Abstract
Microtubule (MT) binding accelerates the rate of ATP hydrolysis in kinesin. To understand the underlying mechanism, using charged-to-alanine mutational analysis, we identified two independent sites in tubulin, which are critical for kinesin motility, namely, a cluster of negatively charged residues spanning the helix 11–12 (H11–12) loop and H12 of α-tubulin, and the negatively charged residues in H12 of β-tubulin. Mutation in the α-tubulin-binding site results in a deceleration of ATP hydrolysis (kcat), whereas mutation in the β-tubulin-binding site lowers the affinity for MTs (K0.5MT). The residue E415 in α-tubulin seems to be important for coupling MT binding and ATPase activation, because the mutation at this site results in a drastic reduction in the overall rate of ATP hydrolysis, largely due to a deceleration in the reaction of ADP release. Our results suggest that kinesin binding at a region containing α-E415 could transmit a signal to the kinesin nucleotide pocket, triggering its conformational change and leading to the release of ADP.
Keywords: ATPase kinetics, kinesin, microtubule, motility, mutant analysis
Introduction
Kinesin is a molecular motor involved in intracellular transport along microtubules (MTs) (Sharp et al, 2000; Hirokawa and Noda, 2008; Gennerich and Vale, 2009). Conventional kinesin consists of two identical heavy chains containing the motor domains responsible for MT binding and ATP hydrolysis (Bloom et al, 1988; Kuznetsov et al, 1988). Kinesin makes discrete 8-nm steps (=the size of a tubulin dimer), with each step tightly coupled to ATP hydrolysis (Hua et al, 1997; Schnitzer and Block, 1997). The mechanism involved in converting the chemical energy of ATP hydrolysis to produce this mechanical step has not yet been understood.
Early studies on the kinesin kinetic mechanism have depicted several key features in its mechano-chemical cycle of ATPase. When kinesin is free in solution, it is in the so-called ‘trapped state,' tightly holding ADP, and it has a very low affinity for MTs. MT binding catalyzes escape from this state, changing the active-site conformation such that the ADP release is accelerated (Hackney, 1988, 1994). In the latter, nucleotide-free state, kinesin binds strongly to MTs with an affinity three-to-four orders of magnitude higher than that in the trapped state (Crevel et al, 1996). Identifying the structural basis regarding how the interaction with MTs induces ADP release is critical to understanding the mechanism of motility, because this kinesin transition from the trapped to the nucleotide-free state is the key event that controls the rate of ATPase hydrolysis and is responsible for force generation (Schief and Howard, 2001; Cross, 2004).
Recent cryo-electron microscopic (cryo-EM) analysis of the kinesin–MT complex revealed the nucleotide-binding pocket in open and closed configurations, depending on the bound nucleotides. The open configuration observed either in the presence of ADP (Hirose et al, 2006; Kikkawa and Hirokawa, 2006) or in the absence of any nucleotides (Hirose et al, 2006; Sindelar and Downing, 2007) was resolved for the first time using the kinesin–MT complex as a specimen, and was clearly different from previous X-ray crystallographic structures of kinesin observed in the absence of MT (Kull et al, 1996; Kikkawa et al, 2001). These structural differences are consistent with the kinetic scheme that suggests that MT binding catalyzes ADP release in kinesin.
To understand the exact molecular mechanism underlying the MT-dependent ADP release, it is crucial to know at which site kinesin and tubulin interact with each other, and via which structural pathway this MT-binding signal is transmitted to the nucleotide pocket for the release of ADP. To this end, we first need to locate the binding sites on both kinesin and tubulin. The cryo-EM structure of the kinesin–MT complex revealed a view of the interface between kinesin and MT, in which the structural elements L7, L8, L11, L12, α4, α5, and α6 of kinesin are located in proximity to H11, the H11–12 loop including H11′, H12, and the C-terminal tail of both α- and β-tubulin (Skiniotis et al, 2004; Hirose et al, 2006; Kikkawa and Hirokawa, 2006; Neumann et al, 2006; Sindelar and Downing, 2007). To further locate the interface at the level of amino acids, Woehlke et al (1997) had conducted alanine-scanning mutagenesis of kinesin and identified several positively charged residues in L7/8, L11, and α4/L12/α5 as MT-interacting kinesin residues. To seek their counterparts in tubulin, we had previously developed an experimental system for mutational analysis of tubulin using a yeast expression system, and we identified the negatively charged residues, E410 and D417, in H12 of β-tubulin as critical residues (Uchimura et al, 2006). However, the cryo-EM structure indicates that these residues might constitute only a part of the MT interface for kinesin. Recent in vivo analysis on human tubulin mutants suggests the possibility that α-tubulin may also be involved in the interaction with kinesin (Keays et al, 2007; Poirier et al, 2007).
In this study, we extended our mutagenesis efforts to the MT outer surface in both α- and β-tubulin, covering nearly the entire area implicated as an interface for kinesin. The result revealed that, in addition to the previously identified residues in β-tubulin, the negatively charged cluster containing E415, E416, E418, and E421 in α-tubulin is also crucial for kinesin motility. Furthermore, ATPase and unbinding force measurements indicated that kinesin–MT interaction probably occurs in two steps: while the initial binding interaction through α-tubulin is proven to trigger ADP release, subsequent binding interaction through α- and β-tubulin locks kinesin firmly on the MT. Using a new technique of tubulin mutational analysis, the study succeeded in dissecting the structure–function relationship of the kinesin–MT interaction in motility for the first time.
Results
Design of charged-to-alanine mutations and mutant phenotypes
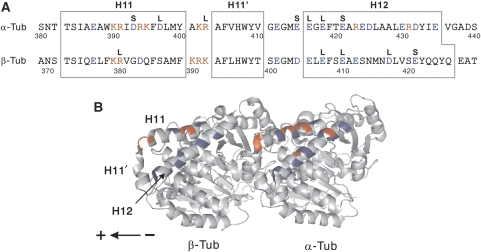
On the basis of the results of structural studies (Skiniotis et al, 2004; Hirose et al, 2006; Kikkawa and Hirokawa, 2006; Neumann et al, 2006; Sindelar and Downing, 2007) and the mutational analysis of kinesin (Woehlke et al, 1997), the charged residues contained in the area of MT spanning H11, the H11–12 loop including H11′, and H12 in both α- and β-tubulin were selected for mutagenesis to alanine (Figure 1). Using the budding yeast, Saccharomyces cerevisiae, we had previously developed an expression system for tubulin dimers composed of single-sequence α- and β-tubulin, Tub1p and Tub2p (Uchimura et al, 2006). Using this expression system, analyses of tubulin mutations were performed.
