Abstract
Mycobacterium tuberculosis, along with other actinobacteria, harbours proteasomes in addition to members of the general bacterial repertoire of degradation complexes. In analogy to ubiquitination in eukaryotes, substrates are tagged for proteasomal degradation with prokaryotic ubiquitin-like protein (Pup) that is recognized by the N-terminal coiled-coil domain of the ATPase Mpa (also called ARC). Here, we reconstitute the entire mycobacterial proteasome degradation system for pupylated substrates and establish its mechanistic features with respect to substrate recruitment, unfolding and degradation. We show that the Mpa–proteasome complex unfolds and degrades Pup-tagged proteins and that this activity requires physical interaction of the ATPase with the proteasome. Furthermore, we establish the N-terminal region of Pup as the structural element required for engagement of pupylated substrates into the Mpa pore. In this process, Mpa pulls on Pup to initiate unfolding of substrate proteins and to drag them toward the proteasome chamber. Unlike the eukaryotic ubiquitin, Pup is not recycled but degraded with the substrate. This assigns a dual function to Pup as both the Mpa recognition element as well as the threading determinant.
Keywords: ARC, degradation, Mpa, proteasome, pupylation
Introduction
In all living organisms, protein degradation is an essential activity that is involved in protein turnover and post-translational quality control, but also fulfills vital regulatory functions (Goldberg, 2003; Sauer et al, 2004). It is carried out by cylindrical complexes that seclude the active sites on their interior, away from general access. Protein substrates are recruited to hexameric ATPase-rings that are associated with the protease cylinder and feed the substrates into the degradation chamber under the expense of energy (Striebel et al, 2009b). Although in eukaryotes and archaea, proteasomes are ubiquitous and represent the main degradation route for cellular proteins (Pickart and Cohen, 2004; Maupin-Furlow et al, 2005), bacteria contain a set of different, but architecturally related degradation complexes, for example Clp, Lon and FtsH proteases (Kress et al, 2009).
Actinobacteria, including the genus Mycobacteria, represent an unusual group, as they have acquired proteasomes by horizontal gene transfer (Tamura et al, 1995; Hu et al, 2006). The 20S proteasomal genes cluster together with other genes of the proteasome degradation pathway, like for example the regulatory ATPase ARC (called Mpa in mycobacteria) (Wolf et al, 1998; Darwin et al, 2005), which are not present in non-proteasome-harbouring bacteria. The function of the acquired proteasomal degradation system in these bacteria is still very poorly understood. However, in vivo relevance of the proteasome system has been shown for the pathogen Mycobacterium tuberculosis (Mtb). Mtb, the causative agent of tuberculosis, is a highly infectious organism with an intracellular life-style. Taken up by macrophages of the lung epithelium, it persists and proliferates in the phagosome. Mutant Mtb strains that lack components of the proteasomal degradation pathway exhibit severe impairment of persistence in infected mice (Darwin et al, 2003; Gandotra et al, 2007; Lin et al, 2009).
It has recently been shown that recruitment of substrates to the mycobacterial proteasome pathway occurs in a manner analogous to ubiquitin-dependent degradation in eukaryotes. The small protein Pup (prokaryotic ubiquitin-like protein), encoded directly upstream of the proteasome, is covalently attached to lysine residues of substrate proteins through its C-terminus, thereby targeting the substrates for degradation through the proteasome pathway (Pearce et al, 2008; Burns et al, 2009). Pup does not have any sequence homology to ubiquitin except a diglycine motif (GGQ) at its C-terminus (Knipfer and Shrader, 1997; Pearce et al, 2008), and unlike the well-structured ubiquitin, Pup adopts a range of extended conformations as shown by NMR analysis (Chen et al, 2009; Liao et al, 2009; Sutter et al, 2009). Despite its C-terminal GGQ motif, mass spectrometry showed that Pup conjugates contain GGE-K isopeptide bonds, suggesting a deamidation step as part of the pupylation pathway (Pearce et al, 2008; Burns et al, 2009). Furthermore, proteasome accessory factor A (PafA) was suggested as a likely component of the conjugation pathway, because pupylated substrates are absent in an Mtb pafA− strain (Pearce et al, 2008). Reconstitution of the complete pupylation pathway in vitro showed that a previously undescribed protein, Dop (deamidase of Pup), encoded directly upstream of Pup deamidates the C-terminal glutamine of Pup and thereby renders it competent for conjugation, which is carried out by the Pup ligase PafA (Striebel et al, 2009a). The function of Dop has recently been confirmed in vivo using an Mycobacterium smegmatis dop− strain, in which pupylation could be restored by exogenous expression of Pup-GGE, but not by expression of Pup-GGQ (Imkamp et al, 2010).
Recognition of Pup occurs via the N-terminal coiled-coil domain of Mpa (Sutter et al, 2009), which makes extensive contacts along the central region of Pup (Chen et al, 2009; Liao et al, 2009; Sutter et al, 2009). The Mpa coiled-coil domain is located on top of a hexameric interdomain comprising a tandem OB-fold that C-terminally connects to the canonical AAA domain (Djuranovic et al, 2009; Wang et al, 2009). Association of proteasomal ATPases with the respective core cylinders is in part mediated by insertion of their C-terminal tails (with a tyrosine in the penultimate position) into binding pockets on the proteasome cylinder face (Smith et al, 2007; Rabl et al, 2008). In analogy to other AAA-proteasome complexes, pupylated substrate proteins have to traverse the central OB-AAA channel to gain access to the proteasome's active sites. Although interaction of Mpa with an open-gate variant of the 20S proteasome has been observed in vitro in the presence of ATPγS by electron microscopy (Wang et al, 2009) and degradation of pupylated substrates by the Mpa–proteasome has been inferred from in vivo data (Pearce et al, 2006, 2008; Burns et al, 2009), degradation of pupylated proteins by the Mpa–proteasome complex has never been shown in vitro.
Here, we have reconstituted the complete bacterial proteasomal degradation pathway, showing that Mpa unfolds and translocates a Pup-GFP model substrate as well as a bona fide proteasomal substrate into the 20S core particle where the substrate and Pup are degraded to peptides. This process involves initial recognition of Pup via the coiled-coil domains of Mpa and subsequent initiation of unfolding via the N-terminal residues of Pup.
Results
Mpa unfolds a Pup-fused model substrate under the expense of ATP
In actinobacteria, ARC (called Mpa in mycobacteria) is suggested as the proteasomal ATPase that recruits pupylated substrate proteins to the 20S core particle for degradation. An mpa-deficient Mtb strain has been shown to accumulate proteasomal substrate proteins and direct physical interaction between Mpa and an open-gate variant of the 20S proteasome has been shown in vitro by negative-stain EM (Pearce et al, 2006, 2008; Wang et al, 2009).
