effects on cellular morphology/dynamics that indirectly affect centrosome clustering.
Materials and methods
Unless otherwise indicated, media and supplements for cell culture were purchased from Gibco-Invitrogen (Carlsbad, CA, USA), plasticware from Corning B.V. Life Sciences (Schiphol-Rijk, The Netherlands), and chemicals from Sigma-Aldrich (St Louis, MO, USA).
Cell lines, culture conditions and chemicals
Wild type, p53−/−, Bax−/− and p21−/− diploid human colon carcinoma HCT 116 cells (kindly provided by Bert Vogelstein), diploid HCT 116 cells stably transfected with a vector encoding the baculoviral caspase inhibitor p35 (Zhang et al, 2006), WT and p53−/− diploid HCT 116 cells transfected with a cDNA coding for the fusion between histone H2B and green fluorescent protein (H2B–GFP, PharMingen, San Diego, CA, USA), as well as WT diploid human colon carcinoma RKO cells were routinely maintained in McCoy's 5A medium (PAA Laboratories GmbH, Pasching, Austria) supplemented with 10% foetal calf serum (FCS) at 37 °C in a 5% CO2 atmosphere. H2B–GFP-expressing cells were cultured in the presence of 20 μg/ml blasticidine. Cells were seeded onto the appropriate supports (6-, 12-, 24- or 96-well plates, 35 or 100 mm Ø Petri dishes) 24 h before the beginning of the experiment. To generate tetraploid clones, parental diploid cells were treated for 48 h with 0.6 μg/ml cytochalasin D or 100 nM nocodazole and then cultured for 2 weeks in drug-free culture medium, followed by staining with Hoechst 33342 (2 μM; Molecular Probes-Invitrogen, Eugene, OR, USA) and cloning of cells characterized by an ∼8n DNA content on a FACSVantage cell sorter (BD Biosciences, San Jose, CA, USA), as previously described (Castedo et al, 2006b). Alternatively, tetraploid clones were isolated by the limiting dilution technique (Castedo et al, 2006b). To prevent drifts in cell composition, clones were split well before complete confluence. For each experiment, at least three different clones for each genotype and/or phenotype were used.
Plasmid transfection and RNA interference
A cDNA encoding for centrin derived from a human testis mRNA library (GenBank accession number BC029515.1) was amplified in a pCMV-sport6 vector (Invitrogen) and then cloned into a pDsRed2-C1 plasmid (Clontech Laboratories, Mountain View, CA, USA) as a BglII restriction fragment, generating a construct for the expression of a centrin-DsRed chimera. The human Mos cDNA (Image Clone 40016104) was purchased from Geneservice (Nottingham, UK) within a pCR-bluntII-TOPO plasmid (Invitrogen). The Mos sequence was then transferred either to the polycystronic expression vector pIRES-hrGFP2 (Agilent Technologies, Santa Clara, CA, USA) as a BamH1–Not1 restriction fragment, or to the pEGFP-C3 plasmid (Clontech Laboratories) as a Xho1–BamH1 restriction fragment upon PCR amplification (primers 5′-TATACTCGAGCCCTCGCCCCTGGCCC-3′ and 5′-TATAGGATCCTCAGCCGAGTTCAGCTTTCA-3′, restriction sites are underlined). In order to generate a non-interferable but functional Mos mutant, the Mos sequence was further modified from within the pIRES-hrGFP2 vector to introduce the silent mutations ATCATA at position 619 (Ile207) and TTGCTA at position 622 (Leu208). Site-directed mutagenesis was performed with the Quikchange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) and the primers 5′-GGACCTGAAGCCCGCGAACATACTAATCAGTGAGCAGGATGTC-3′ and 5′-GACATCCTGCTCACTGATTAGTATGTTCGCGGGCTTCAGGTCC-3′ (mutated nucleotides are underlined), according to the manufacturer's instructions. Briefly, PCR was employed to produce several copies of the entire plasmid including the desired mutations. The reaction mix was then incubated for 1 h at 37°C with the Dpn1 restriction enzyme, which cleaves only methylated DNA (in this case, the parental, non-mutated molecules). Uncleaved (mutated) DNA was then used for transformation of Escherichia coli XL1-blue competent cells (Stratagene).