Figure 1.
Design of mutants and their phenotypes. (A) The positions of the charged residues in the area of the MT spanning H11 and H12, in both α- and β-tubulin, targeted for alanine mutagenesis. Each of the positively (red) or negatively charged (blue) residues was substituted with alanine, which gave rise to 36 mutant strains in total. The resultant phenotypes of yeast mutant cells are marked by the letter L or S, indicating haploid lethal or slow growth, respectively. (B) A ribbon diagram of a tubulin dimer viewed from the putative outside of the MT with its minus end to the right (Lowe et al, 2001). The positions of the charged residues are marked by the same coloring scheme as in (A).
When the mutated tubulins were expressed in the yeast cells (a total of 36 mutant strains), four α-tubulin mutants and four β-tubulin mutants were observed to be haploid lethal, whereas three α-tubulin mutants and one β-tubulin mutant resulted in the slow growth of yeast cells (Figure 1A). The residues in tubulin that are critical for interaction with kinesin are most probably included in these 12 mutations, because kinesin superfamily proteins in budding yeast are known to participate in mitotic spindle function (Hildebrandt and Hoyt, 2000; Wu et al, 2006), and the disruption of interaction between these kinesin superfamily proteins and MTs through tubulin mutation might affect the growth or the viability of yeast cells. Our previous study showed that there is actually a correlation between such yeast mutant phenotypes and the ability of mutant MTs to interact with kinesin (Uchimura et al, 2006). These 12 residues are highly conserved across the species (Little and Seehaus, 1988) and the majority were negatively charged.
Kinesin interacts with MT at two independent sites
We next isolated tubulin from these 12 mutant strains and assessed their ability to serve as tracks for kinesin in the single-molecule motility assay using fluorescently labeled two-headed kinesin (HK560-Cy3; Vale et al, 1996). To prepare MTs from tubulin with lethal mutations, additional tubulin harmless for yeast cells was co-expressed to rescue cell inviability, and this additional tubulin was biochemically separated from the target mutant tubulin in the process of purification (Supplementary Figure S1).
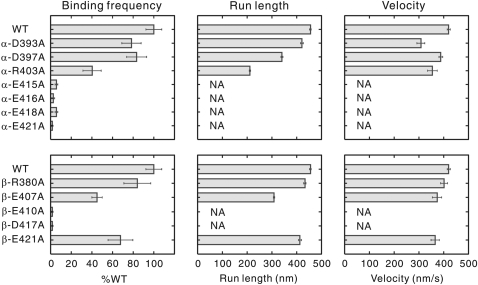
The results of the single-molecule motility assay showed that although kinesin moved processively along the wild-type MTs (run length=455±30 nm; mean±s.e.m.), it scarcely moved along α-E415A, E416A, E418A, E421A, and β-E410A and D417A MTs (Figure 2 and Supplementary Figure S2). The frequency of kinesin–MT interaction showing movement for more than 100 nm on these mutated MTs was lower than 7% of that observed for the wild type. For other six mutants, the effect of mutation on kinesin motility was either moderate or undetectable.
Figure 2.
Binding frequency, run length, and velocity of HK560-Cy3 measured with mutant MTs. The binding frequency, mean run length, and velocity were calculated as described in Materials and methods section. NA stands for data not available and the error bars represent s.e.m. values. The mutants β-E410A, D417A, and E421A were previously characterized in different assay condition (Uchimura et al, 2006). The distributions of the raw data are available in Supplementary Figure S2.
In the molecular structure of a tubulin dimer (Lowe et al, 2001), residues critical for motility are located in two discrete areas (Figure 6A). The critical residues in α-tubulin (E415, E416, E418, and E421) are in a cluster, spanning the H11–12 loop and the N-terminal portion of H12. In contrast, the critical residues β-E410 and D417 are located in H12 of β-tubulin. The latter two residues have been characterized in an earlier study (Uchimura et al, 2006) but these were re-examined here to standardize the experimental conditions with other mutants.
Both α- and β-tubulin contribute to stall force
To investigate in which chemical state of the kinesin–ATPase cycle the interaction was perturbed in these mutants, we next measured the force required to dissociate kinesin from the mutant MTs (unbinding force) in three nucleotide conditions: in the presence of ADP and 5′-adenylylimidodiphosphate (AMP-PNP), and in the absence of nucleotides (Kawaguchi and Ishiwata, 2001; Uemura et al, 2002). In each nucleotide condition, a single-headed kinesin (single-headed heterodimer of Drosophila kinesin; Hancock and Howard, 1998) attached to a polystyrene bead was made to interact with an MT through the use of optical tweezers. An external load was gradually applied to the kinesin–MT complex by moving the stage of the microscope at a constant velocity toward the plus or minus end of the MT until the bead dissociated. The unbinding force, which reflects the stability of kinesin–MT interaction, was calculated by multiplying the magnitude of abrupt bead displacement during detachment with the stiffness of the optical tweezers.
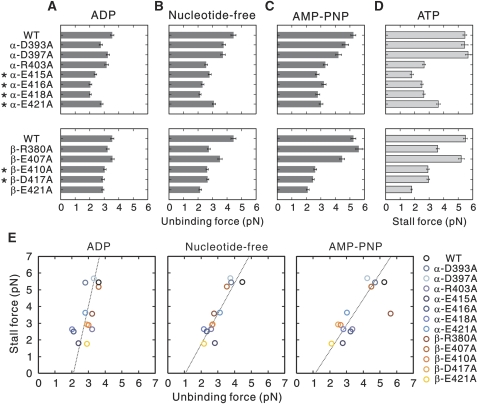
The results revealed that, in the ADP state, kinesin was dissociated from the MTs by smaller forces in α-E415A, E416A and E418A mutants, as compared to that from the wild-type MT (Figure 3A). In several other mutants, including the mutants in β-tubulin, a slight reduction in unbinding force was detected.
Figure 3.
