Abstract
The linker histone H1 has a fundamental role in DNA compaction. Although models for H1 binding generally involve the H1 C-terminal tail and sites S1 and S2 within the H1 globular domain, there is debate about the importance of these binding regions and almost nothing is known about how they work together. Using a novel fluorescence recovery after photobleaching (FRAP) procedure, we have measured the affinities of these regions individually, in pairs, and in the full molecule to demonstrate for the first time that binding among several combinations is cooperative in live cells. Our analysis reveals two preferred H1 binding pathways and we find evidence for a novel conformational change required by both. These results paint a complex, highly dynamic picture of H1–chromatin binding, with a significant fraction of H1 molecules only partially bound in metastable states that can be readily competed against. We anticipate the methods we have developed here will be broadly applicable, particularly for deciphering the binding kinetics of other nuclear proteins that, similar to H1, interact with and modify chromatin.
Keywords: cooperativity, fluorescence imaging, FRAP, in vivo binding, linker histone
Introduction
The H1 or linker histones are stoichiometric architectural components of metazoan chromatin. Through interactions with the nucleosome core and linker DNA, these proteins help organize and compact the chromatin fibre (Thoma et al, 1979; Bednar et al, 1998; Georgel and Hansen, 2001; Brown et al, 2006; Robinson and Rhodes, 2006; Woodcock et al, 2006; Routh et al, 2008). Although H1 was originally viewed as a general transcriptional repressor, more recent studies have identified both negative and positive H1-specific effects on the expression of a growing number of genes (Fan et al, 2005; Happel and Doenecke, 2009). The demonstration that linker histones interact dynamically with chromatin in vivo (Lever et al, 2000; Misteli et al, 2000), continuously exchanging between chromatin sites, leads to the suggestion that this redistribution might function through a network of interacting factors as a mechanism for global or specific gene expression (Misteli, 2001; Brown, 2003; Catez et al, 2006). Evidence supporting this suggestion has begun to accumulate (Meshorer et al, 2006; Yellajoshyula and Brown, 2006), but it remains unclear how the H1 exchange process occurs and how this can lead to chromatin condensation.
The linker histones of higher organisms have a tripartite structure consisting of a central globular core flanked by a long unstructured basic C-terminal tail and a shorter N-terminal extension. Although the C-terminal tail has been reported to be the primary determinant of H1 binding (Hendzel et al, 2004), deletions or point mutations in the globular domain as well lead to severely decreased affinity of the protein for chromatin in live cells (Brown et al, 2006). This suggests that both of these domains contribute to binding, but the details of their cooperation have yet to be worked out or even demonstrated. Three models for H1 binding to chromatin have recently been considered that are consistent with the existing data, but each assigns a different role to the H1 globular and C-terminal domains. In model one (Brown et al, 2006), initial capture of H1 through nonspecific interactions with the intrinsically disordered C-terminal tail promotes efficient specific binding of the globular domain. In model two (Raghuram et al, 2009), the roles of the globular and C-terminal domains are reversed, whereas in model three (Raghuram et al, 2009), both domains participate in the initial capture. In all models, once both domains are stably bound, the newly formed H1–chromatin complex then undergoes a conformational change that induces chromatin compaction.
To directly test these models we have developed and applied a general in vivo method based on fluorescence recovery after photobleaching (FRAP) for measuring cooperation between the different domains of a protein and detecting ensuing conformational changes. FRAP is now regularly used to investigate the dynamics of a wide range of cellular processes. Although most FRAP studies are qualitative, a growing number now quantify FRAP data to extract protein diffusion and binding times (Sprague et al, 2004; Carrero et al, 2004; Houtsmuller, 2005; Beaudouin et al, 2006; Xouri et al, 2007; Mueller et al, 2008). Here, we perform quantitative FRAP on a set of structurally related H10 mutants to show that the H1 globular and C-terminal domains do indeed cooperate when H1 binds, but not in the manner originally anticipated. In particular, our analysis demonstrates that H1 binding is predominantly initiated through the C-terminal domain. After this initial contact is made, we find evidence for a novel conformational change that enables binding of the two DNA-binding sites located on opposite sides of the H1 globular domain: S1 and S2. In the absence of the C-terminal domain, we show that the H1 globular domain is only bound through S1 alone or S2 alone, but never by both sites at the same time. On the other hand, when the C-terminal domain is intact, we show that the H1 globular domain can be bound by both sites at the same time. Our data therefore suggest that binding of the C-terminal domain contracts or reorients chromatin to bring the DNA-binding targets for S1 and S2 together so a single globular domain can bind both targets simultaneously, presumably to form an apposed stem (Bednar et al, 1998). Once the globular and C-terminal domains are bound, the H1 molecule can then enter a unique tightly bound state that is consistent with a second, relatively slow conformational change. Thus, H1-induced compaction is initiated through cooperation between H1 domains rather than by any one domain in particular, highlighting the importance of this type of integrated analysis.
Results
Quantifying the dynamics of the key H1 binding domains through FRAP
To determine the roles of the globular and C-terminal domains of the linker histone, we performed FRAP in previously characterized stable cell lines (Misteli et al, 2000; Brown et al, 2006) expressing GFP-tagged mutant H10 constructs either lacking the C-terminal tail and/or with site-directed mutations in the S1 or S2 binding sites within the globular domain that eliminate their binding affinity (Figure 1). Together the constructs have all eight possible combinations of the important H1 binding regions (S1, S2, and C), with the GFP tag consistently placed on the C-terminal end to enable binding comparisons. We refer to these constructs in bold italics, using WT for the wild type and 3K0 for the triple knockout. For the remaining constructs we use names based on their functional binding regions. For example, S1C indicates an H10 with wild-type S1 and C, but a defective S2 binding site, whereas S1S2 represents an H10 with wild-type S1 and S2, but a deleted C-terminal tail.
Figure 1.
H10 contains three important binding regions: sites S1 and S2 within the central globular domain along with the unstructured C-terminal domain. To determine how these regions cooperatively bind, we examined eight cell lines with GFP-tagged H10 constructs representing all possible combinations of these three binding regions (involving deletions of the C-terminal tail or alanine substitutions of the key residues shown within S1 or S2). We named constructs based on their functional binding regions and signify them with a bold italic font. Although the pattern of fluorescence varied between constructs (see Supplementary Figures S1 and S6 for details), we performed experiments in homogeneous, euchromatic regions of nuclei to exclude the effects of binding at special sites such as nucleoli or the nuclear periphery. A full-colour version of this figure is available at The EMBO Journal Online.