Custom-designed siRNAs duplexes targeting Mos (Mos_1 sense 5′-GCCCGCGAACAUCUUGAUCdTdT-3′; Mos_2 sense 5′-GCCUAAAGCCGACAUUUAUdTdT-3′) and p53 (p53_1 sense 5′-GUGAGCGCUUCGAGAUGUUdTdT-3′; p53_2 sense 5′-GACUCCAGUGGUAAUCUACdTdT-3′) (Gu et al, 2004) were purchased from Sigma-Proligo (The Woodlands, TX, USA). Alternatively, siRNA duplexes for the downregulation of Mos (Mos_3) and HSET (HSET_1) (siGENOME Smart-pool M-003859 and L-004958, respectively) were purchased from Dharmacon (Chicago, IL, USA). As a control, a non-targeting siRNA with an sequence unrelated to both human and murine genomes was used (UNR, sense 5′-GCCGGUAUGCCGGUUAAGUdTdT-3′). HCT 116 cells in 12-well plates were transfected with plasmids at 80% confluence by means of the Attractene® transfection reagent (Qiagen, Hilden, Germany) as recommended by the manufacturer, or with siRNAs at 30–40% confluence by means of the HiPerFect® transfection reagent (Qiagen), as previously described (Hoffmann et al, 2008). After 72 h, transfection efficiency was determined by immunoblotting (see below). When required, cells were transfected first with siRNAs and 24 h later with plasmids.
Cytofluorometric studies
For the simultaneous quantification of DNA content (cell cycle profiling), plasma membrane integrity and mitochondrial transmembrane potential (ΔΨm), live cells were collected and stained with 2 μM Hoechst 33342 (Molecular Probes–Invitrogen), 1 μg/ml propidium iodide (PI, which only incorporates into dead cells) and 40 nM of the ΔΨm-sensitive dye 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3), from Molecular Probes–Invitrogen) (Castedo et al, 2002; Galluzzi et al, 2009). To assess cell cycle distribution, cells were collected, fixed by gentle vortexing in ice-cold 80% (v/v) ethanol (Carlo Erba Reagents, Milano, Italy) and stained with 50 μg/ml PI in 0.1% (w/v) D-glucose in PBS supplemented with 1 μg/ml (w/v) RNAse A (Mouhamad et al, 2007; Mondragon et al, 2009). For the cell count assay, the same amount of FITC-labeled beads (Becton Dickinson) was added to each sample and cells were counted until 5000 beads had passed through the FACS. Cell proliferation was determined by counting the cells every 24 h for 4 days upon the seeding. Cytofluorometric acquisition of blue (Hoechst 33342), red (PI) and green (DiOC6(3)) fluorescence were carried out by means of a FACSCalibur or a FACScan cytofluorometer (BD Biosciences) equipped with a 70 μm nozzle. Statistical analysis was carried out by using the CellQuest™ software (BD Biosciences), upon gating on the events characterized by normal forward scatter and side-scatter parameters.
Videomicroscopy
For videomicroscopy, HCT 116 cells stably transfected with the cDNA encoding for H2B–GFP were grown in 96-well imaging plates (BD Biosciences) under standard culture conditions (37 °C, 5% CO2) and subjected to pulsed observations (every 10 min for up to 48 h) with a BD pathway 855 automated live cell microscope (BD Biosciences). Images were analyzed with the open source software Image J (freely available from the National Institute of Health, Bethesda, MD, USA at the address http://rsb.info.nih.gov/ij/).
Cell proliferation assays
Cell proliferation was measured by the fluorescein-based dye 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) staining. Briefly, 1 × 106 cells were incubated with 2.5 μM CFSE in PBS for 10 min at 37°C. The labelling reaction was stopped by adding an equal volume of FCS and 5 × 104 cells were seeded in 12-well plates. After 24 h, cells were transfected with siRNAs and grown in culture for additional 3–5 days. Cell proliferation was monitored daily on a FACSCalibur cytofluorometer (BD Biosciences) upon excitation at 488 nm and acquisition in the FL1 (green) channel, followed by data analysis by means of the CellQuest software (BD Biosciences).