Unbinding force and stall force of kinesin measured using mutant MTs. (A–C) Unbinding force of single-headed kinesin was measured with load applied toward the MT minus end (A) in the presence of ADP, (B) in the absence of nucleotides, and (C) in the presence of AMP-PNP. The asterisks mark the mutants in critical residues. The distributions of the unbinding forces for each mutant for both minus- and plus-end loadings are shown in Supplementary Figure S3A–C. The stiffness of the optical trap was 0.076 pN nm−1. The error bars represent s.e.m. values. (D) Kinesin stall force for conventional, two-headed kinesin was measured using optical tweezers at trap stiffness of 0.076 pN nm−1. Each bar represents the average stall force with s.e.m. value. The distributions of stall force are shown in Supplementary Figure S3D. (E) The stall force plotted against the unbinding force for minus-end loading measured in three nucleotide conditions. The stall force plotted against the unbinding force for plus-end loading is available in Supplementary Figure S4. Note that the absolute amount of unbinding force depends on the loading rate and, therefore, cannot be equal to the stall force (Kawaguchi et al, 2003). The mutants β-E410A, D417A, and E421A were previously characterized in different assay condition.
In the nucleotide-free and AMP-PNP states, the stability of the kinesin–MT complex was significantly reduced in both α- and β-tubulin mutants (Figure 3B and C). Those mutants showing poor motility in the single-molecule motility assay (Figure 2) were apt to be dissociated from the MTs with smaller forces in the nucleotide-free and AMP-PNP states. However, the opposite case was not true; in nucleotide-free and/or AMP-PNP conditions, the binding of kinesin to the β-R380A and E421A MTs was less stable as compared to that with wild-type MTs, yet these mutant MTs functioned normally as a track for kinesin, at least in terms of kinesin velocity and run length. Taken together, the residues critical for kinesin motility might have some specific roles in the mechanism of kinesin motility by not only increasing the stability of the interaction, but also by transmitting a signal to kinesin through its tertiary structure. For simplicity, only the unbinding force for minus-end loading is shown in Figure 3A–C (for plus-end loading, see Supplementary Figure S3A–C).
We next measured the stall force of kinesin in the presence of ATP, to examine how tubulin mutation affects the force generation of kinesin (Figure 3D and Supplementary Figure S3D). The measurement results for the 12 mutants revealed that kinesin produces a smaller force on those mutant MTs with reduced stability of interaction as compared with the wild-type MTs. When the stall force was plotted against the unbinding force measured in the three nucleotide conditions, it became clear that stall force is related to the unbinding force in the nucleotide-free and AMP-PNP states, but not to that in the ADP state (Figure 3E and Supplementary Figure S4). Each mutation had a very similar effect on the unbinding forces in the nucleotide-free and AMP-PNP states. The only exception was the β-R380A mutant that showed a deviation from the regression line representing the linear relationship between the stall force and the unbinding force in the AMP-PNP state. The most plausible interpretation of this result is that, unlike the other 11 mutants, the residue β-R380 may contribute to the stability of the kinesin–MT interaction only in the nucleotide-free state. In summary, the results indicate that the stall force is governed by the stability of the kinesin–MT interaction in the strong binding state (the nucleotide-free and AMP-PNP states), which are reliant upon both α- and β-tubulin.
α- and β-tubulin have distinct roles in ATPase activation
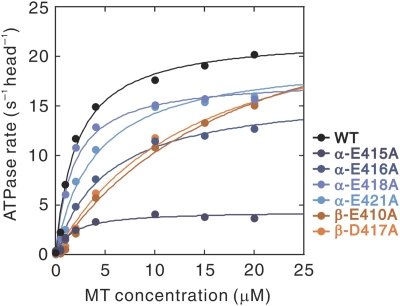
The MT binding to kinesin is known to accelerate ADP release and subsequent ATP binding, thereby accelerating the overall rate of ATP hydrolysis (Kuznetsov and Gelfand, 1986; Hackney, 1988, 1994). To examine how each of the mutations in the residues critical for motility (α-E415, E416, E418, E421, and β-E410 and D417) affects ATP hydrolysis, we measured the MT-activated ATPase of two-headed kinesin (HK560) using the mutant MTs. Although the movement of a single kinesin molecule was scarcely detected on those mutant MTs (Figure 2), they were still capable of activating the ATPase activity of kinesin (Figure 4 and Table 1).
Figure 4.
Steady-state ATPase activity of kinesin (HK560) with mutant MTs. The data shown in this graph are a representative example of the measurements for wild-type and mutant MTs. The maximal turnover rate (kcat) and apparent Michaelis–Menten constant (K0.5MT) were calculated by fitting the data to a hyperbola, and the averages obtained from four to six sets of experiments are shown in Table I.
Table 1. Summary of ATPase measurement and MT gliding assay.
| Construct | MT-activated ATPasea | MT glidinga | ||
|---|---|---|---|---|
| K0.5MT (μM)b | kcat (s−1 head−1)b | Velocity (nm s−1)c | n | |
| WT | 2.3±0.7 | 22.0±0.6 | 324±2 | 250 |
| α-E415A | 1.8±0.1 | 4.5±0.4 | 5±0 | 153 |
| α-E416A | 5.5±0.5 | 17.6±1.9 | 410±3 | 147 |
| α-E418A | 2.3±0.8 | 18.0±0.7 | 292±2 | 240 |
| α-E421A | 4.2±0.9 | 20.7±0.7 | 391±3 | 201 |
| β-E410A | 14.7±3.9 | 26.7±2.4 | 531±3 | 186 |
| β-D417A | 13.7±2.0 | 25.9±1.2 | 540±4 | 166 |
| aBoth ATPase measurement and MT gliding assay were performed using HK560. | ||||
| bMean±s.d. from four to six independent experiments. | ||||
| cMean±s.e.m. | ||||
Interestingly, ATPase measurement revealed a clear difference between the two groups of mutants in α- and β-tubulin. In the former group, the mutation did not significantly affect K0.5MT, whereas their kcat value was reduced to a variable extent depending on the site of mutation. The reduction in kcat value was most prominent in the α-E415A mutant; a five-fold reduction was observed as compared to that of the wild-type MTs. In contrast, mutations in β-tubulin resulted in a lower affinity for kinesin; K0.5MT values for β-E410A and D417A were more than five-fold higher than that for the wild type. In these mutants, their kcat value even increased, opposite to the effect of mutations in α-tubulin. These results demonstrate that it is the interaction mediated through α-tubulin that has a crucial role in activating kinesin ATPase.
The results also suggest that kinesin should still be capable of actively interacting with these mutant MTs if the effective MT concentration is sufficiently high. In fact, when these MTs were tested in the MT gliding assay, all mutant MTs were able to glide on the kinesin-coated glass surface (Table 1). Owing to the simultaneous interactions of multiple kinesin molecules with a single MT, the mutant MTs might be able to maintain continuous contact with a surface coated with kinesin; thus, they displayed a gliding movement over long distances. The gliding speed varied from one mutant to the next, with each velocity nearly proportional to the ATPase rate (kcat).
