Abstract
We have developed an image-analysis and classification system for automatically scoring images from high-throughput protein crystallization trials. Image analysis for this system is performed by the Help Conquer Cancer (HCC) project on the World Community Grid. HCC calculates 12,375 distinct image features on microbatch-under-oil images from the Hauptman-Woodward Medical Research Institute’s High-Throughput Screening Laboratory. Using HCC-computed image features and a massive training set of 165,351 hand-scored images, we have trained multiple Random Forest classifiers that accurately recognize multiple crystallization outcomes, including crystals, clear drops, precipitate, and others. The system successfully recognizes 80% of crystal-bearing images, 89% of precipitate images, and 98% of clear drops.
Electronic supplementary material
The online version of this article (doi:10.1007/s10969-009-9076-9) contains supplementary material, which is available to authorized users.
Keywords: Image analysis, High-throughput protein crystallization
Introduction
Protein crystallization is a difficult step in the structural-crystallographic pipeline. Lacking specific theories that map a target protein’s physico-chemical properties to a successful crystallization cocktail, the structural genomics community uses high-throughput protein crystallization screens to test targets against hundreds or thousands of cocktails. The Hauptman-Woodward Medical Research Institute’s (HWI) High-Throughput Screening Laboratory uses the microbatch-under-oil technique to test 1,536 cocktails per protein on a single plate [9]. Robotic pipetting and imaging systems efficiently process dozens of protein samples (and thus tens of thousands of images) per day. The bottleneck in this process is in the scoring of each image—recognizing crystal growth or other outcomes in an image currently requires visual review by a human expert. To-date, HWI has generated over 100 million images, representing more than 15 million distinct protein/cocktail trials over 12,000 proteins.
We describe here a method developed for automatically scoring protein-crystallization-trial images against multiple crystallization outcomes. Accurate, automated scoring of protein crystallization trials improves the protein crystallization process in several ways. The technology immediately improves throughput in existing screens by removing or reducing the need for human review of images. Multi-outcome scoring (e.g., clear/crystal/precipitate) in particular can speed up crystal optimization by facilitating visualization of a target protein’s crystallization response in chemical space [10]. In the longer term, automated scoring will enable the assembly of millions of protein/cocktail/outcome data points into databases, where data mining tools will turn protein crystallization into a statistical science and lead to rational design of crystallization screens, and potentially result in uncovering principles of protein chemistry.
Crystallization image classification and automated scoring is a two-stage process. In the image analysis stage, the raw image data is first processed into a vector of numeric features. Conceptually, this stage converts thousands of low-information-density image pixels to fewer, high-density features in the vector. Next, during the classification stage, a classifier maps the feature vector to a class, or score. Before new images can be classified, the classifier must first be trained by applying a learning algorithm to a training set of processed and pre-scored images.
The choice of features computed at the image analysis stage sets an upper limit on the success of any classifier built upon it. Image-feature space, like the chemical space of crystallization cocktails, is infinite. Inspired by the incomplete-factorial design of protein crystallization screens, and using previous successes and failures of crystallization image analysis, we have developed a large set of image features: starting with a core set of image-processing algorithms, by varying the parameters of each algorithm factorially, we have created a set of 12,375 distinct features.
This feature-set evolved from our earlier image-analysis work [3, 4], which employed microcrystal-correlation filters, topological measures, the Radon transform, and other tools. Moving from crystal-detection to multi-outcome scoring, it was necessary to expand beyond crystal-specific features, towards analysis of texture. We chose to add grey-level co-occurrence matrices (GLCMs) [5], a set of general-purpose texture descriptors, to our feature set, following [16] and [13]. Alternative texture descriptors used in crystallization image analysis include BlobWorld features [11] and orientation histograms [8].
Crystals, precipitates, and other objects can appear in an image over a range of scales, and in any orientation. Much research in the field therefore uses image analysis methods with explicit multi-scale or rotation-invariance properties. Gabor wavelet decomposition is used by [11] and [8]. Wilson [17] uses Haar wavelets and Fourier analysis. Po and Laine [12] use Laplacian pyramidal decomposition. The BlobWorld features used by Pan et al. [11] incorporate automatic scale-selection. Our features are rotation-invariant, and are computed on multiple low-pass-filtered copies of the original image, though our methods do not formally constitute a hierarchical image decomposition.
Crystals, precipitates, and other reactions can also co-occur in the same image, and so some systems classify local regions or individual objects of an image rather than reasoning on the image globally. Both [18] and [6] use edge-detection to separate foreground objects; individual objects are then analyzed and classified. By contrast, [8] divide the image into overlapping square sub-regions, then analyze and classify each square; objects in the image (crystals, etc.) may span multiple squares. We follow the discrete-object approach in some steps of our analysis, and the sliding-window approach in others. In our work, however, the local analyses are aggregated into global feature vectors prior to classification.
