Abstract
Although some data link archaeal and eukaryotic translation, the overall mechanism of protein synthesis in archaea remains largely obscure. Both archaeal (aRF1) and eukaryotic (eRF1) single release factors recognize all three stop codons. The archaeal genus Methanosarcinaceae contains two aRF1 homologs, and also uses the UAG stop to encode the 22nd amino acid, pyrrolysine. Here we provide an analysis of the last stage of archaeal translation in pyrrolysine-utilizing species. We demonstrated that only one of two Methanosarcina barkeri aRF1 homologs possesses activity and recognizes all three stop codons. The second aRF1 homolog may have another unknown function. The mechanism of pyrrolysine incorporation in the Methanosarcinaceae is discussed.
Keywords: Translation termination, Archeon, Pyrrolysine-utilizing archea, aRF1, Polypeptide release factor
1. Introduction
At the final stage of protein biosynthesis the class-1 release factors (RF1s) recognize stop codons and induce hydrolysis of peptidyl-tRNA in the peptidyl transferase center of the ribosome (reviewed in [1–3]). In eukaryotes a single release factor (eRF1) recognizes all three stop codons, UAA, UAG and UGA. Bacteria have two release factors (RF1 and RF2) that recognize different stop-codon pairs, UAA/UAG and UAA/UGA, respectively. Archaeal class-1 release factors (aRF1s) exhibit a high degree of amino acid sequence similarity with eRF1s and are substantially different from bacterial RFs (Fig. 1). Like eRF1, aRF1 recognizes all three stop codons [4], demonstrating the functional resemblance of aRF1 and eRF1. Thus, archaeal translation termination has common features with eukaryotic termination and their mechanisms are expected to be similar. Most archaea contain only one gene encoding aRF1, but in two species of pyrrolysine (Pyl)-utilizing archaea, Methanosarcina barkeri and Methanosarcina acetivorans, two non-identical class-1 translation termination factors were found (aRF1-1 and aRF1-2), while the other Pyl-containing archaea, Methanosarcina mazei and Methanococcoides burtonii, have only one aRF1 [5].
Fig. 1.
Multiple sequence alignment of the N-terminal domains of release factors from different archaea, eukaryotes and bacteria.
An in-frame UAG codon has been identified in mtmB1 encoding MtmB1, a methylamine methyltransferase participating in methanogenesis from monomethylamine in M. barkeri (reviewed in [6]). The UAG is translated by the novel amino acid Pyl as revealed by the crystal structure of MtmB1 [7]. An in-frame UAG codon is also contained in mtbB1 and mttB1, the genes encoding the di- and tri-methylamine methyltransferases in Methanosarcina spp. (reviewed in [6]), as well as in a number of other open reading frames [8,9]. Examination of the presently sequenced genomes suggests that the existence of Pyl is limited to the Methanosarcinaceae (Methanosarcina spp. and M. burtonii) and to a few bacteria (Desulfitobacterium hafniense [10] and symbiotic δ-proteobacteria [9,11]).
Pyl is co-translationally inserted at UAG codons and as such constitutes the 22nd natural amino acid used in protein synthesis. Pyl has its own tRNAPyl whose CUA anticodon complements the UAG codon [10], and a special aminoacyl-tRNA synthetase (PylRS) that specifically acylates tRNAPyl to form Pyl-tRNAPyl [12,13]. While the molecular principles governing the synthesis of Pyl-tRNAPyl have been worked out in details, the mechanism underlying the recoding of the UGA codon as Pyl sense codon remains poorly understood. Pyl share with selenocysteine (Sec) the property of being inserted at in-frame stop codons (UGA for Sec). Selenocysteine incorporation into protein has been thoroughly examined. For Sec, a RNA stem loop (termed SECIS element) located in the mRNA signals the UGA codon to be recoded to the translation machinery. In addition, an essential tRNASec-specific elongation factor (SelB or EFSec) delivers the Sec-tRNASec at the suppression site, ensuring the successful translation of the in-frame UGA codon as selenocysteine [14–17].
