Abstract
Fragile X-associated Tremor/Ataxia Syndrome (FXTAS) is a neurodegenerative disorder caused by expansion of 55–200 CGG repeats in the 5′-UTR of the FMR1 gene. FXTAS is characterized by action tremor, gait ataxia and impaired executive cognitive functioning. It has been proposed that FXTAS is caused by titration of RNA-binding proteins by the expanded CGG repeats. Sam68 is an RNA-binding protein involved in alternative splicing regulation and its ablation in mouse leads to motor coordination defects. Here, we report that mRNAs containing expanded CGG repeats form large and dynamic intranuclear RNA aggregates that recruit several RNA-binding proteins sequentially, first Sam68, then hnRNP-G and MBNL1. Importantly, Sam68 is sequestered by expanded CGG repeats and thereby loses its splicing-regulatory function. Consequently, Sam68-responsive splicing is altered in FXTAS patients. Finally, we found that regulation of Sam68 tyrosine phosphorylation modulates its localization within CGG aggregates and that tautomycin prevents both Sam68 and CGG RNA aggregate formation. Overall, these data support an RNA gain-of-function mechanism for FXTAS neuropathology, and suggest possible target routes for treatment options.
Keywords: FXTAS, RNA gain-of-function diseases, Sam68
Introduction
Fragile X-associated Tremor/Ataxia Syndrome (FXTAS) is a recently identified neurodegenerative disorder that affects principally older adult males who are carriers of pre-mutation expansions (55–200 CGG repeats) in the 5′-untranslated region (UTR) of the Fragile X Mental Retardation-1 (FMR1) gene (Hagerman et al, 2001; Hagerman and Hagerman, 2004; Ostra and Willemesen, 2009). The major clinical features of FXTAS are progressive intention tremor and gait ataxia, with more variable associated features, including parkinsonism, dysautonomia, anxiety, peripheral neuropathy and cognitive decline (Jacquemont et al, 2003). The neuropathological hallmark of FXTAS is the presence of ubiquitin-positive intranuclear inclusions in both astrocytes and neurons throughout the brain (Greco et al, 2002). It is estimated that nearly 1 in 3000 males has a lifetime risk of developing FXTAS, which would make FXTAS one of the most common single gene causes of tremor, ataxia and cognitive decline among older adults (Jacquemont et al, 2004).
In Fragile X syndrome full mutations (>200 CGG repeats) result in hypermethylation and silencing of the FMR1 gene. In contrast, carriers of the pre-mutation alleles (55–200 CGG repeats) have increased FMR1 mRNA levels but normal, or moderately low, FMR1 protein expression, especially in the upper pre-mutation range (Tassone et al, 2000a, 2000b; Kenneson et al, 2001; Primerano et al, 2002). These observations, coupled with the fact that mRNAs containing expanded CGG repeats accumulate in intranuclear aggregates, suggest a toxic RNA gain-of-function model for FXTAS (Tassone et al, 2004). In support of this hypothesis, a knock-in (KI) mouse model, in which the endogenous eight CGG repeats in the Fmr1 gene has been replaced with an expansion containing ∼100 CGG repeats of human origin, shows ubiquitin-positive intranuclear inclusions and mild neuromotor and behavioural disturbance (Willemsen et al, 2003; Van Dam et al, 2005; Brouwer et al, 2008). Furthermore, sole expression of mRNAs containing 90 CGG repeats outside the context of Fmr1 in a transgenic mouse model is sufficient to recapitulate the neuropathological and molecular features of FXTAS (Hashem et al, 2009). Similarly, heterologous expression of 90 CGG repeats in Drosophila is sufficient to cause neurodegeneration along with formation of neuronal inclusions (Jin et al, 2003). These models show that sole expression of expanded CGG repeats is necessary and sufficient to cause a pathology similar to human FXTAS, and thus indicate that the expanded CGG repeats in RNA are the likely cause of the neurodegeneration in FXTAS.
The FXTAS toxic RNA gain-of-function model show similarities with Myotonic Dystrophies (DM), where expanded CUG or CCUG repeats accumulate in nuclear RNA aggregates that sequester the Muscleblind-like (MBNL1) splicing factor. In DM, partial depletion of the free pool of MBNL1 leads to specific alternative splicing changes, which ultimately result in the symptoms of DM (Ranum and Cooper, 2006; Wheeler and Thornton, 2007). Extending this model to FXTAS, expanded CGG repeats are predicted to sequester specific proteins resulting in loss of their normal molecular functions. Several proteins, including a number of heat-shock proteins, Purα, hnRNP-A2/B1, CUGBP1, MBNL1, lamin-A/C and MBP were found to localize with ubiquitin-positive inclusions in CGG-expressing Drosophila, KI mouse model and FXTAS patients (Iwahashi et al, 2006; Jin et al, 2007; Sofola et al, 2007). However, these proteins were not found to be sequestered by expanded CGG repeats and consequently they are not expected to lose their functions in FXTAS patients.
In this study, we found that in contrast to CUG repeats, expanded CGG repeats accumulate in dynamic intranuclear RNA structures that expand over time. Formation of giant CGG RNA aggregates ultimately results in disorganization of the nuclear lamin structure and cell death. MBNL1 and hnRNP-G proteins were found localized within CGG aggregates but only in the larger inclusions and at late time points after transfection, suggesting these are not the primary defects. In contrast, we identified the Src-Associated substrate during mitosis of 68-kDa (Sam68) protein as the only protein that colocalizes with CGG RNA aggregates at each time point. Sam68 is a nuclear RNA-binding protein involved in alternative splicing regulation (Stoss et al, 2001; Paronetto et al, 2007; Chawla et al, 2009), and its ablation in a mouse knock-out model leads to motor coordination defects (Lukong and Richard, 2008). Sam68 splicing activity, RNA-binding ability and localization are regulated by phosphorylation (Haegebarth et al, 2004; Lukong et al, 2005), and Sam68 interacts with various RNA-binding proteins through several protein–protein interaction domains (Lukong and Richard, 2003). We found that Sam68 is required for subsequent aggregation of MBNL1 and hnRNP G proteins within CGG aggregates. Most importantly, Sam68 is sequestered by expanded CGG-repeat aggregates and thereby loses its splicing-regulatory function. As a consequence, Sam68-regulated splicing is altered in FXTAS patients. Finally, we found that regulation of Sam68 phosphorylation modulates its localization within CGG aggregates. Strikingly, among the various kinase and phosphatase inhibitors tested, we found one, tautomycin, which not only prevents Sam68 colocalization within CGG aggregates, but also abolishes CGG RNA aggregate formation.
Results
Expanded CGG repeats form dynamic nuclear RNA aggregates that expand over time
We first questioned whether pre-mutation-length CGG repeats can form nuclear RNA aggregates in cultured cells. We transfected plasmids expressing either 20, 40, 60 or 100 CGG repeats under the control of a cytomegalovirus (CMV) promoter in various cell lines, and tested the formation of CGG aggregates by RNA fluorescence in situ hybridation (FISH) analysis. We confirmed the specificity of the FISH conditions, and the RNA composition of CGG aggregates, as they were sensitive to RNAse treatment (Supplementary Figure S1). Consistent with an RNA gain-of-function model, expression of 60 or 100 CGG repeats within COS7 cells generated numerous intranuclear CGG aggregates, whereas expression of 20 CGG repeats did not (Figure 1A). Expression of 40 CGG repeats resulted in an intermediate condition with formation of rare small intranuclear aggregates. This is consistent with observations in FXTAS patients in whom it is estimated that ‘normal' CGG polymorphic repeat lengths are 5–45 repeats long, ‘gray zone' alleles contain 45 to 55 repeats and FXTAS patients are defined by pre-mutation allele containing 55–200 CGG repeats (Tassone et al, 2007; Leehey et al, 2008).
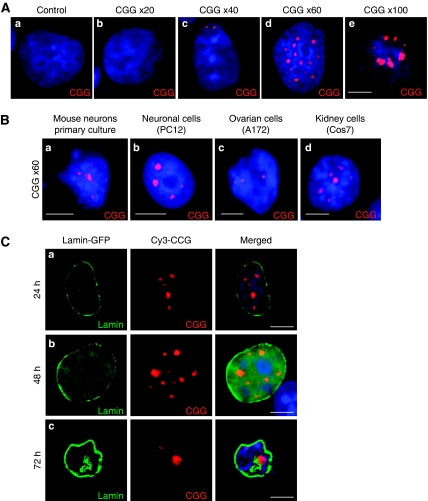
Figure 1.