The computational requirements for computing this feature-set for 100,000,000 images is intractable on most systems. However, since the analysis can inherently be done in parallel, we have made use of a unique computing resource. The World Community Grid (WCG) is a global, distributed-computing platform for solving large scientific computing problems with human impact (http://www.worldcommunitygrid.org). Its 492,624 members contribute the idle CPU time of 1,431,762 devices. WCG is currently performing at 360 TFLOPs, increasing by about 3 TFLOPs per week (Global WCG statistics as of December 18, 2009). Our Help Conquer Cancer project (HCC) was launched on the WCG in November 1, 2007, and Grid members contributed 41,887 CPU-years to HCC to date, an average of 54 years of computing per day. HCC has two goals: first, to survey a wide area of image-feature space and identify those features that best determine crystallization outcome, and second, to perform the necessary image analysis on HWI’s archive of 100,000,000 images.
We have developed three classifiers based on a massive set of images hand-scored by HWI crystallographers [14, 15], with feature vectors computed by the World Community Grid HCC project. Although many works in the literature use a hyperplane-based decision model (e.g., Linear Discriminant Analysis [13], Support Vector Machines [11], our classifiers use the Random Forest decision tree-ensemble method [2]. Alternative tree-based models include Alternating Decision Trees used by [8], and C5.0 with adaptive boosting used by [1].
Materials and methods
Image analysis
Raw image data was converted to a vector of 12,375 features by using a complex, multi-layered image-program running on the WCG. A set of 2,533 features derived from the primary 12,375, and computed post-Grid, augment the feature-set, creating a final set of 14,908 features.
The initial analysis step identifies the circular bottom of the well, approximately 320 pixels in diameter. All subsequent analysis takes place within the 200-pixel-diameter region of interest w at the centre of the well, so as to analyze the droplet interior and avoid the high-contrast droplet edge. The bulk of the analysis pipeline comprises basic image analysis and statistical tools: Gaussian blur (Gσ) with standard deviation σ, the Laplace filter (Δ), Sobel gradient-magnitude (S) and Sobel edge detection (edge), maximum pixel value (max), pixel sum (Σ), pixel mean (μ), and pixel variance (Var). Several groups of features are computed, as described next.
Basic statistics
The first six features computed are basic image statistics: well-centre coordinates x and y, mean μ(w), variance Var(w), mean squared Laplacian 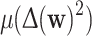 , and mean squared-Sobel μ(S(w)2).
, and mean squared-Sobel μ(S(w)2).
Energy
The next family of features measures the maximum intensity change in a 32-pixel neighbourhood of the image. Let vector 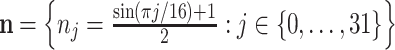 . Then a 32 × 32-element neighbourhood filter is the outer product N = n ⊗ n, and the energy feature of the image is
. Then a 32 × 32-element neighbourhood filter is the outer product N = n ⊗ n, and the energy feature of the image is 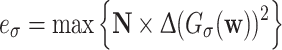 .
.
Euler numbers
These features measure the Euler numbers of the image. Given a binary image, the Euler number is equal to the number of objects in the image minus the number of holes in those objects. The raw image is transformed into binary images b
σ and b
σ,τ by two methods. First, by Sobel edge detection and morphological dilation: b
σ = dilate(edge(Gσ(w)), and second, by thresholding, perimeter-detection, and 3 × 3-pixel majority filtering: 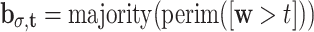 . The resulting features are E
σ = euler(b
σ), and E
σ,t = euler(b
σ,τ).
. The resulting features are E
σ = euler(b
σ), and E
σ,t = euler(b
σ,τ).
Radon-Laplacian features
These features are based on a straight-line-enhancing filter inspired by the Radon transform. Let R be the function mapping images to images, where each output pixel is the maximal sum of pixel values of any straight, 32-pixel line segment centered on the corresponding input pixel. Let 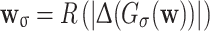 and w
σ,t = [w
σ>t].
and w
σ,t = [w
σ>t].
Two subgroups of Radon-Laplacian features are measured. The global features include global maximum r
σ = max(w
σ), hard-threshold pixel count 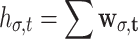 , and soft-threshold pixel sum
, and soft-threshold pixel sum 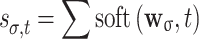 , where, for each pixel value x,
, where, for each pixel value x, 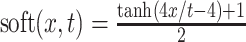 . The blob features are based on foreground objects (blobs) obtained from the binary image w
σ,t.
. The blob features are based on foreground objects (blobs) obtained from the binary image w
σ,t.