While it was initially thought that Pyl insertion mechanism could be modeled on that of Sec, recent data suggest otherwise. A stem loop structure analogous to the SECIS element (and thus termed PYLIS element) was predicted downstream of the in-frame UAG codon in mtmB1 mRNAs [18,19]. However, in contrast to Sec, the presence of the RNA stem loop structure was not critical for the insertion of Pyl into proteins. The PYLIS structure moderately modulated MtmB1 expression in Methanosarcina and did not impact reporter protein expression in an Escherichia coli context [20,21]. Sequence comparison studies showed that PYLIS structure is not conserved in the mtbB1 and mttB1 mRNAs. Lastly, in vitro and in vivo experiments in E. coli demonstrated that elongation factor Tu is capable of interacting and delivering Pyl-tRNAPyl to the UGA suppression site, suggesting that no specialized elongation factor is required for Pyl insertion [22].
In order to gain further insight into UAG recoding mechanism, we have determined stop codon specificity of two M. barkeri aRF1 homologs in an in vitro reconstituted eukaryotic translation system. For these purposes we constructed chimeric proteins which contain the N-terminal domain of M. barkeri aRF1s (responsible for stop-codon decoding) and the MC domains of human eRF1 since the full-length aRF1s from this organism are unable to interact with eukaryotic ribosomes used in translation termination assay. We have shown that only one out of two forms of M. barkeri aRF1s is active in translation termination and recognizes all three stop codons. Taking into account that the frequency of UAG stop-codon usage is substantially decreased in Pyl-encoding genomes, our data suggest that the Methanosarcinaceae may be in a continuing process of the UAG codon reassignment from a stop signal to a sense codon specifying Pyl.
2. Materials and methods
2.1. M. barkeri and Methanococcus maripaludis aRF1 gene constructs
M. barkeri wild-type aRF1-1 and aRF1-2 genes were amplified by PCR using freshly prepared genomic DNA. PCR products were cloned in Topo TA (Invitrogen), sequenced and subcloned into pET15b expression plasmid between BamHI and PstI restriction sites. M. maripaludis aRF1 gene was cloned following the same procedure and subcloned in pET15b between NdeI and XhoI sites.
2.2. Chimeric archaeal/human release factor constructs
The aRF1 gene sequences encoding the N domains of M. barkeri aRF1-1 and aRF1-2 and M. maripaludis aRF1 were PCR-amplified using specific primers. The first primer contained an NdeI site and the second one carried a SalI site in putative boundary of the N and M domains of aRF1 (codons for amino acids 144 and 145, numbered according to human eRF1). The determination of the putative boundaries between the N and M domains of aRF1swas based on the crystal structure of human eRF1 [23] and a multiple alignment of protein sequences of archaeal and human release factors. The resulting PCR products were inserted into NdeI/SalI sites of the pERF4b-Sal plasmid. pERF4b-Sal plasmid with cloned eRF1 gene from Homo sapiens inserted into SalI restriction site of pET23b(+) vector (Novagen) was constructed previously [24]. Thus, three plasmids carrying chimeric genes encoding the archaeal N domain of aRF1s and MC domain of human eRF1 with 6His-tag on the C-terminus were obtained. Mutant forms carrying the N domain of M. barkeri aRF1-2 with amino acid substitutions corresponding to the amino acid sequence of M. barkeri aRF1-1 (K61N or D122V + I124K or D122T + Y123F + I124V; amino acid numbering according to human eRF1) were obtained by PCR mutagenesis as described [25].
2.3. Proteins and ribosomal subunits
The 40S and 60S ribosomal subunits, eIF2, eIF3, eIF4F, eEF1H and eEF2 were purified from rabbit reticulocyte lysate as described (see references in [26]). The eukaryotic translation factors eIF1, eIF1A, eIF4A, eIF4B, eIF5B, eIF5, eRF1, chimeric a/eRF1 were produced as recombinant proteins in E. coli strain BL21 with subsequent protein purification on Ni-NTA-agarose and ion-exchange chromatography (see references in [26]).
2.4. mRNA transcripts
mRNA was transcribed by T7 RNA polymerase on MVHL-stop plasmids, encoding T7 promoter, four CAA repeats, β-globin 5′-untranslated region (UTR), MVHL tetrapeptide followed by one of three stop codons (UAA, UAG or UGA) and 3′-UTR comprising the rest of the natural β-globin coding sequence. MVHL-UAA plasmid was described [26], and MVHL constructs containing UAG and UGA stop codons were obtained by PCR mutagenesis of MVHL-UAA plasmid. For run-off transcription all plasmids were linearized with XhoI.