The H1 constructs exhibit variable expression patterns (see Supplementary Figure S1A for DIC images along with Hoechst staining of heterochromatin). Just as the wild type, all H1 mutants are predominantly within the nucleus and to varying degrees co-localize with heterochromatic foci, although the triple-knockout 3K0, which has severely reduced affinity for chromatin, can also be found at significant levels in the cytoplasm. Unlike the wild type, the mutants also co-localize with nucleoli. As this is most apparent in mutants lacking one or more chromatin binding regions, we attribute this aberrant co-localization to a relatively weak, nonspecific interaction, perhaps with the high concentration of positively charged ribosomal RNA there (which complements the high concentration of negatively charged residues within the H1 molecule). To prevent potential complications arising from localized binding at these and other special sites, we performed all FRAP experiments in fluorescently homogeneous euchromatic regions of the nucleus, avoiding the nuclear periphery as well as the interior and edges of nucleoli (see Supplementary Figure S1B for sample choices of bleach spot locations).
To optimize FRAP for measuring recoveries spanning the wide range of time scales expected for wild-type and mutant H10 molecules lacking binding domains, we developed an improved spatiotemporal implementation. As in standard FRAP experiments, we monitor the fluorescence recovery in a photobleached region of a cell containing a GFP-tagged construct, but we acquire data at speeds exceeding 500 Hz by scanning a single line repeatedly through the centre of the bleached region to produce two-dimensional (2D) spatiotemporal images (Braeckmans et al, 2007; Matsuda et al, 2008) of the FRAP recovery (Figure 2A). We fit the 2D spatiotemporal image with a reaction–diffusion model (Mueller et al, 2008) to determine the diffusion coefficient of each construct and their association and residence times more stringently than traditional 1D fits (Figure 2B and C). From these fits, the bound fraction of each construct can be calculated and decomposed into fast and slow components (Carrero et al, 2004; Sprague et al, 2004; Xouri et al, 2007). To test this procedure, we used it to measure the dynamics of a variety of different proteins, including the rapid dynamics of unconjugated GFP (Supplementary Figure S2A) and the more moderate dynamics of the transcription factor GFP-GR (Figure 2; Table 1, row 1). In all cases, the estimated diffusion and binding times agreed with earlier studies (Braeckmans et al, 2007; Mueller et al, 2008) and/or with independent spot-FRAP experiments (Supplementary Figure S2B). In addition, we performed Monte Carlo simulations of FRAP experiments to verify our fitting routine could accurately detect a wide range of fast and slow binding states (Supplementary Figure S3).
Figure 2.
(A) High-resolution spatio-temporal image of the FRAP recovery for GFP-tagged glucocorticoid receptor (GFP-GR) stably expressed in the nuclei of mouse adenocarcinoma cells (McNally et al, 2000) generated by repeated line scans (yellow double-arrow) that bisect the bleach spot (in dashed black circle), normalized to their pre-bleach value. (B) Instead of fitting the full spatio-temporal image in (A), we fit a subset of the scanned lines that were exponentially distributed in time (more lines earlier in the recovery and less later). This ensured the fast early part of the FRAP recovery was sufficiently sampled to generate an accurate fit. The best-fit surface (shaded green) to the data (blue dots) is shown in 3D, with r representing the radial distance from the centre of the bleach spot. The difference (orange lines) between each data point and the fitting surface is also shown. This yielded D=2.9 μm2/s, ta=7.7 s, and tr=3.1 s, from which we calculated the fraction of bound to free molecules bound/free ∼0.4 using Supplementary equations (S.1) and (S.2). (C) For clarity, the fit from (B) is displayed at select times, with increasing times colour coded from 0 to 40 s, as shown in the bar to the right.
Table 1. Summary of fits for GFP-GR (GR) and all eight H10-GFP constructs.
| FRAP parameters | Calculated parameters | ||||||
|---|---|---|---|---|---|---|---|
| Fast state | Slow state | Bound fraction | Free fraction | Bound/free | |||
| D or Deff (μm2/s) | ta (s) | tr (s) | Slow | Fast | |||
| GR | 2.5±0.3 | 8.5±1.4 | 3.6±0.5 | 30±4% | 0% | 70±4% | 0.4±0.2 |
| 3KO | 6.1±0.2 | — | — | — | 0% | 100% | 0 |
| S2 | 5.3±0.5 | 5.6±2.2 | 1.1±0.3 | 16±6% | 11±7% | 73±9% | 0.4±0.2 |
| S1 | 3.7±0.2 | — | — | — | 39±4% | 61±4% | 0.7±0.1 |
| S1S2 | 3.3±0.3 | 8.8±2.7 | 1.7±0.4 | 16±5% | 39±5% | 45±5% | 1.2±0.3 |
| C | 0.96±0.1 | 17±2.4 | 6.1±0.7 | 26±4% | 61±3% | 13±1% | 6.9±0.7 |
| S2C | 0.52±0.1 | 14±2.2 | 5.7±0.8 | 29±4% | 64±4% | 7±1% | 14±2 |
| S1C | 0.40±0.1 | 17±3.6 | 8.5±1.0 | 33±5% | 62±5% | 5±1% | 20±3 |
| WT | 0.033±0.003 | 270±60 | 100±18 | 28±6% | 71±5% | 0.4±0.1% | 230±30 |
| The spatio-temporal FRAP data were fit for each construct generating estimated parameters for a diffusion coefficient (either pure diffusion D or effective diffusion Deff), and an association time (ta) and residence time (tr). All values are averages of at least 10 fits to single-cell FRAP data taken over 3 or more days. From these, the fraction of molecules bound in fast (effective diffusive) and slow binding states can be calculated using Supplementary equations (S.1) and (S.2). For GR and 3K0, the diffusion coefficient is interpreted as pure diffusion (D). For the other H10-GFP constructs, the diffusion coefficients are always less than the 3K0 pure diffusion coefficient (even after correcting for differences in mass, as described in the Supplementary data), so they are interpreted as effective diffusion. As the 3K0 mutant exhibited negligible binding (see Supplementary data), the binding of the other H10-GFP constructs can be attributed to S1, S2, and C binding regions. | |||||||
We then applied this novel FRAP procedure to measure and compare the binding dynamics of the eight H10 constructs. We began with the H10 wild-type molecule, which recovered in roughly 10 min (Figure 3H). Fitting these FRAP recoveries yielded a diffusion coefficient D=0.033±0.003 μm2/s, an association time ta=270±60 s, and a residence time tr=100±18 s (Table 1, row 9), values consistent with an earlier study (Carrero et al, 2004) of H1 binding in vivo that properly incorporated diffusion into their model of FRAP. The measured association and residence times provide evidence for a slow binding state that can be distinguished from diffusion. In addition to this slow state, there was evidence for a faster binding state. Specifically, the diffusion coefficient was ∼300 times smaller than freely diffusing molecules that are roughly the same size as H1, such as the transcription factor GR (Mueller et al, 2008). This strongly suggests the diffusion is ‘effective' rather than free, in which case the H10 molecule binds iteratively, rapidly hopping from one site to the next, in effect mimicking a slowed diffusive trek (Carrero et al, 2004; Sprague et al, 2004; Xouri et al, 2007). Thus, the wild-type H10 molecule appears to participate in both fast (effective diffusive) and slow binding interactions.