Expanded CGG repeats form intranuclear RNA aggregates. (A) COS7 cells were transfected with a plasmid expressing either no (a), 20 (b), 40 (c), 60 (d) or 100 (e) CGG repeats, transferred to 0.1% serum to block cell divisions and analysed 24 h after transfection by RNA FISH using a (CCG)8x-Cy3 DNA probe. (B) Primary cultures of hippocampal embryonic mouse neurons (a), differentiated PC12 (b), SKOV3 (c) and COS7 (d) cells were transfected with a plasmid expressing 60 CGG repeats and analysed 24 h after transfection by FISH. (C) COS7 cells were co-transfected with a plasmid expressing 60 CGG repeats and a plasmid expressing GFP-tagged lamin-A, and analysed by RNA FISH either 24 (a), 48 (b) or 72 (c) hours after transfection. In all the figures, the magnification is × 630. The scale bars represent 10 μm; nuclei were counterstained with DAPI and one representative experiment from at least three separate experiments is shown.
Next, expression of CGG aggregates was investigated in various cell types. Transfection of constructs containing 60 CGG repeats led to intranuclear CGG RNA aggregate formation in primary cultures of hippocampal embryonic mouse neurons, as well as in various immortalized cell lines such as neuronal (differentiated PC12), ovarian (SKOV3 and SW626) and kidney (COS7)-derived cell lines (Figure 1B). However, no aggregates were observed in A172, U-937, THP1, HeLa, HEK293, NG108-15, IMR-32, Neuro-2a, SH-SY5Y, SK-N-MC or SK-N-SH cell lines (data not shown), confirming a previous report that not all cell lines can support CGG-repeat aggregate formation (Arocena et al, 2005). Tests of colocalization with various nuclear structures indicated that in transfected cells most of the CGG aggregates are associated with nuclear speckles, but not with other structures such as lamin, nucleoli, PML, Gems or Cajal bodies (Supplementary Figure S2A).
Finally, kinetic formation of CGG aggregates was investigated. COS7 cells were transfected with a plasmid expressing 60 CGG repeats and analysed by RNA FISH either 24, 48 or 72 h after transfection. Surprisingly, expanded CGG repeats formed dynamic nuclear structures that expanded over time, resulting in giant inclusions, nuclear lamin architecture disruption and cell death 72–96 h after transfection (Figure 1C). In contrast, expanded CUG (DM1 mutation) or AUUCU (SCA10 mutation) repeats were less toxic and formed stable nuclear aggregates, whose size did not evolve over time (Supplementary Figure S2B). Annexin-V–PE apoptosis tests were negative indicating that the cytotoxicity of CGG repeats is not linked to apoptotic cell death (data not shown), and in agreement with a previous report (Arocena et al, 2005) no ubiquitin-positive aggregates were observed in transfected cells.
CGG aggregates recruit Sam68, then MBNL1 and hnRNP G
To identify which proteins are associated with expanded CGG repeats, we first adopted an in vitro approach. Proteins extracted from mouse brain or COS7-cell nuclei were captured on streptavidin resin coupled to biotinylated in vitro-transcribed RNA composed of 60 CGG repeats, eluted, separated on SDS–PAGE gels and identified by MALDI-TOF analysis. More than 20 proteins were identified (Supplementary Table 1), including a heat-shock protein and several RNA-binding proteins, including MBNL1 and hnRNP-G.
To discard non-specific binding proteins, we tested for colocalization of these candidates with RNA aggregates in COS7 cells transfected with 60 CGG repeats. CGG aggregates expand over time, suggesting that these repeats may recruit different proteins at different time points. Thus, we tested our candidates at 24 and 72 h after transfection (Figure 2A and B). Colocalization of MBNL1 within CGG aggregates increased from 14% at 24 h to 41% at 72 h after transfection. Similarly, colocalization of hnRNP-G increased from 26 to 73% (Figure 2D), suggesting that CGG aggregates form dynamic structures, which constantly recruit proteins.
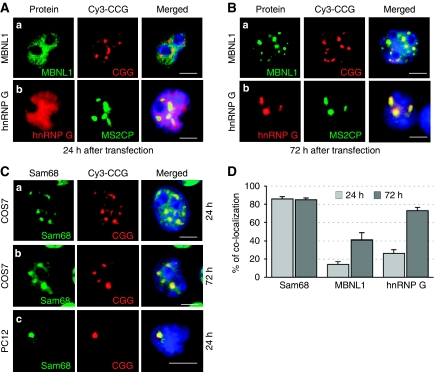
Figure 2.
Sam68 colocalizes with CGG RNA aggregates. (A) COS7 cells were transfected with a plasmid expressing 60 CGG repeats and analysed by FISH/IF using an antibody against MBNL1 (a) 24 h after transfection. None of the antibodies against hnRNP-G that we tested supported FISH conditions. Consequently, endogenous hnRNP-G (b) was analysed by co-transfection of COS7 cells with a plasmid expressing 60 CGG repeats fused to three MS2 tags and a plasmid expressing the GFP-MS2 Coat Protein. The MS2 Coat Protein (MS2CP) possesses a very high and specific affinity for MS2 RNA tags. Endogenous hnRNP-G (b) was detected by IF and the CGG aggregates by localization of the GFP-MS2CP protein, which is bound to the MS2-(CGG)60x RNA. In the absence of MS2-(CGG)60x, GFP-MS2CP was diffuse in the nucleoplasm (data not shown). (B) Similar to panel A but analysed 72 h after transfection. (C) COS7 cells (a, b) or differentiated PC12 neuronal cells (c) were transfected with a plasmid expressing 60 CGG repeats and analysed by FISH/IF using an antibody against Sam68, 24 h (a and c) or 72 h after transfection (b). (D) The percentage of endogenous Sam68, MBNL1 and hnRNP-G colocalized within CGG RNA aggregates in transfected COS7 cells 24 or 72 h after transfection. In all the experiments, three independent transfections totalling a hundred cells were counted, and results are presented as mean±s.d.
By contrast, other in vitro-identified candidates such as SPNR, hnRNP-A1, hnRNP-A2/B, hnRNP-C, hnRNP-D, hnRNP-E and hnRNP-H showed no or very little colocalization with RNA aggregates, and any colocalization observed was only within the giant CGG aggregates that form just before cell death (data not shown). We also observed that neither CUGBP1, nor Purα, colocalized with CGG aggregates in COS7-transfected cells.
These results suggest that MBNL1 and hnRNP-G are not initially recruited by CGG repeats, but join the larger aggregates later on, probably through protein–protein interactions. On the basis of that hypothesis, we searched for proteins that would colocalize with expanded CGG repeats early after transfection, and may capture other RNA-binding proteins through RNA or protein interactions. We screened ∼50 candidates known to bind to RNA or RNA-binding proteins (Supplementary Table 2 and data not shown) and found one, Sam68, that consistently co-localized with CGG RNA aggregates at each time point, including the earliest (Figure 2C and D).
Next, we confirmed by FISH/IF that endogenous Sam68 colocalized with CGG aggregates in neuronal-differentiated PC12 cells (Figure 2C). Similar to COS7 cells, endogenous MBNL1 and hnRNP-G were not recruited within CGG aggregates in PC12 cells 24 h after transfection. We were not able to test Sam68, MBNL1 or hnRNP-G colocalization within CGG aggregates at later transfection time points as PC12 cells are very sensitive to CGG toxicity and die after less than 48 h of CGG expression.
Sam68 is a nuclear protein involved in various aspects of mRNA metabolism and interacts with several RNA-binding proteins, raising question its specificity towards CGG aggregates versus other expanded RNA repeats. COS7 cells were transfected by plasmids expressing either expanded CGG, CUG, CCUG or AUUCU repeats, which are the cause of FXTAS, DM1, DM2 and SCA10 diseases, respectively. We found that 24 or 72 h after transfection, endogenous Sam68 colocalized only with CGG RNA aggregates, but not with other expanded RNA repeats (Figure 3). These results suggest that Sam68 is a specific and early component of CGG aggregates in FXTAS.
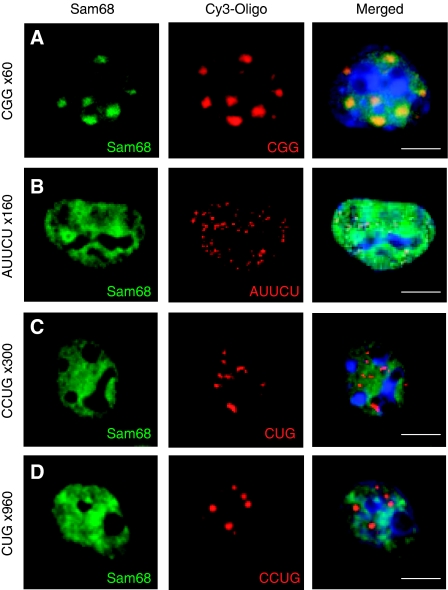
Figure 3.