2.5. Aminoacylation of tRNA
Total calf liver tRNA (Novagen) was purified by size-exclusion chromatography on a Superdex 75 HR column (GE Healthcare) to remove high and low molecular weight contaminants. was aminoacylated by methionine using E. coli methionyl-tRNA synthetase as described [26]. The aminoacylation reaction mixture contained 40 mM Tris–acetate, pH 7.5, 10 mM Mg(OAc)2, 4mM ATP, 0.2 mM L-methionine, 14 KBq/μl [35S]methionine (GE Healthcare), 0.25 mAU280nm/μl purified E. coli methionyl-tRNA synthetase, 0.8 u/μl RNAse inhibitor (RiboLock, Fermentas) and 6–10 μg/ml of purified total calf liver tRNA. Reaction was run for 25 min at 37 °C.
2.6. Pretermination complex assembly and purification
Pretermination complexes were assembled as described [26]. Briefly, 37 pmol of MVHL-stop mRNAs were incubated in buffer A (20 mM Tris–acetate, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2mM DTT) supplemented with 400 u RNAse inhibitor (RiboLock, Fermentas), 1 mM ATP, 0.25 mM spermidine, 0.2 mM GTP, 75 μg total tRNA (acylated with Val, His, Leu and [35S]Met), 75 pmol 40S and 60S purified ribosomal subunits, 125 pmol eIF2, eIF3, eIF4F, eIF4A, eIF4B, eIF1, eIF1A, eIF5, eIF5B each, 200 pmol eEF1H and 50 pmol eEF2 for 30 min and then centrifuged in a Beckman SW55 rotor for 95 min at 4 °C and 50 000 rpm in 10–30% linear sucrose density gradient prepared in buffer A with 5 mM MgCl2. Fractions corresponded to pretermination complexes according to optical density and the presence of [35S]Met were combined, diluted 3-fold with buffer A containing 1.25 mM MgCl2 (to a final concentration of 2.5 mM Mg2+) and used for peptide release assay.
2.7. Peptide release assay
Peptide release assay was run as described [26] with some modifications. Aliquots containing 0.1 pmol of pretermination complexes with an activity of about 10 000 cpm assembled in the presence of [35S]Met-tRNA were incubated at 37 °C for 15 min with different concentrations of release factors (0–120 pmol). Ribosomes and tRNA were pelleted with ice-cold 5% TCA supplemented with 0.75% casamino acids and centrifuged at 4 °C and 14 000×g. The amount of released [35S]Met-containing tetrapeptide, which indicated the efficiency of peptidyl-tRNA hydrolysis, was determined by scintillation counting of supernatants on an Intertechnique SL-30 liquid scintillation spectrometer.
2.8. Sequence analysis
Nucleotide sequences of aRF1 genes were downloaded from the GenBank database (NCBI, NIH). Phylogenetic analysis was done by MEGA package version 4.0 [27]. Phylogenetic tree of aRF1s was constructed by neighbor-joining method using codon-based evolutionary divergence between sequences using HYK85 as implemented in MrBayes [28]. To compare gene pairs and to calculate synonymous and non-synonymous substitutions MUSCLE alignment with default parameters was used [29].
3. Results
3.1. Peptide release activity of the chimeric M. barkeri and M. maripaludis a/eRF1 proteins and the full-length Methanococcus jannaschii aRF1
The activity and codon specificity of the two M. barkeri release factors homologs, aRF1-1 and aRF1-2 were measured in vitro using a fully reconstituted eukaryotic translation system, aminoacylated tRNA and model mRNAs encoding for the MVHL peptide [26]. The coding region of the model mRNAs was terminated by one of the three stop codons (UAA, UGA and UAG). The activity of each release factor was directly measured by the amount of free 35S-labeled MVHL generated [26]. As controls M. maripaludis and M. jannaschii aRF1s of non-Pyl-utilizing archaea were used.