The role of α-E415 in kinesin ATPase activation
The α-E415 residue seems to have a unique role; the impaired motility caused by the mutation at this site was scarcely compensated for by the high density of kinesin molecules used in the gliding assay (Table 1). In stall force measurements, the velocity of kinesin along the α-E415A MT observed at low load was extremely slow compared with the velocity on the other mutant MTs (data not shown). These results suggest the possibility that the α-E415 residue may be a key residue crucial for coupling MT binding and ADP release from kinesin.
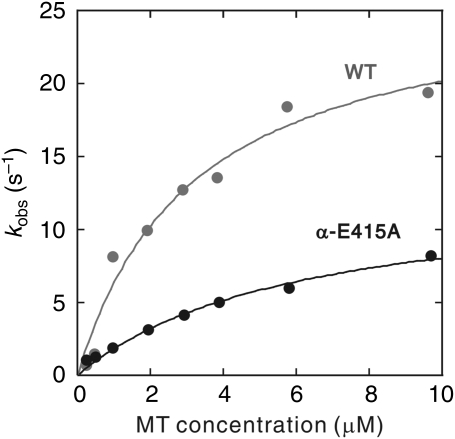
To examine this possibility, the rate of dissociation of mant-ADP was measured by stopped-flow assay, by mixing the HK560 mant-ADP complex with the α-E415A MTs. The rate of ADP dissociation, kobs, was obtained by exponential fitting of the decay in fluorescence. A hyperbolic dependence of kobs on MT concentration gave a value for kmax of 26.6±3.2 and 12.9±1.2 s−1 for wild type and α-E415A mutants, respectively (Figure 5 and Table 2). The dissociation of ADP was indeed slowed down in the α-E415A mutant. The apparent rate constant for kinesin–MT association, calculated by the ratio of kmax/K0.5MT (=kbi(ADP)), decreased roughly four-fold in the mutant, indicating that the initial association of kinesin to the MT was also disrupted in this mutant. The residue α-E415 may be crucial for kinesin to recognize the MT and change its conformation to release ADP from the nucleotide pocket.
Figure 5.
Mictrotubule dependence of mant-ADP release from HK560 kinesin. To determine the ADP release rate, mant-ADP-loaded HK560 was mixed with MTs in a stopped-flow apparatus at post-mixing concentrations of 1 μM HK560, 2 μM mant-ADP, 0.25–10 μM wild-type or α-E415A MTs, and 1 mM ATP. The apparent rate of ADP dissociation was derived by fitting the fluorescence decay by double exponentials, giving fast- and slow-phase rates for ADP release (see Materials and methods section for details). Fast-phase rates (kobs) are plotted as a function of MT concentration. Slow-phase rates were low and independent of MT concentrations, and are omitted from the graphic and analysis. MT dependence of kobs was fit to a hyperbolic model, from which values for parameters kmax and K0.5MT were extracted (Table II).
Table 2. MT dependence of mant-ADP release from HK560.
| Construct | K0.5MT (μM)a | kmax (s−1)a | kbi (μM−1 s−1) |
|---|---|---|---|
| WT | 3.2±0.9 | 26.6±3.2 | 8.3 |
| α-E415A | 6.1±1.0 | 12.9±1.2 | 2.1 |
| aErrors indicate the errors in curve fitting. | |||
When the rate of ADP release (Table 2) and ATPase activity (Table 1) was compared in wild-type MTs (26.6±3.2 and 22.0±0.6 s−1, respectively), it seems that ADP release is the rate-limiting step in the cycle of ATP hydrolysis. However, with α-E415A MTs, their ADP release rate (kmax) was 12.9±1.2 s−1, whereas their ATPase kcat was 4.2±0.9 s−1; the rate of ADP release is not rate limiting in the cycle of ATP hydrolysis. This means that, in a cycle of ATP hydrolysis, in addition to the chemical step of ADP release, other steps in the cycle should also be slowed down, for example, ATP binding, ATP hydrolysis, phosphate release, kinesin dissociation from the MT and so on. Considering the reported result that MT binding accelerates the rate of ATP hydrolysis (Ma and Taylor, 1995, 1997), this step could also be disrupted in the α-E415A mutant.
The value for the kbi ratio (the ratio of the second-order rate constant of MT-activated ATPase (kbi(ATPase)) and that of MT-activated ADP release (kbi(ADP release)) was 1.2 for both α-E415A and wild-type MTs, indicating HK560 is not processive on both types of MTs. This contradicts our observation that, in the single-molecule motility assay, each of the HK560 molecules can move along the wild-type MTs over a distance of several hundred nm (Table 1). This apparent discrepancy may have arisen from the difference in the experimental conditions used for the single-molecule motility assay and that for kinetic measurement.
Discussion
Kinesin–MT interaction through α-tubulin activates the pathway for activation of kinesin ATPase
Using alanine-scanning mutagenesis of yeast tubulin, we have identified charged residues critical for kinesin motility, which are located in two discrete areas of tubulin (Figure 6A). Those MTs with charged-to-alanine mutations at each of the critical residues were further characterized for the stability of their interaction with kinesin (Figure 3), and for their catalytic properties as activators of kinesin ATPase (Figure 4 and Table 1). The results revealed that α- and β-tubulin have distinct roles in the mechanism of motility.
Figure 6.