The first blob feature is the count c σ,t of all blobs in w σ,t. The remaining fourteen blob features u σ,t,j = μ(p j) are means (across all blobs in w σ,t) of geometric properties based on [18]. Per blob, fourteen such properties are measured: the blob area (p 1), the blob/convex-hull area ratio (p 2), the blob/bounding-box area ratio (p 3), the perimeter/area ratio (p 4), Wilson’s rectangularity, straightness, curvature, and distance-extrema range, and distance-extrema integral measures (p 5, p 6, p 7, p 8, p 9), the variance of corresponding blob pixels in Gσ(w) (p 10), the variance of corresponding blob pixels in w σ (p 11), the count of prominent straight lines in the blob (peaks in the Hough transform of w σ) (p 12), values of the first and second-highest peaks in the Hough transform (p 13, p 14), and the angle between first and second-most-prominent lines in the blob (p 15).
Five additional feature subgroups are computed post-Grid. Each summarizes the blob features across all parameter-pairs (σ,t) where w
σ,t contains one or more blobs. The blob means
v
1, …, v
18 measure the means of all u
σ,t,1, …, u
σ,t,15, h
σ,t, s
σ,t, and c
σ,t values. The blob maxs are vectors 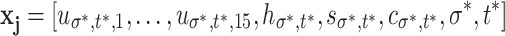 , where, per j,
, where, per j, 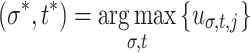 . The blob mins are similarly defined vectors y
j, but with
. The blob mins are similarly defined vectors y
j, but with 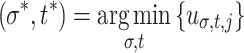 . The first blobless σ and first blobless t are scalar features measuring the lowest values of σ and t for which w
σ,t contains zero blobs.
. The first blobless σ and first blobless t are scalar features measuring the lowest values of σ and t for which w
σ,t contains zero blobs.
Radon-Sobel features
This feature group duplicates the previous set, but substituting the Sobel gradient-magnitude operator for the Laplace filter, i.e., using w σ = R(S(Gσ(w))).
Sobel-edge features
These features are similar to the pseudo-Radon features, with the following differences: they use w
σ = S(Gσ(w)), binary image edge(Gσ(w)) in lieu of w
σ,t, without a threshold parameter t, with blob maxs and mins as vectors of the form 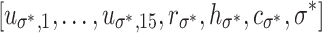 , and without a soft-threshold-sum feature.
, and without a soft-threshold-sum feature.
Microcrystal features
These features are based on the ten microcrystal exemplars of [3]. Let 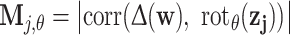 be the product of correlation-filtering some rotation of the exemplar image z
j against the Laplacian of the well image, and let
be the product of correlation-filtering some rotation of the exemplar image z
j against the Laplacian of the well image, and let 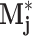 be the pixel-wise maximum of all Mj,θ. Then the peaks of
be the pixel-wise maximum of all Mj,θ. Then the peaks of 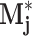 are the points in w that most resemble some rotation of the jth exemplar. Three feature subgroups are calculated. The maximum correlation features
are the points in w that most resemble some rotation of the jth exemplar. Three feature subgroups are calculated. The maximum correlation features 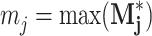 measure the global maxima. The relative peaks features
measure the global maxima. The relative peaks features 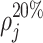 ,
, 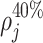 ,
, 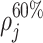 , and
, and 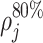 count the number of local maxima in
count the number of local maxima in 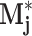 exceeding 20, 40%, etc., of the value of m
j. The absolute peaks features a
j,l (computed post-Grid) approximate the number of local maxima in
exceeding 20, 40%, etc., of the value of m
j. The absolute peaks features a
j,l (computed post-Grid) approximate the number of local maxima in 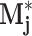 exceeding 26 distinct thresholds l.
exceeding 26 distinct thresholds l.
GLCM features
These features measure extremes of texture in local regions of the image. The GLCM of an image with q grey values, given some sampling vector δ = (δx, δy) of length d, is the symmetric q × q matrix whose elements γ jk indicating the count of pixel pairs separated spatially by δ with grey-values j and k. Haralick et al. define a set of 14 functions on the GLCM that measure textural properties of the image. Our analysis employs the first 13: Angular Second Moment (f 1), Contrast (f 2), Correlation (f 3), Variance (f 4), Inverse Difference Moment (f 5), Sum Average (f 6), Sum Variance (f 7), Sum Entropy (f 8), Entropy (f 9), Difference Variance (f 10), Difference Entropy (f 11), and two Information Measures of Correlation (f 12, f 13). The last Haralick function (Maximal Correlation Coefficient) was discarded due to its high computational cost.
We compute GLCMs and evaluate their functions on every 32-pixel-diameter circular neighbourhood within w, for sampling distances d ∈ {1, …, 25}, and at three grayscale quantization levels q ∈ {16, 32, 64}. For fixed (d, q) and fixed neighbourhood, the range and mean of each f
j are measured across all 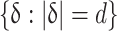 . Feature values are computed by repeating the measurement for all valid neighbourhoods, and recording the maximum neighbourhood mean
. Feature values are computed by repeating the measurement for all valid neighbourhoods, and recording the maximum neighbourhood mean 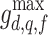 , minimum neighbourhood mean
, minimum neighbourhood mean 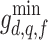 , and maximum neighbourhood range
, and maximum neighbourhood range 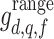 .