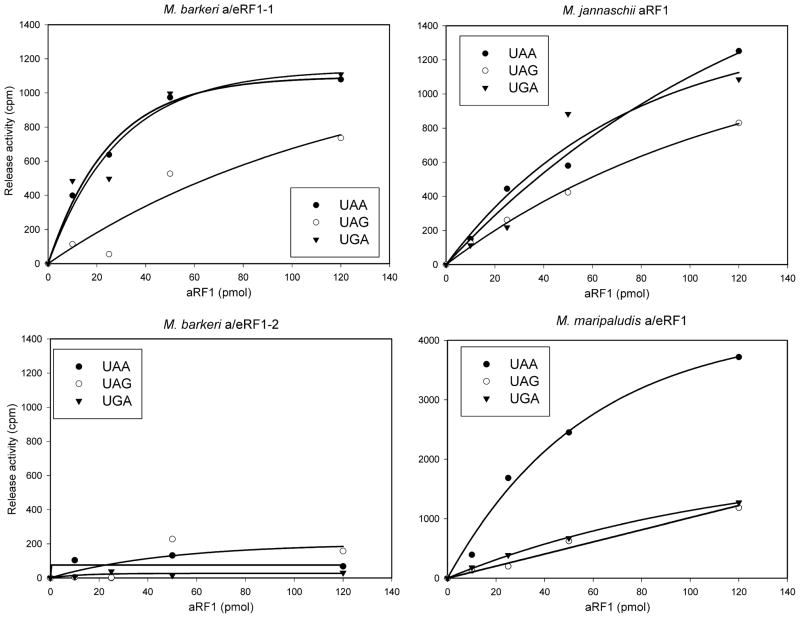
The M. barkeri and M. maripaludis aRF1s turned out to be inactive with any stop codon in the test system probably due to their inability to interact with eukaryotic ribosomal complexes (data not shown). However, the M. jannaschii aRF1 was able to induce peptide release in the presence of all three stop codons (Fig. 2).
Fig. 2.
Release activity of aRF1s from Pyl-utilizing and non-Pyl-utilizing archaea in a reconstituted eukaryotic translation system. Filled circles – activity in the presence of UAA stop codon; empty circles – activity in the presence of UAG stop codon; filled triangles – activity in the presence of UGA stop codon.
In eukaryotes, the N-terminal domain of eRF1 is implicated in the decoding of stop codons as shown by genetic [30] and biochemical [31–33] data. To overcome the lack of interaction of the M. barkeri and M. maripaludis aRF1s with heterologous rabbit ribosomes we constructed chimeric proteins (a/eRF1s) which consisted of the N-terminal domains of aRF1s and the C-terminal part (M and C domains) of human eRF1. These chimeras were designed so that they should have stop codon specificity of the aRF1s and stop-codon- independent ribosome binding properties of human eRF1. This approach has been already successfully used to determine the decoding properties of eRF1s from different species of ciliates [24]. Using this method, we show that only one of the two forms of M. barkeri a/eRF1s, a/eRF1-1 induces peptide release in the presence of all three stop codons, while a/eRF1-2 is inactive with any of the three stop codons (Fig. 2).
We noticed that M. barkeri a/eRF1-1 displays reduced peptide release efficiency when UAG is used as stop codon compared to UAA or UGA. This stop codon preference pattern was also observed for the M. jannaschii aRF1 control, since with this release factor too, UAG stop codon triggered a weaker peptide release activity. M. maripaludis a/eRF1 displayed a different pattern of stop codon preference since the release factor was significantly more active with UAA stop codons than with UGA or UAG. Interestingly, M. maripaludis a/eRF1 had a much stronger release activity than its M. barkeri a/eRF1-1 and M. jannaschii aRF1 counterparts when UAA was used as stop codon.
Alignment of aRF1 sequences (Fig. 1) revealed amino acid substitutions in positions 61 (NIKS motif) and 122–124 (near YxCxxF motif) of non-active M. barkeri aRF1-2 (numeration of amino acids according to human eRF1). These positions have been shown to be essential for eRF1 stop-codon recognition [31,34]. To clarify the influence of these amino acids on decoding activity, three mutants of M. barkeri a/eRF1-2 were constructed with the following amino acid substitutions in the N domain: K61N, D122V + I124K or D122T + Y123F + I124V corresponding to a/eRF1-1 amino acid sequence. However, like their wild-type a/eRF1-2 counterpart, these a/eRF1-2 mutants do not recognize stop codons (data not shown).