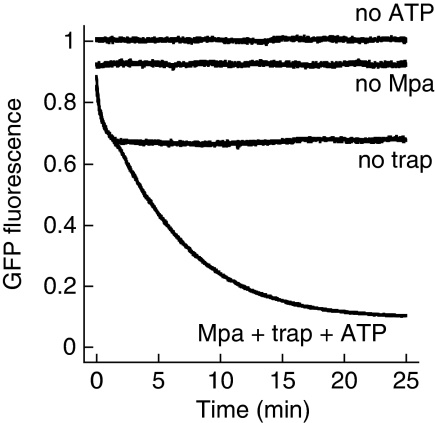
To test the ability of Mpa to unfold pupylated substrates by threading them through its central pore, a model substrate was constructed by fusing Pup N-terminally to GFP. GFP exhibits a strong fluorescence signal in the folded state, but is rendered non-fluorescent upon unfolding. A similar assay has been used to reveal the unfoldase activity of the bacterial ClpA ATPase partner of ClpP (Weber-Ban et al, 1999). Upon mixing of Mpa with Pup-GFP in the presence of ATP, a decrease in GFP fluorescence is observed that is absent when either ATP or Mpa is not added, suggesting an adjustment to equilibrium between Mpa-mediated Pup-GFP unfolding and spontaneous refolding of released, non-native Pup-GFP (Figure 1). Addition of ‘GroEL-trap', a variant of the chaperonin that can bind but not release non-native substrates, interrupts the refolding of GFP by ‘trapping' the unfolded substrates in their non-native state. We, therefore, used a three-fold excess of GroEL-trap over Pup-GFP in the unfolding assay and under those conditions observed a nearly complete loss of Pup-GFP fluorescence, proving that the entire Pup-GFP population is unfolded by the ATP-dependent action of Mpa (Figure 1).
Figure 1.
Mpa unfolds a Pup-GFP model substrate. Pup-GFP (Pup N-terminally fused to GFP) is fully unfolded by Mpa in the presence of GroEL-trap (trap) and ATP. In the absence of GroEL-trap, the fluorescence level reflects the equilibrium between Mpa-driven unfolding and spontaneous refolding of Pup-GFP.
Loops in the Mpa pore mediate unfolding after recognition of Pup at the coiled-coil domains
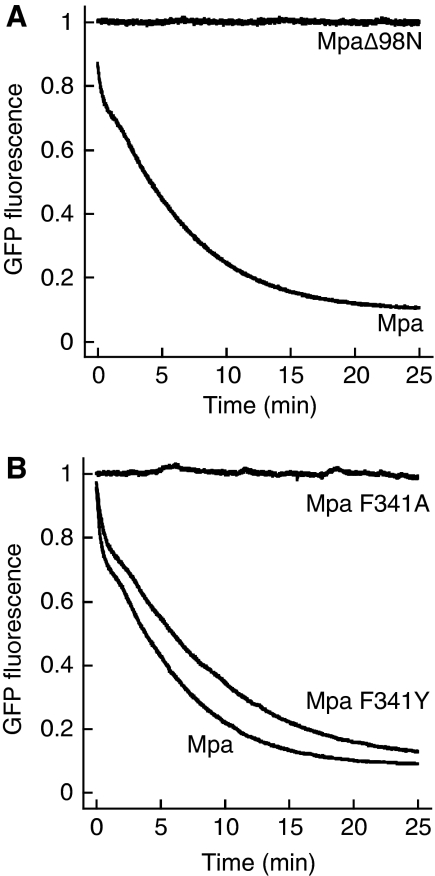
We have shown earlier that the coiled-coil domain of Mpa, but not the interdomain, the AAA domain, or full-length Mpa lacking the coiled-coil domain, can bind the modifier Pup (Sutter et al, 2009). Hence, Mpa lacking the coiled-coil domain (MpaΔ98N) should be unable to recruit and unravel the Pup-GFP model substrate. Indeed, upon mixing of MpaΔ98N with Pup-GFP in the presence of ATP and GroEL-trap, we do not observe any change in the fluorescence signal (Figure 2A), although deletion of the coiled-coil domain does not influence the oligomeric state of Mpa (Zhang et al, 2004; Wang et al, 2009) nor its ATPase activity (Supplementary Figure S1). This shows that the coiled-coil domain is essential for recruitment of Pup-GFP to Mpa.
Figure 2.
Determinants of Mpa-mediated unfolding. (A) Deletion of Mpa's N-terminal coiled-coil domain (MpaΔ98N), which was shown to be the primary recognition site for Pup, results in a complete loss of Pup-GFP unfolding activity in the presence of GroEL-trap and ATP. (B) The conserved pore-loop around F341 of Mpa mediates Pup-GFP unfolding. Mpa F341A can no longer unfold Pup-GFP in the presence of ATP and GroEL-trap, whereas Mpa F341Y retains its unfolding activity.
The common mechanistic feature of AAA ATPases is the conversion of ATP hydrolysis into mechanical activity by coupling distinct conformational states to the different nucleotide states in the nucleotide-binding site (Hanson and Whiteheart, 2005). Several AAA ATPases have been shown to couple their ATPase activity to up- and down-movements of pore loops with a conserved aromatic-hydrophobic-glycine motif (Ar-φ-Gly), which make contact with substrate proteins, exerting a pulling force that leads to unfolding and translocation of the substrate (Wang et al, 2001; Schlieker et al, 2004; Hinnerwisch et al, 2005; Martin et al, 2008). We identified the loop with the conserved motif (FVG in Mpa) by sequence alignment with other members of the AAA-ATPase family (Supplementary Figure S2) and generated two variants replacing the phenylalanine in the conserved motif either with alanine (Mpa-F341A) or with tyrosine (Mpa-F341Y). Whereas the alanine variant can no longer unfold Pup-GFP in the GroEL-trap assay, the tyrosine variant still carries an unfolding activity comparable to wild-type Mpa (Figure 2B).
Interaction of Mpa with the proteasome leads to degradation of a model substrate as well as a bona fide mycobacterial substrate
Protease-interacting ATPase partners unfold the substrate proteins and translocate them in a linear manner into the narrow entrance pores of the respective degradation cylinders (Lee et al, 2001; Reid et al, 2001). As shown earlier, Mpa unfolds a pupylated model substrate, Pup-GFP. As degradation of a pupylated substrate has never been shown in vitro, we now tested, whether Mpa together with the open-gate proteasome (Δ7CP, a core particle in which the α-subunits are truncated by seven N-terminal residues) is sufficient to degrade Pup-GFP. We used the open-gate proteasome, because it was shown by EM that the wild-type proteasome does not interact substantially with Mpa (Wang et al, 2009).