.
Truth data
Truth data was obtained from two massive image-scoring studies performed at HWI: one set of 147,456 images, representing 96 proteins × 1,536 cocktails [14], and one set of 17,895 images specifically containing crystals [15]. A randomly selected 90% of images from these data sets were used to evaluate features and train the classifiers. The remaining 10% were withheld as a validation set.
The raw scoring of each image in the Snell truth data indicates the presence or absence of 6 conditions in the crystallization trial: phase separation, precipitate, skin effect, crystal, junk, and unsure. The clear drop condition denotes the absence of the other six. In combination, these conditions create 64 distinct outcomes. To simplify the classification task, we define three alternative truth schemes: a clear/precipitate-only/other scheme, a clear/has-crystal/other scheme, and a 10-way scheme (clear/precipitate/crystal/phase separation/skin/junk/precipitate & crystal/precipitate & skin/phase & crystal/phase & precipitate), and have trained a separate classifier for each scheme.
Conflicting scores from multiple experts from the 96-protein-study [14] were handled by translating each raw score to each truth scheme, and then eliminating images without perfect score agreement.
Random forests
The random forest (RF) classification model uses bootstrap-aggregating (bagging) and feature subsampling to generate unweighted ensembles of decision trees [2]. The RF model was chosen for its suitability to our task. They generate accurate models using an algorithm naturally resistant to over-fitting. As a by-product of the training algorithm, RFs generate feature-importance measures from out-of-bag training examples, useful for feature selection. RFs naturally handle multiple outcomes, and thus do not limit us to binary decisions, e.g., crystal/no crystal. RFs may also be trained in parallel, or in multiple batches: two independently trained RFs can be combined taking the union of their trees and computing a weighted average of their feature-importance statistics. Finally, earlier, unpublished work suggested that naïve Bayes and other ensembles of univariate models could not sufficiently distinguish image classes; RFs, by contrast, work naturally with multiple, arbitrarily distributed, (non-linearly) correlated features.
For this study, we used the randomForest package [7], version 4.5-28, for the R programming environment, version 2.8.1, 64-bit, running on an IBM HS21 Linux cluster with CentOS 2.6.18-5.
The 10-way classifier
The 10-way classifier was generated in two phases: feature reduction, and classifier training. In feature reduction phase, nine independent iterations of RF were applied. Each iteration trained a random forest of 500 trees on an independently sampled, random subset of images from the training data. Feature “importance” (mean net accuracy increase) measures were recorded for each iteration, and then aggregated using the randomForest package’s combine function. The maximum observed standard deviation in any feature across the nine iterations was 0.08%. From the aggregated results, the 10% most-important (1,492 of 14,908) features were identified (see Fig. 1).
Fig. 1.
Importance measures of the 14,908 features measured during the feature-selection phase of the 10-way classifier training. The 10% highest-scoring features were used to train the three classifiers in this study
In the final training phase, five independent iterations of RF were applied. Each iteration trained a random forest of 1,000 trees on an independently sampled, random subset of images from the training data (see Table 1). The feature-set was restricted to the top-10% subset identified in the first phase. The 1,000-tree-forests from each iteration were combined to create the final 5,000-tree, 10-way RF classifier. This classifier was then used to classify 8,528 images from the validation set, using majority-rules voting.
Table 1.
Number and distribution of image classes in training and validation phases for the 10-way classifier
| Class | In training set | Samples per iteration (feature reduction) | Samples per iteration (final training) | In validation set |
|---|---|---|---|---|
| Clear | 26,160 | 5,000 | 5,000 | 2,887 |
| Precip | 25,951 | 5,000 | 5,000 | 2,897 |
| Crystal | 6,415 | 5,000 | 5,000 | 748 |
| Phase | 5,332 | 5,000 | 5,000 | 564 |
| Precip & crystal | 5,843 | 5,000 | 5,000 | 619 |
| Precip & skin | 3,748 | 3,748 | 3,748 | 466 |
| Phase & crystal | 1,790 | 1,790 | 1,790 | 201 |
| Phase & precip | 384 | 384 | 384 | 45 |
| Skin | 613 | 613 | 613 | 62 |
| Junk | 430 | 430 | 430 | 39 |
The 3-way classifiers
The clear/has-crystal/other classifier-generating process re-used the feature-importance data from the 10-way classifier. The RF was generated in one training phase, using four independent iterations of RF. Each iteration trained a random forest of 1,000 trees on an independently sampled, random subset of images from the training data (see Table 2). The feature-set was restricted to the top-10% subset identified in the first phase of the 10-way classifier (n = 1,492). The 1,000-tree-forests from each iteration were combined to create the final 4,000-tree, clear/has-crystal/other RF classifier. This classifier was then used to classify images from the validation set, using majority-rules voting.
Table 2.