Sam68 colocalization within nuclear RNA aggregates is specific to expanded CGG repeats. COS7 cells were transfected with a plasmid expressing either 60 CGG repeats (A), 160 AUUCU repeats (B), 960 CUG repeats (C) or 300 CCUG repeats (D). Endogenous Sam68 and CGG repeats were analysed 24 h after transfection by FISH/IF.
Sam68 is essential for recruitment of MBNL1 and hnRNP-G within CGG aggregates
Sam68 is recruited earlier than MBNL1 and hnRNP-G, suggesting sequential recruitment of proteins within the CGG aggregates. Furthermore, Sam68 protein contains several protein–protein interaction domains, raising the question whether Sam68 directly recruits hnRNP-G and MBNL1. In agreement with a previous report (Venables et al, 1999), we found by co-immunoprecipitation that Sam68 interacts with hnRNP-G. However, we found no robust interactions between MBNL1 and Sam68 or between MBNL1 and hnRNP-G (Supplementary Figure S3A).
Next, we mapped the domain required for Sam68 colocalization with CGG aggregates. Sam68 protein contains a central KH RNA-binding domain, and N- and C-terminal protein–protein interaction domains. Deletion of the Sam68 N-terminal domain abolished colocalization with CGG aggregates, whereas deletion of the Sam68 RNA-binding domain did not (Supplementary Figure S3B). Similarly, Sam68 paralog proteins, Slm1 and Slm2 (T-Star), which are devoid of the N-terminal extended region of the Sam68 protein, did not colocalize with expanded CGG aggregates 24 h after transfection. The N-terminal part of the Sam68 protein consists of a number of potential protein–protein interaction domains, suggesting that association of Sam68 with CGG repeats might not be direct but mediated through protein–protein interactions.
Finally, we tested whether Sam68 is required for colocalization of MBNL1 and hnRNP-G within CGG aggregates. Transfection of COS7 cells with an shRNA directed against Sam68 greatly reduced (>80%) the expression of endogenous Sam68 (Figure 4C). Importantly, depletion of endogenous Sam68 abolished the colocalization of MBNL1 and hnRNP-G within CGG aggregates (Figure 4 and Supplementary Table 3). These data show that Sam68 protein is required for subsequent recruitment of MBNL1 and hnRNP-G proteins within CGG aggregates.
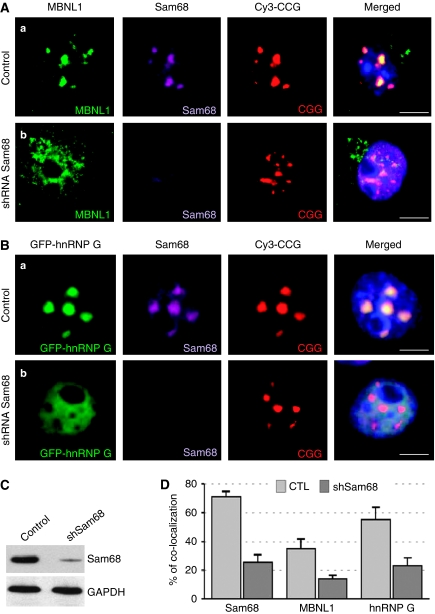
Figure 4.
Sam68 is essential for recruitment of MBNL1 and hnRNP-G within CGG aggregates. (A) COS7 cells were co-transfected with a plasmid expressing 60 CGG repeats and a plasmid expressing either a control LacZ shRNA (a), or a Sam68 shRNA (b), and analysed 72 h after transfection by FISH/IF. Endogenous MBNL1 was detected by IF using an Alexa-488-labelled secondary antibody. Simultaneous detection of Sam68 by IF using a Cy5-labelled secondary antibody confirmed shRNA-mediated depletion of endogenous Sam68. (B) Endogenous hnRNP-G cannot be detected by FISH/IF. Thus, COS7 cells were co-transfected with a plasmid expressing 60 CGG repeats, a plasmid expressing GFP-hnRNP-G and either control (a) or Sam68 (b) shRNA and analysed 72 h after transfection by FISH. (C) shRNA-mediated depletion of endogenous Sam68 was confirmed by western blotting against Sam68. (D) The percentage of endogenous Sam68, MBNL1 and GFP-hnRNP-G colocalized within CGG RNA aggregates 72 h after transfection in COS7 cells transfected with a plasmid expressing either a control or a Sam68 shRNA.
Sam68 colocalizes with CGG aggregates in FXTAS patients
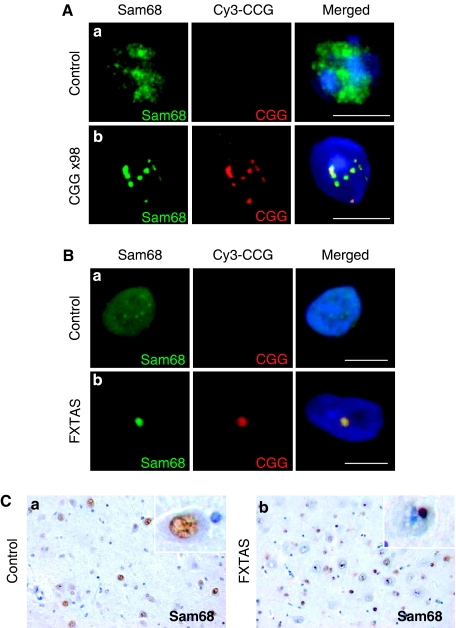
Next, we tested whether Sam68 protein colocalizes with endogenous CGG aggregates. We first analysed the localization of endogenous Sam68 and CGG repeats in the brain sections of a KI mouse model, in which endogenous CGG repeats had been replaced with an expansion of 98 CGG repeats (Willemsen et al, 2003). FISH/IF experiments showed the presence of intranuclear CGG aggregates that colocalized with Sam68 in mice expressing 98 CGG repeats (Figure 5A). By contrast, no RNA aggregates were detected and Sam68 was diffuse throughout the nucleoplasm in control mice (Figure 5A).
Figure 5.
Sam68 colocalizes with endogenous CGG aggregates. (A) Brain sections of mouse expressing either control eight CGG repeats (a) or expanded 98 CGG repeats (b) were analysed by FISH/IF. (B) Similar to panel A. Brain sections (hippocampal area) of age-matched control (a) or FXTAS (b) patients were analysed by FISH/IF. Magnification: × 630. Scale bar, 10 μm. (C) Brain (hippocampal area) sections of age-matched control (a) or FXTAS (b) patients were analysed by immunohistochemistry directed against Sam68. Magnification: × 350.
Then, we investigated the presence of Sam68 and CGG aggregates in FXTAS patients. FISH/IF experiments show that Sam68 consistently colocalized with CGG intranuclear RNA aggregates in the brain sections of FXTAS patients (Figure 5B), but neither Sam68 nor CGG aggregates were found in control brain tissue (Figure 5B). We noted that in FXTAS patients, Sam68 and CGG aggregates were larger and more frequent than in KI mice. This is consistent with the milder neuromotor and behavioural disturbances observed in KI mice as compared with that in FXTAS patients (Willemsen et al, 2003; Van Dam et al, 2005).
Finally, presence of Sam68 aggregates in brain sections of FXTAS patients was confirmed by immunohistochemistry. In brain sections of control patients, Sam68 was diffuse within the nucleoplasm and no detectable Sam68 inclusions were found. In contrast, large intranuclear aggregates of Sam68 were observed in FXTAS patients (Figure 5C), thus confirming that CGG expanded repeats alter Sam68 localization.
Sam68 protein is partially immobilized within CGG expanded repeats
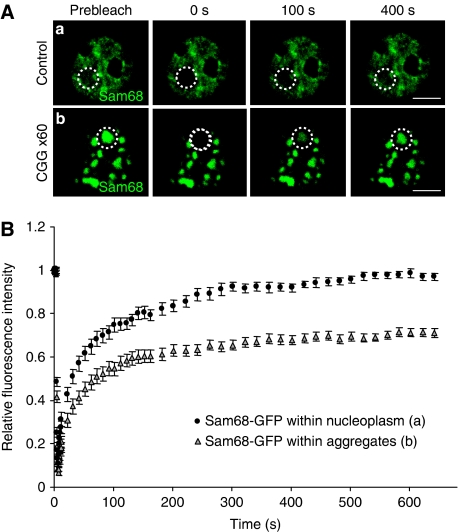
An RNA gain-of-function model for FXTAS predicts that CGG expanded repeats should immobilize Sam68 protein and deplete its molecular activity. To test Sam68 sequestration by CGG repeats, we first analysed its mobility by FRAP (Fluorescence Recovery After Photobleaching) experiments. FRAP of transfected GFP-Sam68 was measured in nuclear regions containing Sam68 aggregates and compared to nuclear regions containing diffuse Sam68 either located within the same nucleus or located in the nuclei of cells not transfected with CGG repeats (Figure 6A). In both cases, nucleoplasmic areas without Sam68 aggregates recovered ∼95% of their initial fluorescence after photobleaching, whereas areas containing Sam68 aggregates recovered only ∼60% of their initial fluorescence (Figure 6B). This shows that a fraction of Sam68 is less mobile in CGG-transfected cells.