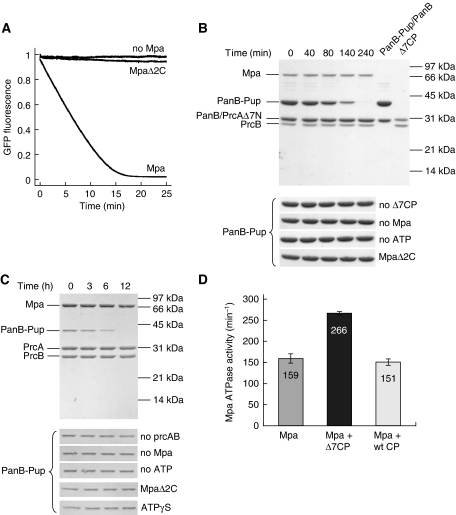
We recorded fluorescence time traces after mixing of Mpa, Δ7CP and the Pup-GFP model substrate in the presence of ATP (Figure 3A). A steep fluorescence decrease is observed, that is absent when Mpa is absent. One possible explanation of the observed fluorescence decrease would be that unfolded Pup-GFP released from Mpa is captured by the open-gate proteasome and degraded. To test this, we repeated the experiment with an Mpa variant (MpaΔ2C) lacking the two C-terminal residues (Pearce et al, 2006) that were shown to insert into binding pockets on the 20S cylinder face for the archaeal proteasome system (Smith et al, 2007; Rabl et al, 2008). For MpaΔ2C a fluorescence decrease is observed that is orders of magnitude slower than the one observed with full-length Mpa (Figure 3A), although MpaΔ2C has the same unfolding activity as full-length Mpa (Supplementary Figure S3). This clearly shows that Mpa is in contact with the core particle during the unfolding of the model substrate and directly feeds the unraveled polypeptide into the degradation chamber. The interaction between Mpa and Δ7CP is further supported by the observation that the basal ATPase activity of Mpa is stimulated by a factor of around 1.7 when Δ7CP is present, whereas no stimulation was observed upon addition of the wild-type core particle (Figure 3D).
Figure 3.
Mpa unfolds and translocates pupylated substrates into the proteasome. (A) The Pup-GFP model substrate is degraded in the presence of Mpa, an open-gate variant of the proteasome (Δ7CP) and ATP. Removal of the two C-terminal residues of Mpa required for interaction with the proteasome (MpaΔ2C) results in a strongly decreased degradation activity. (B) Degradation of the proteasomal substrate protein PanB-Pup by Mpa and Δ7CP in the presence of ATP analysed by SDS–PAGE and Coomassie staining. PanB was modified with Pup using the Pup-ligase PafA. For clarity, PanB-Pup/PanB (lane 6) and Δ7CP (lane 7) are shown. ‘PrcB' refers to the proteasomal β-subunit, ‘PrcAΔ7N' to the proteasomal α-subunit truncated by the seven N-terminal residues. The lower panel shows control reactions performed in the absence of Mpa (No Mpa), ATP (No ATP), Δ7CP (No Δ7CP); or with MpaΔ2C instead of Mpa (MpaΔ2C). (C) Degradation as described in (B) using the wild-type proteasome. As additional control reaction, ATPγS (1 mM) was used instead of ATP. Full-length gels are shown in Supplementary Figure S4. (D) The open-gate proteasome stimulates the Mpa ATPase activity. The basal Mpa ATPase activity is increased around 1.7-fold in the presence of the open-gate proteasome (Δ7CP), whereas no effect is observed for the wild-type proteasome (wt CP). Error bars represent s.d. from three experiments. The Mpa ATPase activity is calculated per hexamer.
In the Pup-GFP model substrate, Pup is connected to the GFP α-amino group through a regular peptide bond. However, in vivo, Pup is conjugated to the ɛ-amino group of substrate lysine residues, resulting in an isopeptide linkage. To also test the degradation of a bona fide Mtb proteasomal substrate, we pupylated the previously identified substrate protein PanB (Pearce et al, 2006) in vitro by using recombinantly produced PanB, deamidated Pup (Pup-GGE) and Pup ligase PafA. After mixing of Mpa, Δ7CP and purified PanB-Pup in the presence of ATP, we drew samples at specific time intervals and analysed them by SDS–PAGE (Figure 3B). The band of PanB-Pup disappears, indicating rapid in vitro degradation. In contrast, PanB is not degraded, showing that only pupylated subunits are extracted from the decameric PanB substrate (Chaudhuri et al, 2003). Control degradation reactions in the absence of either open-gate proteasome, Mpa or ATP show no decrease in the intensity of the PanB-Pup band (Figure 3B). We also observed no significant degradation activity using the MpaΔ2C variant that is impaired in its interaction with the proteasome (Figure 3B). Consistent with the observation by EM that the wild-type proteasome does not interact substantially with Mpa (Wang et al, 2009), we detect only very low activity for the recombinantly produced wild-type proteasome in vitro (Figure 3C; Supplementary Figure S4, also see Discussion below).
Taken together, our results show, for the first time, that the complex between Mpa and the open-gate 20S proteasome degrades pupylated substrate proteins in vitro.
Unfolding by the Mpa–proteasome requires the N-terminal region of Pup
We have proposed earlier that separate regions of Pup that map directly to its primary structure are responsible for distinct functional interactions (Sutter et al, 2009). The flexible C-terminal region is involved in the formation of the isopeptide bond with the substrate protein, and the large central region (residues 21–58, Figure 4A) is responsible for interaction with the Mpa N-terminal coiled-coil domain (Sutter et al, 2009). Consequently, two Pup model substrates where either the N-terminal half of Pup (residues 1–34) or the C-terminal half (residues 35–64) of Pup was fused to GFP (NPup-GFP and CPup-GFP, respectively) are not unfolded by the Mpa ATPase (Supplementary Figure S5).
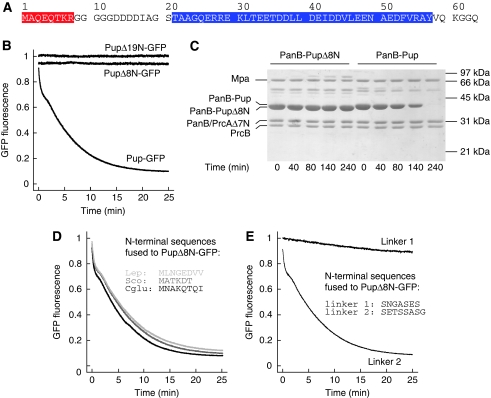
Figure 4.