Figure 3.
FRAP was performed for each of the eight H10 constructs representing all possible combinations of the three key H10 binding regions (S1, S2, and C). For each construct (A–H), a sample fit is shown along with the average bound/free fraction (b/f) obtained from 10–20 such fits taken over 3 or more days. Table I provides the corresponding diffusion coefficients and binding association and residence times. FRAP recoveries from the eight constructs span three orders of magnitude in time (colour coded with increasing time from 0 to 400 s), with the fastest triple-knockout 3KO recovery taking <3 s and the slowest wild-type WT recovery taking over 400 s.
To determine whether the H1 globular and C-terminal domains were responsible for the measured binding states, we then examined the triple-knockout H1 mutant molecule (3K0) with a deleted C-terminal tail and a non-functional globular domain lacking both the S1 and S2 binding sites. As expected, the FRAP recovery of this mutant showed no evidence for binding (Figure 3A). In fact, the full recovery lasted less than 1 s and was well fit by a pure diffusion model, yielding a diffusion coefficient D=6.1±0.2 μm2/s, on par with expectations based on the size of the 3K0 mutant (Table 1, row 2). Although there may be some residual fast binding still present, we estimate this accounts for <1% of the measured wild-type H10 binding (Supplementary data). We therefore conclude that H10 binding is almost entirely due to the C-terminal tail and sites S1 and S2 within the globular domain.
We next wanted to determine the origin of the fast and slow binding sites displayed by the H10 wild type, so we examined mutants lacking selected binding domains. For example, the fast state might be due to the C-terminal tail and the slow state might be due to the globular domain, or vice versa. To test these models, we looked at two mutant H10 molecules, the S1S2 mutant with just the globular domain (Figure 3D) and the C mutant with just the C-terminal tail (Figure 3E). In contradiction to the simple models, however, both mutants exhibited their own fast (effective diffusive) and slow binding states, the latter with association and residence times an order of magnitude faster than the wild-type H10 molecule (Table 1, rows 5 and 6). We therefore conclude that neither the globular nor C-terminal domains by themselves are responsible for the fast and slow states measured in the H10 wild-type molecule.
We then applied this same strategy to analyse the globular domain by itself (the S1S2 mutant), to see whether its fast and slow binding states could be attributed to S1 or S2. Interestingly, the S2 mutant composed of only the S2 binding site exhibited just the slow S1S2 state (Figure 3B), with association time ta=5.6±2.2 s, residence time tr=1.1±0.3 s, and a diffusion coefficient that was the same as the triple-knockout 3K0 mutant (Table 1, row 3). Moreover, the S1 mutant composed of only the S1 binding site exhibited only the fast S1S2 state (Figure 3C), with an effective diffusion coefficient Deff=3.7±0.2 μm2/s (Table 1, row 4). Together these data suggest the effects of sites S1 and S2 within the globular domain are additive in the S1S2 mutant. In other words, the S1S2 effective diffusion coefficient Deff=3.3±0.3 μm2/s is due to site S1 alone, whereas the S1S2 association and residence times ta=8.8±2.7 s and tr=1.7±0.4 s are due to site S2 alone. This implies that S1 and S2 bind independently of each other, at least when the C-terminal domain is not intact. Thus, when the S1S2 mutant is bound, it is either through site S1 alone or through site S2 alone, but never simultaneously through both S1 and S2.
Finally, we wanted to see whether sites S1 or S2 bind independently of the C-terminal tail, so we examined mutant H10 molecules with an intact C-terminal tail, but just one DNA-binding site in the globular domain, either S1 or S2 (the S1C and S2C mutants, respectively). Unlike the S1S2 mutant previously, however, the binding of the S1C mutant could not be decomposed into distinct contributions from its binding regions. Instead the S1C mutant (Figure 3G) had a fast (effective diffusive) binding state not seen in the S1 mutant (with just the S1 binding site) or in the C mutant (with just the C-terminal tail), suggesting the S1 binding site does not bind independently of the C-terminal tail. Similarly, the S2C mutant (Figure 3F) had a fast (effective diffusive) binding state not seen in either the S2 mutant (with just the S2 binding site) or the C mutant, suggesting the S2 binding site also does not bind independently of the C-terminal tail. Reassuringly, however, both the S1C and S2C mutants exhibited the same slow binding state as the C mutant (Table 1, rows 6–8), indicating the C-terminal tail is most likely responsible for this slow binding state. Considering this state is still over 10 times weaker than the wild-type slow state, however, indicates that site S1 and the C-terminal tail by themselves or site S2 and the C-terminal tail by themselves cannot fully bind chromatin. We thus conclude that all three binding regions, sites S1 and S2 within the globular domain as well as the C-terminal tail, are required for functional binding of H10 to chromatin.