Number and distribution of image classes in training and validation phases for the clear/crystal/other classifier
| Class | In training set | Samples per iteration (final training) | In validation set |
|---|---|---|---|
| Clear | 26,160 | 10,000 | 2,887 |
| Has-crystal | 16,763 | 10,000 | 1,879 |
| Other | 81,893 | 10,000 | 9,064 |
The clear/precipitate-only/other classifier was generated by the same process, again re-using the feature-importance data from the 10-way classifier. Training and validation data is summarized in Table 3.
Table 3.
Number and distribution of image classes in training and validation phases for the clear/precipitate-only/other classifier
| Class | In training set | Samples per iteration (final training) | In validation set |
|---|---|---|---|
| Clear | 26,160 | 10,000 | 2,887 |
| Precip-only | 25,951 | 10,000 | 2,897 |
| Other | 34,882 | 10,000 | 3,872 |
Results
Importance measures for the 14,908 image features, calculated during the feature-reduction phase of the 10-way classifier training, are plotted in Fig. 1.
Truth values from the 8,528 images from the 10-way classifier’s validation set were compared against the classifier’s predictions. The resulting confusion matrix is presented in Table 4. An alternative representation is shown in Fig. 2. The terms precision and recall are used in the matrix to measure the accuracy of the classifier on each outcome. For a given outcome X, recall, or true-positive rate, is the fraction of true X images correctly classified as X. Precision is the fraction of images classified as X that are correct. Randomly selected crystal images misclassified as clear and phase are shown in Supplementary Figures S1 and S2, respectively.
Table 4.
Confusion matrix for the 10-way classifier, representing 8,528 classified images from the validation set
| Truth | Machine classification | Total | Recall | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clear | Precip | Crystal | Phase | Precip & crystal | Precip & skin | Phase & crystal | Phase & precip | Skin | Junk | |||
| Clear | 2,746 | 3 | 60 | 53 | 0 | 0 | 0 | 0 | 1 | 24 | 2,887 | 0.951 |
| Precip | 29 | 2,213 | 45 | 77 | 343 | 185 | 1 | 0 | 2 | 2 | 2,897 | 0.764 |
| Crystal | 66 | 9 | 531 | 22 | 96 | 6 | 11 | 0 | 5 | 2 | 748 | 0.710 |
| Phase | 48 | 5 | 21 | 469 | 8 | 8 | 3 | 0 | 0 | 2 | 564 | 0.832 |
| Precip & crystal | 0 | 55 | 84 | 19 | 425 | 33 | 3 | 0 | 0 | 0 | 619 | 0.687 |
| Precip & skin | 1 | 46 | 13 | 6 | 16 | 378 | 0 | 0 | 6 | 0 | 466 | 0.811 |
| Phase & crystal | 3 | 4 | 67 | 48 | 33 | 2 | 42 | 0 | 1 | 1 | 201 | 0.209 |
| Phase & precip | 0 | 13 | 3 | 3 | 20 | 6 | 0 | 0 | 0 | 0 | 45 | 0.000 |
| Skin | 16 | 0 | 4 | 8 | 0 | 0 | 0 | 0 | 32 | 2 | 62 | 0.516 |
| Junk | 9 | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 25 | 39 | 0.641 |
| Total | 2,918 | 2,348 | 831 | 706 | 941 | 619 | 60 | 0 | 47 | 58 | 8,528 | |
| Precision | 0.941 | 0.943 | 0.639 | 0.664 | 0.452 | 0.611 | 0.700 | 0.681 | 0.431 | |||
The element in row i, column j counts the number of images truly belonging to class i, but (mis)classified as j. Diagonal elements count correctly classified images
Fig. 2.