Figure 6.
FRAP analysis uncovers an immobile fraction of GFP-Sam68 within CGG aggregates. (A) Photobleaching was performed 24 h after transfection of COS7 cells with GFP-Sam68 and a plasmid expressing either no CGG repeats (a) or 60 CGG repeats (b). The white circles denote the photobleached regions in the aggregates and the nucleoplasm. Representative images show a single z-section obtained before photobleaching (pre-bleach) and at the indicated time points after photobleaching. (B) Recovery curves of photobleached aggregates and nucleoplasm in cells expressing 0 or 60 CGG repeats are shown as relative fluorescence intensity versus time. In CGG-expressing cells, recovery reached a plateau at ∼60% around 300 s. Each data point is the average of 10 nuclei. The error bars indicate the s.e.m.'s.
Sam68 depletion by CGG repeats affects alternative splicing
Sam68 is a nuclear RNA-binding protein with roles in alternative splicing regulation (Stoss et al, 2001; Paronetto et al, 2007; Chawla et al, 2009). According to the RNA gain-of-function model, titration of free nuclear Sam68 into CGG nuclear aggregates should deplete its functional activity and result in detectable pre-mRNA splicing alterations. To test this hypothesis, we co-transfected constructs encoding expanded CGG repeats with minigenes that recapitulate splicing events directly regulated by Sam68.
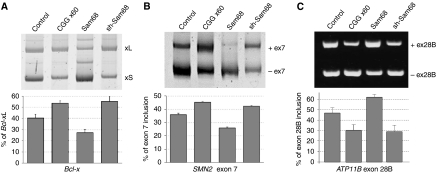
In agreement with a previous report (Paronetto et al, 2007), overexpression of Sam68 with a Bcl-x minigene repressed the formation of the long splicing form, Bcl-xL. We found that overexpression of expanded CGG repeats reproduced a depletion of Sam68 and stimulated the expression of Bcl-xL (Figure 7A). Next, Sam68 paralogues SLM1 and SLM2 are known to regulate the alternative splicing of exon-7 of the survival motor neuron-2 (SMN2) pre-mRNA (Stoss et al, 2004); thus we tested whether Sam68 also regulates SMN2 splicing. We found that overexpression of Sam68 modestly repressed the inclusion of the exon-7 of an SMN2 minigene, while expression of CGG repeats reproduced a depletion of Sam68 and stimulated the inclusion of that exon (Figure 7B). Finally, in a bioinformatic analysis, we identified a novel exon in intron-28 of the human ATP11B gene, which was predicted to be specifically included in the central neural system (Clark et al 2007; Liu et al, 2009). We constructed a minigene in which ATP11B exon-28B and ∼300 bp of its flanking introns were bordered by β-globin exons and co-transfection experiments showed that Sam68 activated its inclusion. In contrast, overexpression of CGG repeats reproduced a depletion of endogenous Sam68 by shRNA and repressed exon-28B inclusion (Figure 7C).
Figure 7.
Alternative splicing is altered in CGG-expressing cells. COS7 cells were co-transfected with a Bcl-x minigene (A), an SMN2 exon-7 minigene (B), or an ATP11B exon-28B minigene (C) and a plasmid expressing either no CGG repeat, or 60 CGG repeats, Sam68 or Sam68 shRNA. Bcl-xL/S, SMN2 and ATP11B splicing isoforms were identified 24 h after transfection by RNA extraction followed by RT–PCR.
hnRNP-G and MBNL1 are also recruited within CGG aggregates, raising the question whether the splicing defects observed in CGG expressing cells are only due to Sam68 sequestration. To test that hypothesis, we used a mutant of Sam68 deleted of its N-terminal domain (Sam68ΔNter), which was no longer recruited within CGG aggregates (Supplementary Figure S3B), but was still able to regulate alternative splicing (Supplementary Figure S4). Co-transfection of Sam68ΔNter rescued the splicing defects caused by expression of CGG repeats (Supplementary Figure S4), suggesting that the splicing alterations observed in CGG-expressing cells were mostly due to sequestration of Sam68.
Alternative splicing is altered in FXTAS patients
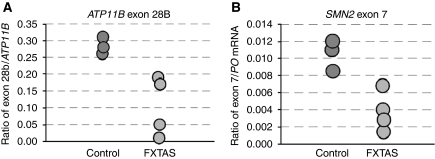
Sam68 has reduced nuclear spatial mobility and splicing regulatory function in CGG-expressing cells; so we tested whether alternative pre-mRNA splicing is altered in human FXTAS patients. RT–PCR analysis of ATP11B alternative splicing in brain samples of control and FXTAS patients showed a significant decrease of exon-28B inclusion from 47±2% in control to 31±8% in FXTAS patients (P<0.005). We confirmed these results by qRT–PCR and found that expression of ATP11B exon-28B is downregulated in FXTAS patients as compared with that in a control (Figure 8A). Similarly, we tested by qRT–PCR the expression of exon-7 of the SMN2 pre-mRNA and found it under-expressed in FXTAS patients (Figure 8B). By contrast, we found no significant differences in the expression of SMN1 exon-7, which is consistent with no alternative splicing regulation of that exon. These results are consistent with alternative splicing being altered in FXTAS patients.
Figure 8.
Alternative splicing is altered in FXTAS patients. (A) qRT–PCR analysis of ATP11B exon-28B in brain RNA samples of three controls and four FXTAS patients. Results are expressed as the ratio between alternative ATP11B exon-28B and constitutive ATP11B exons 21–22. (B) qRT–PCR analysis of SMN2 exon-7 in brain RNA samples from three controls and four FXTAS patients. We were not able to discriminate between SMN1 and SMN2 constitutive exons; therefore, results are expressed as the ratio of the alternative SMN2 exon-7/PO mRNA.
Tyrosine phosphorylation of Sam68 regulates its recruitment within CGG aggregates
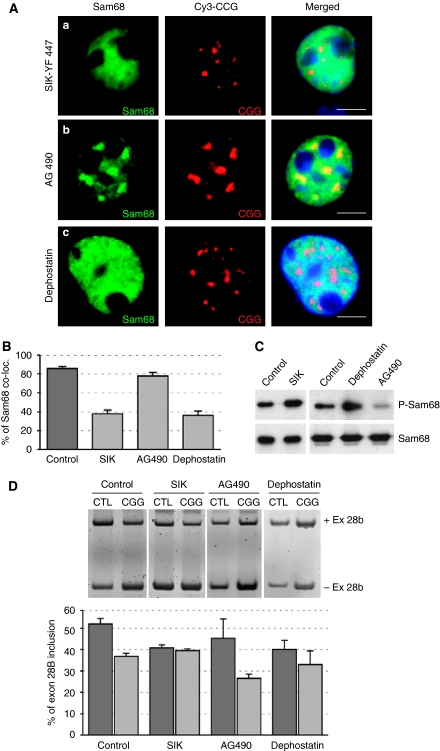
Sam68 is a substrate of the SIK/BRK-tyrosine kinase, which regulates the RNA-binding ability and localization of the Sam68 protein (Derry et al, 2000; Haegebarth et al, 2004; Lukong et al, 2005). We thus tested the effect of Sam68 phosphorylation on the formation of CGG aggregate. We found that expression of SIK/BRK disrupted Sam68 protein localization within CGG aggregates and returned Sam68 to a free nucleoplasmic localization (Figure 9A and B). Sam68 is tyrosine-phosphorylated in response to EGF treatment (Lukong et al, 2005). We found that inhibition of the EGFR tyrosine kinase pathways through tyrphostin/AG490 treatment stimulated the recruitment of Sam68 within CGG aggregates, leading to formation of large aggregates at early time points. By contrast, treatment of transfected cells with dephostatin, a tyrosine phosphatase inhibitor, reduced the recruitment of Sam68 within CGG aggregates (Figure 9A and B). These data suggest a model in which tyrosine phosphorylation of Sam68, mediated by the EGFR-SIK/BRK tyrosine kinase pathway, reduces the recruitment of Sam68 within CGG aggregates (Supplementary Figure S6A).
Figure 9.