The results of our FRAP fits are summarized in columns 1–3 of Table 1 (Supplementary Figures S4 and S5 show sample fits with additional fitting details). From these we calculated the fraction of molecules bound in fast (effective diffusive) and slow binding states using Supplementary equations (S.1) and (S.2). Specifically, the fraction in the slow binding state (Table 1, column 4) is tr/(tr+ta), where ta and tr are the fitted binding association and residence times, respectively (see Supplementary equation (S.1)). The fraction in the fast binding state (Table 1, column 5) is ta/(tr+ta) (1−Deff/D), where Deff is the fitted effective diffusion coefficient of the H1 construct and D is the theoretical pure diffusion coefficient of the H1 construct (see Supplementary equation (S.2)). D can be calculated from the fitted diffusion coefficient D* of the 3K0 mutant: D=D* (m*/m)1/3, where m is the mass of the H1 construct, and m* is the mass of the 3K0 mutant. The fraction not bound in the fast or slow states is the free fraction (Table 1, column 6).
Together, the fast bound fraction, the slow bound fraction, and the free fraction yield the total bound/free fraction (Table 1, column 7). As we discuss in the next section, the bound/free fraction is very useful for comparing and quantifying cooperativity between the various H1 binding regions. To isolate the effects of just S1, S2, and the C-terminal tail, these ratios are reported relative to the triple-knockout 3K0 mutant, which, again, exhibited almost no binding compared to wild-type H10. In Table I, the mutants are ranked from smallest to largest total bound/free fraction. As expected, the predicted ordering shows a satisfying increase in proportion to the number of binding regions present in the H10 mutants.
The results in Table 1 are from FRAP experiments in euchromatin, where fluorescence is homogeneously distributed. Although binding of each mutant was tighter in heterochromatin, we detected a similar ordering of binding affinities there (see Supplementary Figures S6 and S7), suggesting that the overall binding mode was generally conserved from euchromatin to heterochromatin (Supplementary data).
Corroborating FRAP measurements in vitro by salt extractions
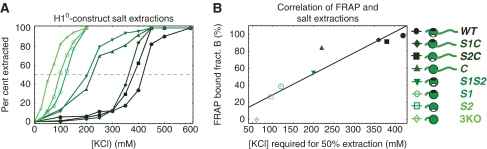
As an independent test of the in vivo FRAP analysis, we performed salt extractions in vitro (Figure 4A). Higher salt concentrations should be required to extract molecules that are more tightly bound to chromatin. The ordering of chromatin affinities for the different constructs that was predicted by the salt extractions showed excellent agreement with the ordering predicted by FRAP (Figure 4B). The two procedures identically ranked six of the eight constructs, with the only discrepancy being an interchange of the sixth and seventh rankings. These results argue that the bound/free fractions measured by FRAP are robust and therefore provide a solid foundation for further comparisons.
Figure 4.
(A) A series of salt-extraction experiments was performed on all eight H1 constructs. Although the wild-type required the largest concentration of salt to fully extract, the 3K0 mutant required the smallest concentration, consistent with its lack of binding domains. The other mutants fell somewhere in between these extremes, with those having fewer binding domains requiring less salt to fully extract than those with more binding domains. On the right the ordering is colour coded from light grey (green online) (3K0) to black (wild-type), along with an illustration of the corresponding construct. (B) A scatter plot relating the bound fraction predicted by FRAP to the salt concentration required for 50% extraction for each construct reveals a strong correlation (fit line) between the two measurements. A full-colour version of this figure is available at The EMBO Journal Online.
Assessing cooperativity between the H1 globular and C-terminal domains
In the absence of the globular domain (the C mutant), the H10 FRAP recovery speeds up considerably and the bound/free fraction drops by a factor of nearly 50. On the basis of this recovery alone, it appears as if the globular domain is the primary determinant of H1 binding (Brown et al, 2006). On the other hand, in the absence of the C-terminal tail (the S1S2 mutant), the FRAP recovery also speeds up considerably, and the bound/free fraction drops by a factor of nearly 200. On the basis of this recovery alone, it appears as if the C-terminal tail is the primary determinant of H1 binding (Hendzel et al, 2004). As Jencks (1981) originally pointed out, however, such conflicting data suggest that neither domain is the primary determinant of binding. Instead, the two domains most likely cooperate to bind, with binding of one facilitating the binding of the other. In this sense, both domains are critical, but only through their mutual cooperation. Thus, removal of either domain results in severely reduced binding.
To test directly for such cooperativity, we developed a systematic procedure based on our measured FRAP bound/free fractions to quantify how much the binding of one H1 domain facilitates the binding of another. Specifically, to determine whether the H1 globular and C-terminal domains cooperate, we need only compare the bound/free fractions of three molecules: the wild-type H10, which has both domains, along with the S1S2 mutant, which only has the globular domain, and the C mutant, which only has the C-terminal domain. As we show in the Supplementary data, if each domain targets independent and separate sites, then the wild-type bound/free fraction is equal to the sum of mutant bound/free fractions (Figure 5A). On the other hand, if the domains cooperate to bind a mutual site, then the wild-type bound/free fraction is greater than the sum of mutant bound/free fractions (Figure 5B). Applying this principle to H10, we find the wild-type bound/free fraction is indeed greater than the sum in mutants S1S2 and C (230>1.2+6.9), indicative of significant cooperation between the globular and C-terminal domains (Figure 5C).
Figure 5.
Proteins like H1 with more than one binding domain can bind their substrates in more than one way. To illustrate, a generic protein with circular and square binding domains X and Y (which could represent the H1 globular and C-terminal domains, for example) can bind its substrate in three ways. In (A) each domain targets an independent set of binding sites. In this case the bound/free fraction in the wild-type (XY) is equal to the sum of bound/free fractions measured in mutants with just the X or Y binding domain (X and Y, respectively), as shown in the bar-plot to the right. Alternatively, in (B), the X and Y domains cooperate to target a mutual site where the binding of one domain tethers the other so it subsequently binds with higher affinity than it would if it were untethered and freely diffusing. In this case, the wild-type bound/free fraction is greater than the sum in mutants, as shown by the grey portion (red online) of the bar-plot to the right. (C) The H10 wild-type bound/free fraction measured by FRAP is significantly greater than the sum of bound/free fractions measured for the globular domain by itself (S1S2=GD) and the C-terminal domain (C) by itself. The difference, shown in grey (red online), represents strong cooperation between these two domains. From the bar-plot, the cooperativity factor γ=27±8 was calculated by dividing the grey portion (red online) by the product of the two white portions (Supplementary equation (S.17)). A full-colour version of this figure is available at The EMBO Journal Online.