Precision/recall plot of the 10-way classifier. Viewed as a row of vertical bar charts, each chart shows the relative distribution of true classes for a given RF-assigned label. Black bars (by width) show the proportions of false-positives. Viewed as a column of horizontal bar charts, each chart shows the relative distribution of RF-assigned labels for a given true class. Black bars (by height) show the proportions of false-negatives. From either perspective, the red bar in each chart shows the proportion of correct classifications, i.e., precision (width) or recall (height)
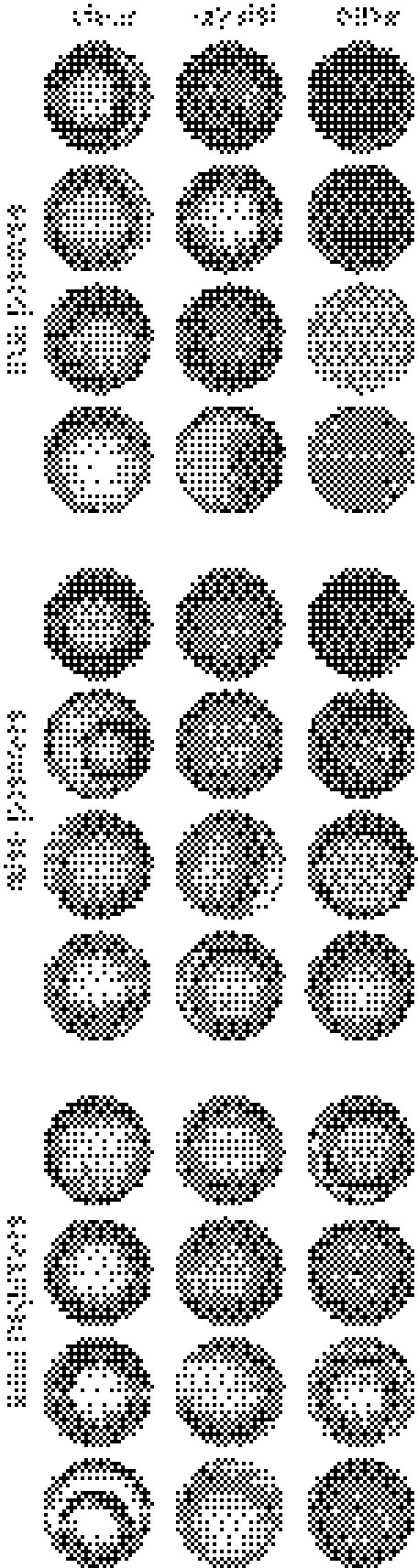
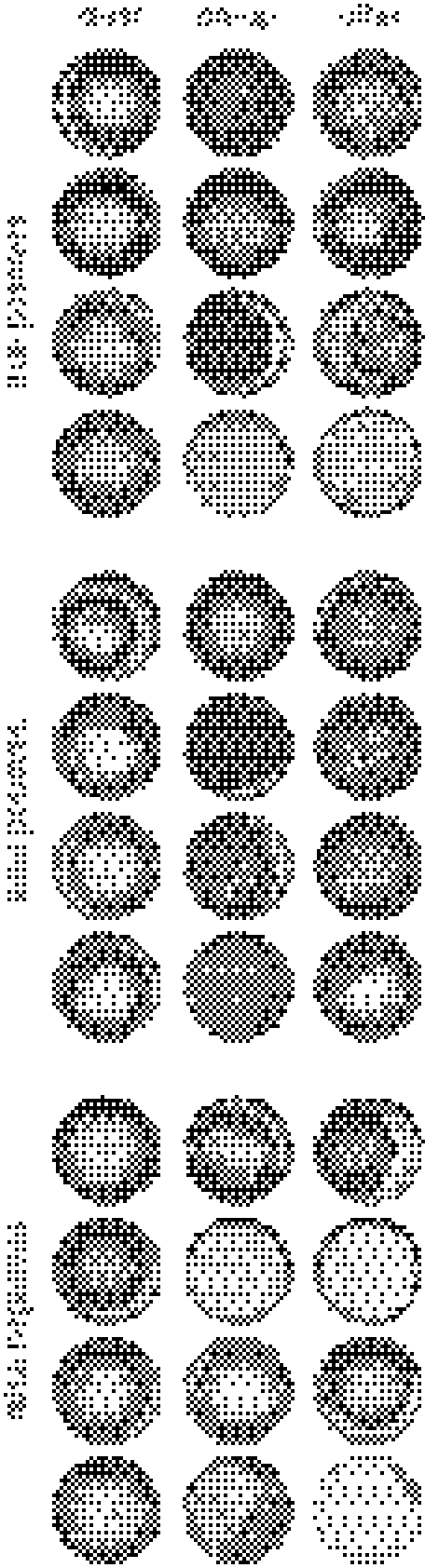
Truth values from the 13,830 images from the clear/has-crystal/other classifier’s validation set were compared against the classifier’s predictions, as were the 9,656 images from the clear/precipitate-only/other validation set. The resulting confusion matrices are presented in Tables 5 and 6, and as precision/recall plots in Figs. 3 and 4. Randomly selected true-positives, false-positives, and false-negatives for each category are presented in Figs. 5 and 6.
Table 5.
Confusion matrix for the clear/crystal/other classifier, representing 13,830 classified images from the validation set
| Truth | Machine classification | Total | Recall | ||
|---|---|---|---|---|---|
| Clear | Has crystal | Other | |||
| Clear | 2,841 | 20 | 26 | 2,887 | 0.984 |
| Has crystal | 99 | 1,507 | 273 | 1,879 | 0.802 |
| Other | 327 | 1,132 | 7,605 | 9,064 | 0.839 |
| Total | 3,267 | 2,659 | 7,904 | 13,830 | |
| Precision | 0.870 | 0.567 | 0.962 | ||
Table 6.
Confusion matrix for the clear/precipitate/other classifier, representing 9,656 classified images from the validation set
| Truth | Machine classification | Total | Recall | ||
|---|---|---|---|---|---|
| Clear | Precip only | Other | |||
| Clear | 2,825 | 7 | 55 | 2,887 | 0.979 |
| Precip only | 22 | 2,571 | 304 | 2,897 | 0.887 |
| Other | 290 | 455 | 3,127 | 3,872 | 0.808 |
| Total | 3,137 | 3,033 | 3,486 | 9,656 | |
| Precision | 0.901 | 0.848 | 0.897 | ||
Fig. 3.