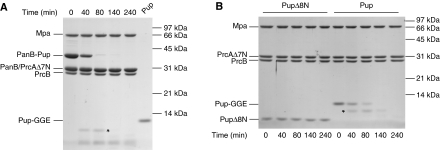
Mpa engages pupylated substrates via the N-terminus of Pup. (A) Primary sequence of Mycobacterium tuberculosis (Mtb) Pup. The region of Pup binding to the coiled-coil domain of Mpa is highlighted in blue; the eight N-terminal residues are highlighted in red. (B) Deletion of the N-terminal 19 or 8 residues of Pup (PupΔ19N-GFP and PupΔ8N-GFP, respectively) abrogates unfolding in the presence of Mpa, GroEL-trap and ATP. (C) PanB conjugated with PupΔ8N is not degraded by Mpa and the open-gate proteasome in the presence of ATP as analysed by SDS–PAGE and Coomassie staining. ‘PrcB' refers to the proteasomal β-subunit, ‘PrcAΔ7N' to the proteasomal α-subunit truncated by the seven N-terminal residues. (D) Fusion of N-terminal Pup sequences from other species to PupΔ8N-GFP restores unfolding by Mpa in the presence of GroEL-trap and ATP. The fused sequences of Corynebacterium glutamicum (Cglu), Streptomyces coelicolor (Sco) and Leptospirillum (Lep) are shown as inset. (E) Generic-linker sequences (shown as inset) were fused to PupΔ8N-GFP, and the respective variants were incubated with Mpa in the presence of GroEL-trap and ATP.
The N-terminal region of Pup is largely unstructured, suggesting that it might serve to reach down into the Mpa pore after recruitment of Pup to the Mpa coiled-coil domains. To test this hypothesis, we generated Pup-GFP variants with deletions at the N-terminus, removing the first 8 or first 19 residues of Pup (PupΔ8N-GFP or PupΔ19N-GFP). Unlike full-length Pup linked to GFP, neither one of the truncated model substrates exhibits a fluorescence decrease when mixed with Mpa and GroEL-trap in the presence of ATP (Figure 4B). To investigate whether deletion of the eight N-terminal residues of Pup also stabilizes a bona fide proteasomal substrate, we purified PupΔ8N and conjugated it to PanB using the Pup-ligase PafA. Upon addition of Mpa and the open-gate proteasome, no significant degradation is observed for PanB modified with PupΔ8N, whereas PanB modified with wild-type Pup is completely degraded (Figure 4C; Supplementary Figure S4). A control experiment showed that PupΔ8N still bound to Mpa in a pulldown assay (Supplementary Figure S6). This suggests that a certain length of the unstructured N-terminal module is required to reach into the Mpa pore, where it can be contacted by the ATP-driven pore loops.
The question remains, however, whether processability is mainly dependent on the length of the Pup N-terminus or whether sequence specificity also has a function. To investigate that question, we created Pup-GFP variants where the eight N-terminal residues were replaced by the N-terminal sequences of Pup proteins from three other organisms (Corynebacterium glutamicum, Streptomyces coelicolor, Leptospirillum sp.). These N-terminal Pup sequences served equally well as the Mtb sequence to recruit Pup-GFP to Mpa for unfolding (Figure 4D). This suggested that the exact sequence of the N-terminal stretch might not be of particular importance, but rather the length of it. Indeed, even when replacing the N-terminal eight residues with a generic-linker sequence (Hennecke et al, 1998) entirely unrelated to the sequence of Pup, the modified targeting tag was as efficient in recruiting GFP to Mpa for unfolding as wild-type Pup (Figure 4E, ‘linker 2'). Taken together, these results show that there is no strict sequence specificity involved in the threading of the Pup-N-terminus into Mpa. However, this does not mean that any sequence will serve this purpose equally well, as is shown by another generic sequence (Figure 4E, ‘linker 1'). A Pup-GFP variant carrying this N-terminus did not exhibit a strong fluorescence signal decrease in the unfolding assay with GroEL-trap and could thus obviously only be engaged poorly into the Mpa pore for unfolding.
Pup is not recycled during in vitro degradation of pupylated substrates
In eukaryotes, processing of polyubiquitinated substrates requires the en bloc removal of the ubiquitin chains, an activity intrinsic to the 19S regulatory particle (Verma et al, 2002; Yao and Cohen, 2002). Ubiquitin is thus recycled and is usually not engaged into the unfolding pore. In fact, it has been shown that in addition to polyubiquitination, an unstructured initiation site must be present in the substrate protein from which the proteasomal ATPases can unravel the substrate (Prakash et al, 2004; Takeuchi et al, 2007). To investigate whether Pup is recycled at the ATPases or whether its N-terminus serves as the unstructured initiation site in this system, leading to degradation of Pup along with the substrate, we performed degradation of pupylated PanB and analysed the reaction by SDS–PAGE (Figure 5A). Upon completion of the degradation reaction, we did not observe recycled Pup, which would be detectable at the concentrations of PanB-Pup used in this experiment (Figure 5A, lane 6). Furthermore, Pup itself is degraded in a proteasome-dependent manner, whereas PupΔ8N is stable over the course of the experiment (Figure 5B). This raises the question how Pup can function as a degradation signal if free Pup is already degraded by the Mpa–proteasome. A competition experiment, in which Mpa and Δ7CP are simultaneously incubated with free Pup and PanB-Pup shows that degradation of free Pup starts only after PanB-Pup has been degraded nearly completely (Supplementary Figure S7).
Figure 5.
Pup is not recycled on degradation of pupylated substrates in vitro. (A) Degradation of PanB-Pup by Mpa (0.3 μM) and open-gate proteasome (0.3 μM) in the presence of ATP analysed by SDS–PAGE and Coomassie staining. For comparison, the expected amount of Pup (Pup-GGE) if Pup were recycled is loaded in lane 6. ‘PrcB' refers to the proteasomal β-subunit, ‘PrcAΔ7N' to the proteasomal α-subunit truncated by the seven N-terminal residues. (B) Degradation of Pup (Pup-GGE) or PupΔ8N (10 μM each) by Mpa (0.2 μM) and open-gate proteasome (0.2 μM) in the presence of ATP analysed by SDS–PAGE and Coomassie staining. The asterisks denote a Pup fragment transiently appearing during degradation (see Supplementary Figures S8 and S9).
In the degradation reactions with PanB-Pup and Pup, a transient band is visible in SDS–PAGE (Figure 5A and B, marked with an asterisk), representing a Pup-fragment as shown by anti-Pup western blot and trypsinization of this band followed by LC-ESI-MS-MS (Supplementary Figure S8 and data not shown). Analysis of this fragment by ESI-MS revealed that Pup is truncated after Tyr58 (Supplementary Figure S9), suggesting that the proteasome preferentially cleaves Pup at this chymotrypsin-specific site.