To quantify the degree of cooperation, a meaningful measure γ is the difference between the wild-type bound/free fraction and the sum of mutant bound/free fractions divided by the product of the mutant bound/free fractions (see Supplementary equation (S.17)). Graphically, this is equivalent to dividing the grey portion (red online) of the bar-plot in Figure 5B by the product of the two white portions. Specifically, the cooperativity factor γ reflects the extent that binding of one domain is enhanced by binding of another domain. For the bound/free fractions associated with the H1 globular and C-terminal domains shown in Figure 5C, this yielded a cooperativity factor γ=27±8. In other words, the binding of one H10 domain (either the globular or C-terminal domain) tethers the other domain such that it subsequently binds with an affinity 27 times greater than it would if it were untethered and freely diffusing. In general, as we derive in the Supplementary data, the cooperativity factor γ is >1 when the binding of one domain is cooperatively enhanced by the binding of another domain, <1 when binding is diminished, and 0 when binding occurs independently at separate sites. Thus, increasingly larger cooperativity factors indicate increasingly stronger cooperative binding enhancements.
This general strategy to quantify the cooperativity between the H1 globular and C-terminal domains can now be applied to any other two groups of H1 binding regions, for example S1 and S2, S1 and C, C-S1 and S2, or C-S2 and S1. As we show in the next section, this enables prediction of the most likely order that different regions of the H1 molecule bind and the occurrence of conformational changes on binding.
Comparing cooperativity to deduce likely H1 binding pathways
To determine which key H1 binding regions cooperate (C, S1, and S2), we compared their corresponding bound/free fractions, as described in the previous section for the globular and C-terminal domains. As Figure 6A shows, the bound/free fraction in S1C was significantly larger than the sum of its parts for S1 and C (20>0.7+6.9). A similar enhancement was observed for S2C (14>0.4+6.9), suggesting binding cooperation both between S1 and C and between S2 and C. In contrast, we found evidence for independent binding of S1 and S2 by themselves, as the bound/free fraction for S1S2 was essentially equal to the sum of its parts S1 and S2 (1.2∼0.7+0.4). This additivity is consistent with earlier findings that suggested the fast binding of S1S2 was due to S1 alone and the slow binding of S1S2 was due to S2 alone.
Figure 6.
Cooperativity analysis for the key H10 binding regions. (A) In the left most bar-plot the bound/free fraction in S1C is significantly greater than the sum of bound/free fractions in S1 and C, indicative of significant binding cooperation between the C-terminal tail and the S1 site. Similarly, the middle bar indicates significant binding cooperation between the C-terminal tail and the S2 site. In constrast, the inset shows no significant binding cooperation between the S1 and S2 sites by themselves. (B) Once two regions cooperatively bind, either C-S2 (left bar) or C-S1 (right bar), the remaining site (either S1 or S2) binds with cooperativity an order of magnitude larger than in the initial cooperative binding step.
To quantify the cooperativity in each of these cases, we calculated the various cooperativity factors γ using Supplementary equation (S.17). This yielded γ=2.6±0.8 for C and S1, γ=2.4±1.6 for C and S2, and γ=0.3±1.5∼0 for S1 and S2. As domains that cooperate more are more likely to bind sequentially, we can begin to rule out possible binding pathways. Specifically, pathways initiated by the sequential binding of S1 and S2 are ruled out as S1 and S2 bind independently on their own (γ∼0). This implies the globular domain by itself cannot initiate binding, leaving pathways that involve the C-terminal tail in the first two steps.
The fact that the bound/free fraction is substantially higher for the C-terminal tail by itself compared with either S1 or S2 by themselves (6.9 in C versus 0.7 in S1 or 0.4 in S2, as shown in Table 1) argues that the C-terminal tail most likely binds in the first step rather than the second, as it can bind well without the other binding regions. In concordance with this, removal of the C-terminal tail eliminated far more binding than removal of S1 alone or S2 alone (bound/free ∼1 in S1S2 versus bound/free ∼14 in S2C and ∼20 in S1C). This leaves two probable binding pathways: C → C-S1 → C-S1-S2 (binding of C followed by S1 and then S2) or C → C-S2 → C-S1-S2 (binding of C followed by S2 and then S1). Both pathways appear to be equally accessible because the cooperativity factor γ for C and S1 is roughly equal to γ for C and S2. This similarity suggests the C-terminal tail provides a common cooperative tethering mechanism in both cases, again implicating its role as binding initiator.
For the two probable pathways we then calculated the cooperativity factor γ for the final binding step (Figure 6B). This yielded γ∼26±14 for S2 and C-S1 (binding of S2 after C and S1 have already bound) and γ∼22±5 for S1 and C-S2 (binding of S1 after C and S2 have already bound). For both pathways, the cooperativity factor is an order of magnitude larger for the final binding step compared with the first. This suggests the stepwise binding of H10 becomes progressively more favourable, leading to a unique and stable final state (with residence time tr∼100 s, Table 1, row 9) in which all three binding regions are simultaneously bound.
Discussion
We have investigated how the C-terminal tail and sites S1 and S2 within the globular domain of H1 contribute individually and cooperatively to chromatin binding in live cells. To do so, we used a novel FRAP analysis to accurately distinguish diffusive kinetics from the fast and slow binding states of H10 mutants having all eight possible combinations of the three key H1 binding regions.
Our H1 FRAP data had a number of reassuring, self-consistent features. Not only did diffusion and binding estimates for the H10 wild type agree with earlier work, but multiple fast and slow binding states also reappeared in H1 mutants with common binding regions (an important verification of the underlying binding model). Furthermore, removal of all three key H10 binding regions eliminated nearly all observed binding. Finally, the predicted H10 mutant bound/free fractions increased with the number of binding regions present in each mutant. This last result was further corroborated by an independent set of in vitro salt extraction experiments. Taken together, the data allowed us to reliably assign quantifiable binding states to the important H1 binding regions for the first time and to analyse their mutual cooperation.