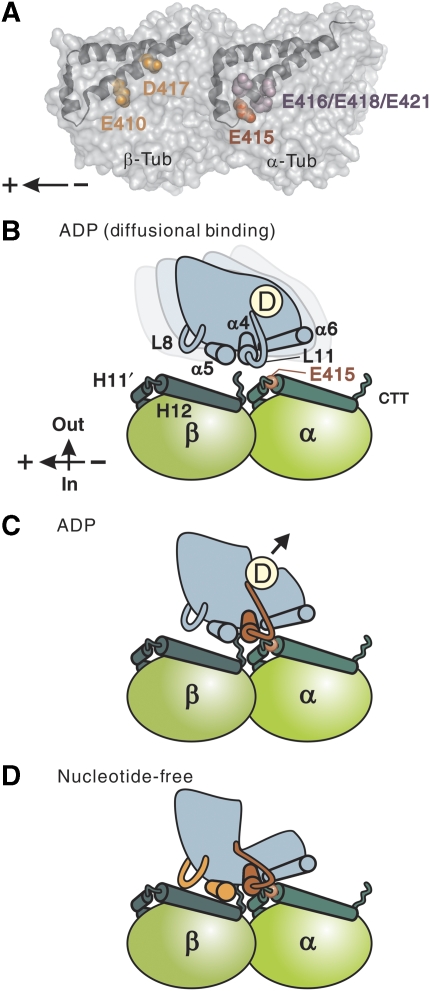
Structural basis for MT-dependent activation of kinesin ATPase. (A) Two independent sites on the MT critical for kinesin motility: the negatively charged residues spanning the H11–12 loop and H12 of α-tubulin (highlighted in red and purple), and the negatively charged residues in H12 of β-tubulin (orange). A tubulin dimer is viewed from the putative outside of the MT tube with its minus end to the right. (B–D) Schematic model for kinesin interaction with MTs in different nucleotide states. In the diffusional binding state (B), kinesin, tightly holding ADP, is in search of the next binding site through diffusion along the MT. Nonspecific electrostatic interaction between the negatively charged C-terminal tail of tubulin (CTT) and the positively charged interface of kinesin contributes to this weak interaction. When the switch II helix/loop of kinesin (red) encounters the H11–12 loop/H12 of α-tubulin (C), kinesin changes its conformation and ejects ADP from the nucleotide pocket. After, or concomitant with ADP release (D), kinesin is further locked on the MT through the interaction between L8/α5 of kinesin (orange) and H12 of β-tubulin. This stable interaction enables kinesin to sustain load.
The unbinding force measurement showed that in the presence of ADP, the interaction of kinesin with the MT was less stable compared with the interaction in the nucleotide-free and AMP-PNP states (Figure 3). The stability of this relatively weak interaction was reduced by the charged-to-alanine mutation most prominently in α-tubulin (α-E415, E416, and E418). In the nucleotide-free and AMP-PNP conditions (Figure 3B and C), the stability of the kinesin–MT complex was reduced in both α- and β-tubulin mutants.
In ATPase measurement, the mutation in α-tubulin resulted mainly in a reduction of kinesin kcat, with little or no effect on MT affinity (K0.5MT), whereas the mutation in β-tubulin resulted in a drastically lower K0.5MT value (Figure 4 and Table 1). The disruption of ATPase activity was most remarkable with the α-E415A mutant, leading to a five-fold reduction in kcat value. These results suggest the possibility that kinesin binding at α-E415 is directly coupled to a chemical reaction in the ATP hydrolysis cycle, most probably to the reaction of ADP release (Hackney, 1988, 1994). The stopped-flow measurement of the rate of ADP dissociation demonstrated that, indeed, MT-dependent ADP release from kinesin was slowed down in the α-E415A mutant (Figure 5 and Table 2).
On the basis of these observations and the results reported in the literature, we propose a model in which the interaction between kinesin and the MT occurs in two sequential steps, with each step requiring a specific binding of kinesin to a distinct tubulin site(s). Owing to the weak electrostatic interaction between the positive charges of the motor head and the negatively charged C-terminal tail of tubulin, a kinesin motor holding ADP will exhibit a diffusional motion along the surface of the MT, along its long axis (Okada and Hirokawa, 2000; Carter and Cross, 2005; Figure 6B). During this diffusional search, an encounter of kinesin head with the α-tubulin-binding site may allow the pair to make specific binding. On formation of the kinesin–ADP–MT complex, structural recognition at α-E415 will initiate a conformational change in kinesin, leading to ADP release from its nucleotide pocket (Figure 6C). Subsequent to this initial interaction at the α-tubulin-binding site, the kinesin head is further locked on the MT through the interaction mediated by α- and β-tubulin-binding sites (Figure 6D), and the resultant kinesin–MT complex is responsible for sustaining load. Force generation is coupled to the entire process of state transition (Figure 6B–D), from the diffusional binding state to the strong-binding, nucleotide-free state (Schief and Howard, 2001; Cross, 2004).
In this model, α-E415 plays a central role in inducing a conformational change in kinesin to release ADP. Among the four negatively charged residues clustered in α-tubulin and identified as critical for kinesin motility, this is the only catalytically critical residue (Table 1). It is noteworthy that, because multiple Glu (E) residues are successively aligned in this area (EEGE), the local charge density around this site is comparable to that of tubulin C-terminal tails. In one possibility, this high density of negative charges, together with the structural flexibility owing to the partial loop structure, may help kinesin in the diffusional binding state find its target and establish specific interaction. Although α-E415 gives kinesin a crucial cue to eject ADP, the acidic residues adjacent to α-E415 may also be important in leading kinesin to the target.
Kinesin motility and MT-dependent ATPase in mutants
Using this model, results of the single-molecule motility assay (Figure 2) and the MT gliding assay (Table 1) can be consistently explained. In the single-molecule motility assay, mutations in the critical residues in α-tubulin may prevent kinesin from switching from the diffusional binding state to the ADP state (Figure 6C), whereas mutations in the critical residues in β-tubulin may make the nucleotide-free state unstable (Figure 6D). In both cases, single kinesin molecules become less capable of initiating interaction with, and/or moving processively along, the mutant MTs, resulting in impaired motility.
However, such perturbation of kinesin–MT interaction by tubulin mutation seems to be surmounted when motility is examined in the MT gliding assay, in which multiple kinesin molecules simultaneously interact with single MTs (Table 1). The observed gliding speed of the mutant MTs was similar to or even higher than that of the wild type, except in the case of the α-E415 mutant, which resulted in very slow movement (5 nm s−1). This result is parallel to that observed in stall force measurements (Figure 3D); single kinesin molecules could exert force on the mutant MTs when they were made to interact with the MTs by manipulation with optical tweezers. Consistent with the results of the MT gliding assay, those mutant MTs were capable of activating MT-dependent kinesin ATPase; the velocity of the MT gliding was nearly proportional to the ATPase rate, kcat (Table 1).
Communication between MT-binding sites and nucleotide pocket
Our model indicates that the interaction at α- and β-tubulin-binding sites has a distinct function in the mechanism of kinesin motility. However, which part of the kinesin motor head would be the binding partner for each binding site in α- and β-tubulin? It is not easy to answer this question on the basis of the results of the structural analyses; first, because an X-ray crystal structure is not available for the kinesin–MT complex and, second, because the cryo-EM map of the kinesin–MT complex has only a limited resolution. Despite a lack of conclusive evidence, the recent cryo-EM map of the kinesin–MT complex has succeeded in depicting, at least in part, a view of the MT interaction site, in which the switch II loop (L11) is extended towards α-tubulin, in proximity to H11′ and the N-terminal segment of H12 (Hirose et al, 2006; Kikkawa and Hirokawa, 2006; Sindelar and Downing, 2007). Such an image is consistent with our results, which showed that both kinesin motility and ATPase were critically dependent upon the α-E415 residue located adjacent to the N-terminus of H12. Interaction at this site could be a starting point in the structural pathway of communication in kinesin connected to its nucleotide pocket.