Taken together, these experiments show that Pup is not recycled from pupylated proteins in vitro but degraded along with the substrate.
Discussion
It has recently been uncovered that mycobacteria and other actinobacteria have evolved an independent solution to proteasomal substrate targeting, termed pupylation (Pearce et al, 2008; Burns et al, 2009), featuring both differences and analogies to the eukaryotic ubiquitination pathway (Striebel et al, 2009a). In this process, the small protein Pup is covalently attached to lysine residues of substrate proteins through its C-terminus. The mechanism of Pup conjugation differs drastically from ubiquitination (Striebel et al, 2009a), but both tags serve as recognition elements targeting the substrates to proteasomal, regulatory binding partners that prepare the substrates for degradation. In M. tuberculosis (Mtb), Pup is recognized by the AAA-ATPase Mpa (called ARC in other actinobacteria) (Chen et al, 2009; Liao et al, 2009; Sutter et al, 2009). In analogy to archaeal and eukaryotic proteasomal ATPases and the bacterial Clp-type ATPases, Mpa is expected to unfold pupylated substrates by threading them through its central pore and to translocate them into the 20S proteasome. Interaction between Mpa and the proteasome had been suggested from the in vivo accumulation of proteasomal substrates in an mpa-knockout strain as well as proteasome-inhibited strains (Pearce et al, 2006, 2008), and was recently confirmed by EM using an open-gate variant of the proteasome (Wang et al, 2009).
In this study, we have reconstituted the unfolding and degradation of pupylated proteins by the Mpa–proteasome system in vitro and established mechanistic properties of this degradation pathway. To obtain a fluorescence-based, real-time readout of substrate processing by the ATPase Mpa, we generated the model substrate Pup-GFP, in which Pup is fused N-terminally to GFP. Making use of the fluorescence-readout and a trapping technique for non-native substrate proteins, we show that Mpa unfolds the Pup-GFP model substrate under the expense of energy (Figure 1). This is the first direct demonstration of an Mpa-unfolding activity. We could further show that unfolding by Mpa leads to efficient proteasomal degradation of Pup-GFP as well as of the bona fide proteasomal substrate PanB-Pup (Figure 3), a substrate in which Pup had been natively conjugated to a lysine residue of PanB by the Pup-ligase PafA.
The entrance to the central cavity of 20S proteasomes is occluded by the N-termini of the α-subunits, which was also observed for the core particle of Mtb (Hu et al, 2006; Lin et al, 2006). For archaea and eukaryotes, it was shown that gate opening requires the interaction with the respective proteasomal ATPase PAN or the Rpt-ATPases. Interestingly, even short peptides with the sequences of the C-termini of PAN or Rpt2/5 (containing the conserved penultimate tyrosine residue) were sufficient to induce the open-gate conformation of the 20S core particle (Smith et al, 2007; Gillette et al, 2008; Rabl et al, 2008). Mpa also contains a conserved penultimate tyrosine (Pearce et al, 2006), and our in vitro degradation assay showed that an Mpa variant truncated by the two C-terminal residues (MpaΔ2C, Figure 3) is strongly impaired in supporting the translocation of substrates into the 20S core particle. Together with in vivo data showing that an Mtb mutant strain carrying the MpaΔ2C-variant led to accumulation of proteasomal substrates (Pearce et al, 2006), this suggests that Mpa uses a similar gate-opening mechanism. Strikingly, actinobacteria that encode Mpa/ARC, but not the proteasomal subunits, for example bifidobacteria and corynebacteria, do not feature the C-terminal extension with the conserved tyrosine.
However, efficient interaction between Mpa and the proteasome und thus efficient substrate degradation in vitro can only be observed if an open-gate variant of the proteasome is used, in which the N-termini of the α-subunits are genetically truncated. It was shown by negative-stain EM that Mpa does not interact substantially with the wild-type proteasome (Wang et al, 2009), which is in agreement with our observation that the Mpa-ATPase activity is stimulated by the open gate, but not the wild-type proteasome (Figure 3D). Also consistent with this, we observed only very slow degradation of PanB-Pup using Mpa and wild-type proteasome after prolonged incubation (Figure 3C; Supplementary Figure S4).
The C-termini of proteasomal ATPases insert into inter-subunit pockets of two neighbouring α-subunits, leading to displacement of a reverse-turn loop with a conserved proline und thus to gate opening (Forster et al, 2003, 2005; Rabl et al, 2008; Yu et al, 2010). The proline is also conserved in bacterial α-subunits; however, the very N-termini are shorter than those from their archaeal and eukaryotic homologs, leading to a clustering of aromatic and hydrophobic residues (MSFPYFI for Mtb, see Supplementary Figure S10). Other residues of archaeal α-subunits that were shown to participate in the binding of the C-termini of PAN (Rabl et al, 2008; Yu et al, 2010) are also partially conserved in bacteria. Strict conservation cannot be expected, as the binding pockets in the α-rings may have evolved differently to allow binding of the cognate C-terminus of the proteasomal ATPase. Taken together, this suggests that the Mpa–proteasome complex likely uses a similar mechanism as other proteasomal systems in regulating gate opening. However, further studies will be necessary to elucidate this and it cannot be ruled out that a factor in addition to the proteasomal ATPase Mpa is needed for efficient proteasomal gate opening in bacteria.
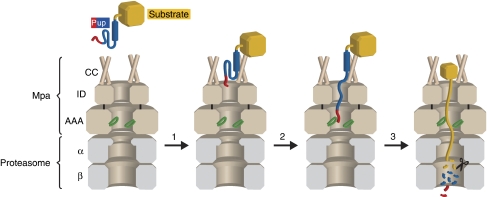
On the basis of our earlier findings (Sutter et al, 2009) and the results presented in this study, we propose the following model for degradation of pupylated substrates (Figure 6): (1) initial recognition of Pup by Mpa, (2) engagement of Pup's N-terminus into the Mpa pore, (3) unfolding, translocation and degradation of the pupylated substrate.
Figure 6.
Proposed mechanism for degradation of pupylated substrates. Step 1: The substrate (yellow) modified with Pup (blue and red) binds to the coiled-coil domains of Mpa (brown, ‘CC') with a KD of 3–4 μM (Sutter et al, 2009). The C-terminal helix of Pup is shown as a blue cylinder, the residues of Pup (21–58) bound by Mpa-CC are shown in blue, and the N-terminal region of Pup is shown in red. Step 2: The N-terminal segment of Pup (red) is engaged by the Ar-φ-Gly-loops (green) in the ATPase domain of Mpa (AAA). ‘ID' refers to the Mpa interdomain. Step 3: Mpa unfolds and translocates the pupylated substrate into the 20S proteasome (grey), where Pup and the substrate are degraded to peptides.