Precision/recall plot of the clear/crystal/other classifier
Fig. 4.
Precision/recall plot of the clear/precipitate/other classifier
Fig. 5.
Randomly selected true-positive, false-positive, and false-negative images from the clear/has-crystal/other classifier’s validation set
Fig. 6.
Randomly selected true-positive, false-positive, and false-negative images from the clear/precipitate-only/other classifier’s validation set. Note that the other category includes precipitates combined with other outcomes (e.g., precip & crystal)
Discussion
Studying the confusion tables of each of the classifiers reveals several trends. Overall, clear drops and precipitates are easily recognized by the classifiers: 98% of all clear drop images are correctly recognized in the simpler classification tasks; this is reduced to 95% with the 10-way classifier. 89% of all precipitate-only images are correctly recognized in the simpler classification task, and this result is also reduced in the 10-way classifier, mainly due to competition with precip & crystal, and precip & skin categories.
Overall, crystals are fairly well detected. 80% of crystals are detected in the simpler classification task. The 10-way classifier has more precise categories, and some accuracy is lost choosing between crystal, precip & crystal, and phase & crystal categories.
The precip & crystal category seems especially attractive to the 10-way classifier, resulting in many misclassifications of precip, phase & precip, and phase & crystal images. Conversely, the phase & precip category was ignored entirely by the classifier: none of the 45 true phase & precip images in the validation set were correctly classified; instead, they were misclassified as mostly precip or precip & crystal. This difficulty is likely caused by two factors. First, the phase & precip category is the rarest category in both the training and validation sets. Second, phase separation seems to introduce a very weak signal in the feature data, whereas precipitate’s signal is very strong. The phase-only outcome is both well-represented in the data, and well-recognized by the 10-way classifier: 83% recall and 66% precision.
The most important outcome to crystallographers is accurately detecting all existing crystals, and improvements to the classifier, the image features, and feature selection must focus here. The 20% false-negative rate for crystals in the clear/has-crystal/other classifier can be dissected somewhat by examining the crystal rows and non-crystal columns of the 10-way classifier’s confusion matrix (Table 4). True crystal images are assigned to non-crystal categories by the classifier at rates of 9% for clear, 3% for phase, and 3% elsewhere. Similarly, true precip & crystal images are assigned to non-crystal categories at rates of 9% for precip, 5% for precip & skin, and 3% for phase. The smaller phase & crystal category is misclassified as phase 24% of the time. The crystal false negatives assigned to clear may be the result of crystals located near the well edge being excluded from the region of interest, or crystals being mistaken for points of contact between the droplet and the plastic well bottom. The majority of crystal false negatives assigned to phase seem to be needle crystals. A deeper look is required at the image features that can better separate the clear, phase, crystal, and phase & crystal categories.
A final note about bias: due to the inclusion of [15] data, both the training and validation sets are enriched for crystal outcomes (11% crystals versus an estimated 0.4% real-world rate). Crystals represent a rare but most important outcome. The additional crystal training data was required in order to sufficiently train the model, but the outcome is a model that will over-report crystals in real-world use, resulting in a decreased precision score, but unchanged recall.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1 Randomly selected crystal images misclassified as clear from the 10-way classifier’s validation set (EPS 3290 kb)
Supplementary Fig. S2 Randomly selected crystal images misclassified as phase from the 10-way classifier’s validation set(EPS 3290 kb)
Acknowledgments
We wish to thank the many members of the World Community Grid, who donated their computing resources to the Help Conquer Cancer project. We also wish to thank IBM and the World Community Grid staff, including Bill Bovermann, Viktors Berstis, Jonathan D. Armstrong, Tedi Hahn, Kevin Reed, Keith J. Uplinger, and Nels Wadycki, for hosting and supporting HCC and Dr. Jerry Heyman and Miso Cilimdzic from IBM. This unique resource provided an essential computing resource for our work. This work would not be possible without data generated at Hauptman-Woodward Medical Research Institute and insight from many discussions during our collaboration with Drs. George T. DeTitta, Edward H. Snell, Joseph R. Luft and their team, Geoff Franks, Thomas Grant, Stacey M. Gulde, Mary Koszelak-Rosenblum, Angela M. Lauricella, Michael G. Malkowski, Raymond Nagel, Stephen A. Potter, Meriem I. Said, Jennifer L. Smith, Max Thayer, and Christina K. Veatch. We acknowledge help from the Jurisica Lab, namely Dr. Kevin Brown and Richard Lu, for helping with computing resources and environment for post-Grid data processing. The authors gratefully acknowledge funding by NIH U54 GM074899, the Canada Foundation for Innovation (Grants #12301 and #203383), the Canada Research Chair Program, the Natural Science & Engineering Research Council of Canada (Grant #104105), and IBM.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- HWI