3.2. Phylogenetic analysis of the release factors from the Methanosarcinaceae family
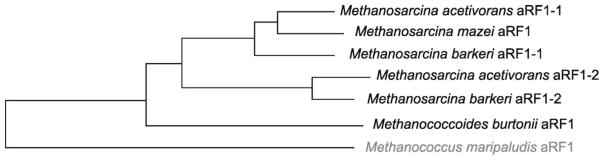
Comparison of coding sequences of aRF1 genes from the Methanosarcinaceae family revealed two observations: (i) higher pair-wise similarity between aRF1-1s of M. barkeri and M. acetivorans and, as well, between aRF1-2s of M. barkeri and M. acetivorans than between aRF1-1 and aRF1-2 within same species and (ii) M. mazei aRF1 belongs to the same phylogenetic clade as M. barkeri and M. acetivorans aRF1-1s (Fig. 3). These results confirm the species phylogenetic tree constructed by Zhang et al. [5]. It also suggests that the duplication of aRF1 gene in genus Methanosarcina occurred before the divergence of M. acetivorans, M. barkeri and M. mazei species. According to this evolutionary scenario M. acetivorans and M. barkeri retained their aRF1-2 while M. mazei genome lost its copy.
Fig. 3.
Phylogenetic tree of aRF1 genes from Methanosarcinaceae, Pyl-utilizing archaeal family, and M. maripaludis aRF1, a non-Pyl-utilizing organism.
Quantitative comparison of gene copies was provided by calculating the ratio of synonymous and non-synonymous substitutions in gene pairs, where M. mazei aRF1 was used as a reference sequence (Table 1). Notably, the number of non-synonymous substitutions in the aRF1-2 forms is higher than in aRF1-1 but this gene still appears to be under negative selection with a relative synonymous/non-synonymous ratio (dn/ds) < 1. Both the M. barkeri and M. acetivorans copies of aRF1-2 genes lack nonsense or frameshift mutations which indicate that the corresponding gene products retained or gained some physiological role.
Table 1.
The number of amino acid substitutions in M. acetivorans and M. barkeri aRF1s compared to M. mazei aRF1.
| Syn/non-syn | |
|---|---|
| M. acetivorans aRF1-1 | 136/27 |
| M. barkeri aRF1-1 | 168/51 |
| M. acetivorans aRF1-2 | 161/222 |
| M. barkeri aRF1-2 | 159/221 |
The first and the second numbers in each cell are the quantity of synonymous and non-synonymous amino acid substitutions, respectively.
3.3. Methanosarcinales genome analysis
It has been shown that UAG is a rare codon in Methanosarcinaceae genomes (less than 5% of all three stop codons compared to UAA and UGA, approximately 45% each) [5]. Using the two directional best BLAST hit approach [35] we have performed a detailed analysis of the stop codon usage in orthologous genes of the order Methanosarcinales. We detected orthologs in four genomes of the Pyl-utilizing archaea in the Methanosarcinaceae family and in one outgroup genome of a non-Pyl-utilizing Methanosaetaceae (Methanosaeta thermophila). While UAG was found in 6–9 orthologous genes in the Methanosarcinaceae, the same codon was found in 160 orthologous genes in the outgroup organism M. thermophila (Table 2). Thus, comparison of stop codon usage in orthologs clearly demonstrates avoidance of UAG stop codon in Pyl-utilizing archaea. It is reasonable to speculate that the majority of UAG codons were changed to UAA stop codons. Indeed the UAG to UAA conversion requires only one substitution and is evolutionary more favorable than the UAG to UGA conversion which requires two nucleotide substitutions. This notion is supported by the fact that first, UAA is used as stop codon in 401–505 orthologs in the Methanosarcinaceae while it is present in only 107 orthologs in the non-Pyl using M. thermophila genome, and second, that the number of orthologs using UGA remains comparable in the Methanosarcinaceae and in M. thermophila (324/431 vs 571, Table 2). Notably, without ortholog analysis we cannot observe UAG stop-codon avoidance. On the whole genome level, taking into consideration poorly conserved and ‘‘newest” (i.e. duplicated, horizontally transferred) genes, UAG is occurred substantially more often, for example, among 3370 genes of M. mazei UAG is used only 125 times.
Table 2.