Initial recognition of Pup takes place at the N-terminal coiled-coil domains of Mpa (Mpa-CC) with a KD of 3–4 μM (determined for Mpa binding to free Pup), a binding event that can occur in the absence of nucleotide (Sutter et al, 2009). Coiled coils are a general feature of proteasomal ATPases (Djuranovic et al, 2009), and it was shown that Mpa-CC contacts the extended middle segment of Pup (residues 21–58, Figure 4A), leaving its N-terminal region unconstrained. Hence, an Mpa variant lacking the coiled-coil domain (MpaΔ98N) is deficient in Pup binding (Sutter et al, 2009) and consequently also in unfolding (Figure 2A).
The second step, engagement of the substrate, requires the N-terminus of Pup. Substrates modified with Pup that was truncated by eight residues from the N-terminus (PupΔ8N) are strongly stabilized against unfolding by Mpa (Figure 4B and C), although these residues do not participate in the initial recognition event (Chen et al, 2009; Liao et al, 2009; Sutter et al, 2009) and PupΔ8N still binds to Mpa in pulldown assays. Fusion of orthologous N-terminal Pup sequences or a generic-linker sequence to PupΔ8N restored unfolding activity by Mpa, indicating little sequence specificity for the initial unfolding event (Figure 4D and E). A similar length dependence has been observed for the bacterial chaperone ClpA in the degradation of bacterial N-end rule substrates (Erbse et al, 2006). It is tempting to speculate that the N-terminal residues of Pup are needed to traverse the Mpa interdomain (Mpa-ID) (Djuranovic et al, 2009; Wang et al, 2009) and contact the conserved Ar-φ-Gly-loops of the Mpa-ATPase domain (Mpa-AAA, Figure 6). This is further supported by the fact that in the partially tagged population of decameric PanB (Chaudhuri et al, 2003), only those PanB subunits are degraded that are covalently fused to Pup, even though the whole complex is tethered to Mpa (Figure 3B). It has been shown for several AAA ATPases that nucleotide hydrolysis leads to up- and down-movements of these pore loops along the pore axis, exerting a pulling force on stretches of substrate they contact, which causes substrate unfolding and translocation (Wang et al, 2001; Schlieker et al, 2004; Hinnerwisch et al, 2005; Martin et al, 2008).
This leads to step three, the processive unraveling and translocation of pupylated substrates into the 20S core particle (Figure 6). An F341A substitution in the conserved pore loop abolishes Mpa unfolding activity (Figure 2B), suggesting that Mpa uses a similar mechanism as described for other AAA proteins. Consistently, a comparable loop mutation of Mpa (V342A) led to in vivo accumulation of a proteasomal substrate (Wang et al, 2009). Importantly, Pup is not recycled in our in vitro assay but degraded along with the substrate and, furthermore, Pup alone is degraded in an Mpa-dependent manner (Figure 5A and B). Nevertheless, the competition experiment with Pup and pupylated substrate (Supplementary Figure S7) suggests that the affinity of Mpa towards pupylated substrates may be higher than to free Pup. Furthermore, the relative cellular levels of free Pup versus pupylated substrates might be rather small. This is, amongst other factors, also influenced by the affinities of the conjugation machinery towards Pup.
The features discussed above highlight important differences to the eukaryotic ubiquitin system, that uses a two-part degradation signal (Schrader et al, 2009). The first part of the degradation signal consists of tethering by polyubiquitin, a process in which the polyubiquitin tag is usually recognized by Ub-receptors at the 19S regulatory particle (Deveraux et al, 1994; Husnjak et al, 2008). However, polyubiquitination is not sufficient for efficient degradation (Thrower et al, 2000; Petroski and Deshaies, 2003), as it was shown that as a second determinant, the substrate needs to contain an unstructured initiation site from which unfolding can occur (Janse et al, 2004; Prakash et al, 2004, 2009; Piwko and Jentsch, 2006; Takeuchi et al, 2007). Thus, the main function of polyubiquitination seems to be tethering of the substrate to the proteasome until engagement for unfolding has occurred. The activity of ubiquitin-ligases and de-ubiquitinases present in the 19S regulatory subunit can fine-tune the time a substrate spends at the proteasome, thereby influencing the likelihood of degradation (Lam et al, 1997; Crosas et al, 2006). Once the substrate is committed for degradation, Rpn 11 catalyses the en bloc removal of poly-Ub, leading to recycling of ubiquitin and degradation of the substrate (Verma et al, 2002; Yao and Cohen, 2002).
In contrast, Pup contains both determinants of the degradation signal on one chain: the extended middle segment functions as a tethering site and localizes the pupylated substrate to the Mpa–proteasome (Chen et al, 2009; Liao et al, 2009; Sutter et al, 2009). The N-terminal residues then serve as the unfolding initiation site, ultimately leading to degradation of the substrate and Pup. This dual function of Pup is intimately connected with its flexible structure (Chen et al, 2009; Liao et al, 2009; Sutter et al, 2009), whereas ubiquitin adopts a stable tertiary fold. Thus, although Pup resembles ubiquitin in that it is a degradation signal conjugated to lysine residues of substrates, it relates mechanistically more to other bacterial degradation signals. These signals usually consist of short N- or C-terminal extensions, like the ssrA-tag, and are sufficient for tethering and initiation of unfolding (Sauer et al, 2004; Moore and Sauer, 2007; Striebel et al, 2009b).
In summary, we have reconstituted the full-proteasomal degradation pathway of M. tuberculosis in vitro and have established several key mechanistic features of this pathway. The results presented here will lay the basis for future studies and provide tools for screening for inhibitors of the proteasome system using the Pup-GFP reporter construct.
Materials and methods
Cloning of constructs
All genes except gfp and groEL were obtained by PCR from M. tuberculosis H37Rv genomic DNA. mpa was cloned with and without a C-terminal His6-tag between the NdeI and BamHI restriction sites into pET20 (Novagen). mpaΔ98N was cloned through NcoI/BamHI into pET16 (Novagen) with a C-terminal His6-tag and deleting the first 98 amino acids of Mpa. The mpa loop variants (F341A, F341Y) were obtained by site-directed mutagenesis from pET20-mpa-his6. The variant mpaΔ2C was obtained by introducing a stop codon after amino acid 607 by site-directed mutagenesis from pET20-mpa.
pup-gfp fusions were generated by cloning pup through BamHI/SacI and gfp through SacI/HindIII restriction sites into a His6-Thioredoxin-TEV fusion vector (Striebel et al, 2009a), resulting in a His6-Thioredoxin-TEV-Pup-GFP construct. The variants Npup-gfp and Cpup-gfp were obtained by fusing residues 1–34 and residues 35–64 of Pup to GFP, respectively. The variants pupΔ8N-gfp and pupΔ19N-gfp were obtained by deleting the first 8 and 19 residues of Pup, respectively. pup-gfp variants replacing the first eight amino acids of Pup were obtained by PCR from pupΔ8N-gfp.