Immunofluorescence microscopy
Immunofluorescence microscopy determinations were performed as previously described (Vitale et al, 2007, 2008). Briefly, cells were fixed in 4% (w/v) paraformaldehyde in PBS, permeabilized with 0.1% SDS and immunostained with antibodies specific for γ- and β-tubulin (both from Sigma-Aldrich). Slides were then incubated with the appropriate Alexa Fluor® conjugates (Molecular Probes–Invitrogen) in the presence of 10 μM Hoechst 33342 (Molecular Probes–Invitrogen), which was used for nuclear counterstaining. Fluorescence and confocal fluorescence images were captured using an IRE2 microscope equipped with a DC300F camera (both from Leica Microsystems GmbH, Wetzlar, Germany) and a TSC-SPE microscope (Leica Microsystems GmbH) equipped with a 63X/1.15 objective (Olympus America, Center Valley, PA, USA), respectively. Signals from different probes were acquired in sequential scan mode and image analysis was performed with the open source software Image J (National Institutes of Health).
Immunoblotting
HCT 116 cells were washed with cold PBS and lysed in a buffer containing 1% NP40, 20 mM HEPES (pH 7.9), 10 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM orthovanadate, 1 mM PMSF, 1 mM dithiothreitol, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 10 μg/ml pepstatin, as previously described (Zermati et al, 2007). Thereafter, protein extracts (50 μg per lane) were separated according to molecular weight on precast 4–12% SDS–PAGE gels (Invitrogen), followed by electrotransfer to Immobilon™ membranes (Sigma-Aldrich) and immunoblotting with antibodies specific for HSET and Mos (both from Santa Cruz Biotechnology, Santa Cruz, CA, USA). Primary antibodies that specifically recognize glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin (both from Millipore-Chemicon International, Temecula, CA, USA) were used as loading control. Finally membranes were incubated with appropriate goat IgG conjugated to horseradish peroxidase (Southern Biotech, Birmingham, AL, USA), followed by chemiluminescence detection with the SuperSignal West Pico® reagent and CL-XPosure® X-ray films (both from Thermo Scientific–Pierce, Rockford, IL, USA).
Cytogenetic analysis and FISH
Chromosome spreads were prepared by conventional procedures (Mouhamad et al, 2007). Briefly, HCT 116 clones were treated with 100 nM nocodazole for 8 h to enrich mitotic cells, then collected and subjected to hypotonic lysis by incubation in 75 μM KCl for 10 min at 37°C. After removal of hypotonic solution, cells were fixed in freshly prepared Carnoy solution (3:1 methanol:acetic acid) and stored at −20°C. Fixed cells were then dropped onto pre-cooled Superfrost Plus glass microscope slides (Thermo Fisher Scientific, Waltham, MA, USA) and dried at room temperature. Chromosomes were stained with 100 ng/ml 4′,6 diamidino-2-phenylindole (DAPI, from Molecular Probes–Invitrogen) and mounted with glass coverslips in Vectashield H-1000 mounting medium (Vector Laboratories, Burlingame, CA, USA). Finally, fluorescence images were visualized and captured using an IRE2 microscope equipped with a DC300F camera (both from Leica Microsystems GmbH). For each experimental condition, 100 cells from three independent experiments were analyzed. For FISH, freshly FACS-purified diploid and sub-tetraploid cells were dropped onto Superfrost Plus polylysine-coated glass microscope slides (Thermo Fisher Scientific) and fixed in situ with 9:1 methanol:acetic acid for 5 min. Thereafter, cells were air-dried overnight and hybridized with a commercial mixture of three probes (Abbott Laboratories, Abbott Park, IL, USA) that detect the centromeric region of chromosome 8 (labelled with FITC—green colour), chromosome 10 (labeled with rhodamine—red colour) and chromosome 18 (labeled with Aqua—blue colour). For each experimental conditions, 100–300 nuclei were surveyed.