Tyrosine phosphorylation reduces Sam68 colocalization within CGG aggregates. (A) COS7 cells were transfected with a plasmid expressing 60 CGG repeats and either co-transfected with a plasmid expressing a constitutively active YF447 mutant of the SIK/Brk kinase (a), or treated with 10 μM of tyrphostin/AG490 (b) or 20 μM dephostatin (c) and analysed by FISH/IF 24 h after transfection. (B) The percentage of endogenous Sam68 colocalized within CGG RNA aggregates in transfected COS7 cells 24 h after transfection. (C) Similar to panel A but phosphorylation of Sam68 was assayed by Sam68 immunoprecipitation followed by western blotting using a Sam68-Y440 phospho-specific antibody. (D) Similar to panel A but COS7 cells were also co-transfected with an ATP11B minigene. Inclusion of exon-28B was quantified 24 h after transfection by RNA extraction followed by RT–PCR.
We confirmed that Sam68 phosphorylation was stimulated by transfection of the SIK/BRK kinase or by inhibition of tyrosine phosphatase pathways through dephostatin treatment. By contrast, inhibition of the EGFR pathway (tyrphostin/AG490 treatment) reduced Sam68 phosphorylation (Figure 9C).
Tyrosine phosphorylation of Sam68 inhibits its splicing activity (Paronetto et al, 2007). We found that transfection of SIK/BRK or dephostatin treatment, both of which induced Sam68 tyrosine phosphorylation (Figure 9C), modified the splicing of ATP11B and mimicked a depletion of Sam68 by CGG or shRNA expression (Figure 9D and data not shown). Interestingly, we noted no cumulative effects on splicing when CGG-expressing cells were co-transfected with SIK/BRK or treated with dephostatin. These data suggest that either ATP11B splicing regulation was saturated or that CGG repeats acted similarly to SIK/BRK or dephostatin, probably through inhibition of Sam68 activity. We observed a similar absence of cumulative effects when CGG repeats and Sam68 shRNA were coexpressed (data not shown), suggesting that CGG repeats acted on splicing mostly through Sam68 depletion.
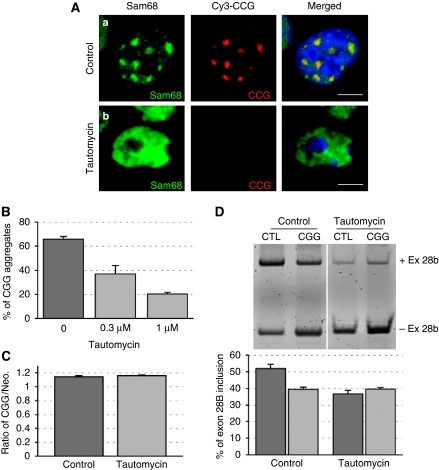
Tautomycin reduces the formation of CGG aggregates
Among the various inhibitors of kinases and phosphatases that we tested, we found one, tautomycin, which not only reduced Sam68 colocalization but also reduced CGG aggregate formation (Figure 10A and Supplementary Table 3). By contrast, tautomycin treatment did not impair the formation of expanded CUG- or AUUCU-repeat RNA aggregates (data not shown).
Figure 10.
Tautomycin abolishes CGG and Sam68 aggregate formation. (A) COS7 cells were co-transfected with a plasmid expressing 60 CGG repeats and either treated with solvent (a) or 1 μM of tautomycin (b), then analysed by FISH/IF 24 h after transfection. (B) The percentage of cells containing CGG RNA aggregates in transfected COS7 cells treated with solvent, 0.3 or 1 μM of tautomycin. (C) Similar to panel A but followed by RNA extraction 24 h after transfection and qRT–PCR quantification of the CGG repeats RNA and of the neomycin mRNA. CGG and neomycin cassettes are expressed from the same pcDNA3.1 plasmid, but under different promoters. (D) Similar to panel A or C but COS7 cells were also co-transfected with an ATP11B minigene. Inclusion of exon-28B was identified 24 h after transfection by RNA extraction followed by RT–PCR.
We confirmed by qRT–PCR that the quantity of RNA containing the expanded CGG repeats was not altered by tautomycin treatment (Figure 10C), suggesting that tautomycin disrupts CGG aggregation without altering their expression.
Finally, we observed that in the presence of tautomycin, ATP11B splicing was no longer altered by expression of expanded CGG repeats (Figure 10D), suggesting that tautomycin reduced the deleterious effects of the CGG repeats on splicing. However and as noted previously (Mermoud et al, 1992; Novoyatleva et al, 2008), treatment with tautomycin alone resulted in splicing changes (Figure 10D), rendering difficult to pinpoint the precise role of tautomycin on the deleterious effects of CGG repeats on splicing.
Discussion
CGG aggregates are dynamic structures that recruit various RNA-binding proteins sequentially
A striking characteristic of CGG expanded repeats is that they form dynamic intranuclear RNA aggregates that enlarge with time, resulting in the formation of giant inclusions, an observation that stands in contrast to the absence of growth over time of expanded CUG (DM1 mutation), CCUG (DM2) or AUUCU (SCA10) repeats. Continuous enlargement of CGG RNA aggregates suggests that these repeats may constantly recruit proteins.
Consistent with that hypothesis, we and others have found various proteins, mainly RNA-binding proteins, to be captured by CGG repeats in vitro (Iwahashi et al, 2006; Jin et al, 2007; Sofola et al, 2007), suggesting that CGG repeats can sequester a large number of proteins through direct RNA–protein interactions, but probably also through indirect protein–protein interactions. When tested in cells, we found that endogenous Sam68, MBNL1 and hnRNP-G colocalized with CGG aggregates. In contrast, other candidate proteins such as Purα, FMRP and CUGBP1 did not colocalize with expanded CGG RNA, whereas hnRNP-A1, hnRNP-A2/B, hnRNP-C, hnRNP-D, hnRNP-E and hnRNP-H presented an intermediary situation with some colocalization, but only at very late time points and within the giant CGG aggregates that form in dying cells. This questions whether these other candidate proteins are recruited specifically at the very last step of CGG aggregate formation, or that they form non-specific aggregates in dying cells. We tested the localization of some of these candidates by FISH/IF in brain sections of control and FXTAS patients.
First, we tested Purα, but could only detect small quantities of Purα within the neuronal intranuclear inclusions in sections from human FXTAS brain as compared with abundant cytoplasmic labelling in the same neurons, which suggests that Purα is still available for its cellular function. This is in agreement with a recent report showing absence of Purα-positive inclusions in a FXTAS mouse model expressing 90 CGG repeats (Hashem et al, 2009). However, Purα was found to colocalize with HSP70 inclusions in CGG-expressing Drosophila and in a single reported human inclusion (Jin et al, 2007). A possible explanation is that Purα, which is cytoplamic, is not found for the most part in CGG aggregates, which are strictly intranuclear in the mammalian cells and FXTAS patients that we analysed (Greco et al, 2006). In contrast, in CGG-repeat-expressing Drosophila, a substantial fraction of the HSP70 inclusions were cytoplasmic and found to colocalize with Purα (Jin et al, 2003, 2007).
Next, we found that MBNL1, which is recruited tardily within CGG aggregates in transfected cells, also colocalizes with CGG inclusions in FXTAS patients (Supplementary Figure S5, Iwahashi et al, 2006). However, we found that the splicing events regulated by MBNL1 are not altered in CGG-expressing cells or in FXTAS patients (Supplementary Figure S5). These results suggest that, while MBNL1 is present within CGG aggregates, it is not immobilized and does not lose its splicing function. We also tested the presence of hnRNP-G within CGG aggregates in FXTAS patients, but none of the anti-hnRNP-G antibodies that we tested resisted the FISH conditions. Thus, whether hnRNP G colocalizes with CGG aggregates in FXTAS patients, remains to be determined.
Finally, we found that, while very late recruited within CGG aggregates, and only in dying cells, both hnRNP-A1 and hnRNP-A2/B1 were present in the CGG aggregates of FXTAS patients (data not shown; Iwahashi et al, 2006). By contrast, little or no hnRNP-A2/B1 aggregates were observed in CGG KI mice (Hashem et al, 2009 and R Willemsen, personal communication), which is consistent with smaller CGG aggregates in mice as compared with that in humans. hnRNP-A1 regulates the alternative splicing of exon-7 of the APP pre-mRNA (Donev et al, 2007), but we found no splicing alterations of APP in FXTAS patients (Supplementary Figure S5), suggesting that, similar to MBNL1, recruitment of hnRNP-A1 within CGG aggregates may not impair its splicing-regulatory function.