PanB-Strep was cloned with a C-terminal Strep-tag through NcoI/EcoRI intp pET-Duet1. For soluble expression of the open-gate 20S proteasome, prcA was truncated by deleting the seven N-terminal amino acids and cloned through NcoI/EcoRI and mature prcB with a C-terminal Strep-tag through NdeI/EcoRV into pET-Duet (Novagen). For expression of inclusion bodies (IBs), wild-type prcA was cloned through NcoI/EcoRI in pETDuet and mature, untagged prcB through NdeI/HindIII into pET20. All constructs were verified by DNA sequencing. Primer sequences are summarized in Supplementary Table I.
Expression and purification of proteins
Pup and PafA-His6 (Striebel et al, 2009a) and GroEL-trap (GroEL D87K) (Fenton et al, 1994) were purified as described. Mpa, PanB-Strep and Pup-GFP were expressed in Escherichia coli BL21 (DE3) from IPTG-inducible plasmids at 30, 20 and 25°C, respectively. The 20S open-gate proteasome was expressed by auto-induction at 25°C. Expression of wild-type α-subunit and mature, untagged β-subunit as IBs was carried out separately in E. coli BL21(DE3) for 5 h at 37°C.
Mpa was purified by an anion-exchange step (Fast Flow Q), a subsequent cation-exchange step (Source 30S) and size-exclusion chromatography (Superose 6) and stored in 50 mM Tris pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 1 mM EDTA, 1 mM DTT. Mpa-His6 variants were purified by Ni2+-affinity chromatography (HiTrap IMAC HP) and size-exclusion chromatography (Superose 6).
Pup-GFP was expressed as His6-Thioredoxin-TEV-Pup-GFP fusion protein and purified by Ni2+-affinity chromatography (HiTrap IMAC HP). After cleavage of the fusion protein with TEV-protease (Invitrogen), His6-Thioredoxin and TEV-protease were removed by Ni2+-affinity chromatography. Pup-GFP was stored in 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA (buffer S). Note that all Pup-GFP constructs contain an additional N-terminal Gly-Ser stemming from the TEV cleavage site.
PanB-Strep was purified by Strep-Tactin affinity chromatography (IBA). The open-gate 20S proteasome was purified by Strep-Tactin affinity chromatography (IBA) and subsequent size-exclusion chromatography (Superose 6) and stored in 50 mM HEPES pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 1 mM EDTA.
The wild-type proteasome was purified from IBs by standard protocols. Briefly, IBs were washed three times by resuspension centrifugation in 50 mM Tris pH 7.5, 1 mM EDTA, 1% (v/v) TritonX-100. The final washing step was carried out without TritonX-100. Then, IBs were solubilized in 100 mM Tris pH 7.5, 8 M urea, 1 mM EDTA and non-solubilized material removed by centrifugation. Subsequently, equal molar amounts of α- and β-subunit were mixed and refolded by dropwise rapid dilution into 200 ml of 50 mM Tris pH 7.5, 1 M arginine, 1 mM EDTA to a final protein concentration of 0.3 mg/ml. After dialysis into 50 mM Tris pH 7.5, 50 mM NaCl, 1 mM EDTA and centrifugation, the soluble proteins were precipitated with 70% ammonium sulphate. The precipitated pellet was dissolved in 50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, and assembled proteasomes were separated from aggregates and monomers by size-exclusion chromatography (Superose 6).
Chromatographic material was purchased from GE Healthcare. Concentrations of proteins were determined by absorption at 280 nm, for GFP at 447 nm. For Mpa and proteasome, the concentrations refer to concentrations of the oligomer (6-mer and 28-mer, respectively), for all others to concentrations of the monomer. All purified proteins were verified by ESI-MS.
Pup-GFP unfolding by Mpa
Unfolding of different Pup-GFP variants was followed by monitoring the fluorescence intensity at 510 nm (excitation at 400 nm). Measurements were carried out on a fluorescence spectrometer (PTI) in buffer R (50 mM Tris pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 20 mM MgCl2, 1 mM DTT) completed with 25 mM phosphocreatine (Sigma) and 1 U/ml creatine phosphokinase (Sigma) at 23°C. Unfolding of Pup-GFP was initiated by mixing 0.5 μM Mpa, 3 μM GroEL-trap and 1 μM Pup-GFP with 5 mM ATP. The fluorescence intensity within one data set was normalized using Fnormalized=F(t)/Finitial.
Pup-GFP degradation by the Mpa proteasome
Pup-GFP degradation was followed as described for the unfolding assay by mixing 0.3 μM Mpa, 0.4 μM open-gate proteasome and 3 μM Pup-GFP with 5 mM ATP.
PanB-Pup degradation by the Mpa proteasome
PanB-Strep (47 μM) was conjugated with Pup by incubation with the Pup-ligase PafA-His6 (1 μM) and Pup-GGE (40 μM) in the presence of ATP (Striebel et al, 2009a). Then, PafA-His6 was removed from the reaction mixture by Ni2+-affinity chromatography and residual Pup-GGE by multiple rounds of ultrafiltration (Amicon spin column, 50000 MWCO). Subsequently, 10 μM PanB/PanB-Pup mixture was incubated with 0.1 μM Mpa, 0.1 μM open-gate proteasome (unless otherwise stated) and 5 mM ATP at 23°C in buffer R supplemented with appropriate amounts of ATP regeneration system. The reaction was terminated at the indicated time points with SDS-sample buffer and analysed by SDS–PAGE and Coomassie-staining. For comparison of PanB-Pup and PanB-PupΔ8N, Mpa and the open-gate proteasome were directly added to the conjugation reaction.
For degradation reactions using the wild-type proteasome, PanB-Pup (1 μM) was incubated with Mpa (0.2 μM) and wild-type proteasome (0.2 μM) in the presence of ATP (5 mM) and ATP regeneration system. The reactions were carried out at 23°C in 50 mM Hepes pH 7.5, 10 mM MgCl2, 1 mM DTT supplemented with 40 mM phosphocreatine and 0.4 U/ml creatine phosphokinase.
ATPase activity of Mpa
The ATPase activity was measured at 23°C in buffer R with a continuous spectrophotometric assay coupled to inorganic phosphate production using 7-methylinosine and the enzyme purine nucleoside phosphorylase (Rieger et al, 1997). Mpa (0.2 μM) was measured in the presence or absence of wild-type or open-gate proteasome (0.5 μM), and the reaction was started by addition of ATP (5 mM).