Clonogenic survival assays
To evaluate clonogenic survival, freshly generated tetraploid and sub-tetraploid cells were stained with 2 μM Hoechst 33342 (Molecular Probes–Invitrogen), FACS-purified on a FACSVantage cell sorter (BD Biosciences), seeded at different concentrations (from 1 to 50 × 103 for well) in 6-well plates, and cultured for up to 10 days under normal conditions. Colonies were then fixed/stained with an aqueous solution containing 0.25% (w/v) crystal violet, 70% (v/v) methanol and 3% (v/v) formaldehyde (Carlo Erba Reagents) and counted (Zhang et al, 2006). Only colonies made of ⩾30 cells were included in the quantification. For each treatment, the surviving fraction (SF) was estimated according to the formula: SF=(number of colonies formed/ number of cells seeded) × plating efficiency (defined as the ratio between the number of colonies formed and the number of cells seeded in control conditions).
In vivo xenograft model
Athymic nu/nu female mice (age=42 days, body weight=20 g, provided by the Institut Gustave Roussy (IGR) in-house animal facility) were used throughout this study in strict compliance with widely accepted ethical guidelines for animal experimentation. Mice were kept in Makrolon® type III wire mesh laboratory cages (Charles River, Boston, MA, USA), under poor germ conditions at 24°C and 50–60% humidity, and were allowed for food and water ad libitum. Light cycle was artificially controlled to provide 14 h of light (from 0630 h to 2030 h). After 4 days of acclimation period, mice were subcutaneously xenografted with 2 × 106 WT or p53−/− tetraploid HCT 116 cells, as previously described (Vitale et al, 2007). Tumour growth was then assessed every 2–4 days with a standard caliper. For ethical reasons, mice were killed before tumours reached a volume of 3400 mm3 or a surface of 250 mm2. Statistical analysis was carried out by means of the SigmaStat package (Systat Software, San Jose, CA, USA). One-way analysis of variance was carried out and statistical significance was determined by means of two-tailed Student's t-test (*P<0.05) in the context of a pairwise comparison procedure.
Tumour recovery
To recover tumour cells, mice were killed by cervical dislocation following the FELASA guidelines. Quickly after killing, tumour tissue was surgically removed and washed in RPMI 1640 culture medium supplemented with 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulphate and 10% FCS. Thereafter, a tissue fragment of ∼125 mm3 was cut into pieces of ∼1 mm3, which were dissociated by incubation with 0.05% trypsin for 45 min at 37°C. Every 15 min the disaggregation of cells was mechanically carried out. Finally, dissociated cells were washed once with RPMI 1640 supplemented with 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulphate and 10% FCS, and seeded into 25 cm2 flasks. Recovered tumour cells were routinely maintained under standard culture conditions (37°C, 5% CO2) in McCoy's 5A medium supplemented with 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulphate, 100 mM HEPES buffer, 1 mM sodium pyruvate and 10% FCS.
Statistical procedures
Unless otherwise specified, all experiments were carried out in triplicate parallel instances and independently repeated at least three times. Data were analyzed with Microsoft Excel (Microsoft, Redmond, WA, USA) and statistical significance was assessed by means of two-tailed Student's t-test (*P<0.05).