The presence of hnRNP-A1 and hnRNP-A2/B1 aggregates only in FXTAS patients is reminiscent of the ubiquitin situation, where no ubiquitin aggregates are found in CGG-transfected cells, while ubiquitin-positive inclusions are a hallmark of FXTAS patients (Greco et al, 2002; Arocena et al, 2005). Overall, these data suggest that accumulation of some proteins, such as MBNL1, hnRNP-A1, hnRNP-A2 and ubiquitinylated proteins, in CGG aggregates of brain from FXTAS patients is a very late-onset event, which may not result in sequestration and loss of function of these proteins.
Sam68 nucleation model
In contrast to late recruited proteins, we found that Sam68 is early and constantly associated with CGG aggregates. Furthermore, we found that depletion of Sam68 abolished the recruitment of MBNL1 and hnRNP-G within CGG aggregates and reduced the formation of giant aggregates. These results suggest that Sam68 is essential for recruitment of novel proteins and the continuous enlargement of the CGG aggregates. Sam68 is involved in various aspects of mRNA metabolism and contains several domains enabling protein–protein interactions, allowing Sam68 to interact with various RNA-binding proteins, including hnRNP-G, hnRNP-A1, Tra2β (SFRS10), Slm1 and Slm2 (T-Star) (Venables et al, 1999, 2000; Paronetto et al, 2007). Co-transfection of these candidates with expanded CGG repeats showed that Slm1, Slm2 and Tra2β colocalized with large CGG aggregates 48 h after transfection. We propose that these proteins can in turn associate with other proteins such as MBNL1 and probably many others, ultimately resulting in massive protein aggregation disastrous for normal cell function and viability (Supplementary Figure S6B). This ‘sequential recruitment' model is consistent with the late colocalization of hnRNP-A, hnRNP-G and MBNL1 in transfected cells, and with the large intranuclear inclusions, cell death and global cerebral and cerebellar atrophy observed in FXTAS patients (Greco et al, 2002, 2006; Tassone et al, 2004).
Such a model of sequential protein recruitment within CGG aggregates implies a founding and nucleating RNA–protein interaction event that would subsequently trap other proteins through indirect RNA–protein or protein–protein interactions. We propose that Sam68 is part of this founding event, as its depletion by shRNA inhibits subsequent protein aggregation and suppresses the formation of giant CGG aggregates. However, Sam68 does not bind directly to CGG RNA (data not shown), and its localization within CGG aggregates requires its N-terminal domain (Supplementary Figure S3), which contains a number of protein–protein interaction domains. These data suggest that association of Sam68 with CGG repeats it not direct but requires an intermediary RNA-binding protein (Supplementary Figure S6B). Characterization of that protein is ongoing (C Sellier, in preparation), but beyond the scope of this paper.
Sam68 protein is partially sequestered and functionally inactivated in FXTAS patients
The RNA gain-of-function model for FXTAS predicts that Sam68 protein should be sequestered by CGG RNA repeats and consequently lose its activity. FRAP analysis showed that there is a significant decrease in the mobile fraction of Sam68 in CGG-transfected cells, suggesting that a fraction of nuclear Sam68 protein is immobilized within Sam68 aggregates. Interestingly, the proportion of Sam68 immobilized in CGG-transfected cells is identical to the fraction of MBNL1 immobilized in CUG-transfected cells (Ho et al, 2005). Through analogy with DM, this shows partial depletion of free Sam68 in CGG-expressing cells, and hence confirms an RNA gain-of-function model for FXTAS.
Sam68 is involved in various aspects of mRNA metabolism, such as alternative splicing regulation, nuclear export, somatodendritic transport, polyadenylation and translation (reviewed by Lukong and Richard, 2003). siRNA depletion of Sam68 impairs neuronal differentiation in cell cultures (Chawla et al, 2009), and its ablation in mouse leads to motor coordination defects (Lukong and Richard, 2008), suggesting that partial Sam68 sequestration and loss of function might be involved in the progressive intention tremor and gait ataxia observed in FXTAS patients.
One of the important functions of Sam68 is in alternative splicing regulation (Stoss et al, 2001; Paronetto et al, 2007; Chawla et al, 2009). We found mis-regulation of pre-mRNA alternative splicing controlled by Sam68 in CGG-transfected cells and in FXTAS patients. Notably, analysis of alternative splicing of ATP11B pre-mRNA showed a splicing mis-regulation similar in FXTAS patients and in Sam68-depleted cells by shRNA transfection. These results are consistent with partial loss of function of Sam68 in CGG-transfected cells and in FXTAS patients. Phospho-type-4 ATPase-11B (ATP11B) is a putative flippase predicted to catalyse aminophospholipid transport and create lipid asymmetry in late secretory and endocytic compartments. Exclusion of exon-28B results in a protein isoform with a distinct C-terminal part. The physiological functions of this splice form remain to be determined.
Next, we tested other splicing events regulated by Sam68 (Chawla et al, 2009). However, we found no significant alterations of alternative splicing of the KTN1, BIN1, DNCIC2, CLASP2 and SGCE2 pre-mRNAs in brain samples of FXTAS patients, possibly due to differences between human patients and murine cells where they were identified as Sam68 targets (Chawla et al, 2009). Furthermore, we found no significant alterations of ATP11B, BIN1, DNCIC2 and CLASP2 alternative splicing in brain samples of Sam68-knockout mice, probably due to the compensatory effects of Sam68 paralogues, Slm1 and Slm2, which are highly expressed in brain (Stoss et al, 2004). We also tested the alternative splicing of Bcl-x and CD44 pre-mRNAs in FXTAS patients (Matter et al, 2002; Paronetto et al, 2007). However, transcripts containing the exon-v5 of the CD44, or corresponding to the Bcl-x Short isoform, were not expressed in the brain samples that we tested.
Finally, we tested the alternative splicing of the SMN1 and SMN2 pre-mRNAs. Loss of the SMN1 gene is responsible for spinal muscular atrophy (SMA), the most common inherited motor neuron disease. The near-identical SMN2 gene does not compensate the deficiency of SMN1 due to skipping of SMN2 exon-7 through alternative splicing. We found that Sam68 weakly regulates the alternative splicing of an SMN2 minigene and inhibits the inclusion of exon-7. However, we found in FXTAS patients decreased expression of SMN2 exon-7 was, which is contradictory with depletion of Sam68 function in FXTAS. SMN2 exon-7 inclusion is robustly stimulated by hnRNP G and Tra2β (Hofmann and Wirth, 2002; Heinrich et al, 2009), which are both late recruited within CGG aggregates in COS7-transfected cells. Whether hnRNP-G and Tra2β are sequestered within CGG aggregates and lose their splicing functions in FXTAS patients remain to be determined. Furthermore, whether decrease of SMN2 exon-7 in FXTAS patients results in lower quantity of SMN protein and is involved in the neuronal death observed in these patients remains also to be determined.
Tautomycin inhibits the formation of CGG aggregates
Sam68 activity is regulated by phosphorylation (Derry et al, 2000; Haegebarth et al, 2004; Lukong et al, 2005), a characteristic that made Sam68 the prototype of the Signal Transducers and Activators of RNA (STAR) family, which transduces information from signalling pathways to mRNA metabolism. Consistent with such a function, we found that tyrosine phosphorylation of Sam68 reduces its recruitment within CGG aggregates and, consequently, the deleterious effects of the CGG repeats on splicing regulation (Supplementary Figure S6A). By contrast, we found that activation of the MAP serine/threonine kinase pathway stimulates Sam68 recruitment within CGG aggregates. However, we failed to detect a direct phosphorylation of Sam68 due to the poor quality of the serine and threonine antibodies that we tested (Supplementary Figure S7).
Strikingly, among the various drugs tested, we found one, tautomycin, which not only prevents Sam68 colocalization with CGG aggregates, but also reduces CGG aggregate formation. We found that tautomycin prevents the deleterious effects of the CGG repeats on alternative splicing regulation, but due to the inherent toxicity of tautomycin, we were not able to assess whether tautomycin also reduces the toxicity of the CGG repeats. By contrast to tautomycin, depletion of Sam68 by shRNA does not alter the formation of CGG aggregates, suggesting that tautomycin may act upstream from Sam68, maybe on the uncharacterized protein that bridges CGG repeats and Sam68 (Supplementary Figure S6B). Tautomycin was first described as an inhibitor of the serine/threonine PP1 phosphatase, and more recently as an inhibitor of the glycogen synthase kinase-3β (GSK-3β) (Adler et al, 2009) and of the Raf1 pathway (Pinchot et al, 2009). Whether formation of CGG aggregates requires the PP1, GSK-3β or Raf pathway remains to be determined.
In conclusion, our results support an RNA gain-of-function mechanism in which Sam68 is partially sequestered within CGG aggregates and consequently loses its regulatory function in neurons from FXTAS patients. Furthermore, our data suggest that CGG aggregates can be dispersed, thus, bringing hope of drug treatments able to reduce CGG aggregate formation in FXTAS patients.