Previous experiments using mutants of the minus-end-directed kinesin, Kar3, have indicated that the interplay between switch I and II might be crucial for passing signals from the MT-binding site to the nucleotide pocket (Yun et al, 2001). The mutations that block the formation of the salt bridge between switch I and switch II resulted in the uncoupling of MT binding from the activation of kinesin ATPase. Interestingly, in their analyses, the charged-to-alanine mutation in R632 in switch II loop (equivalent to K237 in human KIF5B) showed a significant reduction in the turnover rate of MT-activated ATPase (kcat). This result is analogous to our observation that the α-E415A mutant displays a considerable reduction in kcat (Figure 4 and Table 1). Considering this result and the close proximity of R632 in switch II loop (and its equivalent K237 in KIF5B) to the N-terminal portion of α-tubulin H12, this residue could be a binding partner for α-E415. The formation of the salt bridge between the pair may trigger the chain of communication, that is, structural shifts in switch II, switch I, and the P-loop, leading to the opening of the nucleotide pocket (Kull and Endow, 2002; Sindelar and Downing, 2007; Nitta et al, 2008). This hypothesis is worthy of being examined in the future using a combination of kinesin and tubulin mutants.
Parallel with such a plausible model in kinesin, in some of the G-proteins, GDP release is triggered by the binding of the guanine nucleotide-exchange factor (GEF) at the switch II region (Sprang, 1997; Vetter and Wittinghofer, 2001). In general, GEF-induced GDP release from a variety of G-proteins could occur through three different routes: on binding of GEF, GDP held in the nucleotide pocket becomes unstable due to either the displacement of the switch I loop, distortion of the switch II region, or perturbation of the binding site for the nucleotide base, and ultimately, GDP is ejected from the nucleotide pocket. It is possible that MT-dependent ADP release from kinesin could be driven by similar mechanisms (Kull and Endow, 2002; Sindelar and Downing, 2007; Nitta et al, 2008).
With regard to kinesin–MT interaction through β-tubulin H12, our results suggest that interaction at this site becomes effective on ADP release, mainly to increase the mechanical stability of the interaction (Figure 3). This interpretation is consistent with the results of previous cryo-EM studies showing that, upon ADP release, kinesin firmly bound to the MT through the interaction of α4/L12/α5, L8 with β-tubulin H12 (Skiniotis et al, 2004; Hirose et al, 2006).
Our results using the β-tubulin mutants, E410A and D417A, revealed that disruption of kinesin–MT interaction at this site resulted in the acceleration of both MT-gliding velocity and ATPase kcat (Figure 4 and Table 1). This result could be due to the accelerated dissociation of kinesin from the MT after ATP hydrolysis. This observation contrasts with the results of kinetic analysis; the latter demonstrated that the mutation in kinesin L8, which effected a reduction in kcat and K0.5MT values, caused kinesin to become trapped in the MT rigor complex (Klumpp et al, 2003). It is possible that the binding interaction between kinesin L8 and β-tubulin H12 may have a key role in the regulation of kinesin motility; while the loose locking of kinesin to the MT in β-tubulin mutants accelerated the dissociation of kinesin from the MT, the rigid locking of mutant kinesin to the MT rendered kinesin trapped in the MT rigor complex. In summary, both structural and kinetic data seem to be consistent with our model, in which the kinesin–MT interaction through β-tubulin H12 acts as a type of lock to firmly attach kinesin on the MT (Figure 6D).
In the area of MT outside the target of our mutational analysis (Figure 1), there may still be other residues critical for ATPase activation. Recent structural and mutational studies on KIF1A suggested the possibility that the binding interaction of KIF1A L7 with the residues R158 and E159 in H4 of bovine β-tubulin (equivalent to R156 and E157 of yeast β-tubulin, respectively) leads to ADP release from kinesin (Nitta et al, 2008). However, our preliminary experiment showed charged-to-alanine mutation in β-R156 and E157 had little effect on the MT-activated ATPase of HK560 kinesin (data not shown), indicating that these residues are less likely to be binding partners for L7, at least, in HK560 kinesin. It is possible that the key structure in tubulin may differ among kinesin families, depending on the functional demands placed on each species of kinesin; although L7 may have a crucial role in MT-dependent ADP release in KIF1A, its role in HK560 kinesin could be insignificant.
Using mutational analysis of tubulin, this study uncovered the structural basis for MT-dependent activation of kinesin ATPase, which proceeds in two steps. Binding of the residue E415 in α-tubulin to kinesin gives kinesin a structural cue to open its nucleotide pocket, and this conformational change in kinesin is subsequently stabilized by the binding at the β-tubulin-binding site. The property of the intermediate ADP state seems to be unique; it requires a specific binding interaction, yet its mechanical stability is significantly lower than that in the subsequent nucleotide-free state. Such a property should be suitable for the role of mediating the MT-dependent ADP release. This study has paved the way for forthcoming analyses exploring the structural pathway of communication between the MT binding site and the nucleotide pocket.
Materials and methods
Preparation of tubulin mutants and proteins
The plasmids and the yeast mutant strains were constructed as described in Supplementary data and are listed in Supplementary Table SI. Taxol-binding ability was introduced into a β-tubulin gene by site-directed mutagenesis at five amino acids (Gupta et al, 2003), and the gene tub2-A19K-T23V-G26D-N227H-Y270F thus obtained was referred to as TUB2tax. The yeast strain expressing TUB1 and TUB2tax was used as the wild type. Tubulin was purified from the yeast cell lysate by following the previously developed protocol (Uchimura et al, 2006) with modifications (Supplementary data for details). Using the modified protocol, 300–500 μg of tubulin with a purity >95% was purified from 6 l of yeast culture. HK560 was purified and labelled at its C-terminal cysteine residue with Cy3 (Vale et al, 1996; PA23031, GE Healthcare). Polarity-marked MTs were produced using sea urchin sperm axonemes (Tanaka-Takiguchi et al, 1998).