Supplementary Material
Acknowledgments
GK is supported by the Ligue Nationale contre le Cancer (Equipe labellisée), Agence Nationale pour la Recherche, European Commission (Apo-Sys, ChemoRes, ApopTrain), Fondation pour la Recherche Médicale, Institut National du Cancer, Cancéropôle Ile-de-France. MC is supported by the Association pour la Recherche sur le Cancer (ARC). LS is supported by ApopTrain Marie Curie training network of the European Union and FRM, MM and SR-V by FRM, LG by the Apo-Sys consortium of the European Union, OK by EMBO. IV, LS, MJ, MM, OK, LN, AC, SRV, GM, DM, SV, NT, NJ, AV and MC carried out the experiments presented in this article. IV, LG, MC and GK prepared the figures and wrote/edited the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL (2001) Tetraploid state induces p53-dependent arrest of non-transformed mammalian cells in G1. Mol Biol Cell 12: 1315–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M (2006) A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev 20: 2687–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW (2008) Centrosome amplification can initiate tumorigenesis in flies. Cell 133: 1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T (2008) Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henri Harris. J Cell Sci 121(Suppl. 1): 1–84 [DOI] [PubMed] [Google Scholar]
- Caldwell CM, Green RA, Kaplan KB (2007) APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J Cell Biol 178: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll PE, Okuda M, Horn HF, Biddinger P, Stambrook PJ, Gleich LL, Li YQ, Tarapore P, Fukasawa K (1999) Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene 18: 1935–1944 [DOI] [PubMed] [Google Scholar]
- Castedo M, Coquelle A, Vitale I, Vivet S, Mouhamad S, Viaud S, Zitvogel L, Kroemer G (2006a) Selective resistance of tetraploid cancer cells against DNA damage-induced apoptosis. Ann N Y Acad Sci 1090: 35–49 [DOI] [PubMed] [Google Scholar]
- Castedo M, Coquelle A, Vivet S, Vitale I, Kauffmann A, Dessen P, Pequignot MO, Casares N, Valent A, Mouhamad S, Schmitt E, Modjtahedi N, Vainchenker W, Zitvogel L, Lazar V, Garrido C, Kroemer G (2006b) Apoptosis regulation in tetraploid cancer cells. EMBO J 25: 2584–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Ferri K, Roumier T, Metivier D, Zamzami N, Kroemer G (2002) Quantitation of mitochondrial alterations associated with apoptosis. J Immunol Methods 265: 39–47 [DOI] [PubMed] [Google Scholar]
- Colledge WH, Carlton MB, Udy GB, Evans MJ (1994) Disruption of c-mos causes parthenogenetic development of unfertilized mouse eggs. Nature 370: 65–68 [DOI] [PubMed] [Google Scholar]
- Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, Raskind WH, Reid BJ (1995) A p53-dependent mouse spindle checkpoint. Science 267: 1353–1356 [DOI] [PubMed] [Google Scholar]
- D'Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, Whitehead C, Stivala F, Lingle WL, Salisbury JL (2002) Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat 75: 25–34 [DOI] [PubMed] [Google Scholar]
- Doxsey S, Zimmerman W, Mikule K (2005) Centrosome control of the cell cycle. Trends Cell Biol 15: 303–311 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D (2005) Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437: 1043–1047 [DOI] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF (1996) Abnormal centrosome amplification in the absence of p53. Science 271: 1744–1747 [DOI] [PubMed] [Google Scholar]
- Fukasawa K, Vande Woude GF (1995) Mos overexpression in Swiss 3T3 cells induces meiotic-like alterations of the mitotic spindle. Proc Natl Acad Sci USA 92: 3430–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K, Vande Woude GF (1997) Synergy between the Mos/mitogen-activated protein kinase pathway and loss of p53 function in transformation and chromosome instability. Mol Cell Biol 17: 506–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, Bazan NG, Blagosklonny MV, Blomgren K, Borner C, Bredesen DE, Brenner C, Castedo M, Cidlowski JA, Ciechanover A, Cohen GM, De Laurenzi V, De Maria R, Deshmukh M, Dynlacht BD et al. (2009) Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ 16: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D (2009) A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D (2007) Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev 17: 157–162 [DOI] [PubMed] [Google Scholar]