Materials and methods
Plasmids and constructions
Plasmids expressing 20, 40 or 60 CGG repeats were constructed by ligation of oligonucleotides containing 20 CGG repeats in pcDNA3.1. The plasmid expressing 98 CGG repeats was described previously (Arocena et al, 2005). ATP11B exon-28B was amplified using primers Fwd: 5′-AAAAAAAACAATTGCCCTAAATCTTGGTGGCAAATG and Rev: 5′-AAAAAAAACAATTGGTTGTGAGAATATCTTCACAGC, using BAC RP11-36G17 as template, and cloned within the vector pXJ41. Sam68 WT and mutants, Slm1, Slm2 and hnRNP-G expression plasmids were described previously (Venables et al, 2004).
Cell cultures, transfections and treatments
COS7 cells were cultured in Dulbecco's modified Eagle's Medium (DMEM), 10% foetal bovine serum and gentamicin at 37°C in 5% CO2. PC12 were cultured in DMEM, 10% horse serum, 5% foetal calf serum and penicillin at 37°C, 5% CO2. Cells were plated on glass coverslips in a 24-well plate for immunofluorescence and transfected 24 h after plating in DMEM+0.1% foetal bovine serum to block cell divisions, using either FugenHD (Roche) for COS7 cells or Lipofectamine 2000 (Invitrogen) for PC12 cells. PC12 cells were differentiated 6 h after transfection by growing cells in 1% horse serum, 1% foetal calf serum plus 50 ng/ml of NGF (Clinisciences). Primary cultures of hippocampal neurons were obtained from WT E18 mouse and grown into 24-wells plates in 500 μl neurobasal medium (Gibco), 1 × B27 (Gibco), 0.5 mM L-glutamine and penicillin at 37°C, 5% CO2, and were transfected after 4 DIV with Lipofectamine 2000 (Invitrogen).
Cell treatments
Cells were incubated 6 h after transfection in 50 μM PD98059, 4 nM okadaic acid, 40 ng/ml TPA, 10 μM AG490, 20 μM dephostatin or 1 μM tautomycin (Calbiochem).
Patients and brain sections
FXTAS patients have been described previously (case 6, 7 9 and 10 of Greco et al, 2006). KI mouse (72 weeks old, 98 CGG repeats) or human brain sections were deparaffinized two times for 20 min in Histosol Plus (Shandon) and dehydrated as follows: twice in ethanol 100% (5 min), twice in ethanol 95% (5 min), once in ethanol 80% (5 min), once in ethanol 70% (5 min) and rinsed in TBS-Tween 1% before FISH.
FISH combined with immunofluorescence
Glass coverslips containing plated cells or brain sections on slides treated as described above were fixed in 4% paraformaldehyde in PBS (pH 7.4) for 15 min and washed three times with PBS. The coverslips were incubated for 5 min in PBS/0.5% Triton X-100 and washed three times with PBS before pre-hybridization in 40% DMSO, 40% formamide, 10% BSA (10 mg/ml), 2 × SCC for 30 min. The coverslips were hybridized for 2 h in 40% formamide, 10% DMSO, 2 × SCC, 2 mM vanadyl ribonucleoside, 60 μg/ml tRNA, 30 μg/ml BSA plus 0.75 μg (CCG)8x-Cy3 DNA oligonucleotide probe (Sigma). The coverslips were washed twice in 2 × SCC/50% formamide and twice in 2 × SCC. Following FISH, the coverslips were washed twice successively in 2 × SCC/50% formamide, in 2 × SCC and in PBS. The coverslips were incubated overnight with primary anti-hnRNP-G (1/200 dilution; Heinrich et al, 2009), hnRNP-A2/B1 (1/200 dilution, clone DP3B3; Tebu), hnRNP-A1 (1/200 dilution, clone 9H10; Abcam), MBNL1 (1/200 dilution, clone HL1822 3A4-1E9; Sigma) and Sam68 (1/400 dilution, C20 SC-333; Santa Cruz Biotechnology) at 4°C. The coverslips were washed twice with PBS before incubation with a goat anti-rabbit secondary antibody conjugated with Alexa-Fluor 488 (1/500 dilution; Fisher) or Cy5 (1/500 dilution; Interchim) for 60 min. Then, the coverslips were incubated for 10 min in 2 × SCC/DAPI (1/10 000 dilution) and rinsed twice in 2 × SSC before mounting in Pro-Long media (Molecular Probes). Slides were examined using either a simple fluorescence microscope (Leica) or a Leica DM4000 B confocal microscope, equipped with a Leica 100 × HCX Plan Apo CS 1.40 objective, in 1-μm optical sections.
FRAP analysis
COS7 cells were plated in 35-mm glass base dish (Iwaki) and transfected with Fugene HD 24 h after plating. Twenty-four hours after transfection, FRAP experiments were performed using a Leica DM4000 B confocal microscope combined to a heated stage. The cells were maintained for 15 min in growth media at 37°C with no CO2 or humidifier systems; five single scans were obtained followed by five bleach pulses. After photobleaching, images were taken every 10 s for 150 s (post-bleach 1) followed by 40 acquisitions every 20 s (post-bleach 2). Fluorescence intensities were calculated using the Image J software and normalized by total cellular fluorescence intensity. As foci are mobile, an area around the foci was determined for the duration of the experiment.
MALDI analysis
Nuclear extracts were prepared from COS7 cells or mouse brain extract as described by Dignam et al (1983). Nuclear extract was passed over a CGG60x in vitro T7 transcribed and biotinylated RNA (Ambion) bound to a streptavidin agarose column (Invitrogen) in the presence of KCl (100 mM), HEPES (10 mM) and MgCl2 (1 mM). The column was washed with 33 mM MgCl2 and glacial acetic acid. The eluted protein was separated by gel electrophoresis and detected by silver staining. The protein bands were excised, digested and identified using a Reflex IV MALDI-TOF spectrometer (Bruker Daltonics) and the Profound search engine, as described by Argentini et al, 2008.
RNA isolation and PCR
COS cells were cultured as described above in a six-well plate. RNA was isolated 24 h after transfection using a GenElute kit (Sigma). Endogenous ATP11B splicing analysis was performed on total RNA extracted (Trizol reagent) from control or FXTAS patient brain samples. cDNA synthesis reactions were performed with Superscript II (Invitrogen). The primers are described in Supplementary Table 4. PCRs were performed with Taq polymerase (Roche). The conditions for SMN2 and Bcl-x minigenes were described previously (Paronetto et al, 2007; Heinrich et al, 2009). The conditions for ATP11B minigene were 4 min at 94°C; 30 cycles of 40 s at 94°C, 45 s at 60°C and 1 min at 72°C; and a final extension at 72°C for 4 min. The PCR reaction products were analysed on 8% polyacrylamide gels.
Statistical analysis
The percentage of endogenous Sam68, MBNL1 and hnRNP-G colocalized within CGG RNA is expressed as the number of nuclei presenting colocalization/total number of nuclei containing CGG aggregates. Three independent transfections totalling a hundred cells were counted. Results are presented as mean±s.d. The balance between the mRNA levels of the SMN2, Bcl-x and ATP11B splicing variants is calculated as ((mRNA+exon)/(mRNA−exon+mRNA+exon)) × 100. The results are derived from at least three independent experiments. The error bars in the figures indicate the s.e.m.'s. Statistical differences were calculated using t-test.