Single-molecule motility assay and MT gliding assay
The single-molecule motility assay was conducted as previously described (Uchimura et al, 2006), with a couple of modifications. First, MTs were observed by darkfield imaging using an evanescent field (Supplementary Figure S5). Second, MTs were fixed on a glass slide using an anti-tubulin antibody and the ionic strength of the solution was twice increased (x2 MA buffer; 20 mM piperazine-N,N′-bis[2-ethanesulfonic acid] [PIPES], 10 mM Kacetate, 4 mM MgSO4, 2 mM EGTA, 0.2 mM EDTA; pH 6.8). These changes in protocol made the assay much easier in comparison to the original protocol. In brief, a flow chamber assembled from a glass slide and a coverslip was first coated with an anti-tubulin antibody (0.3 mg/ml; T-9026, Sigma) for 3 min, and then washed with 30 μl of x2 MA buffer. Subsequently, 15 μl of yeast MT solution (0.01 mg/ml in x2 MA buffer supplemented with 10 μM Taxol [T-7402, Sigma]) was introduced to the chamber and incubated for another 10 min; in the following process, 10 μM Taxol was consistently included in the solution. The chamber was then incubated with 5 mg/ml casein solution (07319-82, Nacalai Tesque) for 1 min to block non-specific binding of kinesin to the glass surface. Finally, an HK560-Cy3 solution (1 nM in x2 MA buffer) containing 1 mg/ml casein, 1 mM ATP, and an oxygen scavenger (Harada et al, 1990), was introduced into the chamber and motility was observed under a TIRF microscope (Vale et al, 1996). The observation was made at 25°C.
For the MT gliding assay, we adapted a previously-reported method (Howard et al, 1989), with the exception that the standard buffer solution in the original assay was replaced by x2 MA buffer and HK560 was used.
Image analysis of single-molecule motility
The movements of the individual HK560-Cy3 molecules were analyzed using tracking software (G-Track, G-Angstrom). To measure binding frequency, the total number of binding events occurring along a single MT, with its lower limit in the run length set as >100 nm, was counted for 5 min, and this number was divided by the MT length and observation time. For each mutant, the mean value with s.e.m. was calculated from four to ten independent measurements, and was normalized by the mean value for WT (0.53±0.04 μm−1 min−1).
The mean run length was determined by non-linear least squares fitting of the cumulative probability distribution of the data, as previously described (Thorn et al, 2000), with the lower limit of the run set being the same as above. Since the photobleaching rate of Cy3 fluorophore was significantly lower than the detachment rate of kinesin from MT (0.03–0.05 s–1 and 0.93–1.67 s–1, respectively), run length was not corrected for photobleaching. The mean velocity was calculated by fitting the Gaussian distribution with errors representing s.e.m.
Measurements of unbinding force and stall force
For unbinding force measurement, one-headed kinesin heterodimers were prepared as described elsewhere (Hancock and Howard, 1998). For the measurement of stall force, conventional two-headed kinesin was prepared from pig brains. Both unbinding force and stall force measurements were performed according to procedures previously described (Uchimura et al, 2006) with the following two changes being made: (1) the MTs were fixed to the glass slide using an anti-tubulin antibody, and (2) the ionic strength of the solution was increased (x2 MA buffer). This was done to standardize the experimental conditions with the ones used in the single-molecule motility assay.
Steady-state MT-activated ATPase
The steady-state MT-activated ATPase of HK560 was measured using a modified Malachite Green method (Kodama et al, 1986). Reactions were performed in x2 MA buffer containing 0.1 μM HK560, varying concentrations of yeast MTs (0–20 μM), 10 μM taxol, and 2 mM ATP at 25 °C.
Pre-steady state MT-activated ADP release
The kinetics of MT-activated ADP release was measured using the fluorescent ADP analog, mant-ADP (N-methylanthranoyl ADP). Before measurement, HK560 kinesin was incubated with mant-ATP (M-12417, Invitrogen) at a ratio of 1:2 for 60 min on ice to allow the exchange of ADP at the active site with mant-ADP. Pre-steady state mant-ADP release was measured in an SX-20 Spectrometer (Applied Photophysics, Surrey, UK), in which equal volumes (60 μl each) of 2 μM kinesin-mant-ADP are mixed with 0–20 μM MTs and 2 mM ATP (both in x2 MA buffer) at 25°C. An excess amount of ATP is included in the reaction mixture to block rebinding of the mant-ADP to kinesin. Fluorescence was excited at 360 nm and detected after passing through a 395 LP filter. For the data measured using 0.5–10 μM of wild-type MTs, and that with 2–10 μM of α-E415A MTs, each time course of fluorescence intensity was fitted by double-exponential curves using the Pro-Data Viewer software, with the exponent for the fast phase giving the dissociation rate, kobs. For the data measured with 0.25 μM of wild-type MTs and 0.25–1 μM of α-E415A MTs, the time course was fitted by a single exponential. The rates thus calculated showed a hyperbolic dependence on the MT concentration and were fitted according to the following equation: kobs=kmax [MT]/(K0.5MT+[MT]).