Pulldown using Pup as bait
Pulldowns were performed as described (Striebel et al, 2009a) with Pup covalently immobilized on amine-reactive beads in buffer R supplemented with 0.5% (v/v) NP40.
Supplementary Material
Acknowledgments
We thank Wolfgang Kress (ETH Zurich) for making Figure 6, Art Horwich (Yale School of Medicine) for the GroEL-trap plasmid, the Functional Genomics Center Zurich (FGCZ) for MS and Stefan Pitsch for synthesizing 7-methylinosine used in the spectrophotometric ATPase assay. This work was supported by the Swiss National Science Foundation (SNF), the National Center for Excellence in Research (NCCR) Structural Biology program of the SNF, an ETH research grant and a Kekulé fellowship by the ‘Fonds der Chemischen Industrie' to FS. Author Contributions: FS and EWB designed and analysed all experiments and wrote the paper. FS cloned and purified all constructs and performed the experiments. MH performed and analysed part of the experiments under supervision of FS. HS established the Mpa purification protocol.
Footnotes
The authors declare that they have no conflict of interest.
References
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE III (2009) Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem 284: 3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri BN, Sawaya MR, Kim CY, Waldo GS, Park MS, Terwilliger TC, Yeates TO (2003) The crystal structure of the first enzyme in the pantothenate biosynthetic pathway, ketopantoate hydroxymethyltransferase, from M tuberculosis. Structure 11: 753–764 [DOI] [PubMed] [Google Scholar]
- Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ (2009) Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol 392: 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF (2003) The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302: 1963–1966 [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF (2005) Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol 55: 561–571 [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M (1994) A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem 269: 7059–7061 [PubMed] [Google Scholar]
- Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K (2009) Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell 34: 580–590 [DOI] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B (2006) ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439: 753–756 [DOI] [PubMed] [Google Scholar]
- Fenton WA, Kashi Y, Furtak K, Horwich AL (1994) Residues in chaperonin GroEL required for polypeptide binding and release. Nature 371: 614–619 [DOI] [PubMed] [Google Scholar]
- Forster A, Masters EI, Whitby FG, Robinson H, Hill CP (2005) The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell 18: 589–599 [DOI] [PubMed] [Google Scholar]
- Forster A, Whitby FG, Hill CP (2003) The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J 22: 4356–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S (2007) In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med 13: 1515–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN (2008) Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem 283: 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6: 519–529 [DOI] [PubMed] [Google Scholar]
- Hennecke F, Krebber C, Pluckthun A (1998) Non-repetitive single-chain Fv linkers selected by selectively infective phage (SIP) technology. Protein Eng 11: 405–410 [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H (2006) Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol 59: 1417–1428 [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E (2010) Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol 75: 744–754 [DOI] [PubMed] [Google Scholar]
- Janse DM, Crosas B, Finley D, Church GM (2004) Localization to the proteasome is sufficient for degradation. J Biol Chem 279: 21415–21420 [DOI] [PubMed] [Google Scholar]
- Knipfer N, Shrader TE (1997) Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol 25: 375–383 [DOI] [PubMed] [Google Scholar]
- Kress W, Maglica Z, Weber-Ban E (2009) Clp chaperone-proteases: structure and function. Res Microbiol 160: 618–628 [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385: 737–740 [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A (2001) ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell 7: 627–637 [DOI] [PubMed] [Google Scholar]
- Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X (2009) Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J 422: 207–215 [DOI] [PubMed] [Google Scholar]
- Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, Weich N, Nathan C (2006) Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol 59: 1405–1416 [DOI] [PubMed] [Google Scholar]
- Lin G, Li D, de Carvalho LP, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H, Nathan C (2009) Inhibitors selective for mycobacterial versus human proteasomes. Nature 461: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT (2008) Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol 15: 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Gil MA, Humbard MA, Kirkland PA, Li W, Reuter CJ, Wright AJ (2005) Archaeal proteasomes and other regulatory proteases. Curr Opin Microbiol 8: 720–728 [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT (2007) The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem 76: 101–124 [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH (2006) Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J 25: 5423–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322: 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2003) Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell 11: 1435–1444 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE (2004) Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol 5: 177–187 [DOI] [PubMed] [Google Scholar]
- Piwko W, Jentsch S (2006) Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol 13: 691–697 [DOI] [PubMed] [Google Scholar]
- Prakash S, Inobe T, Hatch AJ, Matouschek A (2009) Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol 5: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 11: 830–837 [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell 30: 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BG, Fenton WA, Horwich AL, Weber-Ban EU (2001) ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci USA 98: 3768–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger CE, Lee J, Turnbull JL (1997) A continuous spectrophotometric assay for aspartate transcarbamylase and ATPases. Anal Biochem 246: 86–95 [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA (2004) Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, Bukau B, Mogk A (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11: 607–615 [DOI] [PubMed] [Google Scholar]
- Schrader EK, Harstad KG, Matouschek A (2009) Targeting proteins for degradation. Nat Chem Biol 5: 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL (2007) Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell 27: 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E (2009a) Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol 16: 647–651 [DOI] [PubMed] [Google Scholar]
- Striebel F, Kress W, Weber-Ban E (2009b) Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol 19: 209–217 [DOI] [PubMed] [Google Scholar]
- Sutter M, Striebel F, Damberger FF, Allain FH, Weber-Ban E (2009) A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett 583: 3151–3157 [DOI] [PubMed] [Google Scholar]
- Takeuchi J, Chen H, Coffino P (2007) Proteasome substrate degradation requires association plus extended peptide. EMBO J 26: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, Tanaka K, De Mot R, Baumeister W (1995) The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol 5: 766–774 [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR III, Koonin EV, Deshaies RJ (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH (2001) Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 9: 177–184 [DOI] [PubMed] [Google Scholar]
- Wang T, Li H, Lin G, Tang C, Li D, Nathan C, Darwin KH, Li H (2009) Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa. Structure 17: 1377–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401: 90–93 [DOI] [PubMed] [Google Scholar]
- Wolf S, Nagy I, Lupas A, Pfeifer G, Cejka Z, Muller SA, Engel A, De Mot R, Baumeister W (1998) Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J Mol Biol 277: 13–25 [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419: 403–407 [DOI] [PubMed] [Google Scholar]
- Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y (2010) Interactions of PAN's C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J 29: 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Stoffels K, Wurzbacher S, Schoofs G, Pfeifer G, Banerjee T, Parret AH, Baumeister W, De Mot R, Zwickl P (2004) The N-terminal coiled coil of the Rhodococcus erythropolis ARC AAA ATPase is neither necessary for oligomerization nor nucleotide hydrolysis. J Struct Biol 146: 155–165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.