- Gergely F, Basto R (2008) Multiple centrosomes: together they stand, divided they fall. Genes Dev 22: 2291–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Zacharatos P, Mariatos G, Liloglou T, Kokotas S, Kastrinakis N, Kotsinas A, Athanasiou A, Foukas P, Zoumpourlis V, Kletsas D, Ikonomopoulos J, Asimacopoulos PJ, Kittas C, Field JK (2001) Deregulated expression of c-mos in non-small cell lung carcinomas: relationship with p53 status, genomic instability, and tumor kinetics. Cancer Res 61: 538–549 [PubMed] [Google Scholar]
- Gu J, Zhang L, Swisher SG, Liu J, Roth JA, Fang B (2004) Induction of p53-regulated genes in lung cancer cells: implications of the mechanism for adenoviral p53-mediated apoptosis. Oncogene 23: 1300–1307 [DOI] [PubMed] [Google Scholar]
- Haccard O, Sarcevic B, Lewellyn A, Hartley R, Roy L, Izumi T, Erikson E, Maller JL (1993) Induction of metaphase arrest in cleaving Xenopus embryos by MAP kinase. Science 262: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Hashimoto N, Watanabe N, Furuta Y, Tamemoto H, Sagata N, Yokoyama M, Okazaki K, Nagayoshi M, Takeda N, Ikawa Y, Aizawa S (1994) Parthenogenetic activation of oocytes in c-mos-deficient mice. Nature 370: 68–71 [DOI] [PubMed] [Google Scholar]
- Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T (1996) Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA 93: 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J, Vitale I, Buchmann B, Galluzzi L, Schwede W, Senovilla L, Skuballa W, Vivet S, Lichtner RB, Vicencio JM, Panaretakis T, Siemeister G, Lage H, Nanty L, Hammer S, Mittelstaedt K, Winsel S, Eschenbrenner J, Castedo M, Demarche C et al. (2008) Improved cellular pharmacokinetics and pharmacodynamics underlie the wide anticancer activity of sagopilone. Cancer Res 68: 5301–5308 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW (2009) Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incassati A, Patel D, McCance DJ (2006) Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16 E6. Oncogene 25: 2444–2451 [DOI] [PubMed] [Google Scholar]
- Kalejs M, Ivanov A, Plakhins G, Cragg MS, Emzinsh D, Illidge TM, Erenpreisa J (2006) Upregulation of meiosis-specific genes in lymphoma cell lines following genotoxic insult and induction of mitotic catastrophe. BMC Cancer 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV (1999) A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285: 1733–1737 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C (2007) Mitochondrial membrane permeabilization in cell death. Physiol Rev 87: 99–163 [DOI] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D (2008) Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 22: 2189–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine DS, Sanchez CA, Rabinovitch PS, Reid BJ (1991) Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer. Proc Natl Acad Sci USA 88: 6427–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Li X, Sanchez CA, Paulson TG, Blount PL, Reid BJ (2004) The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res 64: 7629–7633 [DOI] [PubMed] [Google Scholar]
- Margolis RL (2005) Tetraploidy and tumor development. Cancer Cell 8: 353–354 [DOI] [PubMed] [Google Scholar]
- Mondragon L, Galluzzi L, Mouhamad S, Orzaez M, Vicencio JM, Vitale I, Moure A, Messeguer A, Perez-Paya E, Kroemer G (2009) A chemical inhibitor of Apaf-1 exerts mitochondrioprotective functions and interferes with the intra-S-phase DNA damage checkpoint. Apoptosis 14: 182–190 [DOI] [PubMed] [Google Scholar]
- Mouhamad S, Galluzzi L, Zermati Y, Castedo M, Kroemer G (2007) Apaf-1 deficiency causes chromosomal instability. Cell Cycle 6: 3103–3107 [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Abascal F, Diez-Perez J, Carazo JM, Pascual-Montano A (2009) CentrosomeDB: a human centrosomal proteins database. Nucleic Acids Res 37 (Database issue): D175–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Nishizawa M, Furuno N, Yasuda H, Sagata N (1992) Differential occurrence of CSF-like activity and transforming activity of Mos during the cell cycle in fibroblasts. EMBO J 11: 2447–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Sagata N (1995) The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J 14: 5048–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson M, McClements WL, Blair DG, Maizel JV, Vande Woude GF (1980) Properties of a normal mouse cell DNA sequence (sarc) homologous to the src sequence of Moloney sarcoma virus. Science 207: 1222–1224 [DOI] [PubMed] [Google Scholar]