There are several limitations/caveats to our analysis. First, we assume mutant expression does not detrimentally affect cell health. Although the H1 mutants were stably expressed and represent only a small fraction of endogenous H1 levels, the fraction is still significant given the large endogenous pool. In particular, the abnormal co-localization of H1 mutants to nucleoli raises the issue of altered cell metabolism, which could indirectly skew results. Second, we assume the GFP tag does not measurably alter construct binding kinetics. We chose to place the GFP tag on the C-terminal tail of our H1 constructs. Although our wild-type measurements agreed with an earlier study using an N-terminal GFP tag (Carrero et al, 2004), without a direct comparison of multiple tag locations for each construct, there remains some uncertainty about tag interference. Third, as in nearly all work with mutants, we assume isolated binding domains can be used to infer their behaviour in the complete molecule. Complementary methods are therefore important for confirming this reductionist hypothesis. For example, conformational changes induced by DNA binding might be confirmed in vivo using acceptor photobleaching FRET (van Royen et al, 2007) and predictions about preferred binding pathways might be confirmed in vitro using paramagnetic NMR to determine binding intermediates (Iwahara and Clore, 2006). Fourth, our analysis relies on a simplified model that assumes binding occurs homogeneously throughout the FRAP measurement vicinity at a single type of site. In reality binding is heterogeneous, so measurements represent average behaviour. A more sophisticated analysis is therefore required to measure binding at atypical locations. This could involve simulations, like our analysis at heterochromatic foci, or extensions of the underlying model, like those used to investigate localized (Sprague et al, 2006) or mobile binding sites (Braga et al, 2007).
Complex binding of the H10 C-terminal tail
By themselves, both the H10 globular (the S1S2 mutant) and C-terminal domains (the C mutant) exhibited unique fast (effective diffusive) and slow binding states not seen in the wild type. Although the fast and slow states of the globular domain could be attributed to independent binding of sites S1 and S2, the fast and slow states of the C-terminal domain are not so easily decomposed. Considering the high density of positively charged residues within the C-terminal tail, the fast state is likely due to rapid yet weak nonspecific interactions with DNA. On the other hand, the slow state lasts for 10 s and so represents a more specific and stronger interaction. These measured fast and slow states provide in vivo evidence that the C-terminal tail by itself binds chromatin in two steps. This is consistent with models (Hendzel et al, 2004; Th'Ng et al, 2005; Raghuram et al, 2009) that predict DNA binding by an intrinsically disordered C-terminal tail is initiated through fast and weak nonspecific interactions that can then lead to the acquisition of secondary structure within the tail to strengthen the overall bond. This transition to a tighter state could be due to the (ST)/PXK phosphorylation motifs within the C-terminal tail (Th'Ng et al, 2005). These motifs are known to affect DNA binding (Hill et al, 1991) and their numbers and distribution vary between H1 variants. Thus, in contrast to the largely conserved globular domain, these motifs and their interactions may distinguish the role of each H1 variant. In the future it would be interesting to perform an integrated FRAP analysis on these motifs to further elucidate their cooperativity and function within the H1 C-terminal tail.
Two probable H1 binding pathways
To elucidate the H1 binding mechanism, we derived a useful metric for quantifying how cooperative the binding domains of a protein are. This allowed us to compare cooperativity between all possible groupings of the three key H1 binding regions. Of the possible H1 binding pathways, our analysis revealed the two most cooperative (and therefore most likely) pathways were both initiated by the C-terminal tail (Figure 7, step 1). After this initial binding event, our data indicate sites S1 and S2 within the globular domain are favourably tethered such that both subsequently bind with a ∼2.5-fold cooperative enhancement (γ) relative to a freely diffusible H10 molecule (Figure 7, step 2). This demonstrates that binding of a single H10 molecule at a nucleosome is favoured over simultaneous binding of two or three H10 molecules to the three distinct binding sites on the same nucleosome.
Figure 7.
H10 binding model. Step 1: Our cooperative binding analysis of H10 constructs suggests that the majority of initial contact of H10 (dark green) with a nucleosome (grey with red DNA) occurs through the C-terminal tail as the C mutant had a much higher affinity than the S1 or S2 mutants. Step 2: This restricts the search space of the globular domain, allowing either the S1 binding site (black oval) or the S2 binding site (white oval) to subsequently bind their target DNA (black square and white triangle, respectively) at rates that are cooperatively enhanced by γ ∼2.5 times compared with a freely diffusing molecule. Step 3: A conformational change occurs that brings the binding targets for S1 and S2 closer together so that they can be bound by a single globular domain at the same time (not possible in the S1S2 mutant because it lacks the C-terminal tail). This may coincide with the C-terminal tail acquiring structure (shown as a straighter tail) that strengthens the overall bond (the slow state present in the C, S1C, and S2C mutants), although our data cannot conclusively determine when this occurs. Step 4: This further restricts the search space of the globular domain, allowing the remaining binding site to subsequently bind at a rate that is cooperatively enhanced by γ∼25 times compared to a freely diffusing molecule.
Once two regions are bound (either C-S1 or C-S2) the remaining H10 binding region also appears to bind cooperatively with an enhancement factor (γ) that is an order of magnitude larger (∼25 versus ∼2.5) than that between the first and second binding steps. This increase probably reflects even more favourable positioning of the last binding region relative to its binding site once the first two binding regions have been bound (Figure 7, steps 3 and 4). Our data suggest therefore that once the C-terminal domain binds to linker DNA, a progressively more efficient ‘zippering' of the H10 molecule to chromatin generates the final rather tightly bound state (residence time of ∼100 s for the H10 wild-type; Carrero et al, 2004; Phair et al, 2004; Beaudouin et al, 2006; Lele et al, 2006).
With this picture, we can now distinguish between the three current models for how H1 binds chromatin (Raghuram et al, 2009). In model one, binding is initiated by the C-terminal domain at low affinity, followed by binding of the globular domain at high affinity (Brown et al, 2006). In model two, the role of the two domains is reversed, whereas in model three both domains bind simultaneously at low affinity (Raghuram et al, 2009). Of these, only model one is consistent with the two binding pathways we predicted using our procedure (C → C-S1 → C-S1-S2 or C → C-S2 → C-S1-S2).