Stop codon usage for orthologous genes in the Methanosarcinaceae family in comparison with analogous data for Methanosaeta thermophila/order Methanosarcinales.
| UAA | UAG | UGA | |
|---|---|---|---|
| Methanosarcinaceae | |||
| M. acetivorans | 416 | 7 | 415 |
| M. mazei | 445 | 8 | 385 |
| M. barkeri | 505 | 9 | 324 |
| M. burtonii | 401 | 6 | 431 |
| Methanosaetaceae | |||
| M. thermophila | 107 | 160 | 571 |
4. Discussion
4.1. Archaeal release factors are omnipotent
Our data show that archaeal release factors, whether in their native form (M. jannaschii) or as archaeal/eukaryotic chimeras (M. maripaludis and M. barkeri) stimulate hydrolysis of peptidyl-tRNA in a reconstituted eukaryotic translation system and respond to all three stop codons. These data imply that mechanisms of translation termination in eukaryotes and archaea are similar. The comparison of aRF1-1 from M. jannaschii and M. barkeri suggests that termination mechanism is likely to be similar in Methanosarcinaceae and in other non-Pyl-containing archaea.
The role of the second release factor encoded in the M. barkeri and M. acetivorans genome is not known, as this aRF1-2 protein (significantly diverged from the aRF1-1 sequence) is inactive in translation termination. It is pertinent to mention that the M. barkeri genome has two other examples of duplicated translation-related genes; there are two evolutionarily unrelated seryl-tRNA synthetases [36] and lysyl-tRNA synthetases [37].
4.2. Is UAG recoded for Pyl insertion?
The above results now provide the context for a discussion of UAG-directed Pyl incorporation. There are several ways by which UAG could be used as designating Pyl. (i) The first scenario would be similar to Sec, where an intricately executed scheme of recoding is operative involving an RNA structure (the SECIS element) located in the mRNA and a special elongation factor (SelB). This may not be happening for Pyl, as the initial excitement of the PYLIS RNA elements [18] could not be experimentally supported [5], and a new elongation factor was not required for Pyl-tRNA binding [19] or in vivo incorporation [20,21]. (ii) In some organisms stop codons are completely reassigned to a given amino acid; this is mediated by an adjustment of release factor specificity. In Tetrahymena thermophila, where the UAG and UAA codons are recoded as glutamine, the eRF1 only recognizes UGA as stop codon [38]. Similarly, in the ciliate Euplotes aediculatus UGA codon is reassigned to cysteine. In this organism eRF1 has restricted its specificity to UAA and UGA and lost its ability to trigger release at UGA codons [39]. However, as shown above, Methanosarcina aRF1 is omnipotent; thus this scenario is not operative either. Therefore, as in the case of nonsense suppression [40], Pyl insertion most likely results from simple competition between a suppressor aminoacyl-tRNA (in this case Pyl-tRNAPyl) and a release factor (in this case aRF1-1) at ambiguous UAG codons. Early termination results in truncated proteins that are easily disposed off by the protein degradation machinery. On the other hand, the risk of undesired read through of a UAG codon, that is designed to be a stop signal, is often minimized by occurrence of UAA or UGA codons located a short distance downstream of the UAG stop codon [5]. Thus, successful expansion of the genetic code by reassigning a stop codon to a new amino acid (e.g., Pyl) is not dependent on a precise recoding mechanism.
Acknowledgments
We are grateful to Andrey Poltaraus and his colleagues for sequencing a/eRF1 genes. We thank Tatyana Pestova and Chris Hellen for the gift of plasmids encoding initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4G, eIF5, eIF5B, and Anna Yaremchuk and Michael Tukalo for M. jannaschii aRF1. This work was supported by grants from the Presidium of the Russian Academy of Sciences (Program Molecular and Cell Biology), the Russian Foundation for Basic Research (08-04-01091-a to E.A. and 08-04-00375a to L.F.), the National Institute for General Medical Sciences (to D.S.), the National Science Foundation (to D.S.) and the Office of Basic Energy Sciences, DOE (to D.S.).