Supplementary Material
Acknowledgments
We thank Takashi Shimizu (AIST) and Ken Sekimoto (Paris 7) for their invaluable discussions. We also thank Shinsaku Maruta and Masafumi D Yamada (Soka University); Kazuo Sutoh and Takahide Kon (University of Tokyo); and Shuichi Yoshida (TEGA Science) for their kind permission for the use of SX-20 spectrometers; and Yoko Y Toyoshima and Masaki Edamatsu (University of Tokyo) for their generous gift of a kinesin construct. We further thank Research Resources Center and Rie Ayukawa (Brain Science Institute, RIKEN) for the technical support. This study was supported in part by a Grant-in-Aid for Young Scientists (Start-up, No.19870034; to SU) and the Grants-in-Aid for Specially Promoted Research, Scientific Research (A) and the ‘Academic Frontier' Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to SI). YO is a post-doctoral fellow of the Japan Society for the Promotion of Science.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bloom GS, Wagner MC, Pfister KK, Brady ST (1988) Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry 27: 3409–3416 [DOI] [PubMed] [Google Scholar]
- Carter NJ, Cross RA (2005) Mechanics of the kinesin step. Nature 435: 308–312 [DOI] [PubMed] [Google Scholar]
- Crevel IM, Lockhart A, Cross RA (1996) Weak and strong states of kinesin and ncd. J Mol Biol 257: 66–76 [DOI] [PubMed] [Google Scholar]
- Cross RA (2004) The kinetic mechanism of kinesin. Trends Biochem Sci 29: 301–309 [DOI] [PubMed] [Google Scholar]
- Gennerich A, Vale RD (2009) Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol 21: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML Jr, Bode CJ, Georg GI, Himes RH (2003) Understanding tubulin-Taxol interactions: mutations that impart Taxol binding to yeast tubulin. Proc Natl Acad Sci USA 100: 6394–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (1988) Kinesin ATPase: rate-limiting ADP release. Proc Natl Acad Sci USA 85: 6314–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD (1994) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA 91: 6865–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock WO, Howard J (1998) Processivity of the motor protein kinesin requires two heads. J Cell Biol 140: 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Sakurada K, Aoki T, Thomas DD, Yanagida T (1990) Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J Mol Biol 216: 49–68 [DOI] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA (2000) Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta 1496: 99–116 [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y (2008) Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev 88: 1089–1118 [DOI] [PubMed] [Google Scholar]
- Hirose K, Akimaru E, Akiba T, Endow SA, Amos LA (2006) Large conformational changes in a kinesin motor catalyzed by interaction with microtubules. Mol Cell 23: 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ, Vale RD (1989) Movement of microtubules by single kinesin molecules. Nature 342: 154–158 [DOI] [PubMed] [Google Scholar]
- Hua W, Young EC, Fleming ML, Gelles J (1997) Coupling of kinesin steps to ATP hydrolysis. Nature 388: 390–393 [DOI] [PubMed] [Google Scholar]
- Kawaguchi K, Ishiwata S (2001) Nucleotide-dependent single- to double-headed binding of kinesin. Science 291: 667–669 [DOI] [PubMed] [Google Scholar]
- Kawaguchi K, Uemura S, Ishiwata S (2003) Equilibrium and transition between single- and double-headed binding of kinesin as revealed by single-molecule mechanics. Biophys J 84(2 Pt 1): 1103–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, Pinon MC, Dear N, Valdar W, Brown SD, Davies KE, Rawlins JN, Cowan NJ, Nolan P, Chelly J, Flint J (2007) Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell 128: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Hirokawa N (2006) High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J 25: 4187–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N (2001) Switch-based mechanism of kinesin motors. Nature 411: 439–445 [DOI] [PubMed] [Google Scholar]
- Klumpp LM, Brendza KM, Rosenberg JM, Hoenger A, Gilbert SP (2003) Motor domain mutation traps kinesin as a microtubule rigor complex. Biochemistry 42: 2595–2606 [DOI] [PubMed] [Google Scholar]
- Kodama T, Fukui K, Kometani K (1986) The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J Biochem 99: 1465–1472 [DOI] [PubMed] [Google Scholar]
- Kull FJ, Endow SA (2002) Kinesin: switch I & II and the motor mechanism. J Cell Sci 115(Pt 1): 15–23 [DOI] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD (1996) Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380: 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SA, Gelfand VI (1986) Bovine brain kinesin is a microtubule-activated ATPase. Proc Natl Acad Sci USA 83: 8530–8534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SA, Vaisberg EA, Shanina NA, Magretova NN, Chernyak VY, Gelfand VI (1988) The quaternary structure of bovine brain kinesin. EMBO J 7: 353–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M, Seehaus T (1988) Comparative analysis of tubulin sequences. Comp Biochem Physiol B 90: 655–670 [DOI] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E (2001) Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol 313: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW (1995) Kinetic mechanism of kinesin motor domain. Biochemistry 34: 13233–13241 [DOI] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW (1997) Kinetic mechanism of a monomeric kinesin construct. J Biol Chem 272: 717–723 [DOI] [PubMed] [Google Scholar]
- Neumann E, Garcia-Saez I, DeBonis S, Wade RH, Kozielski F, Conway JF (2006) Human kinetochore-associated kinesin CENP-E visualized at 17 A resolution bound to microtubules. J Mol Biol 362: 203–211 [DOI] [PubMed] [Google Scholar]
- Nitta R, Okada Y, Hirokawa N (2008) Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat Struct Mol Biol 15: 1067–1075 [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N (2000) Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA 97: 640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Keays DA, Francis F, Saillour Y, Bahi N, Manouvrier S, Fallet-Bianco C, Pasquier L, Toutain A, Tuy FP, Bienvenu T, Joriot S, Odent S, Ville D, Desguerre I, Goldenberg A, Moutard ML, Fryns JP, van Esch H, Harvey RJ et al. (2007) Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A). Hum Mutat 28: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Schief WR, Howard J (2001) Conformational changes during kinesin motility. Curr Opin Cell Biol 13: 19–28 [DOI] [PubMed] [Google Scholar]
- Schnitzer MJ, Block SM (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature 388: 386–390 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM (2000) Microtubule motors in mitosis. Nature 407: 41–47 [DOI] [PubMed] [Google Scholar]
- Sindelar CV, Downing KH (2007) The beginning of kinesin's force-generating cycle visualized at 9-A resolution. J Cell Biol 177: 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G, Cochran JC, Muller J, Mandelkow E, Gilbert SP, Hoenger A (2004) Modulation of kinesin binding by the C-termini of tubulin. EMBO J 23: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang SR (1997) G protein mechanisms: insights from structural analysis. Annu Rev Biochem 66: 639–678 [DOI] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y, Itoh TJ, Hotani H (1998) Visualization of the GDP-dependent switching in the growth polarity of microtubules. J Mol Biol 280: 365–373 [DOI] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD (2000) Engineering the processive run length of the kinesin motor. J Cell Biol 151: 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura S, Oguchi Y, Katsuki M, Usui T, Osada H, Nikawa J, Ishiwata S, Muto E (2006) Identification of a strong binding site for kinesin on the microtubule using mutant analysis of tubulin. EMBO J 25: 5932–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Kawaguchi K, Yajima J, Edamatsu M, Toyoshima YY, Ishiwata S (2002) Kinesin-microtubule binding depends on both nucleotide state and loading direction. Proc Natl Acad Sci USA 99: 5977–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T (1996) Direct observation of single kinesin molecules moving along microtubules. Nature 380: 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD (1997) Microtubule interaction site of the kinesin motor. Cell 90: 207–216 [DOI] [PubMed] [Google Scholar]
- Wu X, Xiang X, Hammer JA III (2006) Motor proteins at the microtubule plus-end. Trends Cell Biol 16: 135–143 [DOI] [PubMed] [Google Scholar]
- Yun M, Zhang X, Park CG, Park HW, Endow SA (2001) A structural pathway for activation of the kinesin motor ATPase. EMBO J 20: 2611–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.