Interestingly, our data also suggest an unanticipated conformational change induced by the C-terminal tail at an intermediate stage of H10 binding. Specifically, we found that the S1S2 mutant could only be bound by either the S1 site alone or the S2 site alone, but not by both sites at the same time. On the other hand, when the C-terminal tail is intact, as in the H1 wild type, all three regions (S1, S2, and C) can bind simultaneously. Thus, the C-terminal tail appears to be responsible for bringing together or reorienting the DNA-binding targets for S1 and S2 so that they can be simultaneously bound by a single H1 globular domain (Figure 7, step 3). Such a conformational change is again consistent with the concept of an intrinsically disordered C-terminal tail that acquires structure on binding, as already suggested by the fast and slow C-terminal binding states discussed above. In this sense, the slow C-terminal tail binding state may lead to higher-order C-terminal tail structure, and this may then induce the conformational change that brings the DNA-binding targets for S1 and S2 together, although our data cannot conclusively link the timing of these two events. Given that theory predicts S1 targets the curved nucleosomal DNA near the dyad whereas S2 targets the nearby straight linker DNA (Brown et al, 2006), simultaneous binding of S1 and S2 would reduce the angle of incoming/outgoing nucleosomal DNA and promote higher-order chromatin compaction.
In summary, our data reveal in vivo H1 binding is for the most part controlled through cooperation between the globular and C-terminal domains rather than any one domain in particular. This confers many biological advantages. Not only is binding avidity greater than the sum of constituent monovalent affinities, but the net interaction is also more dynamic, with several binding pathways involving partially bound intermediate states (C, C-S1, or C-S2) that can be readily competed against. For example, H1 competitors such as the HMG proteins (Bustin, 1999) could target these partially bound forms of H1 to prevent the globular domain from positioning itself, or they could target the C-terminal tail to prevent H1 from binding in the first place. With evidence for this type of bimodal competition beginning to accumulate (Rochman et al, 2009), our cooperativity analysis could be used to decipher the competitive mechanism by quantifying how specific H1 binding states change in the presence and absence of competition.
Generality of the approach
Our method has been worked out for H10 binding to chromatin, so it could be used to investigate other proteins that are bound to relatively immobile structures inside the cell, including transcription factors, co-activators, or chromatin-remodelers. Of particular interest is the study of chromatin modifying proteins that could have a role in a potential ‘histone code', where it has been suggested that a single protein can apply more than one chromatin modification through the cooperative binding of multiple domains (Ruthenburg et al, 2007). In this case, our method could be used to gauge cooperativity and determine how often different domains are simultaneously bound.
An advantage of our method is that it only requires the measurement of bound/free fractions and therefore can be performed either in vitro or in vivo. As our method quantifies the individual and cooperative binding properties of distinct regions of a molecule, it opens up the intriguing possibility of cataloging the interactions of common binding domains and perhaps explaining the success of specific combinations that appear regularly in related proteins.
Materials and methods
Constructs and cell lines
Plasmid MTH10GFPneo has GFP fused to the C terminus of the coding region for H10 and is under the control of the mouse metallothionein promoter, as described earlier (Gunjan and Brown, 1998; Misteli et al, 2000). For each construct, stable transfectants derived from mouse BALB/c 3T3 were isolated and analysed. Levels of wild-type H10-GFP were shown earlier by HPLC to represent <5% of the total linker histone content (Misteli et al, 2000). As assayed by fluorescence of live cells and the amount of fluorescent material extracted from isolated nuclei, the levels of mutant H10-GFP constructs are <1% of the total linker histone pool.
Imaging conditions
Experiments were performed on a Zeiss 510 confocal microscope with a 100 × /1.3 NA oil-immersion objective. Cells were imaged in LabTek II chambers (Nalgene) kept at 37°C using an air-stream stage incubator (Nevtek, Burnsville, Rochester, NY). Spatio-temporal FRAP recoveries were recorded using the 488 nm line from a 40 mW argon laser operating at 90% laser power with the AOTF set to 0.5% and the pinhole set to 3.4 AU. Experiments were repeated in the same spot to further improve signal-to-noise when needed and repeated once more without an intentional photobleach to correct for observational photobleaching (Mueller et al, 2008). For each construct, data were obtained from 5–10 cells on at least three separate days.
Spatio-temporal FRAP
We determined the spatial profile of the FRAP at each time point in one of two ways. For the slow H10 wild-type recovery, we collected a 2D image (512 pix2=30.7 μm2 in 0.49 s+2 s delay at scan speed 7 and zoom 3) at each time point and then performed a radial average of intensities in concentric regions of the circular bleach profile (single iteration bleach of radius 30 pix=1.80 μm requiring 590 ms). For the other recoveries, we collected only a 1D line scan (128 × 1 pix2=23.03 × 0.18 μm2 in 1.92 ms at scan speed 7 and zoom 4) bisecting the circular bleach profile (single iteration bleach of radius 12.5 pix=2.25 μm requiring 54 ms), and then generated a radial profile by averaging pairs of intensity values at points equidistant from the centre point of the bleach. For the C-terminal tail constructs, a 1 s delay was added between consecutive line scans.
Salt extractions
For each construct, a large prep of crude nuclei was prepared, aliquoted into a series of tubes, and quickly pelleted and then resuspended in buffer containing various concentrations of KCl. These were incubated on ice for 20 min. Nuclei were then pelleted and the supernatant collected. Each pellet was then resuspended in 600 mM KCl and incubated for 20 min on ice so all bound construct was removed. The nuclei were pelleted again and the supernatant collected. Fluorescence in all samples was determined using a BioTek FLx800 multi-detection microplate reader (excitation filter, 485/20 nm; emission filter, 528/20 nm). Values are expressed as the per cent extracted from the first extraction over the total from the 1st and 2nd extractions. Samples from nuclei prepared from cells not expressing H10-GFP yielded a negligible signal.
FRAP analysis
Spatio-temporal images of FRAP recoveries in euchromatin (where fluorescence is distributed homogeneously) were fit to a reaction diffusion model (Mueller et al, 2008) and the resultant parameters were used to determine bound/free fractions (Supplementary equations (S.1) and (S.2) in the Supplementary data). The initial profile was determined by linearly extrapolating from the first 10–20 measured spatial profiles to a time midway through the bleach process. Images were background subtracted, corrected for unintentional photobleaching, and normalized as described earlier (Mueller et al, 2008). Data processing and fits were performed using a custom program written in Mathematica that uses its built-in FindMinimum routine. See the Supplementary data for a description of how FRAP performed at heterochromatin (where fluorescence is distributed heterogeneously) was analysed.