Hauptman-woodward medical research institute
- WCG
World community grid
- HCC
Help conquer cancer
- GLCM
Grey-level co-occurrence matrix
- RF
Random forests
References
- 1.Bern M, Goldberg D, Stevens RC, Kuhn P. Automatic classification of protein crystallization images using a curve-tracking algorithm. J Appl Cryst. 2004;37:279–287. doi: 10.1107/S0021889804001761. [DOI] [Google Scholar]
- 2.Breiman L. Random forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 3.Cumbaa CA, Lauricella A, Fehrman N, Veatch C, Collins R, Luft J, DeTitta G, Jurisica I. Automatic classification of sub-microlitre protein-crystallization trials in 1536-well plates. Acta Crystallogr D. 2002;59:1619–1627. doi: 10.1107/S0907444903015130. [DOI] [PubMed] [Google Scholar]
- 4.Cumbaa CA, Jurisica I. Automatic classification and pattern discovery in high-throughput protein crystallization trials. J Struct Funct Genomics. 2005;6:195–202. doi: 10.1007/s10969-005-5243-9. [DOI] [PubMed] [Google Scholar]
- 5.Haralick RM, Shanmugan K, Dinstein I. Textural Features for Image Classification. IEEE Trans Syst Man Cybern. 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 6.Kawabata K, Takahashi M, Saitoh K, Asama H, Mishima T, Sugahara M, Miyano M. Evaluation of crystalline objects in crystallizing protein droplets based on line-segment information in greyscale images. Acta Crystallogr D. 2006;62:239–245. doi: 10.1107/S0907444905041077. [DOI] [PubMed] [Google Scholar]
- 7.Liaw A, Wiener M. Classification and regression by random forest. R News. 2002;2:18–22. [Google Scholar]
- 8.Liu R, Freund Y, Spraggon G. Image-based crystal detection: a machine-learning approach. Acta Crystallogr D. 2008;64:1187–1195. doi: 10.1107/S090744490802982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/S1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 10.Nagel RM, Luft JR, Snell EH. AutoSherlock: a program for effective crystallization data analysis. J Appl Cryst. 2008;41:1173–1176. doi: 10.1107/S0021889808028938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan S, Shavit G, Penas-Centeno M, Xu DH, Shapiro L, Ladner R, Riskin E, Hol W, Meldrum D. Automated classification of protein crystallization images using support vector machines with scale-invariant texture and Gabor features. Acta Crystallogr D. 2006;62:271–279. doi: 10.1107/S0907444905041648. [DOI] [PubMed] [Google Scholar]
- 12.Po MJ, Laine AF. Leveraging genetic algorithm and neural network in automated protein crystal recognition. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:1926–1929. doi: 10.1109/IEMBS.2008.4649564. [DOI] [PubMed] [Google Scholar]
- 13.Saitoh K, Kawabata K, Asama H, Mishima T, Sugahara M, Miyano M. Evaluation of protein crystallization states based on texture information derived from greyscale images. Acta Crystallogr D. 2005;61:873–880. doi: 10.1107/S0907444905007948. [DOI] [PubMed] [Google Scholar]
- 14.Snell EH, Luft JR, Potter SA, Lauricella AM, Gulde SM, Malkowski MG, Koszelak-Rosenblum M, Said MI, Smith JL, Veatch CK, Collins RJ, Franks G, Thayer M, Cumbaa CA, Jurisica I, Detitta GT. Establishing a training set through the visual analysis of crystallization trials. Part I: approximately 150, 000 images. Acta Crystallogr D. 2008;64:1123–1130. doi: 10.1107/S0907444908028047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snell EH, Lauricella AM, Potter SA, Luft JR, Gulde SM, Collins RJ, Franks G, Malkowski MG, Cumbaa CA, Jurisica I, DeTitta GT. Establishing a training set through the visual analysis of crystallization trials. Part II: crystal examples. Acta Crystallogr D. 2008;64:1131–1137. doi: 10.1107/S0907444908028059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spraggon G, Lesley SA, Kreusch A, Priestle JP. Computational analysis of crystallization trials. Acta Crystallogr D. 2002;58:1915–1923. doi: 10.1107/S0907444902016840. [DOI] [PubMed] [Google Scholar]
- 17.Wilson J. Automated classification of crystallisation images. Euro Pharm Rev. 2007;3:61–66. [Google Scholar]
- 18.Wilson J. Towards the automated evaluation of crystallization trials. Acta Crystallogr D. 2002;58:1907–1914. doi: 10.1107/S0907444902016633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1 Randomly selected crystal images misclassified as clear from the 10-way classifier’s validation set (EPS 3290 kb)
Supplementary Fig. S2 Randomly selected crystal images misclassified as phase from the 10-way classifier’s validation set(EPS 3290 kb)