Supplementary Material
Acknowledgments
We thank the imagery, cell culture and MALDI platforms of the IGBMC; Thomas Cooper for the gift of the DT960 and INSR minigene plasmid; Laura Ranum for the gift of the CCTG300 expression plasmid; Charles Thornton for the gift of the polyclonal MBNL1 antibody; Angela Tyner for the gift of the SIK/Brk constructs; Claudio Sette for the gift of the Bcl-x, shSam68 and Sam68 WT and mutant constructs; Stefan Stamm for the gift of the SMN2 minigene; Stefan Hubner for the gift of the GFP-lamin-A construct; and all members of the French DM Network for fruitful discussion. This work was supported by INSERM AVENIR funding (NCB), ANR GENOPAT grant P007942 (NCB), NIH-NINDS grant NS062411 (RW), NIH Roadmap Initiative DE019583 and AG032119 (PJH), BBSRC grant BB/D013917/1 (DJE), Wellcome Trust grant number WT080368MA (DJE) and an ORSAS international studentship (YL).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adler JT, Cook M, Luo Y, Pitt SC, Ju J, Li W, Shen B, Kunnimalaiyaan M, Chen H (2009) Tautomycetin and tautomycin suppress the growth of medullary thyroid cancer cells via inhibition of glycogen synthase kinase-3beta. Mol Cancer Ther 8: 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argentini M, Strub JM, Carapito C, Sanglier S, Van-Dorsselaer A (2008) An optimized MALDI mass spectrometry method for improved detection of lysine/arginine/histidine free peptides. J Proteome Res 7: 5062–5069 [DOI] [PubMed] [Google Scholar]
- Arocena DG, Iwahashi CK, Won N, Beilina A, Ludwig AL, Tassone F, Schwartz PH, Hagerman PJ (2005) Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG repeat RNA in human cultured neural cells. Human Mol Genet 14: 3661–3671 [DOI] [PubMed] [Google Scholar]
- Brouwer JR, Huizer K, Severijnen LA, Hukema RK, Berman RF, Oostra BA, Willemsen R (2008) CGG repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J Neurochem 107: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G, Lin CH, Han A, Shiue L, Ares M Jr, Black DL (2009) Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol Cell Biol 29: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE (2007) Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol 8: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry JJ, Richard S, Valderrama Carvajal H, Ye X, Vasioukhin V, Cochrane AW, Chen T, Tyner AL (2000) SIK (BRK) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol Cell Biol 20: 6114–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donev R, Newall A, Thome J, Sheer D (2007) A role for SC35 and hnRNPA1 in the determination of amyloid precursor protein isoforms. Mol Psychiatry 12: 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, Hessl D, Becker EJ, Papazian J, Leehey MA, Hagerman RJ, Hagerman PJ (2006) Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS). Brain 1: 243–255 [DOI] [PubMed] [Google Scholar]
- Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, Leehey M, Hagerman PJ (2002) Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain 125: 1760–1771 [DOI] [PubMed] [Google Scholar]
- Haegebarth A, Heap D, Bie W, Derry JJ, Richard S, Tyner AL (2004) The nuclear tyrosine kinase BRK/SIK phosphorylates and inhibits the RNA-binding activities of the Sam68-like mammalian proteins SLM-1 and SLM-2. J Biol Chem 279: 54398–54404 [DOI] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ (2004) The fragile-X premutation: a maturing perspective. Am J Hum Genet 74: 805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ (2001) Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 1: 127–130 [DOI] [PubMed] [Google Scholar]
- Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, Nelson DL (2009) Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum Mol Genet 18: 2443–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, Sperling J, Sperling R, Stamm S (2009) Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J Biol Chem 284: 14303–14315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA (2005) Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci 118 (Part 13): 2923–2933 [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Wirth B (2002) hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet 11: 2037–2049 [DOI] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ (2006) Protein composition of the intranuclear inclusions of FXTAS. Brain 129: 256–271 [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Leehey M, Grigsby J, Zhang L, Brunberg JA, Greco C, Des Portes V, Jardini T, Levine R, Berry-Kravis E, Brown WT, Schaeffer S, Kissel J, Tassone F, Hagerman PJ (2003) Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am J Hum Genet 4: 869–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Leehey MA, Hall DA, Levine RA, Brunberg JA, Zhang L, Jardini T, Gane LW, Harris SW, Herman K, Grigsby J, Greco CM, Berry-Kravis E, Tassone F, Hagerman PJ (2004) Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA 4: 460–469 [DOI] [PubMed] [Google Scholar]
- Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST (2007) Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55: 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST (2003) RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 39: 739–747 [DOI] [PubMed] [Google Scholar]
- Kenneson A, Zhang F, Hagedorn CH, Warren ST (2001) Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet 10: 1449–1454 [DOI] [PubMed] [Google Scholar]
- Leehey MA, Berry-Kravis E, Goetz CG, Zhang L, Hall DA, Li L, Rice CD, Lara R, Cogswell J, Reynolds A, Gane L, Jacquemont S, Tassone F, Grigsby J, Hagerman RJ, Hagerman PJ (2008) FMR1 CGG repeat length predicts motor dysfunction in premutation carriers. Neurology 16: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bourgeois C, Pang S, Kudla M, Dreumont N, Kister L, Sun YH, Stevenin J, Elliott DJ (2009) The germ cell nuclear proteins hnRNP G-T and RBMY activate a testis-specific exon. PLOS Genet 5: e1000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Larocque D, Tyner AL, Richard S (2005) Tyrosine phosphorylation of Sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J Biol Chem 280: 38639–38647 [DOI] [PubMed] [Google Scholar]
- Lukong KE, Richard S (2003) Sam68, the KH domain-containing superstar. Biochim Biophys Acta 1653: 73–86 [DOI] [PubMed] [Google Scholar]
- Lukong KE, Richard S (2008) Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav Brain Res 189: 357–363 [DOI] [PubMed] [Google Scholar]
- Matter N, Herrlich P, König H (2002) Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695 [DOI] [PubMed] [Google Scholar]
- Mermoud JE, Cohen P, Lamond AI (1992) Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res 20: 5263–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AH, Bollen M, Stamm S (2008) Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum Mol Genet 17: 52–70 [DOI] [PubMed] [Google Scholar]
- Ostra BA, Willemesen R (2009) FMR1: a gene with three faces. Biochim Biophys Acta 790: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C (2007) The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol 176: 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchot SN, Adler JT, Luo Y, Ju J, Li W, Shen B, Kunnimalaiyaan M, Chen H (2009) Tautomycin suppresses growth and neuroendocrine hormone markers in carcinoid cells through activation of the Raf-1 pathway. Am J Surg 197: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primerano B, Tassone F, Hagerman RJ, Hagerman P, Amaldi F, Bagni C (2002) Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA 12: 1482–1488 [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA (2006) RNA-mediated neuromuscular disorders. Annu Rev Neurosci 29: 259–277 [DOI] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J (2007) RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron 55: 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoss O, Novoyatleva T, Gencheva M, Olbrich M, Benderska N, Stamm S (2004) p59(fyn)-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1. Mol Cell Neurosci 27: 8–21 [DOI] [PubMed] [Google Scholar]
- Stoss O, Olbrich M, Hartmann AM, Konig H, Memmott J, Andreadis A, Stamm S (2001) The STAR/GSG family protein rSLM-2 regulates the selection of alternative splice sites. J Biol Chem 276: 8665–8673 [DOI] [PubMed] [Google Scholar]
- Tassone F, Adams J, Berry-Kravis EM, Cohen SS, Brusco A, Leehey MA, Li L, Hagerman RJ, Hagerman PJ (2007) CGG repeat length correlates with age of onset of motor signs of the fragile X-associated tremor/ataxia syndrome (FXTAS). Am J Med Genet B Neuropsychiatr Genet 4: 566–569 [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Chamberlain WD, Hagerman PJ (2000a) Transcription of the FMR1 gene in individuals with fragile X syndrome. Am J Med Genet 97: 195–203 [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Loesch DZ, Lachiewicz A, Taylor AK, Hagerman PJ (2000b) Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am J Med Genet 94: 232–236 [DOI] [PubMed] [Google Scholar]
- Tassone F, Iwahashi C, Hagerman PJ (2004) FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS). RNA Biol 1: 103–105 [DOI] [PubMed] [Google Scholar]
- Van Dam D, Errijgers V, Kooy RF, Willemsen R, Mientjes E, Oostra BA, De Deyn PP (2005) Cognitive decline, neuromotor and behavioural disturbances in a mouse model for fragile-X-associated tremor/ataxia syndrome (FXTAS). Behav Brain Res 162: 233–239 [DOI] [PubMed] [Google Scholar]
- Venables JP, Dalgliesh C, Paronetto MP, Skitt L, Thornton JK, Saunders PT, Sette C, Jones KT, Elliott DJ (2004) SIAH1 targets the alternative splicing factor T-STAR for degradation by the proteasome. Hum Mol Genet 13: 1525–1534 [DOI] [PubMed] [Google Scholar]
- Venables JP, Elliott DJ, Makarova OV, Makarov EM, Cooke HJ, Eperon IC (2000) RBMY, a probable human spermatogenesis factor, and other hnRNP G proteins interact with Tra2beta and affect splicing. Hum Mol Genet 9: 685–694 [DOI] [PubMed] [Google Scholar]
- Venables JP, Vernet C, Chew SL, Elliott DJ, Cowmeadow RB, Wu J, Cooke HJ, Artzt K, Eperon IC (1999) TSTAR/ÉTOILE: a novel relative of SAM68 that interacts with an RNA-binding protein implicated in spermatogenesis. Hum Mol Genet 8: 959–969 [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Thornton CA (2007) Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol 20: 572–576 [DOI] [PubMed] [Google Scholar]
- Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, van Unen L, Tassone F, Hoogeveen AT, Hagerman PJ, Mientjes EJ, Oostra BA (2003) The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet 12: 949–959 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.