Supplementary Material
Acknowledgments
We thank Dr Tatiana Karpova for imaging assistance and critical reading of the manuscript and Dr Michael Kruhlak for DIC imaging assistance. We thank Dr Michael Bustin and Dr Michael Seidman for reviewing earlier drafts and Dr Davide Mazza and Dr Ariel Michelman-Ribeiro for discussions. We are grateful to Dr Tom Misteli for assistance in obtaining H1 constructs, discussions, and critical reading of the manuscript. This research was supported in part by the intramural program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Beaudouin J, Mora-Bermudez F, Klee T, Daigle N, Ellenberg J (2006) Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys J 90: 1878–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA 95: 14173–14178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeckmans K, Remaut K, Vandenbroucke RE, Lucas B, De Smedt SC, Demeester J (2007) Line FRAP with the confocal laser scanning microscope for diffusion measurements in small regions of 3-D samples. Biophys J 92: 2172–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga J, McNally JG, Carmo-Fonseca M (2007) A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching. Biophys J 92: 2694–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT (2003) Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol 81: 221–227 [DOI] [PubMed] [Google Scholar]
- Brown DT, Izard T, Misteli T (2006) Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol 13: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol 19: 5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero G, Crawford E, Hendzel MJ, de Vries G (2004) Characterizing fluorescence recovery curves for nuclear proteins undergoing binding events. Bull Math Biol 66: 1515–1545 [DOI] [PubMed] [Google Scholar]
- Catez F, Ueda T, Bustin M (2006) Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol 13: 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan YH, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI (2005) Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123: 1199–1212 [DOI] [PubMed] [Google Scholar]
- Georgel PT, Hansen JC (2001) Linker histone function in chromatin: dual mechanisms of action. Biochem Cell Biol 79: 313–316 [PubMed] [Google Scholar]
- Gunjan A, Brown DT (1998) Effects of H1 histone overexpression on transcription from the MMTV-LTR promoter. Mol Biol Cell 9: 452A [Google Scholar]
- Happel N, Doenecke D (2009) Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431: 1–12 [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th'Ng JPH (2004) The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem 279: 20028–20034 [DOI] [PubMed] [Google Scholar]
- Hill CS, Rimmer JM, Green BN, Finch JT, Thomas JO (1991) Histone—DNA interactions and their modulation by phosphorylation of -Ser-Pro-X-Lys/Arg-motifs. EMBO J 10: 1939–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtsmuller AB (2005) Fluorescence recovery after photobleaching: application to nuclear proteins. Adv Biochem Eng Biotechnol 95: 177–199 [DOI] [PubMed] [Google Scholar]
- Iwahara J, Clore GM (2006) Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440: 1227–1230 [DOI] [PubMed] [Google Scholar]
- Jencks WP (1981) On the attribution and additivity of binding-energies. Proc Natl Acad Sci USA 78: 4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele T, Wagner SR, Nickerson JA, Ingber DE (2006) Methods for measuring rates of protein binding to insoluble scaffolds in living cells: Histone H1-chromatin interactions. J Cell Biochem 99: 1334–1342 [DOI] [PubMed] [Google Scholar]
- Lever MA, Th'Ng JPH, Sun XJ, Hendzel MJ (2000) Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408: 873–876 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Miyawaki A, Nagai T (2008) Direct measurement of protein dynamics inside cells using a rationally designed photoconvertible protein. Nat Methods 5: 339–345 [DOI] [PubMed] [Google Scholar]
- McNally JG, Müller WG, Walker D, Wolford R, Hager GL (2000) The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287: 1262–1265 [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Mistell T (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T (2001) Nuclear structure—protein dynamics: implications for nuclear architecture and gene expression. Science 291: 843–847 [DOI] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408: 877–881 [DOI] [PubMed] [Google Scholar]
- Mueller F, Wach P, McNally JG (2008) Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J 94: 3323–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T (2004) Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol 24: 6393–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram N, Carrero G, Th'ng J, Hendzel MJ (2009) Molecular dynamics of histone H1. Biochem Cell Biol 87: 189–206 [DOI] [PubMed] [Google Scholar]
- Robinson PJJ, Rhodes D (2006) Structure of the ‘30 nm' chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol 16: 336–343 [DOI] [PubMed] [Google Scholar]
- Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu XL, Bubunenko NA, Grigoryev S, Bustin M (2009) The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell 35: 642–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A, Sandin S, Rhodes D (2008) Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA 105: 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Muller F, Pego RL, Bungay PM, Stavreva DA, McNally JG (2006) Analysis of binding at a single spatially localized cluster of binding sites by fluorescence recovery after photobleaching. Biophys J 91: 1169–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Pego RL, Stavreva DA, McNally JG (2004) Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J 86: 3473–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Th'Ng JPH, Sung R, Ye M, Hendzel MJ (2005) H1 family histones in the nucleus—control of binding and localization by the C-terminal domain. J Biol Chem 280: 27809–27814 [DOI] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A (1979) Involvement of histone-H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol 83: 403–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Royen ME, Cunha SM, Brink MC, Mattern KA, Nigg AL, Dubbink HJ, Verschure PJ, Trapman J, Houtsmuller AB (2007) Compartmentalization of androgen receptor protein-protein interactions in living cells. J Cell Biol 177: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan YH (2006) Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14: 17–25 [DOI] [PubMed] [Google Scholar]
- Xouri G, Squire A, Dimaki M, Geverts B, Verveer PJ, Taraviras S, Nishitani H, Houtsmuller AB, Bastiaens PIH, Lygerou Z (2007) Cdt1 associates dynamically with chromatin throughout G1 and recruits Geminin onto chromatin. EMBO J 26: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellajoshyula D, Brown DT (2006) Global modulation of chromatin dynamics mediated by dephosphorylation of linker histone H1 is necessary for erythroid differentiation. Proc Natl Acad Sci USA 103: 18568–18573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.