References
- 1.Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura Y, Ito K. Making sense of mimic in translation termination. Trends Biochem Sci. 2003;28:99–105. doi: 10.1016/S0968-0004(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 3.Poole ES, Askarian-Amiri ME, Major LL, McCaughan KK, Scarlet DJ, Wilson DN, Tate WP. Molecular mimicry in the decoding of translational stop signals. Prog Nucleic Acids Res Mol Biol. 2003;74:83–121. doi: 10.1016/s0079-6603(03)01011-0. [DOI] [PubMed] [Google Scholar]
- 4.Dontsova M, Frolova L, Vassilieva J, Piendl W, Kisselev L, Garber M. Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett. 2000;472:213–216. doi: 10.1016/s0014-5793(00)01466-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280:20740–20751. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 6.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr Opin Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 8.Deppenmeier U, et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between Bacteria and Archaea. J Mol Microbiol Biotechnol. 2002;4:453–461. [PubMed] [Google Scholar]
- 9.Zhang Y, Gladyshev VN. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res. 2007;35:4952–4963. doi: 10.1093/nar/gkm514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 11.Herring S, Ambrogelly A, Gundllapalli S, O’Donoghue P, Polycarpo CR, Soll D. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007;581:3197–3203. doi: 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Soll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci USA. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blight SK, et al. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 14.Bock A, Thanbichler M, Rother M, Resch A. Selenocysteine. In: Francklyn CS, Ibba M, Cusack S, editors. Aminoacyl-tRNA Synthetases. Springer; Georgetown, TX: 2005. pp. 320–327. [Google Scholar]
- 15.Allmang C, Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie. 2006;88:1561–1571. doi: 10.1016/j.biochi.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Squires JE, Berry MJ. Eukaryotic selenoprotein synthesis: mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60:232–235. doi: 10.1002/iub.38. [DOI] [PubMed] [Google Scholar]
- 17.Su D, Hohn MJ, Palioura S, Sherrer RL, Yuan J, Soll D, O’Donoghue P. How an obscure archaeal gene inspired the discovery of selenocysteine biosynthesis in humans. IUBMB Life. 2009;61:35–39. doi: 10.1002/iub.136. [DOI] [PubMed] [Google Scholar]
- 18.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 19.Théobald-Dietrich A, Giegé R, Rudinger-Thirion J. Evidence for the existence in mRNAs of a hairpin element responsible for ribosome dependent pyrrolysine insertion into proteins. Biochimie. 2005;87:813–817. doi: 10.1016/j.biochi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Longstaff DG, Blight SK, Zhang L, Green-Church KB, Krzycki JA. In vivo contextual requirements for UAG translation as pyrrolysine. Mol Microbiol. 2007;63:229–241. doi: 10.1111/j.1365-2958.2006.05500.x. [DOI] [PubMed] [Google Scholar]
- 21.Namy O, Zhou Y, Gundllapalli S, Polycarpo CR, Denise A, Rousset JP, Soll D, Ambrogelly A. Adding pyrrolysine to the Escherichia coli genetic code. FEBS Lett. 2007;581:5282–5288. doi: 10.1016/j.febslet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Théobald-Dietrich A, Frugier M, Giegé R, Rudinger-Thirion J. Atypical archaeal tRNA pyrrolysine transcript behaves towards EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 2004;32:1091–1096. doi: 10.1093/nar/gkh266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor – eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 24.Lekomtsev S, Kolosov P, Bidou L, Frolova L, Rousset JP, Kisselev L. Different modes of stop codon restriction by the Stylonychia and Paramecium eRF1 translation termination factors. Proc. Natl. Acad. Sci. USA. 2007;104:10824–10829. doi: 10.1073/pnas.0703887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seit-Nebi A, Frolova L, Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chavatte L, Kervestin S, Favre A, Lean-Jean O. Stop codon selection in eukaryotic translation termination: comparison of the discriminating potential between human and ciliate eRF1s. EMBO J. 2003;22:1644–1653. doi: 10.1093/emboj/cdg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Emov A, Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 36.Korencic D, Polycarpo C, Weygand-Durasevic I, Soll D. Differential modes of transfer RNASer recognition in Methanosarcina barkeri. J Biol Chem. 2004;279:48780–48786. doi: 10.1074/jbc.M408753200. [DOI] [PubMed] [Google Scholar]
- 37.Ambrogelly A, Korencic D, Ibba M. Functional annotation of class I lysyl-tRNA synthetase phylogeny indicates a limited role for gene transfer. J Bacteriol. 2002;184:4594–4600. doi: 10.1128/JB.184.16.4594-4600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito K, Frolova L, Seit-Nebi A, Karamyshev A, Kisselev L, Nakamura Y. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc Natl Acad Sci USA. 2002;25:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kervestin S, Frolova L, Kisselev L, Jean-Jean O. Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO Rep. 2001;2:680–684. doi: 10.1093/embo-reports/kve156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggertsson G, Soll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988;52:354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]