- Rhodes N, Innes CL, Propst F, Paules RS (1997) Serum starved v-mos-transformed cells are unable to appropriately downregulate cyclins and CDKs. Oncogene 14: 3017–3027 [DOI] [PubMed] [Google Scholar]
- Roumier T, Valent A, Perfettini JL, Metivier D, Castedo M, Kroemer G (2005) A cellular machine generating apoptosis-prone aneuploid cells. Cell Death Differ 12: 91–93 [DOI] [PubMed] [Google Scholar]
- Roy LM, Singh B, Gautier J, Arlinghaus RB, Nordeen SK, Maller JL (1990) The cyclin B2 component of MPF is a substrate for the c-mos(xe) proto-oncogene product. Cell 61: 825–831 [DOI] [PubMed] [Google Scholar]
- Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ (1999) MAPK mediates RAS-induced chromosome instability. J Biol Chem 274: 38083–38090 [DOI] [PubMed] [Google Scholar]
- Sagata N (1997) What does Mos do in oocytes and somatic cells? Bioessays 19: 13–21 [DOI] [PubMed] [Google Scholar]
- Sagata N, Daar I, Oskarsson M, Showalter SD, Vande Woude GF (1989a) The product of the mos proto-oncogene as a candidate ‘initiator' for oocyte maturation. Science 245: 643–646 [DOI] [PubMed] [Google Scholar]
- Sagata N, Watanabe N, Vande Woude GF, Ikawa Y (1989b) The c-mos proto-oncogene product is a cytostatic factor responsible for meiotic arrest in vertebrate eggs. Nature 342: 512–518 [DOI] [PubMed] [Google Scholar]
- Schlegel BP, Starita LM, Parvin JD (2003) Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene 22: 983–991 [DOI] [PubMed] [Google Scholar]
- Senovilla L, Vitale I, Galluzzi L, Vivet S, Joza N, Younes AB, Rello-Varona S, Castedo M, Kroemer G (2009) p53 represses the polyploidization of primary mammary epithelial cells by activating apoptosis. Cell Cycle 8: 1380–1385 [DOI] [PubMed] [Google Scholar]
- Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, Pellman D (2006) Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547 [DOI] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C (2008) The consequences of tetraploidy and aneuploidy. J Cell Sci 121 (Part 23): 3859–3866 [DOI] [PubMed] [Google Scholar]
- Storchova Z, Pellman D (2004) From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 5: 45–54 [DOI] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Taylor SS (2004) Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci 117(Part 26): 6339–6353 [DOI] [PubMed] [Google Scholar]
- Vitale I, Galluzzi L, Vivet S, Nanty L, Dessen P, Senovilla L, Olaussen KA, Lazar V, Prudhomme M, Golsteyn RM, Castedo M, Kroemer G (2007) Inhibition of Chk1 kills tetraploid tumor cells through a p53-dependent pathway. PLoS One 2: e1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Galluzzi L, Criollo A, Vivet S, Castedo M, Kroemer G (2008) Chk1 inhibition activates p53 through p38 MAPK in tetraploid cancer cells. Cell Cycle 7: 1956–1961 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, Deng CX (2006) Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene 25: 7148–7158 [DOI] [PubMed] [Google Scholar]
- Wang XM, Yew N, Peloquin JG, Vande Woude GF, Borisy GG (1994) Mos oncogene product associates with kinetochores in mammalian somatic cells and disrupts mitotic progression. Proc Natl Acad Sci USA 91: 8329–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R, Oskarsson M, Vande Woude GF (1982) Human DNA sequence homologous to the transforming gene (mos) of Moloney murine sarcoma virus. Proc Natl Acad Sci USA 79: 4078–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XY, Grove L, Datta NS, Long MW, Prochownik EV (1999) C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene 18: 1177–1184 [DOI] [PubMed] [Google Scholar]
- Zermati Y, Mouhamad S, Stergiou L, Besse B, Galluzzi L, Boehrer S, Pauleau AL, Rosselli F, D'Amelio M, Amendola R, Castedo M, Hengartner M, Soria JC, Cecconi F, Kroemer G (2007) Nonapoptotic role for Apaf-1 in the DNA damage checkpoint. Mol Cell 28: 624–637 [DOI] [PubMed] [Google Scholar]
- Zhang P, Castedo M, Tao Y, Violot D, Metivier D, Deutsch E, Kroemer G, Bourhis J (2006) Caspase independence of radio-induced cell death. Oncogene 25: 7758–7770 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Kroemer G (2004) Apoptosis and genomic instability. Nat Rev Mol Cell Biol 5: 752–762 [DOI] [PubMed] [Google Scholar]
- Zhou RP, Oskarsson M, Paules RS, Schulz N, Cleveland D, Vande Woude GF (1991) Ability of the c-mos product to associate with and phosphorylate tubulin. Science 251: 671–675 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.